Abstract
Based on tetrapeptide AVPI, we were able to design and synthesize a new simplified scaffold to inhibit the BIR3 domain of the XIAP protein at low micromolar range. The uncomplicated synthesis and the binding activity of the molecule disclosed here represent an attractive alternative to develop new compounds targeting the protein-protein interaction of XIAP/caspase9.
Apoptosis plays a crucial role in the homeostasis and development of living organisms. Deregulation in this mechanism is associated with many diseases including several types of cancer. In the apoptosis pathway, the inhibitors of apoptosis proteins (IAPs) are one of the mechanisms used by tumor cells to evade programmed cell death.1
The XIAP is the most potent caspase inhibitors among IAPs protein family. This protein interacts with initiator capase 9 and executioner caspase 3 and 7 through its BIR3 and BIR2 domains respectively.2 The search for new compounds able to disrupt the XIAP-caspase interaction has attracted the attention of scientific community as a promising strategy for cancer treatment. The natural inhibitor of XIAP is a protein (SMAC/DIABLO) released from the mitochondria into the cytosol in response to apoptotic stimuli. SMAC removes XIAP inhibition of caspase 9 by binding to the BIR3 domain of XIAP through AVPI tetrapeptide present in the N-terminal part of SMAC. This interaction (AVPI/BIR3) has been determined unequivocally by X-ray crystallography.3
Using the AVPI structure, Fesik et al. have performed a comprehensive study to determine which amino acids could be substituted without compromising its binding affinity. The authors have determined the essential amino acids residues to preserve the activity of this tetrapeptide to be the alanine (first amino acid) and proline (third amino acid) 4 (Figure 1).
Figure 1.
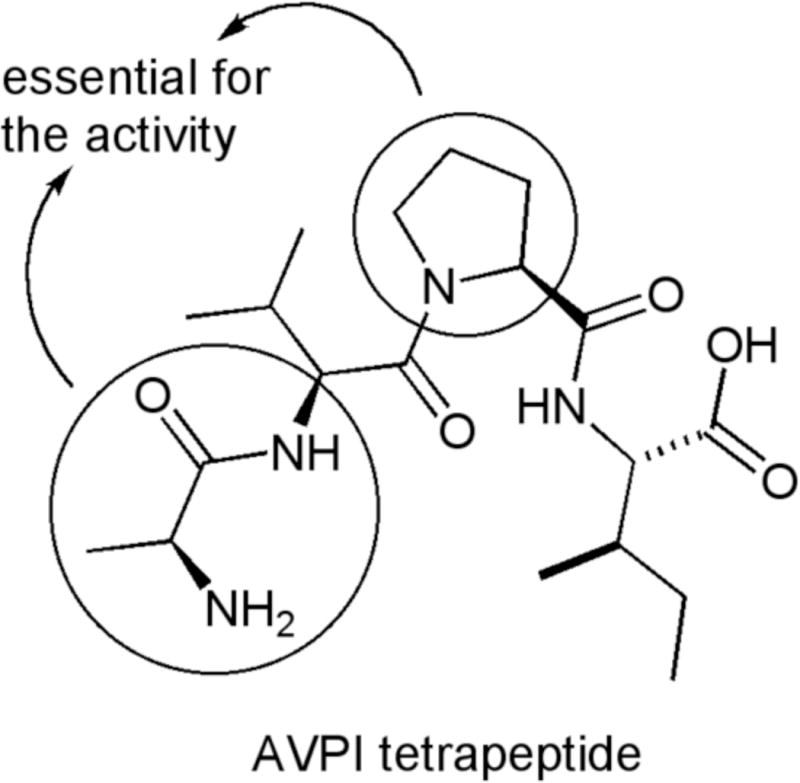
Tetrapeptide from the N-terminal part of SMAC protein.
