Abstract
Genome-wide association studies (GWAS) have identified a region upstream the BIN1 gene as the most important genetic susceptibility locus in Alzheimer's disease (AD) after APOE. We report that BIN1 transcript levels were increased in AD brains and identified a novel 3 bp insertion allele ∼28 kb upstream of BIN1, which increased (i) transcriptional activity in vitro, (ii) BIN1 expression levels in human brain and (iii) AD risk in three independent case-control cohorts (Meta-analysed Odds ratio of 1.20 (1.14–1.26) (P=3.8 × 10−11)). Interestingly, decreased expression of the Drosophila BIN1 ortholog Amph suppressed Tau-mediated neurotoxicity in three different assays. Accordingly, Tau and BIN1 colocalized and interacted in human neuroblastoma cells and in mouse brain. Finally, the 3 bp insertion was associated with Tau but not Amyloid loads in AD brains. We propose that BIN1 mediates AD risk by modulating Tau pathology.
Keywords: Alzheimer, brain, BIN1, Drosophila, Tau
Introduction
Using large-scale Genome-wide association studies (GWAS), we and others have recently identified variants upstream of BIN1 (OMIM *601248) that increase risk of Alzheimer's disease (AD).1, 2, 3 BIN1 is evolutionary conserved from yeast to human and is a member of the Bin1/amphiphysin/RVS167 (BAR) family of genes that have been involved in diverse cellular processes, including endocytosis, actin dynamics and membrane trafficking/tubulation.4 In mammals, BIN1 is widely expressed and produces more than 10 isoforms that differ in their subcellular localization, tissue distribution and protein interactions reflecting its diverse functional roles. Given these multiple functions, it is a challenging task to determine the genetic and molecular mechanisms by which BIN1 affects AD risk. While its role in endocytosis suggests a function for BIN1 in APP metabolism,4 BIN1 might also interact with microtubule-associated proteins like Tau because of its function in regulating cytoskeleton dynamics.4 To address the genetic mechanism by which BIN1 increases AD risk and is involved in the AD pathogenic process, we adopted a multidisciplinary approach, including molecular and functional genetics, cell biology, neuropathology and Drosophila biology.
Materials and methods
mRNA quantification
Total RNA was extracted from frozen frontal cortex brain tissue from the 114 AD5 and 167 control6 samples (see Supplementary Methods section) using a phenol/chloroform protocol (Trizol reagent, Invitrogen, Carlsbad, CA, USA). Total RNA samples from 61 controls (age at death: 80.1±6.2, 42% male, braak stage <2) and 64 AD cases (age at death: 74.7±9.0, 39% male, braak stage >5) were randomly selected for quantification of the expression of BIN1 (0.6 μg per assay) and the housekeeping β-actin or β-glucuronidase genes, (respectively, 0.1 and 0.2 μg per assay), according to the supplier's instructions (Quantigene, Panomics, Fremont, CA, USA) (see Supplementary Methods for additional information).
Genetic analysis of the BIN1 locus
The EADI, GERAD and Belgium-Flanders case-control studies and the imputation methodology are fully described in the Supplementary Method.
We imputed single-nucleotide polymorphisms (SNPs) by using MaCH (http://www.sph.umich.edu/csg/abecasis/mach/index.html) and minimac software (http://genome.sph.umich.edu/wiki/minimac). The reference haplotype data is provided by the MaCH website, which was built for the combined Caucasian populations as part of the 1000 Genomes project. In our data set, all individuals were genotyped on the same platform (the Illumina Human660W-Quad Beadchip, San Diego, CA, USA) and we used 492 941 observed SNP genotypes that passed quality filters as follows: genotyping call rate⩾98%, Hardy–Weinberg equilibrium test P-value ⩾1 × 10−6 and MAF ⩾1%. We first inferred haplotype combinations of each individual using the ‘phase;' option in the MaCH program and then imputed them with minimac. Doses for 7 704 555 SNPs with a MAF >0.01 were available from the French GWA data set using the 1000 Genomes dataset.7 We selected 493 SNPs within the BIN1 locus of interest (chr2:127803530–127903530) and evaluated their associations with AD risk in an additive logistic regression model adjusted for age and gender. A graphic representation (Figure 2a) was then generated with Locuszoom software (http://csg.sph.umich.edu/locuszoom/). Of note, we also used Impute2 package for imputation (mathgen.stats.ox.ac.uk/impute/impute_v2.html) and found very similar results between minor allele frequency estimation between both methods (R2=0.99, data not shown) for SNPs in the BIN1 locus.
Genetic analysis of the LD block of interest in the BIN1 locus
Sequencing of the LD block of interest was performed using genomic DNA extracted from 47 AD cases and 47 controls. Oligonucleotides are available upon request. The Polymorphisms not initially imputed (four SNPs and four Indels, MAF >0.05) were genotyped in a sample of 350 healthy French controls used as a panel reference for imputation in the EAD1 GWAS. After obtaining doses for the eight genetic variations from EADI1 GWA data set, we evaluated their associations with AD risk in an additive logistic regression model adjusted for age and gender.
rs59335482 was also genotyped by direct sequencing in EADI1 (data presented across the manuscript), the Flanders–Belgian population and the brain samples. In order to avoid any genotyping bias, cases and controls were randomly mixed while genotyping and laboratory personnel were blinded to case/control status. All the primer and probe sequences used in the genotyping assays are available upon request. Of note, in line with imputation quality, a 95% concordance was observed between the imputation and direct sequencing genotypes in EADI1. Again, rs59335482 association with AD risk was evaluated in an additive logistic regression model adjusted for age and gender. We used inverse-variance weighting (also known as fixed-effects meta-analysis) to provide meta-analysed, age- and gender-adjusted ORs for association estimates in the EADI1, GERAD1 and Flanders–Belgium studies.
The main characteristics of EADI1, GERAD1 and the Belgian–Flanders case-control studies are described in the supplementary notes.
Drosophila strains, culture conditions and AD-related neurotoxicity assays
UAS-Amph, UAS-AmphP{TRiP.JF02883}attP2 (UAS-AmphKD), GMR-GAL4, Elav-GAL4 (C155) and Act5c-GAL4 lines were obtained from the Bloomington Stock Center. The UAS-Tau (human 2N4R Tau), UAS-Aβ42, Amph5E3 and the Eq-GAL4 lines were a kind gift from GR Jackson, D Crowther, GL Boulianne and H Sun, respectively. Flies were reared under controlled temperature conditions of 25 °C and were fed standard fly medium.
Pictures of external eye morphology were taken at different focus points, with a DP70 camera mounted on an Olympus BX61 (Hamburg, Germany), in a way that an array of images with overlapping depth of fields is obtained. These stacks were focused using ImageJ software with the ‘stack Focuser' plugin (National institutions of Health, Bethesda, Maryland).
To measure eye size female flies were photographed with an Olympus SZX12 stereomicroscope fitted with a XC30 camera. Measurements were performed using AnalySIS FIVE software (Olympus soft Imaging solutions, GmbH, Münster, Germany). A Kruskal–Wallis test was performed followed by a Dunns post-hoc test on Prism 5.0b for MacOS (Graphpad software, La Jolla, CA, USA).
To count notal bristles, flies were mounted on glass slides and photos were taken with the SZX12 stereomicroscope/XC30 camera. Bristles of a well-defined area were counted in a non-automated manner. A Kruskal–Wallis test was performed followed by a Dunns post-hoc test on Prism 5.0b for MacOS (Graphpad software).
