Circulating tumor cells (CTCs)[1] are cancer cells shed from either the primary tumors or metastatic sites. The presence and number of CTCs in peripheral blood can provide clinically significant data on prognosis and therapeutic response patterns, respectively[2]. Thus, as with traditional invasive tumor biopsies that enable gold-standard pathological analysis, CTCs can be regarded as “liquid biopsies” of the tumor, which enable repeated and relatively non-invasive characterization of tumor evolution, especially important during therapeutic interventions. Currently, FDA-cleared CellSearch™ Assay is costly and inefficient in capturing CTCs, and the enriched CTCs are typically contaminated with a large number of white blood cells (WBCs). As a result, the diagnostic value of CTCs has been underutilized. Over the past decade, a diversity of CTC detection technologies[2d, 3] have been developed to overcome the challenges encountered by the immunomagnetic separation-based CellSearch™ Assay.
Different from the existing CTC diagnostic approaches,[2d, 3] we have demonstrated “NanoVelcro” chips[4] that are capable of enriching, identifying and enumerating CTCs in patient blood samples with superb efficiency. First, we pioneered a unique concept of NanoVelcro substrates,[5] by which anti-EpCAM-coated silicon nanowire (SiNW) substrates were utilized to immobilize CTCs in a stationary device setting. We have recently shown that other types of nanostructured substrates (e.g., electrochemically deposited conjugated polymer nano-features[6] and horizontally packed TiO2 nanofibers[7]) also exhibit enhanced affinity for capturing CTCs, demonstrating the general applicability of NanoVelcro substrates. The uniqueness of our approach is the use of nanostructured substrates: There are enhanced local topographic interactions[8] between the anti-EpCAM-coated nanosubstrates and nanoscaled cellular surface components (e.g., microvilli) on a CTC, which are analogous to the working principle of a velcro. Second, by integrating a lithographically patterned NanoVelcro substrate with an overlaid polydimethyl-siloxane (PDMS) chaotic mixer[9] that enhances contact frequency between flow-through CTCs and the substrate, we further improved CTC capture efficiency.[4] Side-by-side analytical validation studies using both artificial and patient CTC samples suggested that the sensitivity of NanoVelcro Chips outperformed[4] that of CellSearch™. Although NanoVelcro Chips allow efficient and reproducible detection of CTCs in patient blood, challenges remain in 1) broadening its general applicability for detecting other types of solid-tumor CTCs that exhibit surface markers other than EpCAM, and 2) enabling isolation of single CTCs for subsequent molecular analyses. To broaden the general applicability of NanoVelcro-based cell affinity assay, we hereby explored a melanoma-specific capture agent[13] (i.e., anti-CD146) to capture circulating melanoma cells (CMCs, a subcategory of solid-tumor CTCs). Further, an increasing number of studies have shown extensive molecular heterogeneity[10] in CTCs from the same types of cancer or even the same patient[11]. Therefore, it is important to develop single-CTC isolation technologies that enable single-cell molecular analyses in order to better understand the origin and role(s) of CTCs in cancer progression.
Improving upon our previously reported NanoVelcro Chips,[4] we introduce a next-generation NanoVelcro Chip (i.e., poly(lactic-co-glycolic acid) (PLGA)-nanofiber embedded NanoVelcro Chip, abbreviation: “PN-NanoVelcro” Chip, Figure 1a and b) capable of not only capturing CMCs with high efficiency, but also enabling highly specific isolation of single CMCs immobilized on the nanosubstrate without contamination by WBCs. Our idea is to replace the non-transparent SiNW substrate in the earlier NanoVelcro Chip[4] with a transparent PN-NanoVelcro substrates, prepared by depositing electrospun PLGA nanofibers[12] (Figure 1c) onto a commercial laser microdissection (LMD) slide, followed by streptavidin-mediated conjugation of a melanoma-specific capture agent[13] (i.e., anti-CD146, Figure 1d). The unique combination of PLGA nanofibers and a LMD slide enables single-CMC isolation by using a highly accurate LMD technique. We first employed melanoma cell lines to optimize and validate the performance of PN-NanoVelcro Chips. In parallel, a 4-color immunocytochemistry (ICC) protocol[14] was established to specifically identify CMCs among nonspecifically captured WBCs. After the CMC capture and ICC melanoma lineage validation, a LMD microscope was applied to cut and harvest single CMCs for subsequent Sanger sequence analysis. To examine the clinical utility of the optimized PN-NanoVelcro Chips, we then performed single-CMC isolation and genotyping using peripheral blood samples collected from two stage-IV melanoma patients, whose melanomas have been previously characterized by conventional PCR-based diagnostics (cobas® 4800, Roche) to harbor a signature oncogenic mutation (i.e., BRAFV600E). Over the past few years, there has been a paradigm shift in the treatment of metastatic melanoma. BRAF inhibitors (e.g., vemurafenib) designed to target BRAFV600E mutation-driven melanomas have demonstrated unprecedented success, eliciting responses in more than 80% of patients and conferring survival benefits.[15] Thus, the BRAFV600E mutation (present in 60% of melanomas) test, developed as a companion diagnostic to the clinical development of the BRAF inhibitor vemurafenib, provides an indispensable predictive biomarker prior to initiating BRAF inhibitor therapy among melanoma patients.