Based on precedents in the literature,5 it is possible to rationalize about some structural features for peptidomimetic derivatives and postulate general “structural guidelines” to design new compounds based on the AVPI structure. As common features, the analogs should contain: (1) an alanine residue or a N-methyl alanine residue, (2) the presence of a rigid core (3) an aromatic residue as a surrogate of the isoleucine, and (4) the molecules should adopt a “U-conformation” for a suitable interaction with the protein (Figure 2). Most of the compounds with biological activity in vitro at nano molar range follow this pattern.
Figure 2.
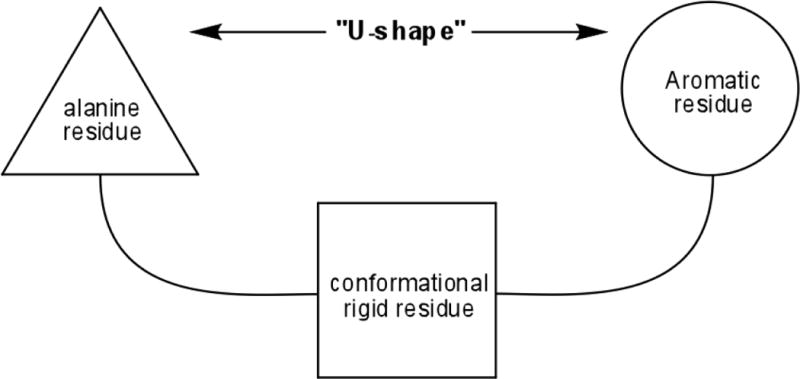
Common structural features found in most of XIAP-BIR3 domain inhibitors.
Structural simplification represents an efficient drug design strategy to shorten synthetic routes while keeping or enhancing the biological activity of complex compounds.6 Combining the molecular simplification concept with the guidelines highlighted previously, we report here a series of simplified compounds inspired by the Smac-AVPI tetrapeptide.
Preserving the alanine residue, we proposed a molecular simplification where the second and third aminoacids were substituted by thiazole ring fused to a carbocycle with different sizes as rigid central core. In this approach, we eliminated one chiral center while at same time conferring more rigidity to the molecule. Finally, different aromatic moieties linked through an amide bond to the rigid portion provided analogs structurally less complex. (Figure 3)
Figure 3.
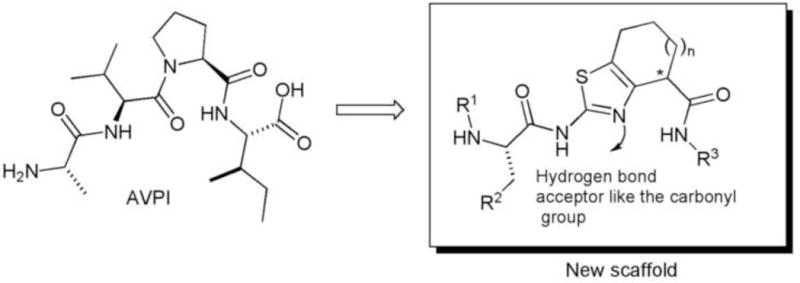
New synthetic scaffold using a thiazole ring fused to a carbocycle as peptide surrogate.
The retrosynthetic analysis of these molecules is depicted in the scheme 1. It is important to mention that the final compounds were predicted to adopt the necessary “U-conformation” for a suitable interaction with the protein based on molecular modeling studies.7
Scheme 1.