Drosophila adult brains were dissected and processed for immunohistochemistry with a mouse monoclonal antifasciclin 2 antibody to visualize mushroom body α-, β- and γ-lobes and the ellipsoid body, as described previously.8 The immunostaining was documented using an Olympus BX61 epifluorescence microscope equipped with a DP70 digital camera controlled with analySIS FIVE software. As a control for staining quality only brains, in which the peduncles could be observed were used in the statistical analysis. Flies were scored blindly for presence of mushroom body β-lobes. Statistical analysis was performed using a Fisher exact test on Prism 5.0b for MacOS (Graphpad software).
To measure dAmph expression levels RNA was isolated from 50 heads of act5c>AmphTRiP, Amph5E3/+, Act5c/+ and AmphTRiP/+. Heads were collected in 1 ml of TRI reagent (Sigma, St Louis, MO, USA) and were grounded with a plastic disposable pestle. Total RNA was isolated by using standard procedures. Complementary DNA was generated from 1 μg of RNA of each sample by using an anchored oligo(dT)18 primer and a hexamer primer according to the instructions of the Transcriptor first-strand complementary DNA synthesis kit; Roche (Meylan, France). Quantitative real-time PCRs were performed on an ABI7000 instrument with qPCR Mastermix Plus for SYBR Green I (Eurogentec, Seraing, Belgium) with primers designed by PrimerExpress software (Applied Biosystems, Foster City, CA, USA). Expression levels of transcripts from the various samples were normalized to the housekeeping genes gadph and Rps13. Primers used: CAGCCCCGACATGAAGGT-forward and CGATCTCGAAGTTGTCATTGATG-reversed for Gadph, GGGTCTGAAGCCCGACATT-forward and GGCGACGGCCTTCTTGAT-reversed for RpS13, TCAGAATCTGCAGGCCAATG-forward and CGCGTCCTTTGGTTAGTTTGAC-reverse for dAmph.
Cell culture and western blot analyses
HEK293 and neuroblastoma SKNSH-SY5Y cell lines were maintained in DMEM supplemented with 10% fetal bovine serum, penicillin and streptomycin at 37 °C in a humidified atmosphere with 5% CO2. Transient transfection was performed using Exgen500 (Fermentas, Villebon sur Yvettes, France) in SKNSH-SY5Y and Fugene-HD (Roche Diagnostics, Rotkreuz, Switzerland) in HEK-293 cells according to the manufacturer's recommendations. The pCNV6XL4-BIN1 isoform 1 (iso.1) vector was purchased from Origene (NM_139343.1) (Rockville, MD, USA). Cell extracts (5–20 μg) were analyzed by SDS-polyacrylamide gel electrophoresis using antibodies listed above. Antibodies directed against human BIN1 (99D), mouse BIN1 (ab27796), human Tau (Tau5) and mouse Tau (mTau5) were from Abcam (Cambridge, UK). A standard ECL detection procedure was then used.
Luciferase reporter assays
Before transfection, cells were plated at 1 × 106 and 1.2 × 106 cells per well in six-wells plate for SKNSH-SY5Y and HEK cells, respectively. Cells were transfected with pGL3promoter (Promega, Madison, WI, USA) plasmids containing or not the sequence (60 bp) surrounding the polymorphisms of interest (wild-type and mutant SDS-page purified oligonucleotides (Eurogentec, Belgium) were directly cloned into plasmids). All the oligonuleotide sequences used for cloning are available upon request. pRL vector (Promega) was used as normalization vector. Luciferase assay was perfomed 48 h after transfection according to promega dual luciferase assay instructions (Promega). Firefly/Renilla Luciferase ratio for all the sequence surrounding the polymorphisms were compared with the ratio measured for pGL3promoter empty vector.
Immunofluorescence
Cells were washed with phosphate-buffered saline (PBS) and fixed in PBS containing 4% paraformaldehyde for 20 min at room temperature. Alternatively, cells were pre-permeabilized with 0.01% of saponin for 3 min before fixation. Cells were permeabilized with 0.25% (v/v) Triton X-100 in PBS for 10 min. After blocking in 1% (w/v) bovine serum albumin, cells were incubated for 2 h at room temperature with primary antibodies (anti-BIN1, 99D; anti-Tau, Tau c-Ter) 1/100 in PBS, 1% bovine serum albumin. Cells were then washed three times with PBS. Appropriate secondary antibodies diluted to 1/400 were used. After washing, coverslips were mounted on slides with Fluoromount (Sigma-Aldrich).
Glutathione-S-transferase (GST) and 6His fusion proteins and pull-down assays
GST-Tagged Tau protein was produced using pGEX vector and expressed in Escherichia coli BL21 cells. Vector for GST-BIN1 neuronal isoform was a gift from Dr Laporte (INSERM U596, Strasbourg, France). In Escherichia coli M15 cells 6His-Tau was expressed using pQE30 vector. Proteins were extracted from bacteria inclusion bodies by treatment with lysosyme, N Sarkosil (0.001%) and Triton X-100 (0.5%) at 4 °C and by sonication on ice. GST fusion proteins were immobilized on glutathione-sepharose beads (Pierce). The GST fusion proteins bound to glutathione-sepharose beads was incubated with HEK293 cells lysates, 1 h at room temperature. Proteins were pull-down by guthlatione beads and loaded on SDS-polyacrylamide gel electrophoresis and analysed by western Blot. Alternatively, GST-BIN1 was incubated with 6His-Tau purified using Ni Sepharose column from His Spin Trap Kit (GE Healthcare, Waukesha, WI, USA) accordingly to the manufacturer.
Coimmunoprecipitation assays
SKNSH-SY5Y cells were washed with PBS and solubilised in ice-cold lysis buffer (10 mM HEPES, pH 7.4; 140 mM NaCl; 0.5% NP40; 1 × complete protease inhibitor mixture; 1 × PhosSTOP, Roche Applied Science). After centrifugation at 11 000 g for 10 min at 4 °C, supernatants were incubated overnight at 4 °C with the indicated antibodies (anti-BIN1, 99D; anti-Tau, Tau-5) and protein-G. The following day, antibodies were precipitated and washed three times with lysis buffer and then were resuspended in loading buffer and resolved by SDS-polyacrylamide gel electrophoresis. Alternatively, protein samples from synaptosomal protein fraction (0.5 mg) were precleared with 50 μl of magnetic beads (protein G coated Dynabeads, Invitrogen) during 30 min of agitation (1200 r.p.m.). Primary antibodies (anti-BIN1, 99D; anti-Tau, mTau-5) were bound to protein G-coupled beads according to the manufacturer's instructions (1 μg of antibody per 10 μl of bead slurry). Precleared lysates were incubated with 50 μl antibody-bead complexes overnight (1200 r.p.m., 4 °C). After four washes with RIPA buffer, beads were eluted with SDS polyacrylamide gel electrophoresis sample buffer and analysed with western blotting.
