Summary
The surface protein Pfs47 mediates Plasmodium falciparum evasion of the Anopheles gambiae complement-like immune system.
Plasmodium falciparum transmission by Anopheles gambiae mosquitoes is remarkably efficient, resulting in a very high prevalence of human malaria infection in sub-Saharan Africa. A combination of genetic mapping, linkage group selection, and functional genomics was used to identify Pfs47 as a P. falciparum gene that allows the parasite to infect A. gambiae without activating the mosquito immune system. Disruption of Pfs47 greatly reduced parasite survival in the mosquito and this phenotype could be reverted by genetic complementation of the parasite or by disruption of the mosquito complement-like system. Pfs47 suppresses midgut nitration responses that are critical to activate the complement-like system. We provide direct experimental evidence that immune evasion mediated by Pfs47 is critical for efficient human malaria transmission by A. gambiae.
The mosquito Anopheles gambiae is the natural vector of the human malaria parasite Plasmodium falciparum in large regions of Africa. There is compelling evidence, mostly from murine models, that A. gambiae can mount robust and effective antiplasmodial responses. For example, midgut epithelial cells activate nitration reactions in response to Plasmodium infection that limit parasite survival (1, 2), by modifying the parasite and promoting lysis by thioester-containing protein 1 (TEP1), a key component of the mosquito complement-like system (3). However, several studies indicate that disrupting the complement-like system has a modest (4, 5) or no (6) effect when A. gambiae is infected with P. falciparum.
The A. gambiae L3‐5 strain was selected to be refractory (R) to Plasmodium cynomolgi (simian malaria), but also eliminates most other Plasmodium species including P. falciparum strains from the New World, and forms a melanotic capsule (i.e., deposition of melanin, a black insoluble pigment) around the dead parasites. In contrast, this strain is highly susceptible to infection with some African P. falciparum strains, such as NF54, 3D7, and GB4 (7, 8). Some parasite lines from malaria-endemic areas where A. gambiae is the natural vector are able to evade the mosquito immune system (8). Co-infection experiments reveal that the immune response (or lack thereof) to a P. falciparum strain did not affect the fate of other parasites present in the same mosquito midgut (8); suggesting that parasite survival is determined by genetic differences between P. falciparum strains (8). In this study, quantitative trait locus (QTL) mapping, linkage group selection, and functional genomics were used to identify the first P. falciparum gene that promotes infection by modulating the host immune system.
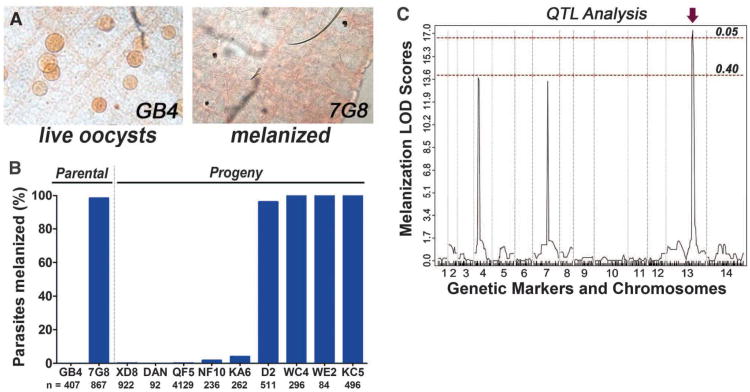
We took advantage of the phenotypic difference between A. gambiae R infected with two P. falciparum lines—7G8 from Brazil (97–100% melanized) and GB4 from Ghana (0–3% melanized) (8) (Fig. 1A)—that have been previously subjected to a genetic cross (9). We phenotyped nine cloned progeny lines, five of them had the GB4 phenotype and survived well (0-5% melanization), while four had the 7G8 phenotype and were mostly melanized (98–100% melanization) (Fig. 1B) (10). The presence of two distinct phenotypes in the progeny suggested a monogenic trait. Repeated attempts to phenotype 16 additional progeny lines failed, because most clonal lines had lost the ability to generate mature gametocytes. QTL mapping, using a previously reported linkage map (9) and the phenotypes obtained, identified three logarithm of odds (LOD) peaks (Fig. 1C), but only one of them, located in chromosome 13 (Chr13), was significant (P < 0.05). The boundaries of the recombination sites for this locus were precisely mapped and defined a 172-kb region coding for 41 genes (FS1; TS1).
Fig. 1.
Survival of the parental and progeny Plasmodium falciparum lines in refractory (R) mosquitoes and quantitative trait locus (QTL) mapping of the melanization phenotype. (A) P. falciparum GB4 and 7G8 parasites in the midgut of R mosquitoes. (B) Melanization phenotype of the parental and progeny lines of the GB4 × 7G8 genetic cross in R mosquitoes. (C) Logarithm of odds scores (LOD) of genome-wide QTL analysis of the melanization phenotype. Red dotted lines, statistical significance thresholds at P = 0.05 and P = 0.40; arrow, significant QTL.
