Abstract
Comparative studies have examined the expression and function of homologues of the Drosophila melanogaster pair rule and segment polarity genes in a range of arthropods. The segment polarity gene homologues have a conserved role in the specification of the parasegment boundary, but the degree of conservation of the upstream patterning genes has proved more variable. Using genomic resources we identify a complete set of pair rule gene homologues from the centipede Strigamia maritima, and document a detailed time series of expression during trunk segmentation. We find supportive evidence for a conserved hierarchical organisation of the pair rule genes, with a division into early- and late-activated genes which parallels the functional division into primary and secondary pair rule genes described in insects. We confirm that the relative expression of sloppy-paired and paired with respect to wingless and engrailed at the parasegment boundary is conserved between myriapods and insects; suggesting that functional interactions between these genes might be an ancient feature of arthropod segment patterning. However, we find that the relative expression of a number of the primary pair rule genes is divergent between myriapods and insects. This corroborates suggestions that the evolution of upper tiers in the segmentation gene network is more flexible. Finally, we find that the expression of the Strigamia pair rule genes in periodic patterns is restricted to the ectoderm. This suggests that any direct role of these genes in segmentation is restricted to this germ layer, and that mesoderm segmentation is either dependent on the ectoderm, or occurs through an independent mechanism.
Keywords: Segmentation, Pair rule gene, Pattern formation, Arthropod, Chilopod, Evolution
Highlights
-
•
We use genomic resources to identify 17 Strigamia pair rule gene homologues.
-
•
Sixteen of these are expressed in periodic patterns before the appearance of morphological segments.
-
•
We allocate genes to primary and secondary tiers in the hierarchy based on detailed time series data.
-
•
Expression of pax3/7 and slp homologues is conserved at the parasegment boundary across arthropods.
-
•
Segmental expression is restricted to ectoderm, suggesting mesoderm segmentation is secondary.
Introduction
The genetic dissection of the mechanism of segmentation in the fruit fly, Drosophila melanogaster, has laid the foundation for a growing body of comparative research on other arthropods (Nusslein-Volhard and Wieschaus, 1980; Peel et al., 2005). The Drosophila work identified a number of genes involved in segment pattern formation, and divided them into four functional categories based on their mutant phenotypes. These categories are maternal factors, gap genes, pair rule genes and segment polarity genes. Work over the years has pieced together the hierarchy of interactions between these genes, and shown how this hierarchy is capable of reproducibly generating precise patterns (Akam, 1987; Jaeger et al., 2004; Pankratz and Jaeckle, 1993).
Homologues for most of the Drosophila segmentation genes can be identified throughout the arthropods. The key segment polarity genes wingless, engrailed, hedgehog and Cubitus interruptus show highly conserved expression at the parasegment boundary in a range of arthropod species, including chelicerates and myriapods, as well as insects (Damen, 2007; Farzana and Brown, 2008). Thus, the parasegment is likely to be an ancient feature of arthropod segmentation. However, the extent of conservation in the gene regulatory network that acts upstream of the segment polarity genes is not yet clear (Peel et al., 2005).
Besides Drosophila, there are two other insects for which extensive functional data are available on the role of pair rule gene homologues: the red flour beetle Tribolium castaneum (Choe and Brown, 2007, 2009; Choe et al., 2006) and the honeybee Apis mellifera (Wilson and Dearden, 2012). In the honeybee, transcripts for a number of the pair rule gene homologues are deposited into the oocyte and have acquired novel roles in early developmental patterning, which obscure any later roles in segmentation in RNAi experiments (Wilson and Dearden, 2012). Therefore, the main work able to address the role of these genes in segment pattern formation is in Tribolium. There are a number of differences in the roles of pair rule genes between Tribolium and Drosophila. For example, the beetle homologues of the pair rule genes hairy, odd-paired (opa) and fushi-tarazu (ftz) appear to have no essential role in Tribolium trunk segmentation, or at least give no detectable RNAi phenotype (Aranda et al., 2008; Choe et al., 2006).
One aspect of segment patterning that is largely conserved between Tribolium and Drosophila is the division of the pair rule genes into primary and secondary tiers, based on their functional position in the hierarchical cascade. Primary pair rule genes are upstream in the cascade and are regulated by other factors (maternal coordinate factors and gap genes), whereas the secondary pair rule genes are downstream in the cascade and regulated by the primary pair rule genes. In Drosophila, even-skipped (eve), runt and hairy are considered the primary pair rule genes; sloppy-paired (slp) and paired (prd) are considered the secondary pair rule genes. ftz and odd-skipped (odd) were originally considered to be secondary, but a more recent and thorough analysis of their cis-regulatory architecture has shown that in some respects they qualify as primary pair rule genes (Schroeder et al., 2011). In Tribolium, eve, runt, and odd are found to be primary pair rule genes, and slp and prd to be secondary pair rule genes. Thus, not only do many of the Tribolium pair rule gene homologues play a role in segmentation, they are also found to occupy similar levels in the gene regulatory hierarchy. (Choe et al., 2006).
In non-insect arthropods there is indirect evidence that the functional division of the pair rule network into primary and secondary levels is conserved. Importantly, the timing of expression of the pair rule genes in segment patterning reflects the functional division of the hierarchy. That is to say, the alternate expression of the primary pair rule genes is established first in the process, and the periodic patterning of the secondary pair rule genes appears afterwards. Therefore in arthropods where functional tools are not available, indirect evidence for the functional division can still be obtained from the temporal order in which the genes are expressed during segment pattern formation. This has been done for spider (Damen et al., 2005) and millipede segmentation (Janssen et al., 2011). A consistent finding across studies is the early expression, and where tested, upstream function of eve, runt and odd homologues during segment patterning; and the late expression, and where tested, downstream function of prd and slp homologues. This suggests that the functional division between primary and secondary pair rule genes, and at least some of the genes that occupy these categories, may be a conserved feature of arthropod segmentation.
A second aspect of segmental patterning conserved between Tribolium and Drosophila concerns the role of the secondary pair rule genes, prd and slp, in the regulation of the key segment polarity genes, wingless (wg) and engrailed (en) (Choe and Brown, 2009). In both insects, the prd homologue is expressed in both wg- and en-positive cells, and thus overlaps the parasegment boundary, whereas the slp homologue is restricted to wg-positive cells. The same relationship is observed between these four genes in the pill millipede Glomeris marginata (Janssen et al., 2011), and the available data suggests that it holds for a pax3/7 homologue and en in the grasshopper Schistocerca americana (Davis et al., 2001). This striking conservation of relative expression suggests an ancient origin for the regulatory module in which these four genes act, even accepting that conserved transcriptional output cannot be taken to imply that the underlying transcriptional networks are also conserved (Ludwig et al., 2000; Romano and Wray, 2003).
We have tested whether these features of the pair rule gene network are also conserved in another ancient branch of the myriapods – the Chilopoda. Our study species is the geophilomorph centipede, Strigamia maritima. Previous work on Strigamia has characterised a number of the major features of segmentation (Brena and Akam, 2012; Chipman et al., 2004b; Kettle et al., 2003). At the time of germ band formation, a large population of progenitor cells forms a disc of unsegmented tissue at the posterior of the embryo. As the germ band elongates, this posterior disc narrows and segments emerge from it sequentially in anteroposterior order. Because segments are added in a temporal sequence, the progression of segment patterning can be visualised in each embryo as a sequence from posterior to anterior.
Expression of some of the pair rule gene homologues has already been examined in Strigamia – two even-skipped homologues (Sm-eve1 and Sm-eve2); one member of the odd-skipped gene family (Sm-odr1); and two members of the hairy/deadpan family (originally named Sm-hes1 and Sm-hes4) (Chipman and Akam, 2008; Chipman et al., 2004a). This work has shown that there are two phases of patterning during the major stage of trunk segmentation. In the first phase, dynamic patterns of gene expression resolve to a series of concentric rings in the posterior disc, defining a double segment repeat. In the second phase, the double segment pre-pattern resolves to a single segment repeat by the splitting and/or intercalation of expression domains. The region where this segmental resolution occurs, in between the posterior disc and the segmented germ band, is called the transition zone. The domain of dynamic expression is restricted to a population of cells surrounding, but largely anterior to, the proctodeum and lying within the first resolved ring of expression. We refer to this territory as the peri-proctodeal region.
We set out to test four hypotheses about the degree of conservation of the pair rule gene homologues in segment pattern formation. First, are the pair rule genes in Strigamia hierarchically organised into primary and secondary tiers, as has been described in other arthropods? Second, are the relative expression domains of key genes conserved during the specification of the parasegment boundary? In particular, is the spatial registration of slp and pax3/7 homologues conserved in relation to the abutting wg- and en-expressing cells? Third, is there evidence from the relative expression of eve, runt, odd and hairy homologues that the upstream tiers of segment patterning are more divergent? Fourth, is the expression of the Strigamia pair rule genes restricted to the ectoderm germ layer? And if not, are any of these genes expressed in patterns that suggest an early role in mesoderm segmentation? We have addressed these hypotheses by examining the spatial and temporal dynamics of expression of a complete set of Strigamia pair rule homologues during segment patterning.