Synaptosomal preparation
Preparation of synaptosomal fractions from mouse brain was performed as previously described.9 One snap frozen hemisphere was homogenized in 10 ml homogenization buffer (0.32 M Sucrose, 1 mM EDTA, 1 mg ml−1 bovine serum albumin, 5 mM HEPES pH 7.4, phosphatase (PhosStop, Pierce, Thermoscientific, Rockford, IL, USA) and protease (Complete, Roche inhibitors) using a glass-teflon douncer (10 strokes, 600 r.p.m., 4 °C). After centrifugation (10 min; 3000 g, 4 °C), the supernatant, containing cytosolic fraction and synaptosomes, was centrifuged again (12 min, 14 000 g, 4 °C) resulting in a pellet containing the synaptic vesicles. The latter was resuspended in 0.733 ml Krebs-Ringer buffer (140 mM NaCl, 5 mM KCl, 5 mM glucose, 1 mM EDTA, 10 mM HEPES, pH 7.4, phosphatase (PhosStop, Pierce, Thermoscientific) and protease (Complete, Roche inhibitors). After adding 0.6 ml Percoll (Sigma) and centrifugation (20 000 g, 2 min, 4 °C), synaptosomal fraction appeared as an opaque floating band, which was collected and pelleted in 1 ml of Krebs Ringer buffer (20 000 g, 30 s, 4 °C). The final pellet was dissolved in 0.5 ml RIPA buffer (Sigma) for further analysis.
Immunohistochemistry
Formalin-fixed brain samples were obtained from the Lille Neurobank and included seven AD patients (two Braak II, three Braak IV and three Braak VI) and two controls. Forty micrometer sections from the hippocampal formation were processed for BIN1 immunohistochemistry using the anti-human BIN1 antibody (99D, Millipore, Billerica, MA, USA). For double labelling, another BIN1 antibody was also used (sc30099, Santa Cruz, Santa Cruz, CA, USA). Other antibodies were anti-neurofilaments (SMI31, Merck, Whitehouse Station, NJ, USA), Iba1 (WAKO) and GFAP (sc33673, Santa Cruz). Secondary antibodies were alexa Fluor 488 anti-mouse, alexa Fluor 488 anti-rabbit, alexa Fluor 568 anti-mouse, alexa Fluor 568 anti-rabbit (Invitrogen). Sections were counterstained with DAPI. Brain sections were analyzed on a TCS SP Leica (Lasertechnik GmbH, Jena, Germany) confocal microscopy.
In the brain sample from Manchester, the proportion of tissue area (%) occupied by Aβ40 and Aβ42 were quantified in immunohistochemically stained sections from brodmann area 8/9 of the frontal cortex, as previously reported.5 Tau load (proportion of tissue area (%)) was determined after immunostaining for phosphorylated Tau using a standard procedure employing monoclonal antibody AT8 (Innogenetics, Gent, Belgium), as primary antibody.10
Endophenotype analyses
We extracted BIN1 mRNA levels measured in lymphoblastoid cell lines from the public MTAB-198 database (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-198). These lymphoblastoid cell lines were derived from 109 caucasian HapMap 3 individuals and mRNA levels were measured using a whole-genome expression array (Illumina Sentrix WG-6, Version 2).5 rs59335482 imputation was successfully performed as described in the imputation section for 98 individuals (info measure=0.94) and association study of rs59335482 with BIN1 lymphoblastoid cell lines mRNA levels were performed using a general linear model.
Comparison of BIN1 mRNA amounts in the brain of AD cases and controls, and the association study of rs59335482 with BIN1 cerebral mRNA levels were performed using a general linear model adjusted for RNA degradation level (28 s/18 s ratio). Association studies of rs59335482 with Tau, Aβ40 and Aβ42 score were performed using a general linear model. All the comparisons were also performed using a non-parametric Wilcoxon test and the results did not change (data not shown).
Results
BIN1 expression is increased in AD brains when compared with controls
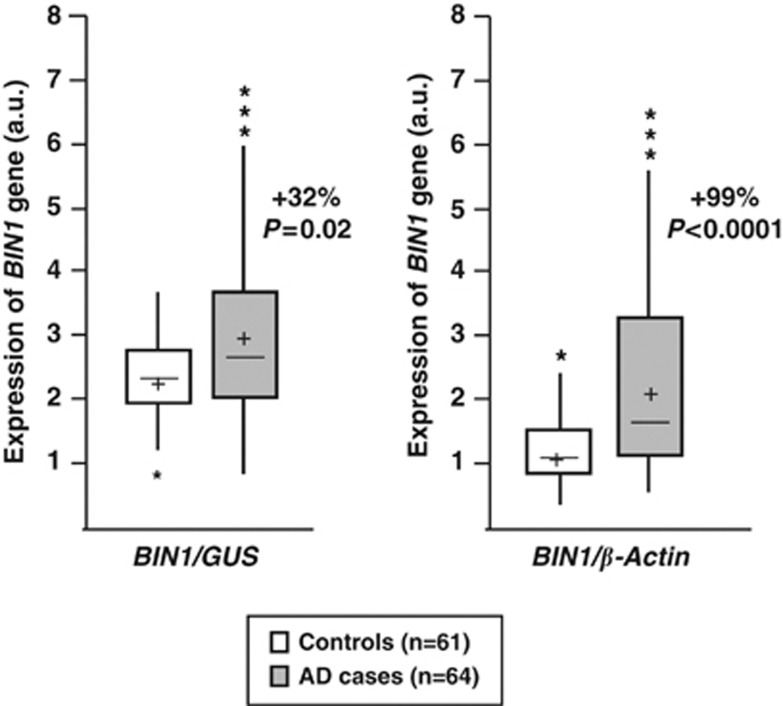
We first analysed BIN1 expression in the human brain and observed increased BIN1 transcript levels in brains of AD cases when compared with controls (Figure 1). From this observation, we hypothesized that altered BIN1 expression might constitute the genetic mechanism by which BIN1 increases AD risk. This hypothesis is in line with the fact that the Index signal rs744373 observed in the initial publication of Seshadri et al.3 is located 28 kb upstream of the BIN1 coding region.
Figure 1.
BIN1 mRNA levels in AD and control brains. All values of BIN1 mRNA are shown as arbitrary units (a.u.) following normalisation by GUS (β-glucuronidase) or β-actin mRNA quantification. mRNA quantifications were carried out in triplicate in all individuals. cross: mean of BIN1 expression in cases and controls; middle line: median; upper horizontal line: inclusion of 75% of the individuals; lower horizontal line: inclusion of 25% of the individuals. *Individuals exhibiting extreme values (out of the global distribution).