Figure 1.
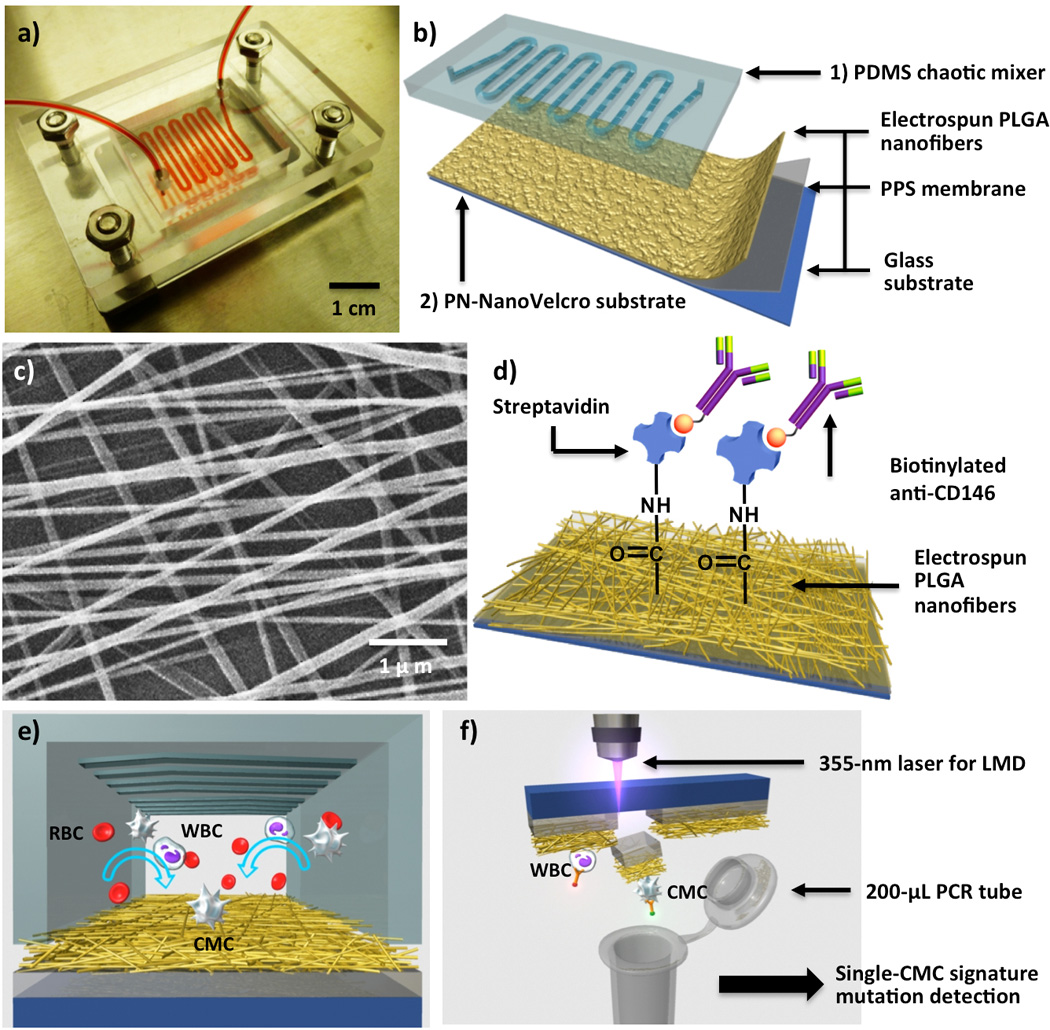
a) Photograph of PN-NanoVelcro Chip for capture and isolation of single circulating melanoma cells (CMCs). b) A PN-NanoVelcro Chip is composed of 1) an overlaid PDMS chaotic mixer and 2) a transparent PN-NanoVelcro substrate. c) SEM image showing the morphologies of electrospun PLGA nanofibers on a PN-NanoVelcro substrate. d) NHS chemistry is employed to covalently anchor streptavidin for conjugation of biotinylated anti-CD146, a melanoma specific-capture agent. e) Conceptual illustration of the operation mechanism of PN-NanoVelcro CMC Chip. f) Graphic representation depicting the LMD-based single CMC isolation in action.