Materials and methods
Embryo collection, fixation and staging
Embryos were collected in the field from a population near Brora, Scotland and fixed as described previously (Chipman et al., 2004b). Embryos were staged by counting the total number of morphologically visible leg-bearing segments, either in whole embryos or in flat-mount preparations.
Gene identification and cloning
A genome and adult and embryonic transcriptomes have recently been assembled for Strigamia maritima (genome release Smar_1.0 http://www.ncbi.nlm.nih.gov/assembly/322118/). Pair rule gene homologues were identified by similarity searches against these resources, and orthology of the genes was confirmed by reciprocal similarity searches. To clone genes, gene-specific primers were designed against the genomic or transcriptomic sequence, and products amplified by standard PCR from embryonic cDNA. Genes were cloned into a pGEM-T Easy vector (Promega). The clones of ftz and twist were a gift of C. Brena, the clones of slp and opa1 were a gift of V. Hunnekuhl and the clone of wg was a gift of L. Hayden. An annotated gene set for Strigamia is provided at EnsemblMetazoa (http://metazoa.ensembl.org/Strigamia_maritima/Info/Index). The Ensembl IDs of the pair rule and segment polarity gene homologues examined in this paper are provided in Supplementary Table 1.
Phylogenetics
Phylogenetic analysis of the genes was carried out using Phylemon2 (Sanchez et al., 2011). The multiple sequence alignment was performed on protein sequences using MUSCLE (Edgar, 2004). The gene trees were built by maximum likelihood analysis in PhyML (Guindon and Gascuel, 2003) using the LG substitution model with 100 bootstrap replicates. We used empirical frequencies, had four substitution rate categories and a proportion of invariant sites, and estimated the gamma distribution parameter. Trees were visualised in the programme FigTree v1.3.1 (Rambaut, 2009). For clarity, any nodes with less than 70/100 bootstrap support were collapsed to polytomies using the programme TreeCollapserCL v3.2 (Hodcroft, 2012).
Whole mount in situ hybridisation
Single and double colorimetric in situ hybridisation reactions were carried out on whole mount embryos as described previously (Chipman et al., 2004b; Chipman and Stollewerk, 2006). After staining, the embryo was dissected away from the yolk with fine tweezers and flat-mounted under a cover slip on a microscope slide. Bright field or DIC images of embryos were taken on a Zeiss Axiophot compound microscope with a Leica DFC 300 FX camera. Fluorescent images and optical sections were taken using a Leica TCS SP5 confocal microscope. The photos were adjusted using Adobe Photoshop (version CS5).
A note on terminology
The term “pair rule” has been used in three different ways in the literature. The original sense of the term was a functional categorisation based on the loss-of-function mutant phenotype (Nusslein-Volhard and Wieschaus, 1980). Second, the term was used to describe genes expressed in a double segment pattern, even if the loss-of-function phenotype was not consistent with the classical pair rule phenotype (Choe et al., 2006). Third, the term has been used as a convenient ‘category-level name’ for the set of genes in other species that are orthologous (one-to-one or one-to-several) to the set of genes historically identified in Drosophila under that name (Damen et al., 2005; Janssen et al., 2011). This third use only implies homology of the genes themselves between the taxa of interest, and explicitly makes no statement on homology at the level of expression or function of the genes. In this paper, we use the term “pair rule gene” in this third sense.
Results
A complete set of pair rule gene homologues identified and cloned in Strigamia
Based on current genomic and transcriptomic resources, we believe that we have identified and cloned all the Strigamia homologues of the eight canonical Drosophila pair rule genes. In total 17 genes are identified and characterised: one homologue each of runt (Sm-run), sloppy-paired (Sm-slp) and fushi-tarazu (Sm-ftz); two homologues of the paired/gooseberry/gooseberry-neuro class of genes (Sm-pax3/7-1 and Sm-pax3/7-2); two homologues of odd-paired (Sm-opa1 and Sm-opa2); three homologues each of even-skipped (Sm-eve1, Sm-eve2 and Sm-eve3) and odd-skipped (Sm-odr1, Sm-odr2, Sm-odr3); and four hairy/deadpan homologues (Sm-h1, Sm-h2, Sm-h3, Sm-h4). Phylogenetic analysis confirms that there is no ambiguity about the many-to-many orthology of the genes examined, and suggests that, with the exception of hairy, where there are multiple gene family members in Strigamia, these are the result of lineage-specific duplications within the arthropods (Fig. S1). In general, it is not possible to define one-to-one orthologous relationships between the genes. For example, the myriapod family of eve genes form a separate clade from the insect family, but relationships within the myriapod clade are otherwise unresolved (Fig. S1A).
Phylogenetic analysis identifies opa2 as a fast-evolving, divergent sequence (Fig. S1B). opa2 appears not to be expressed in embryos – transcripts are not identified in the embryonic transcriptome, and the gene failed to amplify from embryonic cDNA (results not shown). However, reads for this gene are present in the adult transcriptome, showing that opa2 is expressed in adult tissue. The high divergence of the sequence is consistent with the gene adopting a specialised or novel role in adults.
There are multiple runt-related genes in other arthropods, for example runt, lozenge, runxa and runxb in Drosophila (Bao and Friedrich, 2008). Analysis of the amino acid sequence of the single Strigamia runt homologue confirms its orthology to the arthropod runt genes. It contains the canonical runt DNA-binding domain and WRPY motif (results not shown). However, there is no good evidence for a one-to-one orthologous relationship between the Strigamia runt gene and any other arthropod runt-related genes (Fig. S1C).
Three odd homologues are identified in Strigamia by the presence of conserved zinc-finger domains and their high similarity to odd family members from other arthropods (results not shown). Phylogenetic analysis shows no strong evidence for any one-to-one orthology between the Strigamia genes and the insect odd/sob/bowl class of genes, nor between them and any chelicerate odd family members (Fig. S1D). We therefore refer to these genes as odd-skipped-related1, 2 and 3 (odr1, odr2 and odr3).
Two putative homologues of the Drosophila prd gene are identified in Strigamia. The Drosophila pair rule gene prd, and the closely related gooseberry genes, are the Drosophila Pax3/7 family members. There are four major families of Pax genes in bilaterians – Pax1/9, Pax2/5/8, Pax4/6 and Pax3/7 (Balczarek et al., 1997). Phylogenetic analysis shows that the two Strigamia homologues form a well-supported Pax3/7 clade to the exclusion of the other Pax families (Fig. S1E). However, orthology relationships within the clade are not well resolved. We therefore refer to these genes in Strigamia as pax3/7-1 and pax3/7-2.
slp is a Fox family transcription factor. There are 17 subclasses in the Fox family (Mazet et al., 2003). In Drosophila melanogaster, there are three members of the FoxG subclass, slp1, slp2 and forkhead domain 19B (Lee and Frasch, 2004; Mazet et al., 2003). Phylogenetic analysis shows that there is only one member of the FoxG subclass in Strigamia, which we refer to as slp (Fig. S1F).
The Strigamia orthologue of ftz is identified by conserved residues in the homeodomain (Fig. S1G) and by its genomic position in a conserved Hox cluster – it lies between the orthologues of Sex combs reduced and Antennapedia (genome release Smar_1.0).
Phylogenetic analysis of hairy, deadpan, enhancer of split and other basic helix-loop-helix (bHLH) genes was carried out by L. Duncan and P. Dearden as part of the Strigamia maritima genome annotation (unpublished data). It identified four members of a hairy/deadpan clade in Strigamia. These genes are distinct from genes in the enhancer of split clade, and from other bHLH genes. Therefore we refer to these as hairy1, 2, 3 and 4 (h1, h2, h3 and h4). h1, h2 and h3 are linked in a 30 kb genomic cluster, whereas h4 is on a separate genomic scaffold. Four hairy/enhancer of split (HES) related genes were reported previously (Chipman and Akam, 2008). Two of these, both isolated as cDNA clones, correspond to two of the genes defined in the genome annotation: hes1 becomes h4, and hes4 becomes h3. The other two (hes2 and hes3) were previously recovered only as short fragments from a PCR screen. They show no close match at the nucleotide level to any sequence in the Strigamia genome or transcriptome assemblies. They may be derived from contaminating PCR fragments in the original study.
A hierarchy of primary and secondary pair rule genes in Strigamia
We report here the expression of 16 pair rule gene homologues during Strigamia trunk segmentation (Figs. 1 and S2). All the embryos examined are at stage 4.2 or early 4.3, during the major phase of segment addition; stage 4.2 embryos have between ≈14 and ≈27 leg-bearing segments (Brena and Akam, 2012). Note that our descriptions are restricted to the segmenting tissue of the embryo, including the peri-proctodeal territory, but they exclude the tissues of the proctodeum itself and the immediately adjacent tissue. These comprise a unique set of cell populations, including those associated with the future gut. They will be described in detail elsewhere.