Insertion/deletion rs59335482, potentially involved in BIN1 overexpression in AD
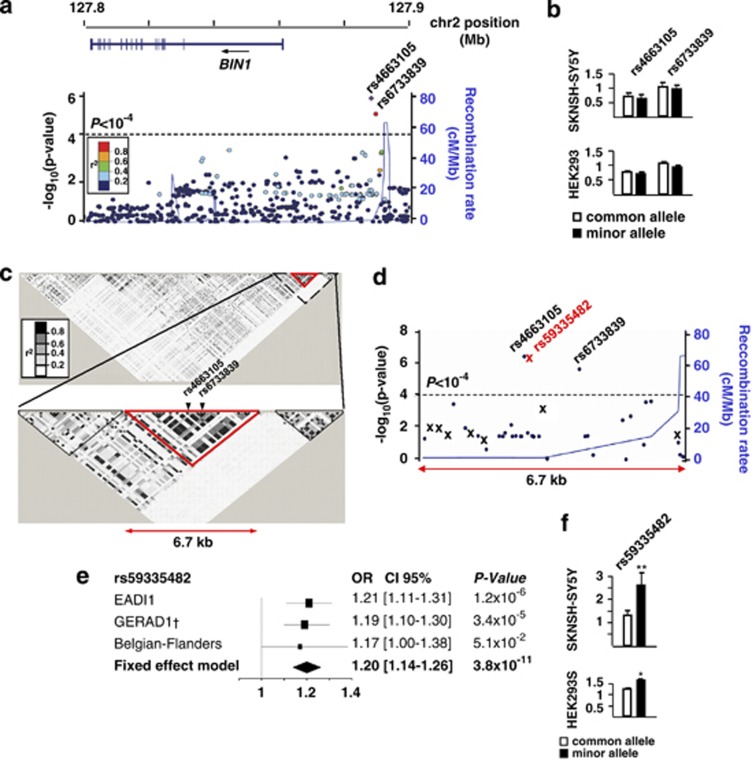
To further investigate this possibility, we fine-mapped the BIN1 risk locus by performing imputation of genotypes in our French EADI12 cohort using the 1000 Genomes data set (http://www.1000genomes.org). We analysed 493 SNPs and after Bonferroni correction (P<10−4), we observed two SNPs, rs4663105 and rs6733839, associated with AD risk ((Figure 2a). We therefore investigated these two SNPs for their potential to modify transcription in vitro using luciferase reporter assays. However, none of them showed evidence for allele-specific differences in transcriptional activity in both SKNSH-SY5Y and HEK293 cells (Figure 2b). To further fine-map the BIN1 locus, we performed linkage disequilibrium (LD) block analysis by using Haploview software (MIT, Cambridge, MA, USA) and by applying Gabriel's criteria to define these LD blocks. This indicated that rs4663105 and rs6733839 were encompassed in a small 6.7 kb LD block (from rs11680911 to rs6431223) (Figure 2c; Supplementary Table S1). This LD block also includes rs744373, which is the index signal observed in the initial publication of Seshadri et al.3 These data suggested that this LD block might harbour (a) functional risk variant(s). We therefore fully sequenced this 6.7 kb region in 47 AD cases and 47 controls and observed eight new polymorphisms including four SNPs and four insertion/deletion variants (Indel) (Figure 2d and Supplementary Table S2). These polymorphisms were then genotyped in a sample of 338 French controls, which was used as a reference panel for imputation in the entire GWAS EADI1 data set (Supplementary Table S2). Among these eight new variants, only Indel rs59335482 (an insertion of three C bases), was associated with a higher AD risk (respectively, odds ratio (OR)=1.21, 95% Confidence interval (1.13–1.38), P=1.3 × 10−6) when applying our initial Bonferroni criteria correction (P<0.0001). We finally attempted to replicate the rs59335482 insertion association with AD risk in the GERAD1 GWAS and in an independent Flanders–Belgian population. This resulted in a genome-wide significant meta-analysed OR of 1.20 (1.14–1.26) (P=3.8 × 10−11) (Figure 2e; Supplementary Table S3). We then addressed the possibility that this Indel variant may affect transcriptional activity and observed that the rs59335482 insertion allele was associated with an increase in luciferase activity in both neuroblastoma SKNSH-SY5Y (+101%) and HEK293 (+33%) cells (Figure 2f). Accordingly, we finally observed the rs59335482 insertion allele was associated with an increase in BIN1 mRNA expression in the brain (Table 1). To corroborate this observation, we also imputed rs59335482 in 98 Hapmap 3 individuals for whom BIN1 RNA level was measured in lymphoblastoid cell lines.11 We observed that the rs59335482 insertion allele was again associated with an increase in BIN1 mRNA levels (P=0.01; Table 1). Together these results strongly support that functional(s) polymorphism(s) that increase BIN1 expression in the brain, may increase AD risk.
Figure 2.
Genetic analysis of the BIN1 locus. (a) Locuszoom view of the imputation analysis in the EADI1 cohort showing the genomic positions of the rs4663105 and rs6733839 SNPs with P<10−4. (b) Measure of luciferase activity in HEK293 and SKNSH-SY5Y cells for the rs4663105 and rs6733839 SNPs. Cells were transfected with the pGl3 promoter vectors containing the common or minor allele of each polymorphism. pGl3 promoter empty vector has been used as reference. The graphs represent the average of three independent experiments. Histograms indicate the means±s.d. (c) LD mapping showing an LD block (from rs11680911 to rs6431223) of 6.7 kb including the most strongly associated SNPs rs4663105 and rs6733839. (d) Locuszoom view of the imputation analysis in the EADI1 cohort showing all polymorphisms identified by sequencing of the 6.7 kb region in 47 AD cases and 47 controls. X denotes the eight new polymorphisms identified by sequencing. The Indel rs59335482 was significantly associated with AD risk with P<10−4. (e) Association of rs59335482 with AD risk in EADI1, GERAD1 and the Flanders–Belgian population. P-values and ORs with the associated 95% confidence interval have been calculated under an additive model using logistic regression models adjusted for age, gender and centers when necessary. (f) Measure of luciferase activity as in b with pGl3 promoter vectors containing the common or minor allele of the rs59335482. The graphs represent the average of three independent experiments. Histograms indicate the means±s.d. *P<0.05; **P<0.001.
Table 1. Association of rs59335482 with BIN1 mRNA level in brain and in lymphoblasts (Hapmap 3).
| rs59335482 | DD | ID | II | P-value |
|
|---|---|---|---|---|---|
| Model 1 | Model 2 | ||||
| BIN1 mRNA level | |||||
| Whole (n=125) | 1.7±1.4 | 2.3±2.2 | 2.6±1.8 | 0.02 | 0.01 |
| Controls (n=61) | 1.2±0.5 | 1.3±0.6 | 1.7±0.7 | 0.17 | 0.07 |
| Cases (n=64) | 2.3±1.9 | 3.2±2.7 | 3.2±2.7 | 0.15 | 0.11 |
| Hapmap3a (n=98) | 10.8±0.9 | 11.1±0.7 | 11.3±0.7 | 0.02 | 0.01 |
Abbreviations: D, deletion; I, insertion. Analysis was performed using a non parametric Wilcoxon test following recessive or additive models (respectively model 1 and model 2).
Denotes populations imputed for the rs59335482.
