Summary
Several mechanisms that increase the rate of mutagenesis across the entire genome have been identified; however, how the rate of evolution might be promoted in individual genes is unclear. A majority of the genes in bacteria are encoded on the leading strand of replication1–4. This presumably avoids the potentially detrimental head-on collisions that occur between the replication and transcription machineries when genes are encoded on the lagging strand1–4. We identified the ubiquitous (core) genes in Bacillus subtilis and determined that 17% of them are on the lagging strand. We found a higher rate of point mutations in the core genes on the lagging strand compared to those on the leading strand, with this difference being primarily in the amino acid changing (nonsynonymous) mutations. We determined that overall, the genes under strong negative selection against amino acid changing mutations tend to be on the leading strand, co-oriented with replication. In contrast, based on the rate of convergent mutations, genes under positive selection for amino acid changing mutations are more commonly found on the lagging strand, indicating faster adaptive evolution in many genes in the head-on orientation. Increased gene length and gene expression levels are positively correlated with the rate of accumulation of nonsynonymous mutations in the head-on genes, suggesting that the conflict between replication and transcription could be a driving force behind these mutations. Indeed, using reversion assays, we show that the difference in the rate of mutagenesis of genes in the two orientations is transcription-dependent. Altogether, our findings indicate that head-on replication-transcription conflicts are more mutagenic than co-directional conflicts and that these encounters can significantly increase adaptive structural variation in the coded proteins. We propose that bacteria, and potentially other organisms, promote faster evolution of specific genes through orientation-dependent encounters between DNA replication and transcription.
Concurrent DNA replication and transcription leads to conflicts that stall replication, especially on the lagging strand, where the two machineries meet head-on (Supplementary Figure 1) 5–8. Presumably, to avoid these encounters that can delay replication, bacteria have co-oriented the majority of their genes with replication by encoding them on the leading strand1,2. However, it is unclear why, even in species with a strong orientation bias, like B. subtilis, 25% of all genes and 6% of all essential genes remain on the lagging strand1,2.
We investigated the relationship between gene orientation and mutagenesis, by analyzing the rate of mutations of ubiquitous (core) genes from five clonally-divergent strains of B. subtilis (Supplementary Table 1). To be defined as a core gene, the gene must be present in all five strains and have at least 95% nucleotide identity, and at least 95% of the length coverage, to exclude the highly diverse genes affected by non-homologous gene shuffling9. Among the 759 core genes, 132 (17%) were encoded on the lagging strand (Supplementary Table 2). Out of the 148 core genes that were determined to be essential10, only 6 (4%) were on the lagging strand, consistent with the previously described strong orientation bias in the essential genes1,2.
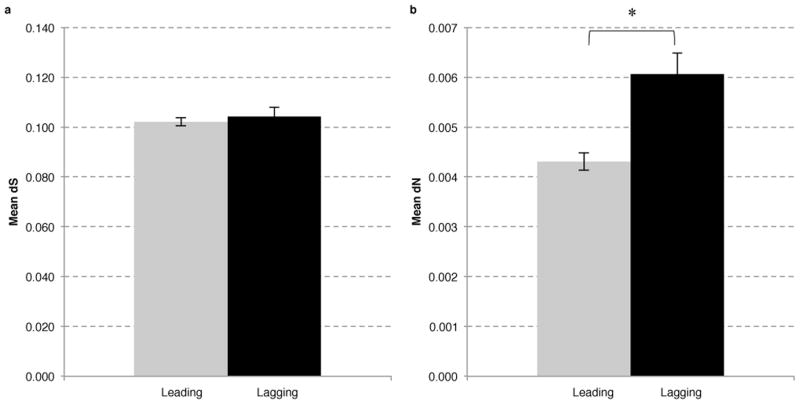
We compared the rates of silent or synonymous (dS), and amino acid (aa) changing or nonsynonymous (dN) mutations, of the core genes on the leading and lagging strands. The rate of synonymous mutation on the lagging is marginally (2%) higher than on the leading strand (Figure 1A). In contrast, the rate of nonsynonymous mutations of the lagging strand genes is 42% higher than the genes on the leading strand (Figure 1B, p<0.0001), without a significant number of outliers (Supplementary Figure 2). The relative increase of dN in the genes on the lagging compared to the leading strand was irrespective of the dS level of the corresponding genes (Supplementary Figure 3). We found a similar trend when we analyzed the mutagenesis rate in all leading or lagging strand genes, i.e. besides the core genes, on the two strands (Supplementary Table 3). However, among the core genes that were previously identified as essential, we did not detect a difference in structural variability between the two strands (Supplementary Figure 4).