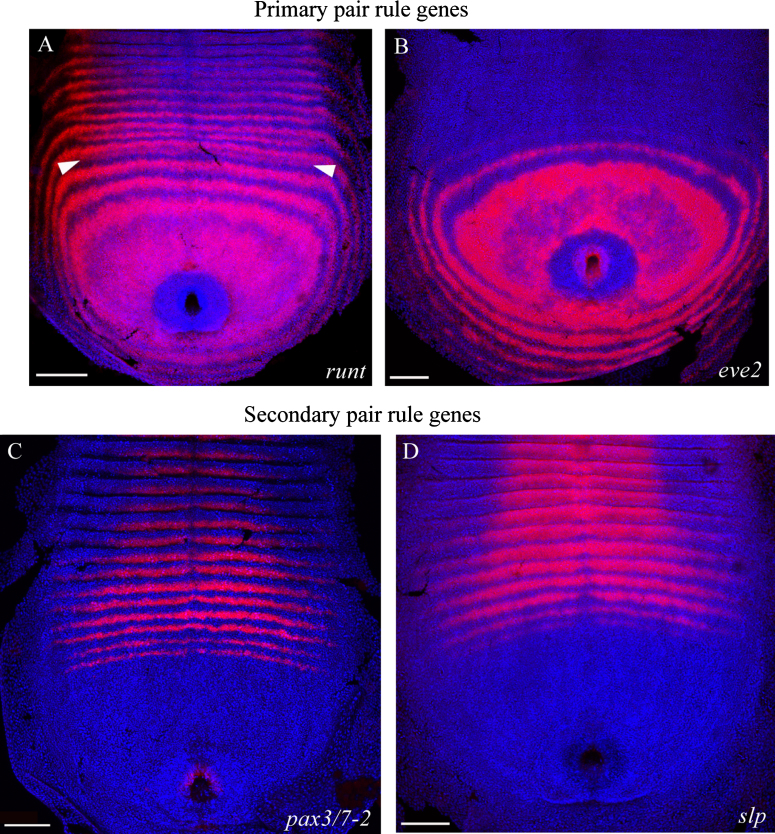
Fig. 1.
Examples of primary and secondary pair rule genes in Strigamia. (A) Expression of the primary pair rule gene runt in the posterior disc and segmenting germ band of a stage 4 Strigamia embryo with 23 leg-bearing segments (lbs). runt initiates as a dynamic domain in the peri-proctodeal region and resolves to a series of concentric rings at double segment periodicity. These broad stripes then resolve to a single segment repeat through the intercalation of a shorter stripe between two broad stripes (white arrowheads). (B) Expression of eve2 at a similar stage, 17 lbs. eve2 initiates in a similar pattern to runt, but is inactivated after one or two rings have resolved from the dynamic domain. It never undergoes resolution to a single segment periodicity. (C) Expression of the secondary pair rule gene pax3/7–2, 28 lbs. (D) Expression of slp, 33 lbs. pax3/7-2 and slp initiate at a more anterior position in the germ band, and are only ever expressed at single segment periodicity. Anterior is to the top. Scale bar is 100 μm. For similar data on the complete set of primary and secondary pair rule genes, see Supplementary Fig. 2.
With the exception of opa2, all of the genes studied are expressed in the tissue of the posterior disc and/or the forming germ band, before the appearance of morphological segments. Therefore, these genes may play some role in segment patterning. All of the genes are activated before the first expression of the segment polarity gene homologues en and wg (Fig. 3). This is consistent with a hierarchical role upstream of parasegment boundary specification.
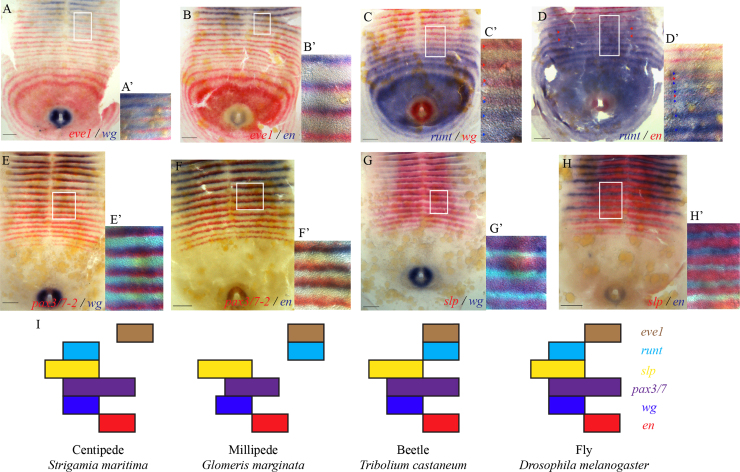
Fig. 3.
Conserved aspects of gene expression at the parasegment boundary. (A) Expression of eve1 (red) and wg (blue) on a stage 4 embryo with 21 lbs. (A′) eve1 and wg stripes are out of phase. (B) eve1 (red) and en (blue), 21 lbs. (B′) en overlaps the anterior margin of eve1, but with an anterior offset. (C) runt (blue) and wg (red), 30 lbs. (C′) wg stripes appear directly overlapping runt. wg transcripts appear at the same time that runt transcripts are clearing from cells. (D) runt (blue) and en (red), 17 lbs. (D′) runt and en stripes are out of phase. (E) pax3/7-2 (red) and wg (blue), 20 lbs. (E′) The first stripe of wg appears overlapping with the anterior one or two cell rows of the pax3/7-2 stripe, and maintains this registration until morphological segmentation occurs. (F) pax3/7-2 (red) and en (blue), 28 lbs. (F′) The first stripe of en appears overlapping with the posterior one or two cell rows of the pax3/7-2 stripe. (G) slp (red) and wg (blue), 33 lbs. (G′) The first stripes of wg are overlapping with the stripes of slp expression. (H) slp (red) and en (blue), 29 lbs. (H′) Expression of en and slp are out of phase. Panels are higher magnification DIC images of the white boxed areas. Blue arrowheads in C′ and D′ indicate runt stripes, and red arrowheads indicate wg or en stripes respectively. Lighter colours in C′ indicate the gradual appearance of wg transcripts at the same time that runt transcripts are clearing. Embryos are flat-mounted preparations. Anterior is to the top. Scale bar is 100 μm. (I) Schematic comparison of the relationships of expression domains of key genes at the parasegment boundary between insects and myriapods. In Drosophila and Tribolium, the mechanism of wg and en regulation is different in alternate parasegments, and the relationship of expression domains of some genes is also different in alternate parasegments. For Drosophila the summary of relative gene expression is shown for the parasegment boundaries between odd-numbered en stripes and even-numbered wg stripes. But for Tribolium, the expression data is shown for the opposite parasegment registration, that is to say, even-numbered en and odd-numbered wg stripes. Anterior is to the left. Modified from Janssen et al., 2011.
From an analysis of the expression dynamics, there is good evidence that the Strigamia pair rule genes are hierarchically organised into primary and secondary tiers. A subset of the genes is activated early in the segmentation process and initiates at a double segment periodicity in the posterior disc (Figs. 1A, B and S2A–K). This set of genes includes all the Strigamia homologues of eve, runt, odd and hairy. The activation of these genes early in the progression of the pattern is consistent with them operating upstream in the regulatory network. We shall refer to them as primary pair rule genes. Most of these genes resolve to a single segment repeat by the intercalation or splitting of expression domains in the transition zone (Fig. 1A). However, two of them (eve2 and odr1) are inactivated before this, and never make the transition to single segment periodicity (Figs. 1B and S2D).
In contrast, a second set of genes is first activated at a more anterior position in the germ band. These are pax3/7-2 and slp (Fig. 1C, D). We designate these as secondary pair rule genes because their later activation in the development of the pattern is consistent with them occupying a downstream tier in the network. They show no evidence of being activated in a double segment pre-pattern. Rather, they initiate at single segment periodicity, coincident with or shortly after, the transition of the primary pair rule genes to a single segment repeat (Fig. S2M and O). Their expression never appears before the primary pair rule genes undergo this transition.
Therefore, there is clearly a division into two classes of genes that initiate at different periodicities and at different points in the progression of pattern formation. This is consistent with a hierarchical organisation of these gene classes in the regulatory network. It is striking that homologous genes occupy the same relative positions in the hierarchy as in other arthropods examined to date.
Three genes do not fit comfortably into either of these categories, pax3/7-1, opa1 and ftz (Fig. S2P, Q and R). pax3/7-1 is not expressed in the ectoderm before morphological segmentation, but is restricted to a subset of mesodermal cells (Fig. S2P and S4N). This is addressed in more detail in a following section.
Transcription of opa1 initiates in the posterior disc and transcripts persist through the transition zone and into nascent morphological segments (Fig. S2Q). However, opa1 expression shows no modulation in relation to the forming segment pattern; any appearance of modulation is actually caused by cell rearrangements during the folding in of furrows at morphological segmentation.