Variation of Amph expression, that is, the BIN1 Drosophila ortholog modulates Tau neurotoxicity in vivo
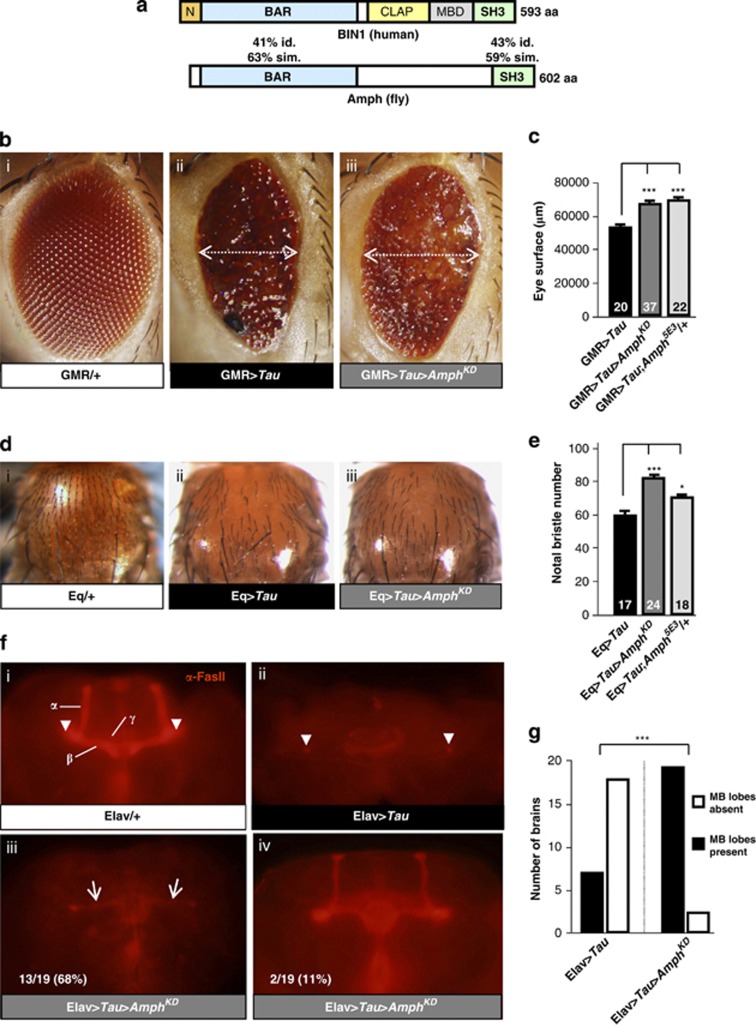
Next, in order to define how altered BIN1 expression might be involved in AD pathogenesis, we used Drosophila genetics to explore in vivo the link between altered expression levels of the BIN1 ortholog Amph and AD-relevant neurotoxicity readouts. Amph is strongly homologous to BIN1 (Figure 3a and Supplementary Table S4) and, like BIN1, Amph functions in membrane morphogenesis in muscle and neuronal tissues.11, 12, 13 We first investigated if altered expression of Amph could modulate Aβ42 neurotoxicity in Drosophila. We used a model, in which neuronal expression of the Aβ42 peptide leads to rough eyes and neurodegeneration characterized by intraneuronal accumulation of non-amyloid Aβ42.14 We checked that knockdown of Amph and amph null allele decrease and abolish respectively Amph expression at the RNA and protein levels (Supplementary Figures 1a–c). We checked also as negative controls that altered Amph expression did not affect external eye morphology on its own (Supplementary Figures 1d–e). Altered Amph expression did not modify the Aβ42-induced rough eye phenotype (Supplementary Figure 2a).
Figure 3.
Amph modifies Tau neurotoxicity in Drosophila. (a) Schematic showing evolutionary conservation of the protein domains of Drosophila Amph and human BIN1 isoforms (iso.). BAR, BIN1-amphiphysin-Rvs167; CLAP, clathrin and AP-2 binding; MBD, Myc-binding domain; SH3, Src homology 3. PI, phosphoinositide binding region. (b) External eye morphology. (i) GMR/+ control (outcrossed eye specific GMR-GAL driver) showing normal regular eye morphology; (ii) GMR>Tau (Tau control, eye specific overexpression of Tau) showing severely reduced eye size and external roughness; (iii) GMR>Tau>AmphKD (simultaneous eye-specific overexpression of Tau and knockdown of Amph) showing increased eye size compared with (ii). (c) Quantification of eye size in flies overexpressing Tau alone (GMR>Tau) and simultaneous decreased Amph expression by either RNAi-mediated knockdown (GMR>Tau>AmphKD) or by removal of one genomic copy of Amph (GMR>Tau;Amph5E3/+). (d) Notal bristle number. (i) Eq/+ (control, outcrossed notal bristle specific Eq-GAL driver) showing normal number of bristles; (ii) Eq>Tau (Tau control, notal bristle specific overexpression of Tau) showing severely reduced number of bristles; (iii) Eq>Tau>AmphKD (simultaneous bristle specific overexpression of Tau and knockdown of Amph) increased number of bristles compared to (ii). (e) Quantification of notal bristle number in flies overexpressing Tau alone (Eq>Tau) and simultaneous decreased Amph expression by either RNAi-mediated knockdown (Eq>Tau>AmphKD) or removal of one genomic copy of Amph (Eq>Tau;Amph5E3/+). (f) Mushroom body morphology. (i) Immunohistochemical α−FasII labeling in whole-mount brains showing normal mushroom body morphology in a control brain (Elav/+, outcrossed neuron-specific Elav-GAL4 driver). α-, β- and γ-lobes and peduncles (arrowheads) are indicated; (ii) Neuron-specific Tau overexpression (Elav>Tau) ablates mushroom body lobes while peduncles can still be observed in most cases (arrowheads); (iii) Neuron-specific Tau overexpression and simultaneous Amph knockdown (Elav>Tau>AmphKD) reveals thin β-lobes in 13/19 brains (arrows); (iii) Elav>Tau>AmphKD shows morphologically normal mushroom bodies in 2/19 brains. (g) Quantification of the presence of mushroom body (MB) lobes in flies panneuronally overexpressing Tau alone (Elav>Tau) and simultaneous decreased Amph expression by RNAi-mediated knockdown (Elav>Tau>AmphKD). Histograms in c and e indicate means±s.e.m. *P<0.05; ***P<0.001.
Next, we investigated if altered Amph expression could alter human Tau neurotoxicity in Drosophila. Eye-specific Amph overexpression did not significantly alter the size of the Tau-induced rough eye although the number of ommatidia was significantly reduced due to increased ommatidial fusions (P<0.0001; Supplementary Figures 2b and c). Knockdown of Amph suppressed Tau-toxicity, leading to a 30% increase in eye surface compared with flies solely overexpressing Tau (Figures 3b and c). Nearly identical results were obtained in Tau overexpressing flies heterozygous for an Amph null allele (Figure 3c and Supplementary Figure 1). We checked as a negative control that altering Amph expression in the eye did not affect human Tau transgene expression levels (Supplementary Figure 1c). In order to further confirm a link between Amph and Tau toxicity, we also measured Tau neurotoxicity in the notal bristles of adult flies.15 The Drosophila notum carries a multitude of bristles, which are sensory organs, connected at the base with the dendrite of a sensory neuron. Silencing Amph suppressed the Tau-induced bristle loss phenotype resulting in 37% more notal bristles compared with Tau overexpression alone (Figures 3d and e). Similar results were obtained for the heterozygous Amph null allele (Figure 3e). We also checked as negative controls that modulating Amph levels in a wild-type background did not induce phenotypes that could explain the genetic interactions (Supplementary Figures 1d and e). Finally, we looked at the Drosophila mushroom bodies, which like the hippocampi of mammals, are crucial for learning and memory. Panneuronal overexpression of human Tau leads to a selective ablation of the mushroom bodies.16 In most Tau overexpressing flies we observed residual staining of the two peduncles but not of α- or β-lobes (Figure 3f and g). In contrast, in the majority of Tau overexpressing flies in which Amph was knocked down, thin β-lobes could be clearly discerned and two brains even displayed an almost wild-type mushroom body morphology (Figure 3f). Together, these results are in accordance with the hypothesis that increased BIN1 expression in the brain might be deleterious and indicated that loss of Amph was able to suppress Tau-induced neurotoxicity in Drosophila.