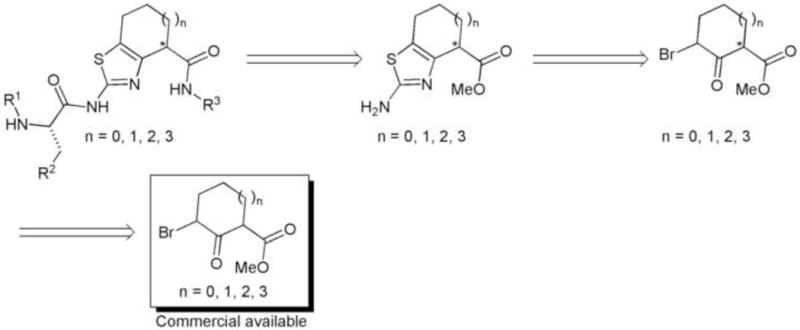
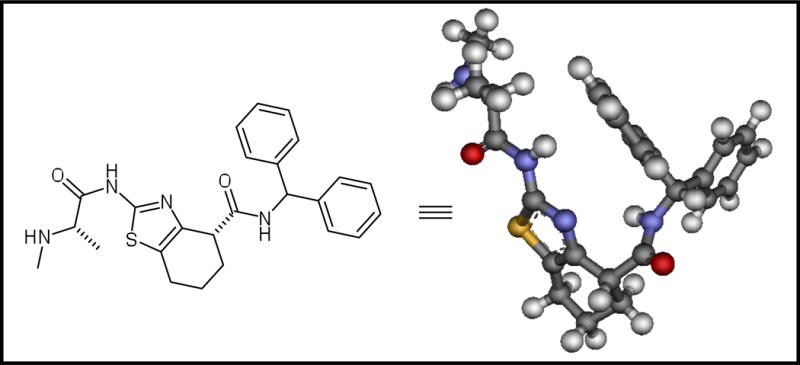
Retrosynthesis and conformational analysis of the proposed compounds.
After docking analysis of the molecules containing different sizes in the central core with the BIR3 domain of the XIAP protein (RCSB PDB ID 2jk7), one of the suitable candidates to initiate the synthesis was the compound containing the 7 member ring carbocycle.8 Furthermore, examples containing a 7 member ring fused to a 5 member ring have been reported in the literature with excellent biological activity.5b,7
The synthesis started with the bromination of the commercially available compound methyl-2-oxo-1-cycloheptanecarboxilate (1).10 The product 2 was used without purification in the thiazole formation assisted by microwave irradiation affording the fused bicyclic compound 3. The peptide coupling with the amino acids was achieved using standard conditions (DIC/HOAt/CH2Cl2) leading to compound 4 in good yields (85%–95%). The methyl ester 4 was hydrolyzed using LiOH and the product 5 was used in amide formation using different commercial available amines. Finally, deprotection of the Boc group present in the amino acid residue was performed using a solution of 10%TFA (v/v) in CH2Cl2 providing the first set of molecules. (Scheme 2)
Scheme 2.
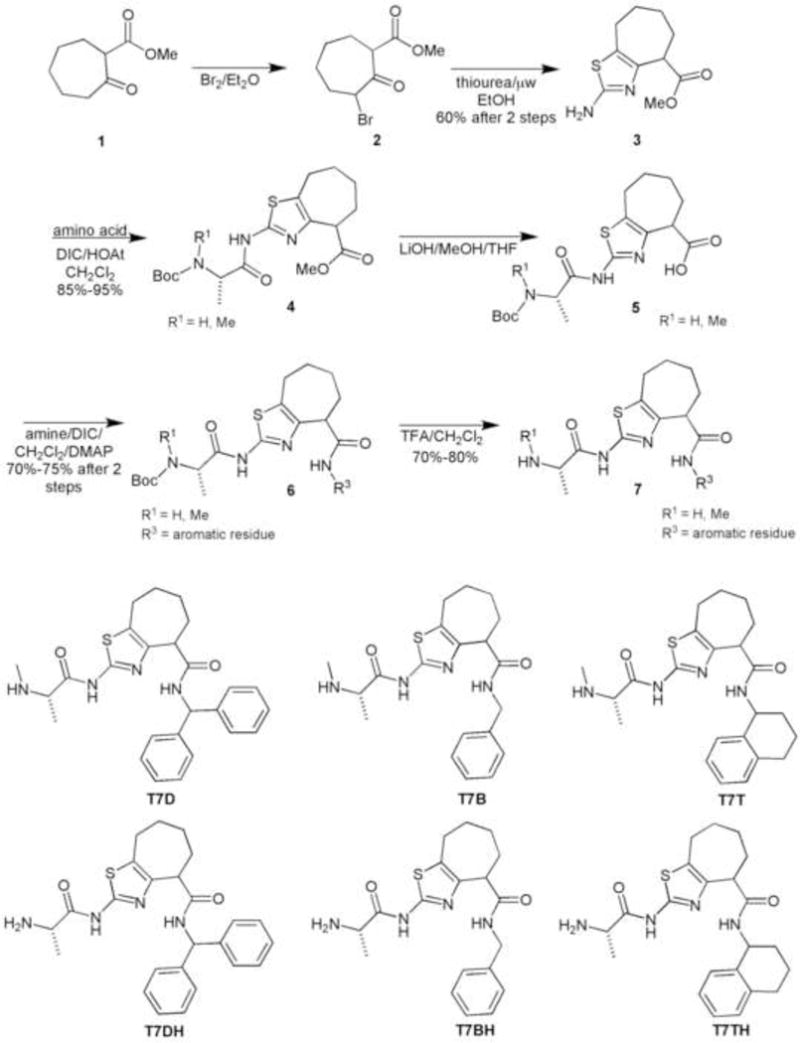
Synthesis of the thiazole ring fused to a 7 member ring carbocycle.