The expression of the ftz orthologue exhibits a double segment modulation. It initiates as broad double segment stripes at the anterior margin of the peri-proctodeal region. In the transition zone these resolve into alternating long and short segmental stripes. However, it does not show any dynamic expression in the peri-proctodeal region itself (Fig. S2R). This suggests that ftz is entrained by the upstream pair rule patterning system, but is not involved in the primary pattern generation.
Map of relative expression domains for the Strigamia pair rule genes
We have determined the registration of the primary and secondary pair rule genes, with respect to each other and to the segment polarity genes, using double in situ hybridisation. These data are summarised in an expression map (Fig. 2A, further supporting data in Fig. S3).
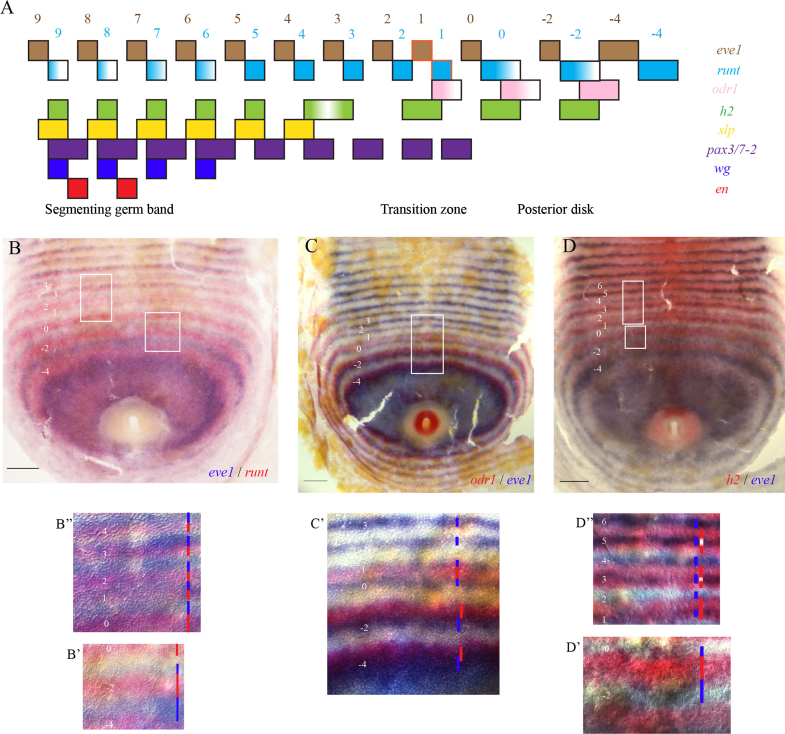
Fig. 2.
Schematic time series of expression of the Strigamia pair rule genes during the generation of periodic patterns. Panel A summarises the expression of pair rule and segment polarity genes during the progression of segment pattern formation in Strigamia. Each row represents the expression of a single gene along the anteroposterior axis from the posterior disc up to the specification of the first two parasegment boundaries. Anterior is to the left. Effectively, this presents a time series running from right to left. The relative horizontal position of domains summarises the degree of overlap, or otherwise, of the expression of the respective genes. For eve1 and runt, an orange outline highlights the first appearance of stripes intercalating between the double segment stripes in the transition zone. Colour fading inside the boxes represents gradual clearing of the transcripts from cells. Brown and blue numbering indicates the relative position of eve1 and runt stripes respectively. Stripe number 1 is the first intercalating stripe (see below). Panels B–D present examples of the data from which the map in panel A was generated. For a more complete dataset, see Supplementary Fig. 3. (B) Expression of eve1 (blue) and runt (red) on a stage 4 embryo with 18 lbs. (C) Expression of odr1 (red) and eve1 (blue), 22 lbs. (D) Expression of h2 (red) and eve1 (blue), 17 lbs. Panels below are higher magnification DIC images of the white boxed areas in B, C, D. Coloured bars in panels are schematic representations of the accompanying stripes of gene expression, indicating stripe width and inter-stripe spacing. (B′) highlights boxed area on right hand side, (B″) on left hand side in B. (D′) highlights the lower, (D″) the upper boxed area in D. The stripe numbering indicates the relative position of a stripe in the pattern. The first intercalated stripe of expression is annotated as stripe 1. This is the first stripe that marks the transition to single segment periodicity. For stripes anterior to stripe 1, these are numbered in posterior-to-anterior progression 2, 3, 4 etc. The stripes posterior to stripe 1 are at double segment periodicity, and are thus numbered backwards in twos, in anterior-to-posterior order 0,−2, −4 etc. up to and including the first double segment ring which is clearly demarcated from the dynamic domain. Importantly, the numbering only pertains to one of the two genes shown on the double stain. In the bottom right-hand corner of each primary image, the two genes stained are indicated. The gene on the right-hand side of the separator defines the gene to which the stripe numbering annotated on the image applies. In all cases this gene is either runt or eve1. Anterior is to the top. Scale bar is 100 μm.
The map confirms previous work showing that there are two phases of periodic patterning – a double segment pre-pattern and the resolution of this to a single segment repeat. None of the genes that we have examined provide any evidence for single segment patterning occurring before cells enter the transition zone. Also, the summary shows that although the primary pair rule genes are expressed in similar patterns, they are expressed in different phases of the pattern repeat, and thus have distinct relationships with one another (Fig. 2A).
Fig. 2B, C, and D show three representative examples comparing the expression of runt, odr1 and h2 with eve1. For eve1 and runt, at the anterior margin of the peri-proctodeal region, the dynamic pattern resolves into adjacent, space-filling stripes of runt and eve1 that are at least three or four cell rows wide (Fig. 2B, B′). These stripes then narrow to around two cell rows, such that runt continues to abut eve1 at its anterior margin, but an inter-stripe territory expressing neither gene appears posterior to the runt stripe, anterior to the next eve1-expressing cells. During the resolution to a single segment repeat, the intercalating stripes appear within this gap, and when fully resolved have the same relationship to one another as the stripes that derive from the narrowing of the primary stripes (Fig. 2B″).
For odr1 and eve1, the pattern resolves initially into two overlapping stripes with a small anterior offset of odr1 (Fig. 2C, C′). This pair of stripes is then separated from the next pair by non-expressing cells. Moving more anteriorly, the offset becomes more pronounced, such that they mature into largely non-overlapping stripes, with odr1 abutting the anterior margin of eve1. In the transition zone, the intercalating stripe of eve1 appears at the anterior border of the odr1 stripe, and not overlapping with odr1. odr1 is then inactivated.
For h2 and eve1, during the double segment phase of the pattern, the expression dynamics are almost identical to those of runt and eve1, as described above (Fig. 2D, D′). In this phase, h2 and runt are almost directly overlapping (Fig. S3G). However, h2 has a different mode of transition to single segment periodicity. For eve1 and runt, it seems that the intercalating stripes appear de novo in between the primary stripes (Fig. S2A, G). The h2 primary stripes, on the other hand, appear to split by the repression of expression in cells within the stripe. The intercalating stripe of eve1 is activated inside the h2-positive domain, approximately in the middle but with a posterior offset (Fig. 2D, D″). The h2 transcripts are repressed in these eve1-positive cells. This repression leaves the anterior and posterior rows of h2-expressing cells intact, and separated from each other by non-expressing cells, thus producing segmental stripes.
A further major inference that can be drawn from the expression comparison is on the relative expression of different members of a gene family with respect to one another. Strigamia has three members each of the eve and odr gene families, and four members of the hairy family. We examined the expression of each gene with respect to a common reference gene, runt or eve1, in order to determine the registration of the family members to one another.
Each eve gene is activated early, in the peri-proctodeal region, but subsequently inactivated at a different position along the anteroposterior axis, so each is expressed in a distinct subset of the overall pattern. In each phase of the pattern, all three eve genes are expressed in the same spatial relationship with runt (Fig. S3A, B, C). Therefore, from the resolution of the first double segment ring, we can infer that all the eve family members are expressed in phase with one another. That is to say, in phases of the pattern where two or three of the genes are expressed, the transcripts are in the same cells and the stripes directly overlap.
The same logic can be applied to the odr family and the same conclusion drawn. Each odr gene is inactivated at a different anteroposterior position, but all three are expressed in the same spatial relationship with respect to eve1 (Fig. S3D, E, F).
The situation is different for the hairy family. h2 and h3 have distinct spatial relationships with runt, and are therefore not expressed in phase with one another (Fig. S3G, H). Unfortunately, because the signal from the h1 and h4 probes is so weak, it has not been possible to obtain registration information from double stains with these probes.
Finally, the distinct expression domains of the Pax3/7 gene family warrant comment (Fig. S2L and P). The two members of this family in Strigamia are orthologous to three genes in Drosophila – prd, gooseberry and gooseberry-neuro (see above). pax3/7-1 is activated later than pax3/7-2 in the ectoderm, and only in medial cells that are likely to be neurectoderm. This neurectodermal expression of pax3/7-1 resembles that of gooseberry-neuro in Drosophila, whereas the expression of pax3/7-2 is most similar to the later segmental expression of paired, and of gooseberry. However, the one-to-one orthology of genes in this family remains uncertain.