Figure 1. Highly conserved “core” genes show a higher rate of nonsynonymous mutations on the lagging compared to the leading strand.
The mean values for the core genes in relation to diversity are presented. The dS (A) and dN (B) values are plotted for the leading (gray) and lagging (black) strands. Error bars represent the standard error of the mean. A statistically significant difference in dN between the two strands is detected with a p<0.0001 (*). Analysis of statistical significance was performed using the z-test for dS and dN values (see methods).
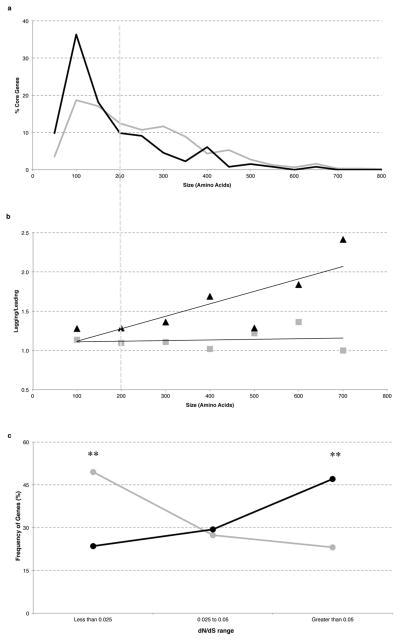
The observed difference between the rates at which leading and lagging strand genes vary, may be due to orientation-dependent encounters between replication and transcription. It was previously suggested that transcript length contributes to severity of conflicts based on the observation that gene operons oriented head-on tend to be of a relatively small size4. Interestingly, we found that the vast majority (82%) of core genes on the lagging strand are not grouped together, and are organized as single genes rather than operons (Supplementary Figure 5). Moreover, there is a significant bias for genes on the lagging strand to be significantly shorter on average, with the average gene size being 48% larger on the leading strand (681 vs. 459 bp). In particular, genes coding for proteins with a length of larger than 200 aas are underrepresented on the lagging strand, with only 26% of genes (34 out of 132 genes) being of this size category, whereas 48% of genes exceed 200 aas on the leading strand (Figure 2A, p<0.0001). These data are consistent with selection for genes on the lagging strand to be shorter, potentially to decrease the frequency of head-on conflicts.
Figure 2. For head-on genes, a significant positive correlation exists between increased length, mutagenesis, and positive selection.
(A) The percentage of genes in each size category for lagging (black) and leading (gray) strands within increasing windows of 50 aas. (B) The fold difference between either dN (black) or dS (gray) for lagging compared to the leading strand genes in seven size categories (R2=0.65 for dN and 0.02 for dS, over length). (C) dN/dS ratios for genes bigger than 200 aas on the leading strand (gray) and lagging strand (black) (**p<0.005). p values were determined using a 2×2 χ2 test.
Because gene size likely correlates with conflict level, we analyzed the relationship between nonsynonymous mutation rates and gene length. Indeed, there was a significant positive correlation between increased length and increased dN on the lagging compared to the leading strand (Figure 2B), with the relative difference in dN being most apparent amongst the genes coding for proteins >200 aas. In this size category (303 genes on the leading and 34 genes on the lagging strand), we also find a positive correlation between transcript abundance and dN, with the mean expression values being three-fold higher in the group with higher dN levels compared to those with the lower dN levels only in the lagging strand genes (Supplementary Figure 6). Together, the positive correlation between increased mutation rates with gene length, as well as expression levels, suggest that replication-transcription conflicts are likely responsible for the increased mutation rates in the lagging strand genes.