The PN-NanoVelcro Chip is composed of two functional components (Figure 1a and b), including 1) an overlaid PDMS chaotic mixer[4] (Figure 1e) and 2) a transparent PN-NanoVelcro substrate, which were assembled together using a plastic chip holder (using 4 parallel-oriented screws). A 22-cm-long microchannel with embedded herringbone features[9] for increasing cell–substrate contact frequency was fabricated into the PDMS chaotic mixer (Figure 1e). A custom-designed eletrospinning system[7] (see supporting information) was employed to deposit PLGA nanofibers (130.6 ± 62.7 nm in diameter, Figure 1c, also see supporting information) onto a LMD slides (Leica, with a pre-deposited 1.2-µm-thick poly(phenylene) sulfide, PPS membrane) to generate transparent PN-NanoVelcro substrates. NHS chemistry was then employed to activate carboxylic acid groups on the surfaces of PLGA nanofibers for covalent conjugation of streptavidin (Figure 1d). A syringe pump and syringes were utilized to introduce cell suspensions or blood samples, ethanol for fixation and permeabilization and ICC agents into PN-NanoVelcro Chips with controlled flow rates. Prior to the cell-capture studies, biotinylated anti-CD146 (8 µg mL−1, in 500-µL PBS with 1% (w/v) BSA) was freshly grafted onto the PN NanoVelcro substrates.
To optimize and validate the performance of PN-NanoVelcro Chips, we prepared suspensions of the M229 human melanoma cell line[16] (100 cells mL−1 in DMEM medium) as a model system. We first examined how different eletrospinning times affect the CMC-capture efficiency. Under a 1.0-mL h−1 flow rate,[3e, 4, 9] an optimal deposition time of 3 h (Figure 2a, corresponding to a 3-µm-thick PLGA-nanofiber membrane, confirmed by profilometer) led to ca. 87% capture efficiency. By applying 3-µm-thick PLGA-nanofiber membranes in the devices, we then determined that a slower flow rate of 0.5 mL h−1 (Figure 2b) helps to achieve further improved cell-capture performance. We analyzed the spatial distribution (Figure 2c) of substrate-immobilized cells at different locations of the 22-cm serpentine microchannel. At a flow rate of 0.5 mL h−1, 68% of the cells were captured in the first 8 cm of the 22-cm-long microchannel, suggesting that such a channel length is sufficient to achieve the desired cell-capture performance. Further, four control studies using 1) PN-NanoVelcro Chips without anti-CD146, 2) PN-NanoVelcro Chips without herringbone microfeatures,[9] 3) anti-CD146-coated PLGA films (3-µm thick, no nanofeatures), and 4) anti-CD146-coated SiNW substrates[4] were carried out in parallel with CD146-coated PN-NanoVelcro Chips. The results summarized in Figure 2d suggest that the capture agent, microfluidic chaotic mixer[9] and nanostructures play indispensable roles in achieving the superb cell-capture performance. We also tested the dynamic range of PN-NanoVelcro Chips using a series of artificial CMC samples that were prepared by spiking DMEM medium and healthy donors’ blood with DiO-stained M229 cells at densities of 10, 50, 100, 200, 500 and 1,000 cells mL−1. The results (Figure 2e) show that the devices exhibit sufficient performance for clinical samples that usually have a CMC density of few to hundreds cells mL−1. Finally, we tested the general applicability and specificity of PN-NanoVelcro Chips for capturing CD146-positive melanoma cells. Three CD146-positive melanoma cell lines[16] (M229, M229R – a vemurafenib-resistant sub-line derived from M229, and M202) were studied in parallel with two CD146-negative cancer-cell lines (Jurkat leukemia and PC3 prostate cancer cells) and freshly isolated human WBCs. Summarized results in Figure 2f suggest that PN-NanoVelcro Chips were capable of specifically capturing melanoma cells, and its background to non-specifically immobilize other types of cells can be mitigated via the subsequent ICC protocol and fluorescence LMD technique.