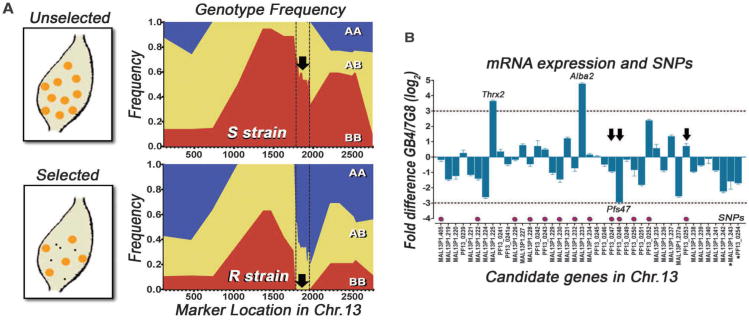
Linkage group selection analysis, a method that allows de novo location of loci encoding selectable phenotypes of malaria parasites (11), was used to obtain independent confirmation of the locus in Chr13. The un-cloned recombinant progeny from the original genetic cross was used to generate gametocytes and infect either the R strain or a permissive susceptible (S) A. gambiae G3 line in which both parental parasite lines survive. Individual oocysts were isolated, subjected to whole genome DNA amplification, and genotyped for multiple markers along Chr13. Oocysts derive from the diploid ookinete stage and can be homozygous for the African GB4 (AA) or Brazilian 7G8 alleles (BB), or heterozygous (AB). In the S strain, the BB genotype is highly abundant in the central region of Chr13, reaching a frequency of > 90% (Fig. 2A; FS2, TS2), which was already observed in the progeny clones from the genetic cross (9) and is not due to selection by the mosquito, because both parental strains survive in the S strain (7). In the R strain, we identified a well-defined region, indicated by the dotted line, in which the BB genotype is under strong negative selection and is totally absent (0%) (Fig. 2A). In contrast, prevalence of the BB genotype in the same chromosomal region is 55% in the S strain (P < 0.00001; χ2 test), which does not exert selective pressure on the parasite. It is noteworthy that the two markers that define this region (Fig. 2A, dotted lines) are the same as those that limit the 172-kb region identified by QTL analysis. Although 50 additional individual oocysts dissected from the R strain were genotyped, no oocyst with the BB genotype was detected for any of the markers within the region under strong selection (FS3). The locus could therefore not be narrowed down any further.
Fig. 2.
Linkage group selection, mRNA expression, and coding region sequence analysis. (A) Genotype frequency of homozygous African (AA; blue), Brazilian (BB; red), or heterozygous (AB, yellow) markers along Chr. 13 in individual oocysts dissected from S or R mosquitoes. (black arrow, region with BB under extreme negative selection in R strain). (B) Relative mRNA expression of candidate genes in GB4/7G8 Plasmodium falciparum ookinete stage. Magenta dots, genes with non-synonymous single nucleotide polymorphisms (SNPs) between GB4 and 7G8; Arrows, SNPs shared between GB4-3D7 and 7G8-SL strains.
Gene expression analysis of the 41 candidate genes in the ookinete stage identified three genes with large differences in expression (8-fold or higher) between the parental lines: Pfs47 (PF13_0248), thioredoxin 2 (MAL13P1.225), and the nucleic acid binding protein ALBA2 (MAL13P1.233) (Fig. 2B; TS3) (P < 0.0001; t-test). Thioredoxin 2 and ALBA2 also had differences in expression between the parental lines in the gametocyte stage (TS3). Sequencing the coding regions of the 41 candidate genes identified non-synonymous single nucleotide polymorphisms (SNPs) between the parental lines in 13 genes (Fig. 2B, magenta dots; TS4). Some non-synonymous SNPs in three of these genes, four SNPs in Pfs47, and one in Pf48/45 (PF13_0247) and ethanolamine-phosphate cytidylyltransferase (PF13_0253) also correlate with parasite survival in two other P. falciparum strains (Fig. 2B, black arrows; TS4). The GB4 SNP alleles are shared with the NF54 and 3D7 strains that also survive, and the 7G8 SNP alleles with the SL strain that is melanized by the R strain (7). Five genes were selected as top candidates for detailed genetic analysis based on large differences in gene expression and/or on polymorphisms that correlates with survival in other strains (TS5).