We examined the ratio of dN to dS, to evaluate the relative strength of negative selection against structural changes in the genes coding proteins >200aas in length. A significantly higher proportion of the genes on the leading strand (about half) are in the very low dN/dS range of <.025 when compared to those on the lagging strand (p=0.004, Figure 2C). Furthermore, there are 25 genes on the leading strand but none on the lagging strand that lack any structural variation at all. Thus, genes that are under especially strong selection against nonsynonymous changes are heavily biased to be co-oriented with replication. In contrast, almost half of the genes on the lagging strand had a relatively higher dN/dS range of >.075, significantly more than that on the leading strand (p=.002). To determine whether the increased dN/dS on the lagging strand genes could be in part due to selection for nonsynonymous mutations, we looked for the presence of convergent amino acid mutations, which is strong evidence for positive selection and adaptive evolution11–13. Amongst the genes encoding proteins >200 aas, convergent mutations were detected in 24% of the core genes on the lagging strand in contrast to only 11% of the genes on the leading strand (P<0.04) (Table 1 and Supplementary Table 4), indicating faster adaptive evolution of the corresponding genes.
Table 1.
Distribution of genes with convergent amino acid mutations in lagging and leading strands.
| Strand | Number of genes* | Genes with convergent amino acid mutations | 2×2 χ2 P-value |
|---|---|---|---|
| Lagging | 34 | 8 | 0.04 |
| Leading | 303 | 34 |
Genes that encode proteins with length of 200 or more amino acids.
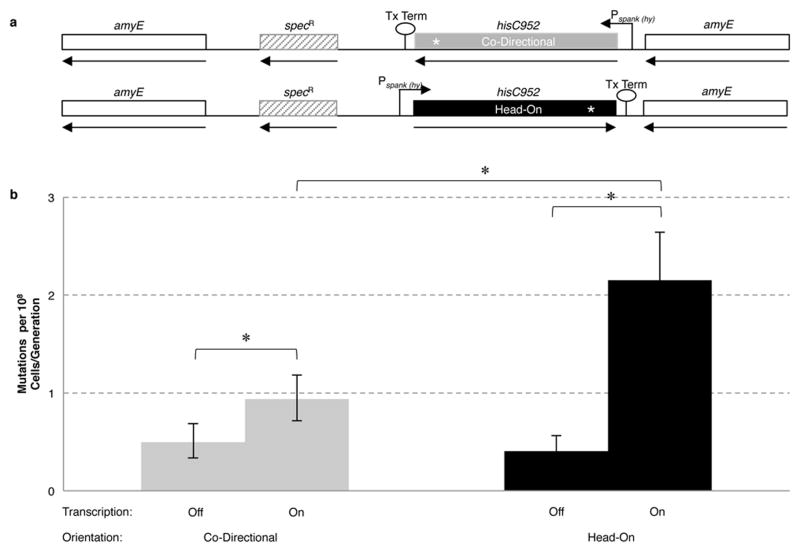
We next determined whether replication-transcription conflicts are responsible for the increased mutation rates in the lagging strand genes experimentally, using classic reversion assays. The positive correlation between increased gene length as well as expression levels with increased dN in the lagging strand genes strongly suggests that conflicts are driving the mutagenesis of the head-on genes (Figure 2B and Supplementary Figure 6). We engineered strains auxotrophic for histidine biosynthesis, and integrated an ectopic copy of the histidine synthetase (hisC) gene with a premature stop codon at the 318th position, under the control of the IPTG inducible promoter Pspank(hy) onto the chromosome, in either the head-on or co-directional orientation (Figure 3). Reversion assays and fluctuation analyses were performed in the presence and absence of transcription, for both orientations (see methods). In the absence of transcription, the orientation of the gene did not impact mutation rates (Figure 3 and Supplementary Table 6). When activated, transcription led to an increase in the mutation rate in both orientations, but the increase was much more prominent for the head-on orientation (Figure 3 and Supplementary Table 6). The dependence of increased mutation rates on both transcription and orientation indicates that head-on replication-transcription conflicts lead to increased mutagenesis. These data demonstrate that in the absence of transcription, mutagenesis rates of the two strands are not significantly different.