Figure 2.
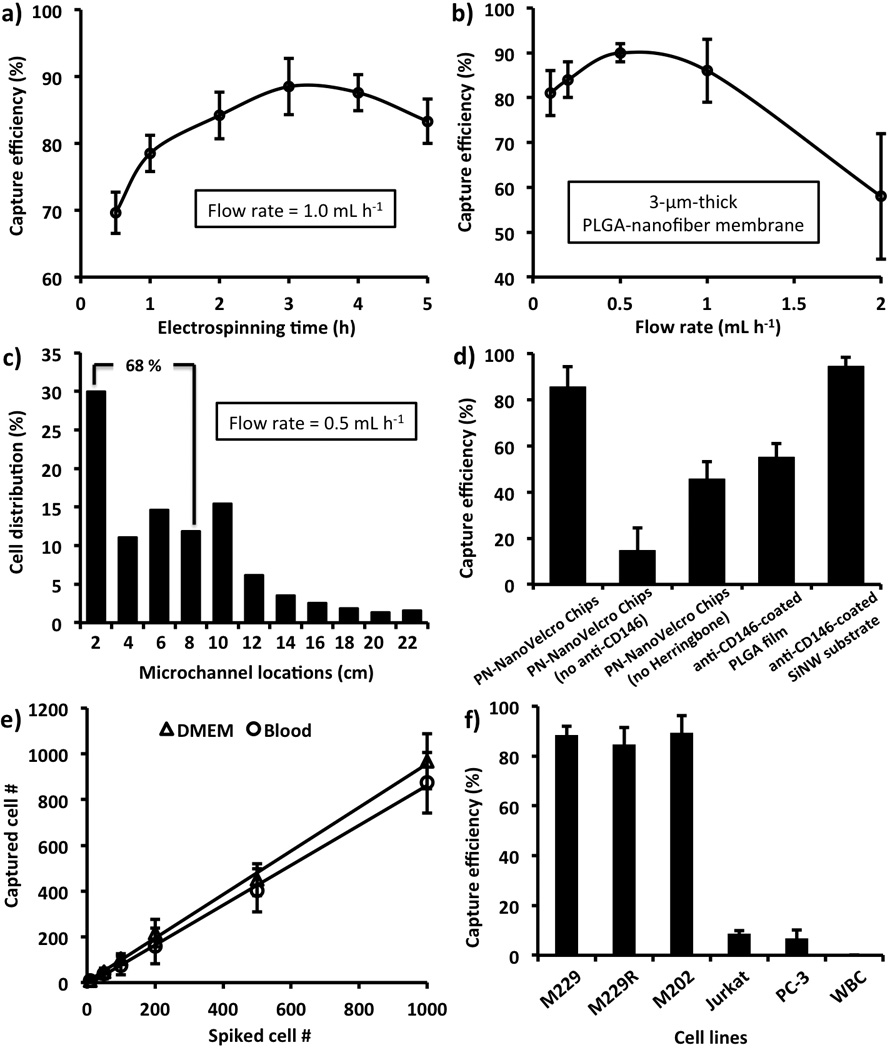
Optimization and validation of PN-NanoVelcro Chips using M229 melanoma cells (Error bars show standard deviations, n ≥ 3): a) Capture efficiency obtained as the function of electrospinning deposition times of 0.5, 1.0, 2.0, 3.0, 4.0 and 5.0 h). b) Based on the optimal electrospinning time of 3h, capture efficiencies were measured at flow rates of 0.1, 0.2, 0.5, 1.0 and 2.0 mL h−1. c) Spatial distribution of substrate-immobilized cells along the serpentine microchannel at the flow rates of 0.5 mL h−1. d) Comparison of cell-capture performance between anti-CD146-coated PN-NanoVelcro Chips with four different control systems, i.e., 1) PN-NanoVelcro Chips without anti-CD146, 2) PN-NanoVelcro Chips without herringbone microfeatures, 3) anti-CD146-coated PLGA films (3-µm thick, no nanofeatures), and 4) anti-CD146-coated SiNW substrates. e) Capture efficiency at different cell numbers ranging from 10 to 1000 cells mL−1 in both DMEM medium and whole blood. f) Capture efficiency using suspensions of melanoma cells (M229, M229R and M202), T-lymphocyte (Jurkat), prostate (PC3), and WBCs.