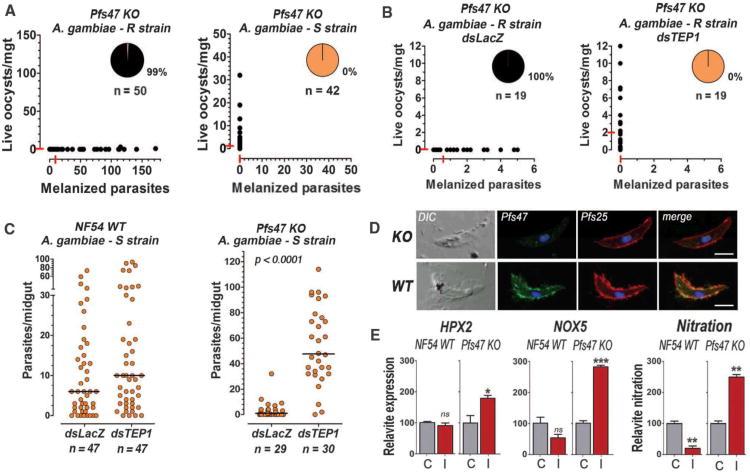
Two top candidate genes, Pfs47 and Pfs48/45, code for members of the 6-cysteine protein family that are expressed on the gametocyte surface. Previous gene disruption experiments in the NF54 line revealed that Pfs48/45 is critical for gamete fertility (12). Pfs47 is expressed in female gametocytes but is not essential for P. falciparum fertilization, although its homolog in Plasmodium berghei is required for female gamete fertility (12, 13). The intensity of infection with the Pfs48/45 knockout (KO) line (NF54 genetic background) in the R strain was low, probably due to reduced fertility, but those parasites that invaded the midgut had a similar phenotype as wild-type (WT)NF54parasites (8) and only 3% were melanized (FS4); indicating that Pfs48/45 is not required to evade the mosquito immune system. In contrast, Pfs47 KO (NF54 genetic background) parasites develop and invade the midgut but are eliminated by R mosquitoes (99% melanization) (Fig. 3A); however, although Pfs47 KO parasites are not melanized by S mosquitoes (Fig. 3C), the infection level in the S strain (median of 1 oocyst/midgut) is much lower than that in An. stephensi mosquitoes (60 oocysts/midgut median) (Fig. S5). Notably, this An. stephensi strain has been selected to be highly permissive to P. falciparum infection (14).
Fig. 3.
Phenotype of NF54 wild type (WT) or Pfs47 knockout (KO) on different mosquitoes. (A) Number of melanized (x-axis) and live (y-axis) KO parasites in R and S mosquitoes. Each dot represents an individual midgut. Medians are indicated by the red lines. (B) Effect of TEP-1 silencing on KO infection in R mosquitoes. (C) Effect of silencing TEP1 on WT and KO infection in S mosquitoes. (D) Immunofluorescence staining of WT and KO ookinetes with Pfs47 (green) and Pfs25 (red) (scale bar = 5μm). (E) Midgut mRNA expression of HPX2 and NOX5, and midgut nitration using ELISA, 24h after S females were infected with WT or KO parasites (I = infected) or fed uninfected blood (C = control).
To determine whether Pfs47 interacts with the mosquito immune system, the A. gambiae complement-like system was disrupted by silencing TEP1. Reducing TEP1 expression completely reversed melanization of Pfs47 KO parasites in the R strain (Fig. 3B). In the A. gambiae S strain (G3), neither NF54 WT nor Pfs47 KO parasites were melanized (Fig. 3C); however, while TEP1 silencing had no significant effect on infection with NF54 WT parasites (Fig. 3C), it dramatically increased both the intensity (P < 0.0001 Mann-Whitney test) and prevalence of infection (P < 0.001; χ2 test) of Pfs47 KO parasites (Fig. 3C). This indicates that Pfs47 is necessary for P. falciparum parasites to evade two well-characterized immune responses mediated by TEP1 in A. gambiae: killing followed by melanization in the R strain and parasite lysis without melanization in the S strain.
Pfs47 protein is present on the surface of WT NF54 ookinetes, the stage that invades the midgut, but is absent in Pfs47 KO parasites (Fig. 3D). The expression of HPX2 and NOX5, two enzymes that mediate midgut nitration in response to P. berghei infection and promote TEP1 activation (2) was evaluated in S mosquitoes. HPX2 and NOX5 were not induced by NF54 WT parasites and nitration levels were lower than in uninfected controls (Fig. 3E). In contrast, Pfs47 KO parasites induced expression of HPX2 and NOX5, and a robust nitration response, indicating that Pfs47 may prevent TEP1-mediated lysis by suppressing midgut epithelial nitration responses (Fig. 3E).