Figure 3. Increased rates of mutagenesis in the head-on orientation are transcription-dependent.
(A) Diagram of the amyE locus containing reporter co-directionally (HM420) or head-on (HM419) to replication. (B) Mutation rates based on reversion assays for hisC952 when the gene was oriented head on or co-directionally to replication, in the presence (transcription on) or absence (transcription off) of IPTG (*p<0.05). Error bars represent 95% confidence intervals. We confirmed by sequencing that the transcription-dependent mutations were indeed occurring in the hisC allele at amyE (Supplementary Table 9). YB955 did not show an increase in hisC952 reversions with IPTG treatment (data not shown).
We analyzed the functional categories of the core genes on the lagging strand and found enrichment in genes for: 1) sporulation, 2) iron binding, 3) transcription regulation, and 4) cellular homeostasis (Figure 4). In general, the majority of these genes represent a variety of stress responses. For example, the transcription regulation genes include ECF-type sigma factors (SigV and SigM), which are known to be the most variable of the sigma factors and are involved in responding to extracytoplasmic functions, as well as master regulators of biofilm formation (SinI and SinR), which control the transition from a free-swimming lifestyle to a stress-tolerant, biofilm lifestyle. We propose that these particular functions are kept relatively variable, possibly to produce population heterogeneity that can more easily respond to rapid changes in the environment.
Figure 4. Functional categories of leading and lagging strand genes.
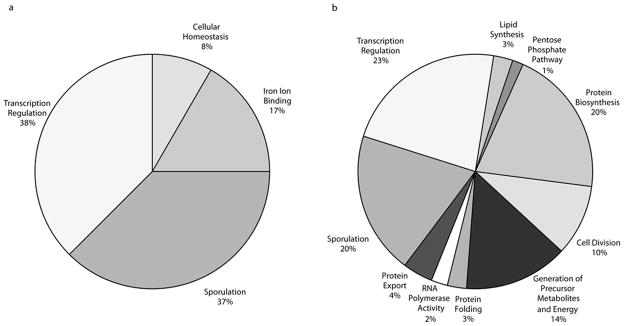
Pie charts presenting significantly (p < 0.05) overrepresented functional categories (as determined by medium stringency using DAVID23) of protein products from lagging (A) and leading (B) strand of replication for B. subtilis strain168. The genes categorized for the lagging strand are: abrB, acuA, acuB, ahpC, ahpF, antE, cotM, cspD, cueR, def, fer, gerPF, hpr, ispG, katA, lexA, mrgA, nasE, sacY, sda, sigM, sigV, sinI, sinR, spoIISB, sspB, sspD, sspI, sspK, sspL, sspM, sspO, tnrA, ycnC, ydbP, ydgJ, yhdK, yjbI, ynzD, ypoP, ytzE, yutI, yuxN, ywoH.
Taken together, our data indicate that genes positioned on the lagging strand evolve at a significantly higher rate than those encoded on the leading strand, and that the mutagenic nature of the lagging strand is due, at least in part, to the conflicts between replication and transcription. The increased rate of mutagenesis in the genes on the lagging strand is likely to be detrimental for many, especially essential genes, and is probably responsible for the strong bias for these genes to be co-orientated with replication. Two models were proposed previously to explain the underlying cause of the bias against head-on orientation of genes: 1) the detrimental effect of increased mutagenesis, by Mirkin and Mirkin, and 2) increased rate of gene transcript truncations, by Rocha and Danchin1,14. Experimental work from B. subtilis, E. coli and S. cerevisiae has shown that genomic instability (which may increase the rate of mutations) is a potential consequence of head-on conflicts8,15,16. Indeed, our findings show that the bias for the co-directional orientation of genes in bacteria is at least partially determined by negative selection against increased rate of nonsynonymous mutations, consistent with the Mirkin and Mirkin model.