These compounds were synthesized to identify which amino acid residue (R1) and aromatic moieties (R3) would provide convenient substitution patterns for this scaffold. Their binding affinities to XIAP-BIR3 domain were evaluated using competitive fluorescence polarization BIDING assay (FPA).
Despite the preliminary docking result, which indicated the thiazole ring fused with a 7 member carbocycle as a promising candidates, these molecules did not exhibit relevant activity.11
Focusing our attention in the central part of the proposed scaffold, we synthesized another group of molecules containing a 5 member ring in the rigid portion. This modification was envisioned to confer more rigidity to system and at same time probe any unfavorable steric interactions in the binding pocket of the protein.
The synthesis of these compounds followed the same procedure as described above although the starting material was the commercially available compound cyclopetanone-2-carboxylic acid methyl ester. (Figure 4)
Figure 4.
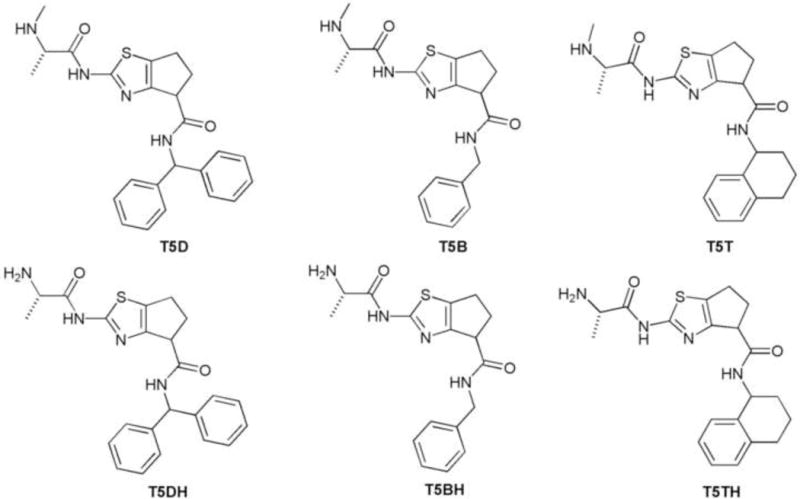
Set of molecules containing thiazole ring fused to 5 member ring carbocycle.
The results obtained in the fluorescence polarization assay (FPA) experiment prompted us to make some considerations about the structure and activity relationship (SAR) of this set of molecules. (Figure 5)
Figure 5.