To examine the clinical utility of PN-NanoVelcro Chips, we performed single-CMC isolation and genotyping on blood samples collected from two stage-IV melanoma patients whose metastatic tumors harbor clinically confirmed BRAFV600E mutation. For comparison, an artificial sample was prepared by spiking 50 M229 cells (with know BRAFV600E mutation) into a 1.0-mL blood sample collected from a healthy donor. After flowing blood samples through PN-NanoVelcro Chips, a 4-color ICC protocol for parallel staining of FITC-labeled anti-Mart1, TRITC-labeled anti-HMW-MAA, DAPI, and Cy5-labeled anti-CD45 was established to identify CMCs (DAPI+/Mart1+/HMW-MAA+/CD45-, 40 µm > diameter > 10 µm) among nonspecifically captured WBCs (DAPI+/Mart1−/HMW-MAA−/CD45+, diameter < 10 µm) and cellular debris immobilized on PN-NanoVelcro substrates. After performing ICC, the overlaid microfluidic chaotic mixer was removed, and the transparent PN-NanoVelcro substrate with immobilized CMCs were mounted onto a fluorescent microscope (Nikon Ni) for identification, registration and enumeration of CMCs. 45, 43 and 36 CMCs were captured and identified from the 1.0-mL artificial control sample, patient #1 and #2 blood samples, respectively. Subsequently, a fluorescent microscope (Leica LMD7000) equipped with a 355-nm LMD setup was employed to cut (Figure 3b) the PLGA nanofibers along with the supporting PPS membrane located underneath the pre-registered CMCs, allowing the collection of a single CMC into a 200-µL PCR tube for whole genome amplification and sequence analysis. Among the 45, 43, and 36 captured CMCs, 30, 24 and 18 CMCs were isolated by LMD for individual analyses.
Figure 3.
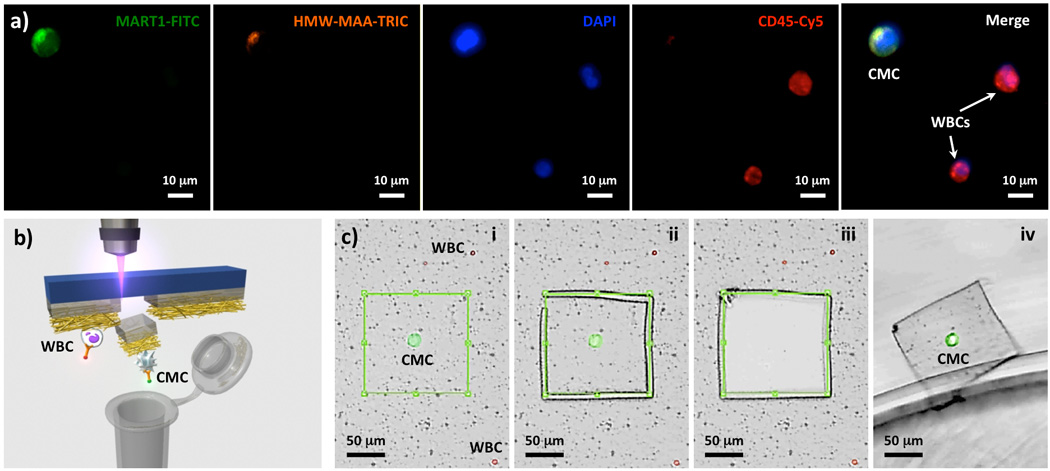
a) Typical micrographs of a CMC and WBCs immobilized on a PN-NanoVelcro Chip. A 4-color immunocytochemistry (ICC) protocol for parallel staining of FITC-labeled anti-Mart1, TRITC-labeled anti-HMW-MAA, DAPI and Cy5-labeled anti-CD45 was established for identification of substrate-immobilized CMCs (DAPI+/Mart1+/HMW-MAA+/CD45-, 40 µm > diameters > 10 µm) from nonspecifically captured WBCs (DAPI+/Mart1−/HMW-MAA−/CD45+, diameters < 10 µm) and cellular debris. b) and c) Micrograph images recording stepwise operation of LMD-based single CMC isolation: i) identification of a CMC, ii) laser-dissecting the CMC, iii) and iv) releasing the CMC from the substrate, followed by harvesting into a 200-µL PCR tube.