The other side of the coin, however, in contrast to the essential genes, is that the increased mutation rate could also be the reason for the head-on orientation of certain core genes, i.e. head-on orientation could be promoted by positive selection. The action of positive selection is apparent from the high rate of convergent evolution in the proteins coded by genes on the lagging strand – a strong indication of their adaptive significance. In contrast to the nonsynonymous changes, synonymous mutations are generally (but not always) considered to be fitness-neutral in nature and their accumulation over time is expected to be driven largely by genetic drift. Thus, accumulation of synonymous mutations could be a relatively slow process compared to positive selection for the adaptive changes, possibly explaining the low level of accumulation of the synonymous mutations in the lagging strand genes, compared to the nonsynonymous mutations. Though it is possible that there is a stronger negative selection against synonymous mutations in the lagging strand genes due to codon usage bias in highly expressed genes, we did not see a major difference between codon usage in the two strands’ genes (Supplementary Figure 7).
It is unclear how the phenomenon, described here, extends to other Gram-positive bacteria or to Gram-negative organisms such as E. coli and Salmonella. A systematic investigation of orientation, transcription and rates of evolution in core genes has not been performed in these organisms, and previous studies looking at some aspects of these questions have produced contradictory results depending on methodology 17–22.
Replication-transcription conflict-mediated mutagenesis could be used by many organisms, including eukaryotes, as a universal strategy to link gene expression and evolution rate under selection. Genes on the lagging strand can evolve at a relatively fast rate because of head-on conflicts. Thus, a simple switch in orientation, using short sequence homologies and recombination dependent mechanisms, could facilitate evolution in specific genes in a targeted way. Investigating the major targets of conflict-mediated mutagenesis is likely to reveal far reaching biological insights into adaptation and evolution of organisms.
Methods
Strains
For the bioinformatics analysis, fully assembled genomes of 5 B. subtilis strains were downloaded from NCBI Genbank. Those strains are: B. subtilis subsp. subtilis str. 168, B. subtilis BSn5, B. subtilis subsp. subtilis str. RO-NN-1, B. subtilis subsp. spizizenii TU-B-10 and B. subtilis subsp. spizizenii str. W23 (Supplementary Table 1).
All experiments were performed using isogenic derivatives of Bacillus subtilis strain YB955 (ile, leu, his, met) (Supplementary Table 7). To construct a strain with an exogenous copy of histidine synthetase oriented head-on to replication hisC952 was first amplified from genomic DNA from YB955 using the primers HM386 (AAA AGT CGA CAA GGA GGT ATA CAT TTG CGT ATC AAA GAA CAT TTA AAA C) and HM396 (AAA GCA TGC TCC TTA TGA ATG GGA CTT ATA AAA TTT CAG CTA AAA TGG). The PCR fragment was digested with SalI and SphI and ligated into the integration vector pDR111 to generate the plasmid pHM392. This plasmid was introduced by double crossover into YB955 at amyE to generate strain HM19 (leuC427 metB5 hisC952 amyE::{Pspank(hy)-hisC952 Head-On spc}). To construct a strain with the hisC952 allele oriented co-directionally with replication a fragment containing hisC952 and Pspank(hy) was amplified off of pHM392 using primers HM392 (AAA GAA TTC TAA CTC ACA TTA ATT GCG TTG C) and HM393 (AAA GGA TCC TGG CAA GAA CGT TGC TCG AGG). This fragment was digested with EcoRI and BamHI, inverted and ligated into pDR111 to generate plasmid pHM394. This plasmid was introduced by transformation into strain YB955, generating strain HM420 (leuC427 metB5 hisC952 amyE::{ Pspank(hy)-hisC952 Co-Directional spc}). See supplementary table 8 for a list of the plasmids. Candidate spectinomycin resistant transformants were screened for disruption of the amyE locus via starch agar plate assay. Genomic DNA was isolated from amylase deficient transformants and the chromosomal region containing amyE was amplified using primers HM205 (TCC AAA CTG GAC ACA TGG AA) and HM207 (AAA GAG GCG TAC TGC CTG AA). The resulting PCR fragments were sequenced to confirm integration of hisC952 in the proper orientation.