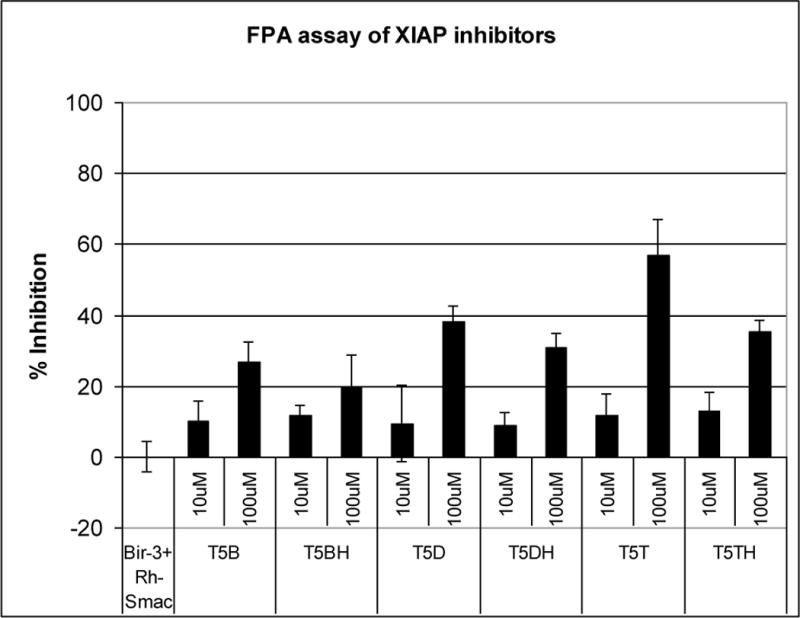
FPA result obtained with the thiazole fused to a 5 member ring carbocycle derivatives.
As shown in Figure 5, all the compounds with the N-methyl alanine residue exhibited better activity than those containing the alanine. On the other hand, the compounds with 1,2,3,4 tetrahydronaphthalene moiety showed better results when compared with others with different aromatic substitution patterns.
The promising Ki = 37μM obtained with the compound T5T12 has provided valuable information about this scaffold, which includes:(1) the amino acid residue must contain an N-methyl group; (2) the aromatic moiety should be the 1,2,3,4 tetrahydronaphthalene; and (3) probably because of unfavorable steric interactions, the thiazole ring in the central core should be fused with a 5 member ring.
Another important aspect for the activity of the small molecules targeting the BIR3 domain is the correct configuration of the chiral centers present in the molecule. This is not unexpected, considering that the binding pocket is a chiral environment and many examples support the importance of the correct diasteroisomer.5b,9,13
In our initial studies we wanted to determine the suitable substitution pattern and also the convenient central core for BIR3-binding activity. Nonetheless, the chiral aspect of the molecules was not considered. As can be observed, only one of the three chiral centers (the amino acid residue) present in the structure is defined while the other two are in the racemic form. Thus, the compound T5T exists in a diasteroisomeric mixture of four compounds. (Figure 6)
Figure 6.
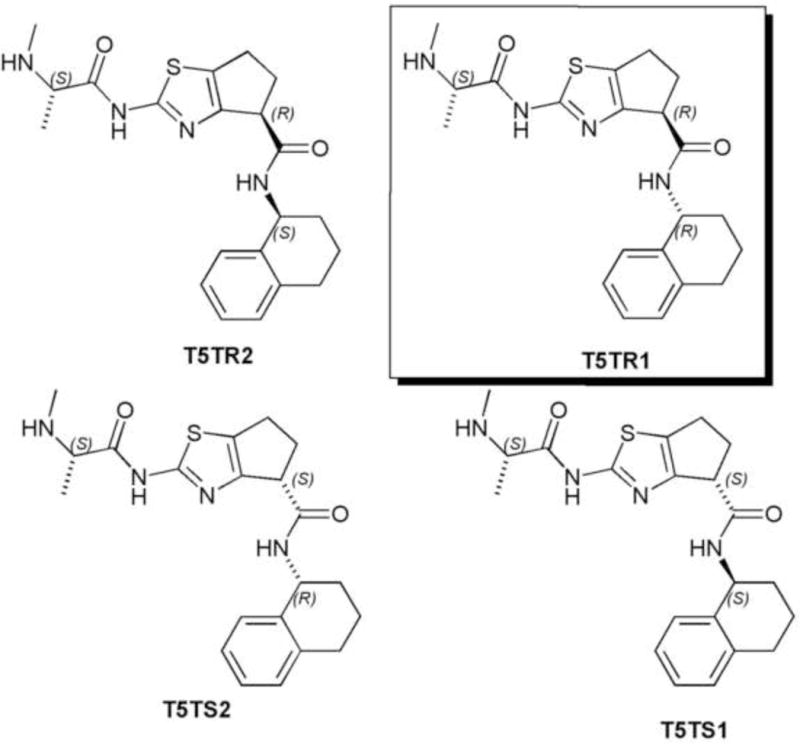
Diasteroisomers present in the T5T mixture.