BIN1 interacts with Tau
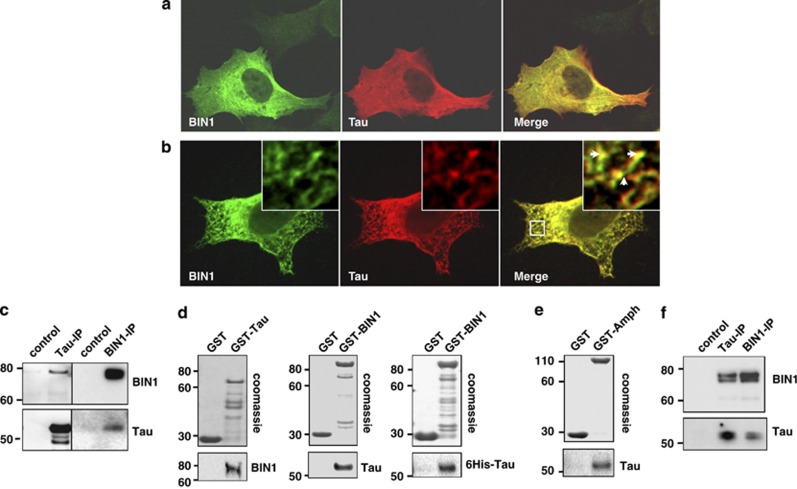
In order to verify if BIN1 and Tau were able to physically interact, we first compared their respective subcellular localizations in human neuroblastoma SKNSH-SY5Y cells, transiently transfected with neuronal BIN1 isoform (iso.1) and Tau (1N4R). As shown in Figure 4a, BIN1 and Tau were highly expressed throughout the cell and exhibited strong cytosolic staining. In conditions of pre-permeabilization with 0.01% of saponin, which removes an important part of the cytosol and leaves the cytoskeleton intact, BIN1 and Tau showed a strong intracellular costaining and colocalization (Figure 4b). We thus explored the possibility that BIN1 and Tau (1N4R) form a complex by performing reciprocal immunoprecipitations with antibodies directed against BIN1 (99D) and Tau (Tau-5). Figure 4c shows that BIN1 coimmunoprecipitated with Tau in cells overexpressing the two proteins.
Figure 4.
Characterisation of a link between BIN1 and Tau. (a) Representative confocal images showing the subcellular distribution of BIN1 and Tau. SKNSH-SY5Y cells were transiently transfected with BIN1 (iso.1) and Tau (1N4R). After 24 h, cells were fixed in paraformaldehyde and staining with anti-BIN1 (green) (99D) and anti-Tau (red) (Tau c-Ter) antibodies. (b) Representative confocal images showing the intracellular distribution of BIN1 and Tau. SKNSH-SY5Y cells were transiently transfected as in a and pre-permeabilized with 0.01% of saponin before fixation to remove an important part of cytosol. Arrows denote colocalization staining between BIN1 and Tau. (c) SKNSH-SY5Y cells were transiently transfected with Tau (1N4R) and BIN1 (iso.1) or control empty plasmid (control). After 24 h, cells extractswere immunoprecipitated (IP) with anti-BIN1 (99D) or anti-Tau (Tau-5) antibody. Precipitated proteins were resolved by SDS-PAGE and visualized with anti-BIN1 (99D) and anti-Tau (Tau c-Ter) antibodies using True-Blot system. (d) GST-Tagged protein or GST alone was incubated with lysate from HEK293 cells overexpressing Tau (1N4R) or BIN1 (iso.1). Alternatively, GST-BIN1 was incubated with 6His-Tau purified protein. Pull-downs were resolved by SDS-polyacrylamide gel electrophoresis and visualized with Coomassie or by western blot using anti-Tau (Tau c-Ter) and anti-BIN1 (99D) antibodies. (e) GST-Amph or GST alone was incubated with lysate from HEK293 cells overexpressing Tau (1N4R). Pull-downs were resolved by SDS-polyacrylamide gel electrophoresis and visualized with Coomassie or by western blot using anti-Tau (Tau c-Ter) antibody. (f) Synaptosomes fraction were extracted from mouse brain. Precleared lysates were incubated for immunoprecipitation with anti-Tau (mTau-5), anti-BIN1 (99D) antibody or with protein G-coupled beads alone (control). Pull-downs were resolved by SDS-polyacrylamide gel electrophoresis and visualized by western blot using True-Blot system.
To validate the BIN1-Tau interaction, we used GST pull-down assays using either recombinant GST-Tagged Tau (GST-Tau (1N4R)) or GST-Tagged BIN1 proteins. GST-Tau pull-down assays with lysates of the HEK293 cell line transiently transfected with BIN1, revealed that BIN1 coprecipitated with GST-Tau but not with GST alone (Figure 4d). Similarly, Tau coprecipitated with GST-BIN1 from lysates of HEK293 cell line overexpressing Tau. The direct BIN1-Tau interaction was validated by GST-BIN1 pull-down assays with recombinant 6xHis-Tagged Tau (6His-Tau (1N4R)) purified protein. Of note, using GST pull-down assays, we showed that Drosophila Amph was also able to interact with human Tau (Figure 4e), indicating that the modulation of the Tau-neurotoxicity readouts by Amph in Drosophila models may be explained by a direct Amph-Tau interaction.
Finally, to confirm BIN1-Tau interaction at the physiological level we performed a reciprocal immunoprecipitations using wild-type mouse brain homogenates. As BIN1 was described to be enriched in nerve-terminals,8 we performed reciprocal reciprocal immunoprecipitations using synaptosomal fractions. We observed that endogenous BIN1 and Tau coimmunoprecipitated confirming this interaction at a physiological level (Figure 4f).
Insertion/deletion rs59335482 is associated with an increase of Tau pathology in AD brains
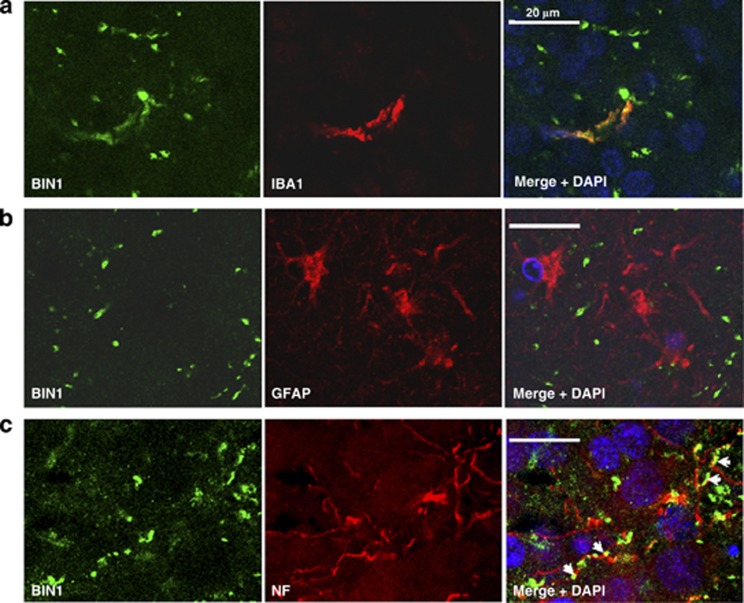
Next, to better define the link between BIN1 and Tau in the AD context, we performed immunohistochemical analysis in the brain of AD cases. BIN1 expression was rarely observed in microglia, was not detectable in astrocytes (Figures 5a and b) but was mainly present in neurons where it followed the neurofilament labelling in axons (Figure 5c). This observation in the human brain was consistent with a previous report showing BIN1 expression in axonal segments in rat brain.17 On the other hand, we observed no consistent colabelling between neurofibrillary tangles and BIN1 (data not shown), although the functional rs59335482 insertion risk allele was associated with Tau loads but not with Aβ40 and Aβ42 loads in the brains of AD patients (Table 2). Taken together, these observations suggest that increased BIN1 expression might modulate Tau pathology in pre-pathological stages.
Figure 5.
Representative confoncal images of BIN1 staining in AD brains. Representative confoncal images of BIN1 staining in hippocampus from AD patients (Braak VI). (a) Anti-IBA1 and (b) anti-GFAP antibodies have been used as microglia and astrocytes markers, respectively. (c) Representative confocal images of BIN1 staining in AD brains. BIN1 staining followed the neurofilament (NF) labeling in axons (arrows).