A commercial whole genome amplification[11] (WGA kit, Sigma/Aldrich) was employed to amplify genomic DNA from each CMC, and the resulting WGA DNA was then subjected to specific PCR amplification in the presence of BRAF exon 15 (containing the BRAFV600 codon)-specific primers.[16b] The double-amplified gDNA was then subjected to Sanger sequencing (Figure 4). We were able to detect BRAFV600E mutation from the M229-based control sample and CMCs isolated from both patient #1 and #2 blood samples. The BRAFV600E identified in the CMCs are consistent with those observed in their tumor biopsies. As a negative control, WBCs isolated from the two melanoma patients showed no BRAFV600E mutation. It is noteworthy that the Sanger sequencing data obtained for the BRAFV600E mutation in single CMCs (Figure 4) displayed a strong signal-to-noise ratio. In contrast, various levels of sequencing noise and BRAFV600E zygosity are often encountered from sequencing of traditional melanoma biopsies as a result of standard tissue fixation and tissue heterogeneity (“contamination” with stromal and immune cell types).
Figure 4.
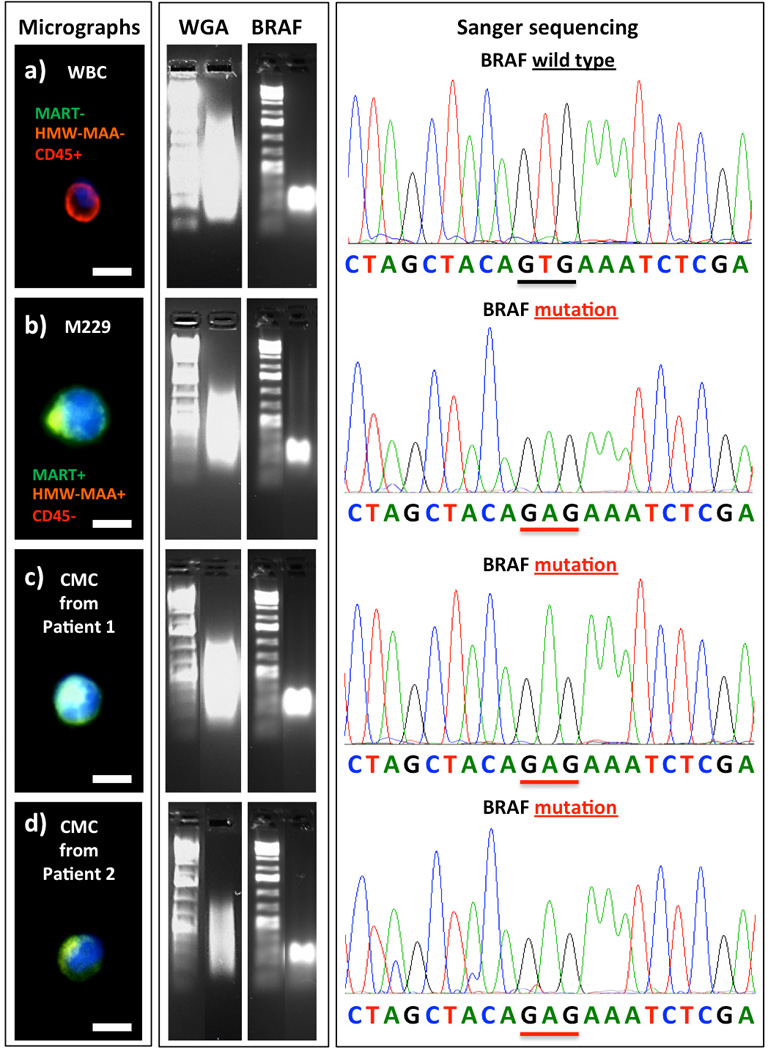
Left: Fluorescent micrographs of a negative control WBC, a M229 melanoma cell isolated from control blood sample, and two CMCs isolated from patient #1 and #2. Middle: Results from whole genome amplification (WGA) and PCR amplification using a BRAF-specific primer. Right: Sanger sequencing of the individually isolated WBC, M229 cell and the two CMCs. (Scale bar: 15 µm)
In conclusion, the continuous development of NanoVelcro CTC Assay, including replacement of the original non-transparent SiNW NanoVelcro substrate by a new transparent PN-NanoVelcro substrate to enable laser-assisted microdissection, has facilitated single-tumor cell isolation in addition to its outstanding CMC-capture performance. Furthermore, we were able to extend the applicability of this new PN-NanoVelcro Chip beyond epithelial cancers to specifically detect and isolate CMCs. Our proof-of-concept study validated the feasibility of performing a streamlined process starting from CMC detection, isolation, and all the way to single-CMC genotyping for a key melanoma drug target, the BRAFV600E mutation. Most importantly, the BRAFV600E mutation detected in single CMCs matched that detected in the patients’ tumor biopsies, supporting a positive correlation between CTCs and their tumor origin.