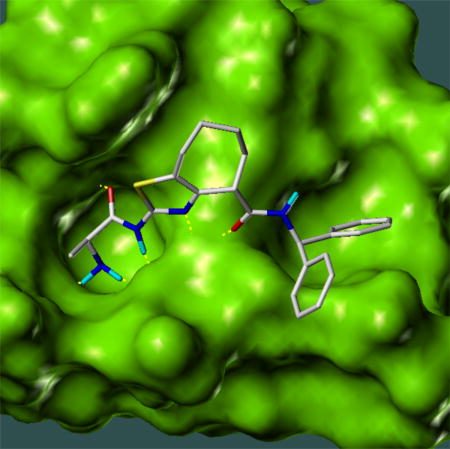
To determine which diasteroisomer was responsible for the inhibitory activity, we performed a docking study of these different diasteroisomers. The result obtained indicated that the only diasteroisomer suitable for the interaction with the XIAP-BIR3 domain is T5TR1 with the configuration S,R,R, highlighted in the Figure 6. The docked conformation of this diasteroisomer is shown in Figure 7.
Figure 7.
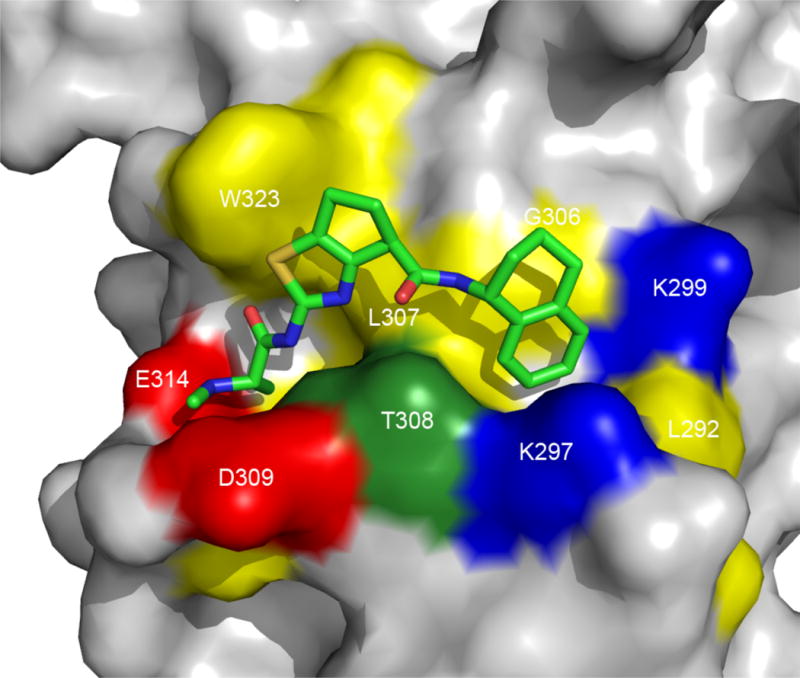
The docked conformation of the diasteroismer T5TR1 (shown in Fig. 6) in XIAP BIR3 domain (RCSB PDB ID 2jk7) as predicted by molecular docking calculation using the program SYBYL 8.0.