Table 2. Association of rs59335482 with pathological hallmark lesions in AD brains.
| rs59335482 | DD (n=31) | ID (n=30) | II (n=6) | P-value |
|
|---|---|---|---|---|---|
| Model 1 | Model 2 | ||||
| Pathological hallmarks of AD | |||||
| Tau Loads | 2.5±2.7 | 4.6±4.2 | 5.7±3.2 | 0.03 | 0.009 |
| Aβ40 loads | 3.2±3.2 | 3.9±4.2 | 4.1±3.8 | 0.70 | 0.40 |
| Aβ42 Loads | 10.3±4.3 | 9.7±3.9 | 8.4±3.3 | 0.55 | 0.40 |
Abbreviations: AD, Alzheimer's disease; D, deletion; I, insertion.
Analysis was performed using a non parametric Wilcoxon test following recessive or additive models (respectively model 1 and model 2).
Discussion
GWASs in common disorders such as AD identify a large number of novel risk-increasing loci without predetermined ideas about their functions. As a consequence, our understanding of the pathophysiological pathways involved in AD might be significantly revised in the light of the new general picture of AD genetics. However, to gain a detailed understanding of the functional link between these genetic determinants and AD is extremely challenging as it not only requires the identification of the risk gene and its functional risk variant(s), but also its role in the pathophysiological process. The novel BIN1 AD risk locus clearly illustrates this point as the genetic and molecular mechanisms, by which it affects AD pathophysiology are completely unknown. In this context, the objective of this multidisciplinary study was to pave the way for our understanding of BIN1 involvement in the AD pathophysiological process.
First, we obtained evidence that a deregulation of BIN1 expression is likely pathological: (i) BIN1 is over-expressed in the brain of AD cases; (ii) a functional variant, rs59335482, associated with increased transcriptional activity in vitro could mediate AD risk by increasing BIN1 cerebral expression in vivo; (iii) loss of the Drosophila BIN1 ortholog Amph was able to suppress Tau-induced neurotoxicity in line with a deleterious role of increased BIN1 expression in the AD brain.
Second, our data further strongly suggest that BIN1 modulates AD pathogenesis at the level of the Tau pathway. We were able to identify associations between Tau and BIN1 at the biochemical level (in vitro and in vivo), the genetic, and neuropathological level.
In conclusion, we found that genetically determined increased BIN1 expression increases AD risk and that this interferes at the level of Tau biology. However, even if our study allowed us to shed light on the molecular mechanism by which BIN1 might modulate AD pathophysiology, there are several caveats to be taken into account.
At the genetic level, even if rs4663105 and rs6733839 are the best associated SNP in terms of P-value, our results seem to indicate that they are likely not functional. In this context it is important to keep in mind that P-value (the level of association) is dependent on the sample size, the magnitude of association (OR) but also the allele frequency. Furthermore, as the GWAS-defined genes are generally associated with a low magnitude of association (OR≈1.20), slight variations in sample size and allele frequencies in particular can have strong consequences when calculating P-values. In others words, although SNPs can exhibit similar ORs, P-values can fluctuate and are not enough to discriminate for genetic variants explaining a locus association. This clearly points out the difficulty to exhaustively characterize functional polymorphisms responsible for a locus association with a disease risk.
Importantly, rs59335482 likely explains a part of the BIN1 signal detected by the GWASs. As rs59335482 was in almost complete LD with rs744373 (D′=0.98, r2=0.94), for example, the signal reported as associated with AD risk in the initial paper of Seshadri et al.,3 the two polymorphisms likely reflect the same signal. Of note, rs744373 association with AD risk in EADI1, GERAD1 and in the Flanders–Belgian case-control study resulted in a genome-wide significant meta-analysed OR of 1.17 (1.11–1.24) (P=4.7 × 10−8) (Supplementary Figure 3). In contrast, rs59335482 was in incomplete LD with rs4663105 (D′=0.98, r2=0.56) and rs6733839 (D′=0.94, r2=0.47) probably due to the difference between MAF of rs59335482 (0.27) and rs4663105 (0.40) and rs6733839 (0.39). However, we cannot exclude that in addition to rs59335482, there are likely multiple functional alleles in the BIN1 locus to be characterized and responsible for its association with AD risk. Nevertheless, our data indicate that rs59335482 explains the original rs744373 association and thus at least partly explains the BIN1 association with AD risk. In addition, the functional genetic data indicating increased BIN1 expression are in agreement with increased BIN1 expression in AD brains and with rs59335482 allele specific increased expression data in human brains and lymphoblasts.
At the biological level, the exact pathogenic mechanism(s) linking Tau, BIN1 and the AD pathophysiological process remain(s) to be determined. For instance, it is possible that BIN1 modulates (i) microtubule stability, (ii) Tau phosphorylation/aggregation or (iii) neurofibrillary tangle formation. The two first points are supported by previous reports showing that BIN1 can stabilize T-tubule structure in muscles18 or have a role in protein phosphorylation.19, 20 The last possibility seems less probable as we did not detect any colocalisation of BIN1 with neurofibrillary tangles. This observation might indicate that BIN1 does not have a role in late stages of the Tau-related pathology development and might affect Tau aggregation/oligomer formation in earlier disease stages. Of note, several GWAS have been published on tauopathies such as Parkinson's disease, progressive supranuclear palsy or frontotemporal dementia.21, 22, 23 None of them reported BIN1 as a locus reaching genome-wide significance. Even if we can not exclude that these studies did not have enough statistical power to detect a potential association of BIN1 with these diseases, this could imply that BIN1 might be involved in an AD-specific pathological process linking amyloid and Tau pathology.
In conclusion, we found that genetically determined increased BIN1 expression increases AD risk and that this interferes with Tau biology. If confirmed, BIN1 would be the first genetic risk factor for AD linked to the ‘Tau pathway'.