Conserved core of pax3/7-2 and slp expression domains during parasegment boundary specification
Our observations support the hypothesis that the en/wg juxtaposition (i.e. the parasegment boundary) is the primary patterning boundary in each Strigamia segment, as it is in other arthropods. Fig. 2 shows that each new en stripe appears posteriorly adjacent to a wg stripe, and does not overlap with it. At the mid-stage in trunk segmentation depicted here, en expression is delayed with respect to wg, such that wg stripes appear two or three segments ahead of en (Fig. S3J). This temporal relationship changes at different stages of the segmentation process (data not shown). Even so, abutting stripes of wg and en are consistently established in each segment before any sign of morphological segmentation. When the segmental furrow forms, it lies immediately behind the en stripe. Later this furrow will carry the en-expressing cells down into it (Eibner, 2010) (L. Hayden and W. Arthur, in prep).
Fig. 3 compares the relative expression domains of some key genes at the point of parasegment boundary specification in Strigamia with the data available in other arthropods. This comparison shows that the expression domains of the secondary pair rule genes slp and pax3/7-2 have a conserved registration with the domains of wg and en at the parasegment boundary.
Each new stripe of wg appears overlapping with the anterior one or two cell rows of a pax3/7-2 stripe, and maintains this registration until morphological segmentation occurs (Fig. 3E, E′). Conversely, new stripes of en appear overlapping with the posterior one or two cell rows of the pax3/7-2 stripe, and similarly maintain this registration (Fig. 3F, F′). For technical reasons it has not been possible to monitor the expression of all three genes simultaneously, but it is clear that pax3/7-2 is co-expressed in some, if not all, of both the wg- and en-positive cells, and thus overlaps the parasegment boundary.
Each new stripe of wg also overlaps with a stripe of slp expression (Fig. 3G, G′). Within the limitations of double colorimetric in situ, it appears that all wg-positive cells are encompassed within the slp domain. In contrast, the expression of en and slp appear to be out of phase, with no or very little overlap (Fig. 3H, H′).
These observations confirm the conservation of expression of the key secondary pair rule genes at the parasegment boundary across arthropods (Fig. 3I).
Evolutionary flexibility of expression of primary pair rule genes
The eve, runt and hairy homologues show a greater degree of divergence in the spatial relationships of their expression than the secondary pair rule or segment polarity gene homologues (Fig. 3).
Examining the registration of eve1 at the parasegment boundary, we observe that the wg stripes are out-of-phase with the eve1 stripes (Fig. 3A, A′). The appearance of the first en stripe is partially overlapping with the anterior margin of the eve1 stripe (Fig. 3B, B′). It appears that the en stripe is anteriorly offset, such that some en-expressing cells are outside of the eve domain. This spatial relationship between eve1 and en differs from that in both Glomeris, where there is no overlap, and in Drosophila and Tribolium, where the en stripe is entirely within the eve domain (Fig. 3I) (Janssen et al., 2011; Lawrence et al., 1987; Patel et al., 1994).
For runt, the first stripes of en and wg initiate in a region of the germ band where runt transcripts are beginning to clear from cells (Fig. 3C, D). The first stripes of wg expression initiate directly on top of the runt stripes (Fig. 3C, C′). The first stripes of en appear in between the segmental stripes of runt, and do not overlap (Fig. 3D, D′). At the mid-segmentation stage examined here, soon after the activation of two stripes of wg or en, runt transcripts have completely disappeared from this region of the germ band. Comparing this pattern with other arthropods (Fig. 3I), the spatial relationship of runt with respect to eve and to the parasegment boundary has diverged between flies and beetles, and between centipedes and millipedes.
Furthermore, in the double segment phase of segment patterning, the registration of the runt homologue and at least two of the hairy homologues has diverged between fly and centipede. In Drosophila, the double segment stripes of hairy and runt are expressed out-of-phase. In the double segment phase of Strigamia however, runt and h2 are expressed in phase; and runt and h3 stripes are partially overlapping (Fig. S3G, H). Overall, these observations confirm earlier suggestions from other arthropods that the upstream tiers of segment patterning are more evolutionarily flexible (Peel et al., 2005).
Periodic expression of Strigamia pair rule genes is restricted to the ectoderm
From an analysis of nuclear-stained embryos in cross-section, it is clear that there are two distinct cell layers in the posterior disc (Fig. 4). It is most likely that the surface layer is the ectoderm, and the deep layer is the mesoderm. To test this, we examined the expression of the Strigamia homologue of the mesodermal marker gene twist (Fig. 4D). At the stage examined, the surface cells never express twist, consistent with an ectodermal identity. Except in the immediate vicinity of the proctodeum itself, the deep cells posterior to the transition zone are also twist-negative. However, deep cells in the transition zone do express twist. These twist-expressing cells are initially loosely organised, but more anteriorly they line up into rows that come to lie directly underneath the ectodermal segments. More anteriorly still, these cells organise themselves into coelomic sacs. The twist-negative deep cells of the posterior disc are continuous with the twist-positive cells that will form the mesoderm in the segmented germ band. Therefore on the basis of the behaviour and topology of the cells, as well as the expression of a mesodermal marker gene, we conclude that these cells are indeed mesodermal precursors.
Fig. 4.
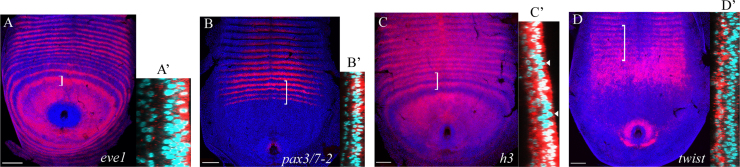
Expression of pair rule genes in the ectoderm and mesoderm. In all panels, the side panels show a higher magnification, orthogonal section along the anteroposterior axis of the bracketed area in the main panel. (A) Expression of eve1 in the posterior disc and germ band of a stage 4 embryo with 20 leg-bearing segments. (A′) eve1 expression is restricted to the ectoderm. Transcripts appear to be predominantly apically localised. (B) pax3/7-2 expression, 28 lbs. (B′) pax3/7-2 expression is restricted to the ectoderm. (C) h3 expression, 23 lbs. (C′) The striped expression of h3 is restricted to the ectoderm. White arrowheads indicate the ectodermal inter-stripe regions. h3 has an additional uniform expression domain in the mesoderm. (D) twist expression, 18 lbs. (D′) twist is restricted to the mesoderm. The twist-positive cells line up into rows and arrange themselves underneath the segmented ectoderm. In the overview and accompanying cross-section images, the RNA is stained in red and shown overlaying the nuclear stain. In the overview images, nuclei are shown in dark blue. In the cross-section images the nuclei are shown in cyan, which gives a clearer distinction of the two germ layers and the expression of the RNA in relation to them. Embryos are flat-mounted preparations. Anterior is to the top. In cross-sections, the ectoderm is oriented on the right, and the underlying mesodermal cells to the left. Scale bar is 100 μm (overviews only).
For all the pair rule gene homologues, the expression of the genes in dynamic and periodic patterns, in the posterior disc and transition zone, is restricted to the ectodermal cell layer (Figs. 4A, B and S4). Labelling of the mesoderm is much weaker, probably at background levels, and this labelling is not modulated into double or single segment patterns. This suggests that any direct role of these genes in segment pattern formation is restricted to the ectoderm.
There are two genes that have additional domains of expression in the mesoderm, h3 and pax3/7-1 (Figs. 4C and S4N). h3 is expressed uniformly throughout the mesodermal layer of the posterior disc and continues into the mesoderm of the morphological segments. However, the dynamic and striped domains of expression are restricted to the ectoderm (Fig. 4C). pax3/7-1 is never expressed in the ectoderm before the appearance of morphological segments. It is expressed in mesodermal precursor cells throughout much of the posterior disc (Fig. S4N). As furrows form to demarcate the segments, pax3/7-1 is inactivated in the mesodermal cells, and reinitiates in a specific row of ectodermal cells at the posterior of each segment. This expression is restricted to the ventral neurectoderm territory. This pattern is distinct from that of all other pair rule genes. Importantly, when viewed in XZ or YZ cross-sections, the mesodermal expression of both h3 and pax3/7-1 shows no evidence of modulation into double or single segment patterns. Expression appears to be uniform in intensity. Any appearance of segmental modulation in surface views (XY) is actually caused by cell rearrangements, as the ectodermal furrows fold inwards and the mesodermal cells organise into rows.
In summary, the data support the hypothesis that any direct role of the pair rule gene homologues in the generation of periodic patterning is restricted to the ectoderm. This implies that mesoderm segmentation is either induced by signals from the overlying ectoderm, or occurs through an independent developmental mechanism.