To verify experimentally that T5TR1 is indeed the diasteroisomer responsible for XIAP BIR3 binding, we conducted synthesis of the four diasteroisomers using the same conditions as depicted in the scheme 2. However, in the coupling with the aromatic moiety, the commercially available enantiopure compounds (R) and (S)-1,2,3,4 tetrahydronaphthalene were used. The diastero isomeric mixture from the coupling with (R)-1,2,3,4 tetrahydronaphthalene was possible to separate in a regular (silica gel) chromatographic column, affording the compounds T5TR1 and T5TR2. In the docking analysis, the isomers from the coupling with (S)-1,2,3,4 tetrahydronaphthalene (T5TS1 and T5TS2) did not exhibit interaction with protein. We therefore tested these compounds as diasteroisomeric mixture (T5TSM). (Figure 8)
Figure 8.
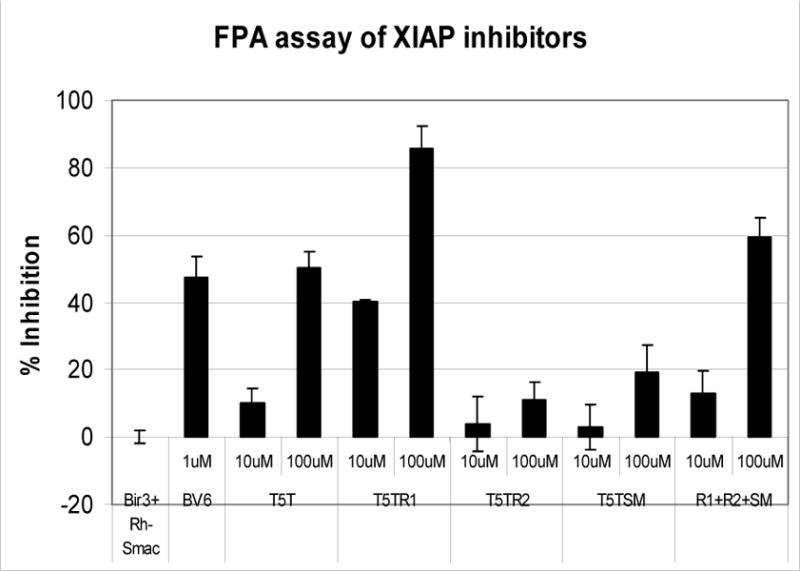
FPA result obtained with diasteroisomers of compound T5T.
As a control experiment we have used the known compound BV6, which was determined to have a Ki value of 0.90 μM for BIR3 domain in our FPA assay.14 The Ki obtained for T5TR1 was 5.58 μM while the other disteroisomers did not exhibit significant binding activity, supporting the docking result and demonstrating that T5TR1 is indeed the active diasteroisomer responsible for XIAP BIR3 binding (Figure 9).
Figure 9.
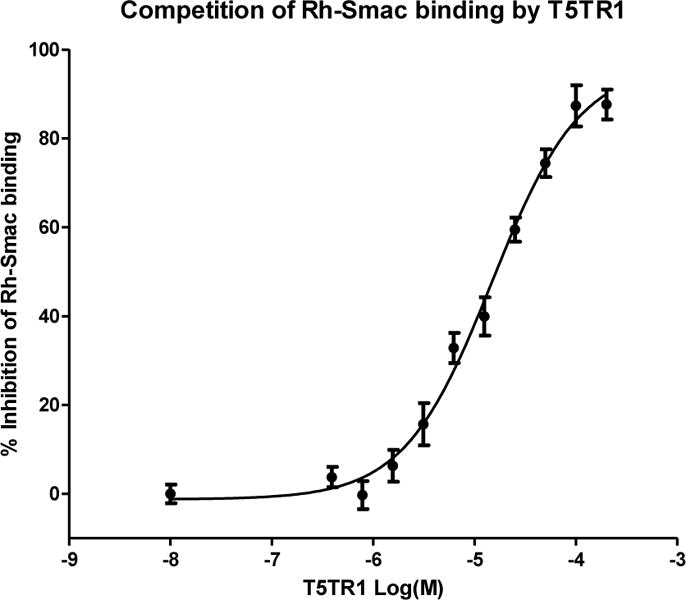
Titration curve to determine the Ki value of compound T5TR1.