Detection of common orthologous genes
In order to identify the core genes shared between all 5 strains of B. subtilis, we used B. subtilis subsp. subtilis str. 168 as the reference genome and extracted homologs from other 4 strains with nucleotide sequence identity and length coverage values of ≥ 95%. We excluded the annotated pseudogenes and genes with either internal stop codon or non-ACGT characters.
Identification of leading and lagging strand genes
We considered the origin of replication to be positioned between ribosomal protein L34 and chromosomal replication initiation protein (DnaA), whereas the terminus region before replication terminator protein (Rtp) for all 5 strains (Kunst et al. 1997). Depending on these locations we defined the genes present in leading strand and lagging strand for each strain. For analysis, we considered those genes which are present in the same strand (either leading or lagging) for all five strains. In total we found 759 core polymorphic genes, 627 in leading strand and 132 in lagging strand (Supplemantary Table 2).
Molecular evolutionary analysis
Alignment of every gene set was performed by clustalW24. The rates of nonsynonymous (dN) and synonymous (dS) mutations for each gene were computed by the ratio of number of nonsynonymous and synonymous changes to total number of nonsynonymous and synonymous sites respectively, using the mutation-fraction method25. To assess the statistical significance between any two sample sets, Z-test was applied to dS and dN values26.
Detection of convergent mutations was performed by Zonal Phylogeny Software27. Zonal Phylogeny reconstructs an unrooted protein phylogram based on nucleotide sequences where multiple alleles differentiated by synonymous changes are collapsed into single protein variant, followed by the mapping of phylogenetically unlinked repeated mutations at specific amino acid positions (i.e. convergent amino acid mutations). DnaSPv5 was used to calculate the 2×2 χ2 statistics28.
Media and growth conditions
For all experiments cells were grown at 37°C, shaking at 260 RPM. Liquid cultures of Bacillus subtilis were grown in rich medium (LB) supplemented with spectinomycin (50 ug/ml) where appropriate. Escherichia coli was grown in LB supplemented with ampicillin at 100 ug/ml. To determine viabilities B. subtilis cells were plated on solid agar plates containing Spizizen’s Minimal Medium (0.2 mg/ml ammonium sulfate, 1.4 mg/ml monobasic potassium phosphate, 0.6 mg/ml dibasic potassium phosphate, 0.1 mg/ml sodium citrate dihydrate, 0.02 mg/ml magnesium sulfate heptahydrate, 100 ug/ml glutamic acid, 5 ug/ml glucose), supplemented with isoleucine, methionine, leucine and histidine at 50 ug/ml. To select for reversion to prototrophy, cells were plated on solid agar plates containing Spizizen’s Minimal Medium supplemented with isoleucine, methionine and leucine at 50 ug/ml.
Reversion assays
Fluctuation assays were performed to determine the mutation rate. Strains HM419 and HM420 were grown overnight in LB supplemented with spectinomycin at 50 ug/ml. Saturated cultures were diluted 1:104 into 2 ml LB medium and 2 ml LB medium supplemented with 1 mM IPTG. For these experiments 12 parallel cultures for each strain and condition were grown for 5 hours at 37°C with shaking at 260 rpm. When cells reached saturation, 100 ul of culture was withdrawn, diluted 1:106, and spread onto solid agar plates containing Spizizen’s Minimal Medium supplemented with all required amino acids to determine the number of viable cells present in the culture at plating. The remainder of the culture was pelleted by centrifugation, re-suspended in Spizizen’s Minimal Salts and spread onto solid agar plates lacking histidine. Revertant colonies were counted after 48 hours of growth at 37°C. The experiment was repeated three times on different days.