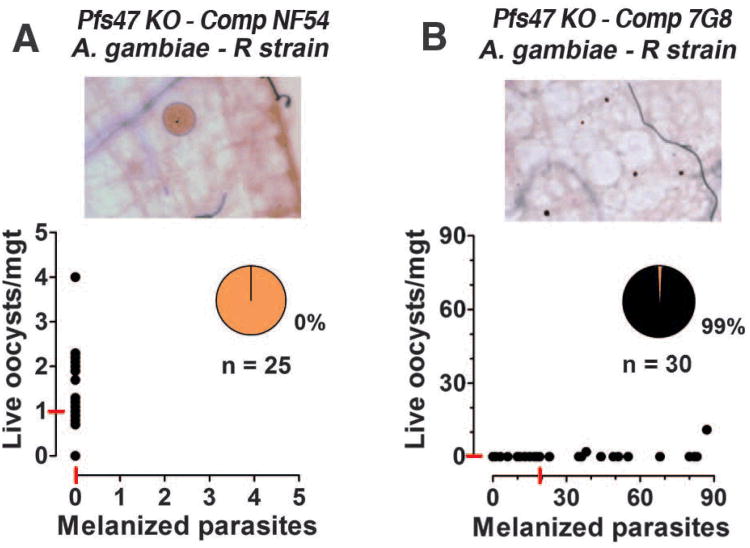
Finally, we confirmed the importance of Pfs47 for parasite survival by complementing the Pfs47 KO line with different Pfs47 alleles (FS6-9). As expected, the NF54 allele of Pfs47 reversed the melanization phenotype (0% melanization) in the R strain when the complemented parasites were kept under sustained drug pressure, confirming that this allele of Pfs47 is sufficient to evade the immune system (Fig. 4A). A reversal to a mixed live/melanization phenotype was observed when the drug pressure was reduced (FS10). In contrast, complementation with the 7G8 allele failed to rescue parasites in the R strain, as 99% melanization was observed (Fig. 4B).
Fig. 4.
Effect of complementing Pfs47 knockout (KO) parasites with the Brazilian (7G8) and African (NF54) alleles of Pfs47. Infectivity of Pfs47 KO parasites complemented with the (A) NF54 or (B) 7G8 Pfs47 alleles in the Anopheles gambiae R strain.
Together, our findings identify Pfs47 as an essential survival factor for P. falciparum that allows the parasite to evade the immune system of A. gambiae, a major mosquito vector in Africa. However, other parasite genes may also be involved in this process. Interestingly, Pfs47 is a highly polymorphic gene with a marked population structure in field isolates and exhibits extreme fixation in non-African regions of the world (15, 16). Our findings suggest that the population structure of Pfs47 may be due to adaptation of P. falciparum to the different Anopheles vector species present outside of Africa. The fact that the 7G8 allele of Pfs47 is sufficient to evade the TEP1 complement-like system in S mosquitoes but not in the R strain indicates that there are also genetic differences in the vector that determine compatibility with parasites that express specific Pfs47 alleles. It appears that Pfs47 evolved a function in P. falciparum that increases parasite survival in A. gambiae mosquitoes and may be responsible, at least in part, for the very high rates of malaria transmission in hyperendemic regions in Africa. Disruption of the immunomodulatory activity of Pfs47 may prove to be an effective strategy to reduce malaria transmission to humans.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, NIAID, National Institutes of Health. We thank Karen Hayton and Thomas Wellems for advice with the genetic cross; André Laughinghouse and Kevin Lee for technical support; and Brenda Marshall for editorial assistance.
Footnotes
References and Notes
- 1.Kumar S, Gupta L, Han YS, Barillas-Mury C. J Biol Chem. 2004;279:53475. doi: 10.1074/jbc.M409905200. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira GdA, Lieberman J, Barillas-Mury C. Science. 2012;335:856. doi: 10.1126/science.1209678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blandin S, et al. Cell. 2004;116:661. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 4.Garver LS, Dong Y, Dimopoulos G. PLoS Pathog. 2009;5:e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitri C, et al. PLoS Pathog. 2009;5:e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohuet A, et al. EMBO Rep. 2006;7:1285. doi: 10.1038/sj.embor.7400831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins FH, et al. Science. 1986;234:607. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 8.Molina-Cruz A, et al. Proc Natl Acad Sci U S A. 2012;109(28):E1957. doi: 10.1073/pnas.1121183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayton K, et al. Cell Host Microbe. 2008;4:40. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supporting online materials are available on Science Online.
- 11.Carter R, Hunt P, Cheesman S. Int J Parasitol. 2007;37:285. doi: 10.1016/j.ijpara.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 12.van Dijk MR, et al. Cell. 2001;104:153. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 13.van Schaijk BC, et al. Mol Biochem Parasitol. 2006;149:216. doi: 10.1016/j.molbiopara.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Feldmann AM, Ponnudurai T. Med Vet Entomol. 1989;3:41. doi: 10.1111/j.1365-2915.1989.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 15.Anthony TG, Polley SD, Vogler AP, Conway DJ. Mol Biochem Parasitol. 2007;156:117. doi: 10.1016/j.molbiopara.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Manske M, et al. Nature. 2012;487:375. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.