Although the Ki value for the compound T5TR1 was approximately 7 times less potent compared to the control BV6, the uncomplicated synthesis (6 steps) and the structural simplicity of our designed scaffold make it an interesting starting point to develop new analogs.
Precedents in the literature have in their structures between 4 and 5 chiral centers (as in the case of the original AVPI peptide), and in some examples the synthesis has up to 10 chemical steps to achieve the final product.5c,11
In conclusion, we have designed a simplified analog using the AVPI tetrapeptide as template. The expedient synthetic route (6 steps) combined with binding activity of this new compound (T5TR1) represents an alternative to other more complex compounds attacking the XIAP BIR3 domain. This lead compound presents the structural novelty of a thiazole ring fusioned to 5 member ring carbocycle (2-amino-5,6-dihydro-4H-cyclopenta[d]thiazole-4-carboxylic acid) as a peptide surrogate. New analogs inspired by this scaffold are undergoing evaluation in our laboratory.
Acknowledgments
This work was supported by the NIH PO1 grants CA055164, U01-CA113318.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Reed J. Nat Rev Drug Disc. 2002;1:111. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 2.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mol Cell. 2003;11:519. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Hermann J, Wu JC, Fesik SW. Nature. 2000;408:1004. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 4.Oost TK, Sun C, Armstrong RC, Assad A-S, Betz S, Deckwerth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg S, Shoemaker A, Tomaselli K, Zou H, Fesik SW. J Med Chem. 2004;47:4417. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 5.a) Park CM, Sun C, Olejniczak ET, Wilson A, Meadows R, Betz S, Elmore S, Fesik SW. Bioorg Med Chem Lett. 2005;15:771. doi: 10.1016/j.bmcl.2004.11.010. [DOI] [PubMed] [Google Scholar]; b) Sun H, Coleska ZN, Yang CY, Xu L, Liu M, Tomita Y, Pan H, Yoshika Y, Krajewski K, Roller P, Wang S. J Am Chem Soc. 2004;126:16686. doi: 10.1021/ja047438+. [DOI] [PubMed] [Google Scholar]; c) Sun H, Coleska ZN, Yang CY, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller P, Stuckey J, Wang S. J Am Chem Soc. 2007;129:15279. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wist AD, Gu L, Riedl S, Shi Y, McLendon GL. Bioorg Med Chem. 2007;15:2935. doi: 10.1016/j.bmc.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Wender PA, Verma VA, Paxton TJ, Pillow TH. Acc Chem Res. 2008;41:40. doi: 10.1021/ar700155p. [DOI] [PubMed] [Google Scholar]
- 7.The structure was minimized using AM1 semi-empirical method in the program HyperChem 7.5 for windows.
-
8.Docking result for the thiazole ring fused to a 7 member ring carbocycle using SYBYL 8.0 and the XIAP-BIR3 domain crystal structure with RCSB PDB ID 2jk7.
- 9.Zobel K, Wang L, Varfolomevv E, Franklin MC, Elliott LO, Wallweber HJA, Okawa DC, Flygare JA, Vucic D, Fairbrother WJ, Deshayes K. ACS Chem Biol. 2006;8:525. doi: 10.1021/cb600276q. [DOI] [PubMed] [Google Scholar]
- 10.Hedberg C, Kallstrom K, Brandt P, Hansen LK, Andersson PG. J Am Chem Soc. 2006;128:2995. doi: 10.1021/ja057178b. [DOI] [PubMed] [Google Scholar]
- 11.These compounds did not exhibit activity at concentration of 100μM.
- 12.The Ki value was calculated according to; Coleska-N Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Anal Biochem. 2004;332:261. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, Coleska ZN, Qin D, Sun H, Yang CY, Bai L, Qiu S, Wang Y, Ma D, Wang S. J Med Chem. 2009;52:593. doi: 10.1021/jm801101z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Elliot LO, Wallweber HJA, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. Cell. 2007;131:669. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]