Discussion
We have shown that, with the exception of pax3/7-1 and opa2, all Strigamia homologues of the Drosophila pair rule genes are expressed in the ectodermal cell layer of the posterior disc and/or forming germ band before the formation of morphological segments. Most are expressed in periodic patterns and their expression precedes the activation of the segment polarity genes wg and en, consistent with a role in the generation of segmental pattern.
It might be argued that most genes would show such periodic patterns in the posterior disc and transition zone, in response to a much smaller subset of transcription factors that are actually necessary for segment patterning. To assess this argument, we examined the expression of a sample of 25 other Strigamia genes that we and our colleagues had cloned as mesodermal or neural markers for use in other studies, and which were not known to be involved in arthropod trunk segmentation from studies in other arthropods. None of these 25 is expressed in a dynamic pattern in the peri-proctodeal region; and the majority (18/25) is not expressed before the appearance of morphological segments (data not shown). Therefore, the expression dynamics of the Strigamia pair rule gene set is distinctive, providing suggestive evidence that many if not most of these genes play a role in segmentation.
As we have no tools for gene manipulation in Strigamia, these expression data provide our best proxy for inferring gene function. However, we must be cautious. The patterned expression of a gene during a process does not mean that it plays any necessary role in that process. For example, the striped expression of ftz and hairy in Tribolium is consistent with a function in segmentation, but it is known that RNAi knockdown of these genes gives no detectable trunk segmentation phenotype (Aranda et al., 2008; Choe et al., 2006). In these cases, regulatory linkage into the segmentation gene network might reflect a retained vestige of an ancestral function that is no longer necessary, or a role in coupling the segmentation network to other processes (e.g. possibly Hox gene regulation, in the case of ftz). Such patterns may even have arisen by chance, persisting because they are not deleterious. That being said, we interpret our data as suggesting that the set of transcription factors encoded by the pair rule gene homologues has been involved in segment pattern generation since at least the origin of the major living arthropod lineages.
Conservation of hierarchical network structure
In the two major insect models Tribolium and Drosophila, in at least one representative of the chelicerates, the spider Cupiennius salei, and now in two myriapods, the millipede Glomeris marginata and the centipede Strigamia maritima, there is evidence for a hierarchical organisation of the pair rule genes (Choe et al., 2006; Damen et al., 2000, 2005; Ingham and Gergen, 1988; Ingham, 1988; Janssen et al., 2011; Schroeder et al., 2004). In particular, eve and runt homologues are identified as primary pair rule genes, expressed early and where tested functioning upstream; slp and pax3/7 homologues as secondary pair rule genes, expressed later and functioning downstream, in all species examined. This feature of the network organisation may be more stable than the exact regulatory linkages between individual genes, which have clearly diverged in different insect lineages – and of which we as yet know essentially nothing in non-insect arthropods.
An ancestral arthropod patterning system for specifying the parasegment boundary
In both insects and myriapods, a pax3/7 homologue is expressed across the parasegment boundary in both wg- and en-positive cells; and a slp homologue is expressed only in wg-positive cells (Choe and Brown, 2009; Janssen et al., 2011). Therefore in Strigamia pax3/7 and slp homologues are expressed in the right cells and at the right time to have a function in regulating wg and en during parasegment boundary specification, but this remains to be tested.
Within the insects, the functional data indicates a mixed picture. RNAi experiments in Tribolium confirm that these genes have a role in segment patterning, and are required for the correct establishment of parasegmental stripes of wg and en (Choe and Brown, 2007). However, the regulatory interactions between genes that underlie the execution of this patterning function have diverged from Drosophila (Choe and Brown, 2009). This is consistent with the growing evidence that developmental systems drift is pervasive – the same or similar phenotypic results are frequently accomplished through alternative developmental pathways (True and Haag, 2001).
Given our data in Strigamia, we propose that the involvement of slp and prd in a regulatory module that sets the borders of wg and en stripes is a stable and ancient feature of segment patterning, but that the involvement of other genes and the exact linkages between genes in the module is more evolutionarily labile, and will have diverged in different lineages. It is important to study these questions in other non-insect arthropods, particularly crustaceans and chelicerates, in order to build a more complete catalogue of those parts of the segmentation gene regulatory network which are stable, and those which are more flexible. This comparative dataset will be very useful for testing hypotheses on how the location and connectedness of regulatory modules in a network affect its susceptibility to evolutionary change.
Evolutionary flexibility of primary pair rule genes
In contrast to the high degree of conservation of the secondary pair rule and segment polarity genes at the parasegment boundary, the expression of the primary pair rule genes, during double segment patterning and at the parasegment boundary, appears to be more evolutionarily flexible (Figs. 2 and 3).
The expression of eve1 shows a mixture of conserved and divergent aspects. At the parasegment boundary, eve1 is expressed out-of-phase with wg stripes, and partially overlapping with en stripes at its anterior margin (Fig. 3A, B). The observation that the overlap between the anterior margin of eve1 and the first en stripe is only partial is significant. This is because, assuming there are no differences in expression between eve transcripts and protein, the presence of en-expressing cells outside of the eve1 domain suggests that eve1 cannot be responsible for regulating the entire en stripe. This is different from the situation found in Drosophila and Tribolium, where all en-positive cells are encompassed within the eve stripe (Lawrence et al., 1987; Patel et al., 1994), but more consistent with the pattern described in Glomeris, where all en-positive cells are outside the eve domain (Janssen et al., 2011). It suggests that the role of eve in setting the borders of the en stripes might have diverged or been lost in myriapods.
In comparison with other arthropods, the expression of runt with respect to the other examined genes at the parasegment boundary is particularly flexible. Its relative expression domain is divergent between flies and beetles, and between millipedes and centipedes (Choe and Brown, 2009; Janssen et al., 2011). During double segment patterning, the registration of runt with respect to two of the hairy homologues is also divergent between Strigamia and Drosophila (Fig. S3G, H). This evolutionary flexibility in spatial registration might reflect corresponding differences in its functional relationship with other pair rule genes.
It is known from Drosophila and mammalian systems that runt exhibits context-dependent transcriptional regulation (Collins et al., 2009; Swantek and Gergen, 2004; Walrad et al., 2011). It can act as a transcriptional repressor or activator, depending on its interactions with different cofactors. Given the deep conservation of the Runx family of transcription factors across metazoans (Sullivan et al., 2008), it is highly likely that this biochemical potential of runt is also conserved in non-insect arthropods. On this basis, one possible hypothesis for the flexibility of runt is that its dual regulatory properties enable it to occupy many different positions in the transcriptional network. Also, the biochemical simplicity of switching between being an activator and repressor through an exchange of cofactors may have facilitated its fast evolution.
Restriction of dynamic and periodic expression to the ectoderm
None of the pair rule gene homologues that we have examined is expressed in a dynamic or periodic pattern in the mesoderm. With the exception of pax3/7-1 and h3, which are expressed in non-periodic mesodermal domains, the rest of the genes are either never expressed in the mesodermal cells of the posterior disc and transition zone, or expressed at such low levels as to be indistinguishable from background.
In Drosophila, pair rule genes are expressed in the mesoderm, and it is known that periodic stripes of eve and slp are necessary for correct patterning of the mesoderm into cardiac, somatic and visceral fates (Azpiazu et al., 1996; Riechmann et al., 1997). On the other hand, most of Drosophila segmentation is complete before gastrulation, and it is known that this is a derived condition within the arthropods. Tribolium is more typical for the arthropods, where segments are generated after gastrulation, in a sequential manner from a growth zone with both ectodermal and mesodermal progenitor cells. In Tribolium, eve and odd homologues are expressed in the mesoderm of the growth zone before morphological segmentation (Sarrazin et al., 2012), but their function in these cells is unknown.
In general mesoderm segmentation is not understood in Tribolium, but there are a couple of suggestive observations. First, the re-activation of twist in segmental bands in the growth zone mesoderm occurs just anterior to the appearance of newly formed en stripes – and thus in a region where wg and hedgehog are being transcribed, and might have begun signalling to surrounding cells (Handel et al., 2005). However, it is not known whether this temporal correlation reflects a causal link between ectodermal signalling and mesoderm segmentation. Second, it is known that in a dorsalised Tribolium embryo with no mesoderm, ectoderm segmentation still occurs (da Fonseca et al., 2008). This supports the hypothesis that ectoderm segmentation is autonomous in Tribolium.
There are no genes examined to date in Strigamia that are expressed in a periodic pattern in the mesoderm before such patterns are established in the ectoderm. If this holds true as more genes are studied, it is suggestive evidence that mesoderm segmentation might be induced by signals from the overlying ectoderm. There is supporting evidence from Drosophila and the amphipod crustacean Parhyale hawaiensis that mesoderm segmentation is secondary and at least partially dependent on the ectoderm (Azpiazu et al., 1996; Frasch, 1999; Hannibal et al., 2012). In all it suggests that primary segmentation of the ectoderm may be the ancestral arthropod condition. It is an interesting area for future work to investigate how mesoderm segmentation occurs in other sequentially segmenting arthropods.