Supplementary Material
Footnotes
This research was supported by the National Institutes of Health (R21 CA151159 and R33 CA157396). Dr. F. Charles Brunicardi, Dr. Antoni Ribas, Dr. Roger S. Lo and Dr. Hsian-Rong Tseng are members of UCLA Josson Comprehensive Cancer Center. They acknowledge JCCC’s generous support and promotion of the teamwork.
Contributor Information
Shuang Hou, Email: hrtseng@mednet.ucla.edu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA, Web: http://www.tseng-lab.com.
Libo Zhao, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
Qinglin Shen, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA; Department of Applied Physics and Department of Oncology Surgery, Wuhan University, Wuhan, PRC.
Juehua Yu, Department of Surgery, University of California, Los Angeles.
Charles Ng, Division of Hematology and Oncology, Department of Medicine, Department of Surgery, and Department of Molecular and Medical Pharmacology, University of California, Los Angeles.
Xiangju Kong, Division of Dermatology, Department of Medicine, University of California, Los Angeles.
Dongxia Wu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
Min Song, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
Xiaohong Shi, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
Xiaochun Xu, CytoLumina Technologies Corp., 21038 Commerce Point Dr., Walnut, CA 91789, USA.
Wei-Han OuYang, CytoLumina Technologies Corp., 21038 Commerce Point Dr., Walnut, CA 91789, USA.
Rongxian He, Department of Applied Physics and Department of Oncology Surgery, Wuhan University, Wuhan, PRC.
Xing-Zhong Zhao, Department of Applied Physics and Department of Oncology Surgery, Wuhan University, Wuhan, PRC.
Tom Lee, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
F. Charles Brunicardi, Department of Surgery, University of California, Los Angeles.
Mitch André Garcia, Email: mgarcia@cytolumina.com, CytoLumina Technologies Corp., 21038 Commerce Point Dr., Walnut, CA 91789, USA.
Antoni Ribas, Email: aribas@mednet.ucla.edu, Division of Hematology and Oncology, Department of Medicine, Department of Surgery, and Department of Molecular and Medical Pharmacology, University of California, Los Angeles.
Roger S. Lo, Email: rlo@mednet.ucla.edu, Division of Dermatology, Department of Medicine, University of California, Los Angeles.