Acknowledgments
The work was made possible by the generous participation of the control subjects, the patients, and their families. This work was supported by the National Foundation for Alzheimer's disease and related disorders, the Institut Pasteur de Lille, the Centre National de Génotypage, Inserm, FRC (fondation pour la recherche sur le cerveau) and Rotary. This work has been developed and supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant (Development of Innovative Strategies for a Transdisciplinary approach to ALzheimer's disease). FH was funded by the Alzheimer's Association (Grant IIRG-06-25487) and the ‘Ligue contre la Maladie d'Alzheimer (LECMA; Grant 09705). MG and LVB are PhD fellows of the Institute for the Promotion of Innovation by Science and Technology (IWT) in Flanders. CB received funding from the fondation pour la recherché médicale. AM received funding from the INSERM and the Nord-Pas de Calais Regional Council. JC was funded by the MEDIALZ Project (Grant 11001003) financed by ERDF (European Regional Development Fund) and Conseil Régional Nord Pas de Calais. FL received funding from the university of Lille II and the Nord-Pas de Calais Regional Council. BD received support from the Foundation for Alzheimer Research (SAO/FRMA) in Belgium, the Foundation for Research Flanders in Belgium (FWO). Research on Drosophila dementia models at INSERM U744 is supported by grants from the Région Nord/Pas-de-Calais and European Regional Development Funds ERDF (N0 11005007), France, to B D and from the Fondation Plan Alzheimer, France, to J-C L and BD. PC was supported by Interuniversity Attraction Poles program of the Belgian Science Policy. We thank the TRiP at Harvard Medical School for providing transgenic RNAi fly stocks and/or plasmid vectors used in this study. ID is supported by the Fonds de la Recherche Scientifique (FNRS), Belgium. DM and ID were sponsored by the IWT-Flanders. The research at the Antwerp site was in part supported by the international Consortium of Centers of Excellence in Neurodegeneration, the Interuniversity Attraction Poles program of the Belgian Science Policy Office, Foundation for Alzheimer Research (SAO/FRMA), a Methusalem Excellence Grant of the Flemish Government, the FWO, the University of Antwerp and the Special Research Fund of the University of Antwerp, the Antwerp Medical Research Foundation and Neurosearch, Belgium. KS and KB received a postdoctoral fellowship and RV a senior clinical investigator mandate of the FWO. Acknowledged are Karolien Bettens for her contributions to the genetic analyses of BIN1 and Nathalie Le Bastard, Sylvie Slaets and Jill Luyckx for the biomarker analyses in CSF. Also acknowledged are the contributions by the personnel of the Departments of Neurology and Memory Clinics of the Hospital Network Antwerp Middelheim and Hoge Beuken and the University Hospitals Leuven Gasthuisberg, the VIB Genetic Service Facility and the Biobank of the Institute Born-Bunge. The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France and the joint French Ministry of Research/INSERM ‘Cohortes et collections de données biologiques' programme. Lille Génopôle received an unconditional grant from Eisai. This study incorporated summary results from the GERAD1 genome-wide association study. GERAD1 Acknowledgements: Cardiff University was supported by the Wellcome Trust, Medical Research Council (MRC), Alzheimer's Research Trust (ART) and the Welsh Assembly Government. ART supported sample collections at the Kings College London, the South West Dementia Bank, Universities of Cambridge, Nottingham, Manchester and Belfast. The Belfast group acknowledges support from the Alzheimer's Society, Ulster Garden Villages, N.Ireland R&D Office and the Royal College of Physicians/Dunhill Medical Trust. The MRC and Mercer's Institute for Research on Ageing supported the Trinity College group. The South West Dementia Brain Bank acknowledges support from Bristol Research into Alzheimer's and Care of the Elderly. The Charles Wolfson Charitable Trust supported the OPTIMA group. Washington University was funded by NIH grants, Barnes Jewish Foundation and the Charles and Joanne Knight Alzheimer's Research Initiative. Patient recruitment for the MRC Prion Unit/UCL Department of Neurodegenerative Disease collection was supported by the UCLH/UCL Biomedical Centre. LASER-AD was funded by Lundbeck SA. The Bonn group was supported by the German Federal Ministry of Education and Research (BMBF), Competence Network Dementia and Competence Network Degenerative Dementia, and by the Alfried Krupp von Bohlen und Halbach-Stiftung. The GERAD1 Consortium also used samples ascertained by the NIMH AD Genetics Initiative. The KORA F4 studies were financed by Helmholtz Zentrum München; German Research Center for Environmental Health; BMBF; German National Genome Research Network and the Munich Center of Health Sciences. The Heinz Nixdorf Recall cohort was funded by the Heinz Nixdorf Foundation (Dr jur G Schmidt, Chairman) and BMBF. Coriell Cell Repositories is supported by NINDS and the Intramural Research Program of the National Institute on Aging. We acknowledge use of genotype data from the 1958 Birth Cohort collection, funded by the MRC and the Wellcome Trust which was genotyped by the Wellcome Trust Case Control Consortium and the Type-1 Diabetes Genetics Consortium, sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development and Juvenile Diabetes Research Foundation International.
Dr Lambert reports being a consultant to Genoscreen.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Mann D, Richard F, Tian J, Shi J, Thaker U, et al. Is there a relation between APOE expression and brain amyloid load in Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2005;76:928–933. doi: 10.1136/jnnp.2004.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr C, Lambert JC, Sazdovitch V, Amouyel P, Chartier-Harlin MC, Mohr M, et al. Neuropathological epidemiology of cerebral aging: a study of two genetic polymorphisms. Neurobiol Aging. 2001;22:227–235. doi: 10.1016/s0197-4580(00)00227-x. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Grenier-Boley B, Harold H, Zelenika D, Chouraki V, Kamatani Y, et al. Genome-wide haplotype association study identifies the FRMD4A gene as a risk locus for Alzheimer's disease Mol Psychiatry 2012. in press. [DOI] [PMC free article] [PubMed]
- Meunier B, Quaranta M, Daviet L, Hatzoglou A, Leprince C. The membrane-tubulating potential of amphiphysin 2/BIN1 is dependent on the microtubule-binding cytoplasmic linker protein 170 (CLIP-170) Eur J Cell Biol. 2009;88:91–102. doi: 10.1016/j.ejcb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Nagy A, Delgado-Escueta AV. Rapid preparation of synaptosomes from mammalian brain using nontoxic isoosmotic gradient material (Percoll) J Neurochem. 1984;43:1114–1123. doi: 10.1111/j.1471-4159.1984.tb12851.x. [DOI] [PubMed] [Google Scholar]
- Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, et al. Transcriptomic and genetic studies identify IL-33as a candidate gene for Alzheimer's disease. Mol Psychiatry. 2009;14:1004–1016. doi: 10.1038/mp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis PA, Chow BM, Stewart BA, Iyengar B, Campos AR, Boulianne GL. Drosophila Amphiphysin is a post-synaptic protein required for normal locomotion but not endocytosis. Traffic. 2001;2:839–850. doi: 10.1034/j.1600-0854.2001.21113.x. [DOI] [PubMed] [Google Scholar]
- Razzaq A, Robinson IM, McMahon HT, Skepper JN, Su Y, Zelhof AC, et al. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther DC, Kinghorn KJ, Miranda E, Page R, Curry JA, Duthie FA, Gubb DC, Lomas DA. Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer's disease. Neuroscience. 2005;132:123–135. doi: 10.1016/j.neuroscience.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Yeh PA, Chien JY, Chou CC, Huang YF, Tang CY, Wang HY, et al. Drosophila notal bristle as a novel assessment tool for pathogenic study of Tau toxicity and screening of therapeutic compounds. Biochem Biophys Res Commun. 2010;391:510–516. doi: 10.1016/j.bbrc.2009.11.089. [DOI] [PubMed] [Google Scholar]
- Kosmidis S, Grammenoudi S, Papanikolopoulou K, Skoulakis EM. Differential effects of Tau on the integrity and function of neurons essential for learning in Drosophila. J Neurosci. 2010;30:464–477. doi: 10.1523/JNEUROSCI.1490-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MH, David C, Ochoa GC, Freyberg Z, Daniell L, Grabs D, et al. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BS, Toussaint A, Muller J, Laporte J. Defective membrane remodeling in neuromuscular diseases: insights from animal models. PLoS Genet. 2012;8:e1002595. doi: 10.1371/journal.pgen.1002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumi A, Aizaki H, Suzuki T, DuHadaway JB, Prendergast GC, Komuro K, et al. Reduction of hepatitis C virus NS5A phosphorylation through its interaction with amphiphysin II. Biochem Biophys Res Commun. 2005;336:572–578. doi: 10.1016/j.bbrc.2005.08.142. [DOI] [PubMed] [Google Scholar]
- Pineda-Lucena A, Ho CS, Mao DY, Sheng Y, Laister RC, Muhandiram R, et al. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J Mol Biol. 2005;351:182–194. doi: 10.1016/j.jmb.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.