Acknowledgements
This work was supported by a Wellcome Trust Ph.D. studentship to J. Green and by the Marie Curie training network “Evonet”. We thank members of the Akam group for assistance with embryo collection; Carlo Brena, Vera Hunnekuhl, Cornelius Eibner, Luke Hayden and Wallace Arthur for sharing clones and information prior to publication; Peter Dearden and Andrew Peel for comments on the manuscript prior to publication.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found in the online version at 10.1016/j.ydbio.2013.06.017.
Appendix A. Supporting materials
Supplementary Fig. S1.
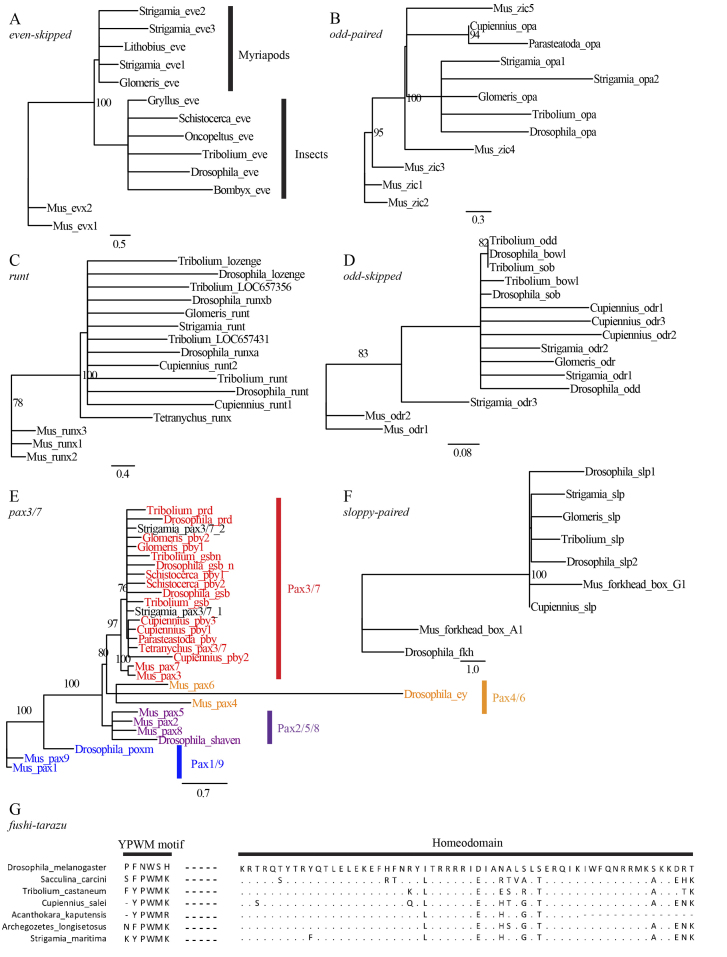
Phylogenetic analysis of Strigamia pair rule gene homologues. Phylogenetic trees for homologues from various arthropods and the mouse of (A) even-skipped, (B) odd-paired, (C) runt, (D) odd-skipped, (E) paired and related pax genes, (F) sloppy-paired. (G) Protein alignment of fushi-tarazu orthologues from various arthropods. Values at nodes are bootstrap support values (out of 100 bootstrap replicates). Nodes with less than 70/100 support are collapsed to polytomies. Abbreviations for genes: eve, even-skipped; evx, even-skipped homeobox; opa, odd-paired; zic, zinc finger protein; odd, odd-skipped; odr, odd-skipped-related; bowl, brother of odd with entrails limited; sob, sister of odd and bowl; prd, paired; gsb, gooseberry; gsb-n, gooseberry-neuro; pby, pairberry; pax, paired box; ey, eyeless; poxm, pox-meso; slp, sloppy-paired; fkh, forkhead. Abbreviations for animals: Bombyx, Bombyx mori (silkmoth); Cupiennius, Cupiennius salei (spider); Drosophila, Drosophila melanogaster (fly); Glomeris, Glomeris marginata (millipede); Gryllus, Gryllus bimaculatus (cricket); Lithobius, Lithobius atkinsoni (lithobiomorph centipede); Mus, Mus musculus (mouse); Oncopeltus, Oncopeltus fasciatus (milkweed bug); Parasteatoda, Parasteatoda tepidariorum (spider); Schistocerca, Schistocerca americana (grasshopper); Strigamia, Strigamia maritima (centipede); Tetranychus, Tetranychus urticae (mite); Tribolium, Tribolium castaneum (beetle).
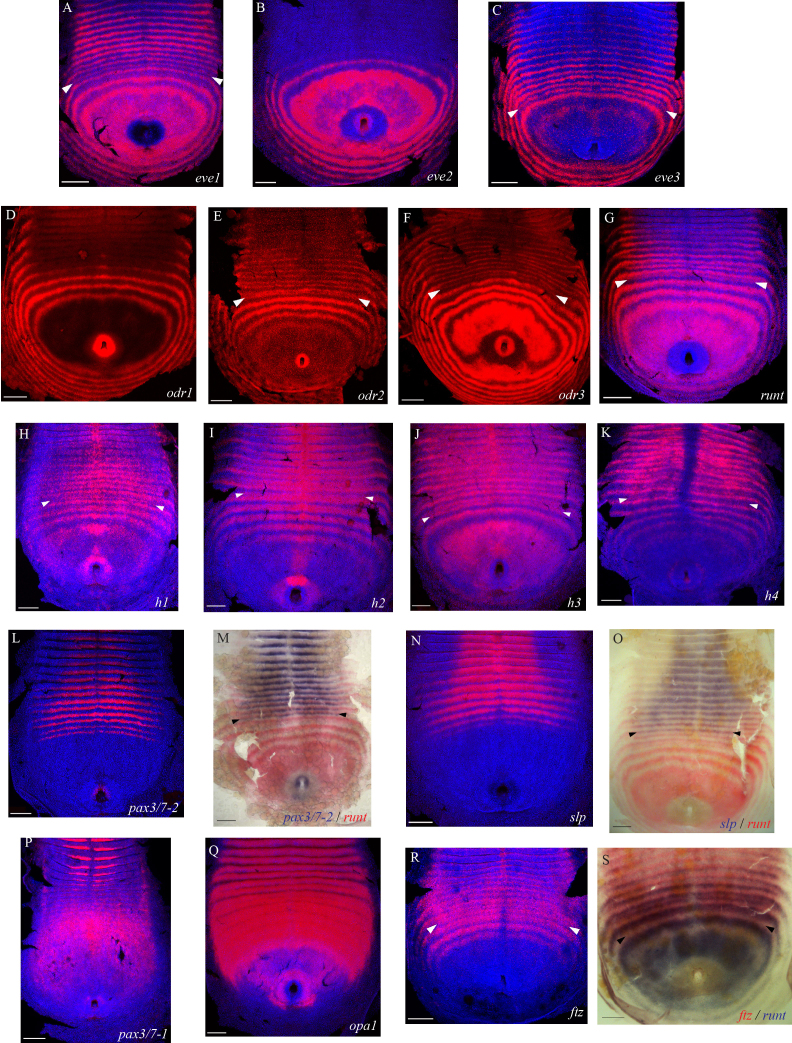
Supplementary Fig. S2.
Expression of pair rule gene homologues during Strigamia trunk segmentation. (A) Expression of eve1 in the posterior disc and germ band of a stage 4 Strigamia embryo with 21 leg-bearing segments. (B) eve2, 17 lbs. (C) eve3, 21 lbs. (D) odr1, 25 lbs. (E) odr2, 18 lbs. (F) odr3, 18 lbs. (G) runt, 23 lbs. (H) h1, 14 lbs. (I), h2, 17 lbs. (J) h3, 23 lbs. (K) h4, 16 lbs. (L) pax3/7-2, 28 lbs. (M) Double stain for pax3/7-2 (blue) and runt (red), 20 lbs. The first stripe of pax3/7-2 appears overlapping with the posterior cell rows of the first intercalating runt stripe (black arrowheads). (N) slp, 33 lbs. (O) slp (blue) and runt (red), 19 lbs. The first stripe of slp appears overlapping with the first primary stripe of runt, after two secondary runt stripes have intercalated (black arrowheads). (P) pax3/7-1, 22 lbs. (Q) opa1, 25 lbs. (R) ftz, 18 lbs. (S) ftz (red) and runt (blue), 24 lbs. ftz transcripts first appear at the anterior margin of the peri-proctodeal region, anterior to the first demarcated ring of runt (black arrowheads). White arrowheads indicate the transition from double to single segment periodicity, through the intercalation of new stripes in between the primary stripes (A, C, E, F, G, H, J, K), or the splitting of broad, primary stripes into alternating long and short stripes (I, R). (A, B, C, G, H, I, J, K, L, N, P, Q, R) RNA is stained in red and nuclei are stained in blue. The expression pattern is overlain on the nuclear stain. (D, E, F) RNA expression is shown on its own, with no nuclear stain overlay. This is to better visualise the weaker, new stripes of expression that intercalate between the primary stripes, which are otherwise obscured against the background of the nuclear stain. Embryos are flat-mounted preparations. Anterior is to the top. Scale bar is 100 μm.