Confirmation of reversions in the hisC gene at amyE
To determine if reversions were occurring in the copy of hisC at amyE, twenty-eight colonies, twenty-four head-on and four co-directional, from plates with IPTG were sequenced. The revertant colonies were grown up in selective (histidine- spectinomycin+) media and genomic DNA was prepared. The amyE region was amplified via PCR with primers HM205 (TCC AAA CTG GAC ACA TGG AA) and HM207 (AAA GAG GCG TAC TGC CTG AA). The PCR products were sequenced using the primer HM396 (AAA GCA TGC TCC TTA TGA ATG GGA CTT ATA AAA TTT CAG CTA AAA TGG). Changes at position 952 were analyzed (Supplementary Table 9)
Calculation of mutation rates
Mutation rates were calculated using the Ma-Sandri-Sarkar Maximum Likelihood method29,30, with the aid of the Fluctuation Analysis Calculator. Statistical significance was determined using a standard two-tailed t-test.
Supplementary Material
Acknowledgments
We would like to acknowledge the work of others that has contributed to our understanding of the topic which we were unable to reference here due to size limitations. We thank the anonymous reviewers that helped improve this manuscript during the revision process. We thank Drs. Bonita Brewer, Joseph Mougous, Ferric Fang, Matthew Parsek, and Samuel Miller for critical reading of the manuscript. We thank Dr. Christopher Merrikh for comments on the manuscript and technical help with sequencing of revertants. We thank Dr. Eduardo Robleto for the auxotrophic YB955 strain. H.M. was supported by start-up funds from the department of Microbiology at the University of Washington. S.M.W. was supported by the University of Washington Biophysics Training Grant.
Footnotes
Author Contributions
H.M., E.S., S.P., and S.M.W. designed experiments, S.P., S. C., and S.M.W. carried out the bioinformatics analysis and the experiments, H.M., E.S., S.P., S. C., and S.M.W. analyzed the data. H.M. and E.S. wrote the paper.
References
- 1.Rocha EP, Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic acids research. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha EP, Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nature genetics. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 3.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 4.Price MN, Alm EJ, Arkin AP. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic acids research. 2005;33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nature reviews. Microbiology. 2012;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiology and molecular biology reviews : MMBR. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Septenville AL, Duigou S, Boubakri H, Michel B. Replication fork reversal after replication-transcription collision. PLoS genetics. 2012;8:e1002622. doi: 10.1371/journal.pgen.1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay S, et al. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12412–12417. doi: 10.1073/pnas.0906217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, et al. Essential Bacillus subtilis genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 12.Christin PA, Weinreich DM, Besnard G. Causes and evolutionary significance of genetic convergence. Trends in genetics : TIG. 2010;26:400–405. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 14.Mirkin EV, Mirkin SM. Mechanisms of transcription-replication collisions in bacteria. Molecular and cellular biology. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS genetics. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA repair. 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veaute X, Fuchs RP. Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science. 1993;261:598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- 18.Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maliszewska-Tkaczyk M, Jonczyk P, Bialoskorska M, Schaaper RM, Fijalkowska IJ. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12678–12683. doi: 10.1073/pnas.220424697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczepanik D, et al. Evolution rates of genes on leading and lagging DNA strands. Journal of molecular evolution. 2001;52:426–433. doi: 10.1007/s002390010172. [DOI] [PubMed] [Google Scholar]
- 21.Lin CH, Lian CY, Hsiung CA, Chen FC. Changes in transcriptional orientation are associated with increases in evolutionary rates of enterobacterial genes. BMC bioinformatics. 2011;12(Suppl 9):S19. doi: 10.1186/1471-2105-12-S9-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juurik T, et al. Mutation Frequency and Spectrum of Mutations Vary at Different Chromosomal Positions of Pseudomonas putida. PloS one. 2012;7:e48511. doi: 10.1371/journal.pone.0048511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, et al. Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic acids research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y, Gojobori T. The Phylogenetic Handbook. Vol. 1. Cambridge University Press; 2003. pp. 283–311. [Google Scholar]
- 27.Chattopadhyay S, Dykhuizen DE, Sokurenko EV. ZPS: visualization of recent adaptive evolution of proteins. BMC bioinformatics. 2007;8:187. doi: 10.1186/1471-2105-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- 30.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.