Hsian-Rong Tseng, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770, USA.
References
- 1.a) Bernards R, Weinberg RA. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]; b) Pantel K, Brakenhoff RH. Nat. Rev. Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]; c) Pantel K, Alix-Panabieres C. Trends Mol. Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]; d) Kaiser J. Science. 2010;327:1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]; e) Criscitiello C, Sotiriou C, Ignatiadis M. Curr. Opin. Oncol. 2010;22:552–558. doi: 10.1097/CCO.0b013e32833de186. [DOI] [PubMed] [Google Scholar]
- 2.a) Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]; b) Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Clin. Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]; c) Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI. Clin. Cancer Res. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]; d) Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Am. J. Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J. Chromatogr. A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]; c) Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Biomed. Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]; e) Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Tan SJ, Lakshmi RL, Chen P, Lim WT, Yobas L, Lim CT. Biosens. Bioelectron. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]; h) Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal. Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Dickson MN, Tsinberg P, Tang ZL, Bischoff FZ, Wilson T, Leonard EF. Biomicrofluidics. 2011:5. doi: 10.1063/1.3623748. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Br. J. Cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]; l) Riethdorf S, Pantel K. Ann. N. Y. Acad. Sci. 2010;1210:66–77. doi: 10.1111/j.1749-6632.2010.05779.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng H-R. Angew. Chem. Int. Ed. Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng H-R. Angew. Chem. 2011;123:3140–3144. [Google Scholar]
- 5.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng H-R. Angew. Chem. Int. Ed. Engl. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng H-R. Angew. Chem. 2009;121:9132–9135. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekine J, Luo SC, Wang S, Zhu B, Tseng H-R, Yu HH. Adv. Mater. 2011;23:4788–4792. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Adv. Mater. 2012;24:2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 8.a) Fischer KE, Aleman BJ, Tao SL, Daniels RH, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Curtis ASG, Varde M. J. Natl. Cancer. Inst. 1964;33:15. [PubMed] [Google Scholar]; c) Liu WF, Chen CS. Adv. Drug Deliv. Rev. 2007;59:1319–1328. doi: 10.1016/j.addr.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 10.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, He W, Zeng L, Xing M, Wu R, Jiang H, Liu X, Cao D, Guo G, Hu X, Gui Y, Li Z, Xie W, Sun X, Shi M, Cai Z, Wang B, Zhong M, Li J, Lu Z, Gu N, Zhang X, Goodman L, Bolund L, Wang J, Yang H, Kristiansen K, Dean M, Li Y. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Shin HJ, Lee CH, Cho IH, Kim YJ, Lee YJ, Kim IA, Park KD, Yui N, Shin JW. J Biomater Sci Polym Ed. 2006;17:103–119. doi: 10.1163/156856206774879126. [DOI] [PubMed] [Google Scholar]; b) Xin X, Hussain M, Mao JJ. Biomaterials. 2007;28:316–325. doi: 10.1016/j.biomaterials.2006.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kim SJ, Jang DH, Park WH, Min BM. Polymer. 2010;51:1320–1327. [Google Scholar]
- 13.Anti-CD146 is a melanoma-specific capture agnet used in CellSearch™ circulating melanoma cell assay. See its clinical studies in: Rao C, Bui T, Connelly M, Doyle G, Karydis I, Middleton MR, Clack G, Malone M, Coumans FA, Terstappen LW. Int. J. Oncol. 2011;38:755–760. doi: 10.3892/ijo.2011.896.
- 14.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Bollag G, Hi-th P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KYJ, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D/'Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Hersey P, Kefford R, Lawrence D, Puzanov I, Lewis KD, Amaravadi RK, Chmielowski B, Lawrence HJ, Shyr Y, Ye F, Li J, Nolop KB, Lee RJ, Joe AK, Ribas A. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, Sazegar H, MacConaill LE, Barretina JG, Kehoe SM, Attar N, von Euw E, Zuckerman JE, Chmielowski B, Comin-Anduix B, Koya RC, Mischel PS, Lo RS, Ribas A. J. Transl. Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Shi H, Kong X, Ribas A, Lo RS. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, Solit DB. Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