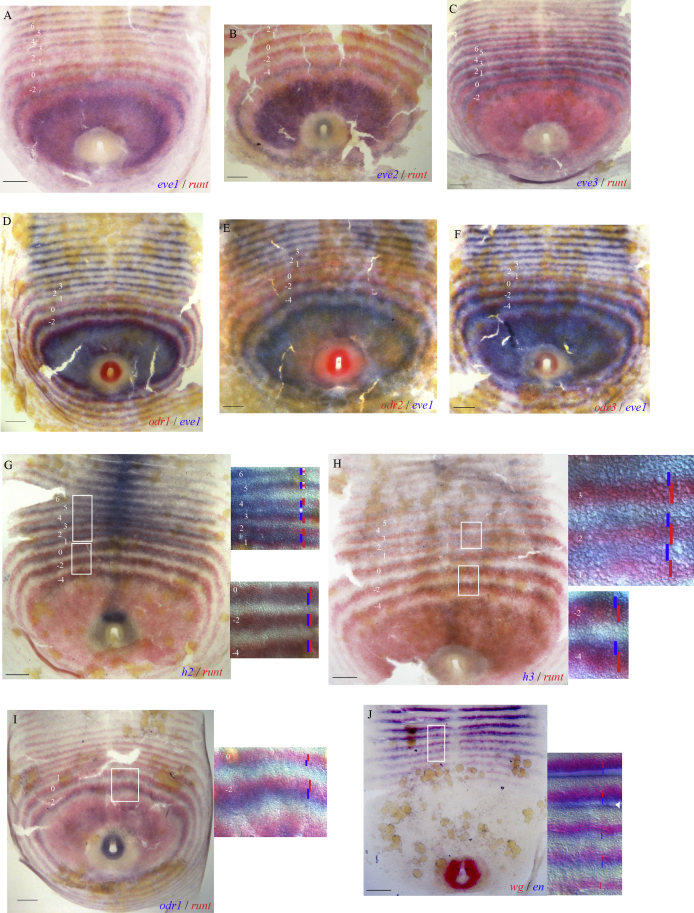
Supplementary Fig. S3.
Registration of the pair rule genes during the progression of pattern formation. (A) Expression of eve1 (blue) and runt (red) on a stage 4 embryo with 18 lbs. (B) eve2 (blue) and runt (red), 19 lbs. (C) eve3 (blue) and runt (red), 19 lbs. (D) odr1 (red) and eve1 (blue), 23 lbs. (E) odr2 (red) and eve1 (blue), 19 lbs. (F) odr3 (red) and eve1 (blue), 19 lbs. (G) h2 (blue) and runt (red), 24 lbs. (H) h3 (blue) and runt (red), 19 lbs. (I) odr1 (blue) and runt (red), 24 lbs. (J) wg (red) and en (blue), 29 lbs. Panels are higher magnification DIC images of the white boxed areas in G, H, I and J. Coloured bars in panels are schematic representations of the accompanying stripes of gene expression, indicating stripe width and inter-stripe spacing. Stripe number as explained in Fig. 2. Embryos are flat-mounted preparations. Anterior is to the top. Scale bar is 100 μm.
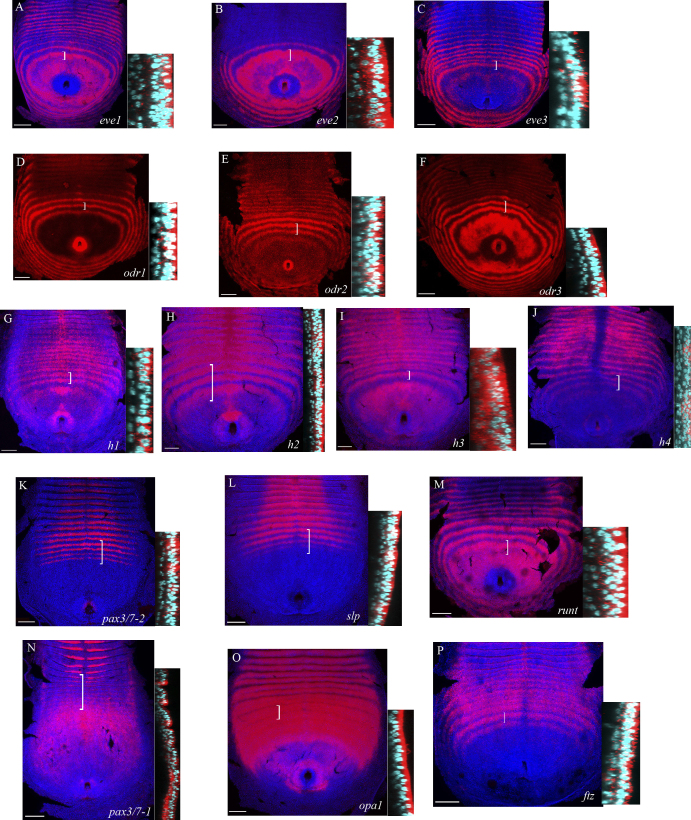
Supplementary Fig. S4.
Expression of pair rule genes in different germ layers. (A) Expression of eve1, 17 lbs. (B) eve2, 17 lbs. (C) eve3, 21 lbs. (D) odr1, 25 lbs. (E) odr2, 18 lbs. (F) odr3, 18 lbs. (G) h1, 14 lbs. (H) h2, 28 lbs. (I) h3, 23 lbs. (J) h4, 16 lbs. (K) pax3/7-2, 28 lbs. (L) slp, 33 lbs. (M) runt, 20 lbs. (N) pax3/7-1, 22 lbs. (O) opa1, 25 lbs. (P) ftz, 18 lbs. The side panels show a higher magnification, orthogonal section along the anteroposterior axis of the bracketed area in the main panel. In the overview and accompanying cross-section images, the RNA is stained in red and shown overlaying the nuclear stain. In the overview images, nuclei are shown in dark blue. In the cross-section images the nuclei are shown in cyan, which gives a clearer distinction of the two germ layers and the expression of the RNA in relation to them. Embryos are flat-mounted preparations. Anterior is to the top. In cross-sections, the ectoderm is oriented on the right, and the underlying mesodermal cells to the left. Scale bar is 100 μm (overviews only).
Supplementary Table 1
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Aranda M., Marques-Souza H., Bayer T., Tautz D. The role of the segmentation gene hairy in Tribolium. Dev. Genes Evol. 2008;218:465–477. doi: 10.1007/s00427-008-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N., Lawrence P.A., Vincent J.P., Frasch M. Segmentation and specification of the Drosophila mesoderm. Genes Dev. 1996;10:3183–3194. doi: 10.1101/gad.10.24.3183. [DOI] [PubMed] [Google Scholar]
- Balczarek K.A., Lai Z.C., Kumar S. Evolution and functional diversification of the paired box (Pax) DNA-binding domains. Mol. Biol. Evol. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- Bao R., Friedrich M. Conserved cluster organization of insect Runx genes. Dev. Genes Evol. 2008;218:567–574. doi: 10.1007/s00427-008-0244-x. [DOI] [PubMed] [Google Scholar]
- Brena C., Akam M. The embryonic development of the centipede Strigamia maritima. Dev. Biol. 2012;363:290–307. doi: 10.1016/j.ydbio.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Chipman A.D., Akam M. The segmentation cascade in the centipede Strigamia maritima: Involvement of the Notch pathway and pair-rule gene homologues. Dev. Biol. 2008;319:160–169. doi: 10.1016/j.ydbio.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Chipman A.D., Arthur W., Akam M. A double segment periodicity underlies segment generation in centipede development. Curr. Biol. 2004;14:1250–1255. doi: 10.1016/j.cub.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Chipman A.D., Arthur W., Akam M. Early development and segment formation in the centipede, Strigamia maritima (Geophilomorpha) Evol. Dev. 2004;6:78–89. doi: 10.1111/j.1525-142x.2004.04016.x. [DOI] [PubMed] [Google Scholar]
- Chipman A.D., Stollewerk A. Specification of neural precursor identity in the geophilomorph centipede Strigamia maritima. Dev. Biol. 2006;290:337–350. doi: 10.1016/j.ydbio.2005.11.029. [DOI] [PubMed] [Google Scholar]
- Choe C.P., Brown S.J. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev. Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C.P., Brown S.J. Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Dev. Biol. 2009;325:482–491. doi: 10.1016/j.ydbio.2008.10.037. [DOI] [PubMed] [Google Scholar]
- Choe C.P., Miller S.C., Brown S.J. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. USA. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A., Littman D.R., Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca R.N., von Levetzow C., Kaischeuer P., Basal A., van der Zee M., Roth S. Self-regulatory circuits in dorsoventral axis formation of the short-germ beetle Tribolium castaneum. Dev. Cell. 2008;14:605–615. doi: 10.1016/j.devcel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Damen W.G.M. Evolutionary conservation and divergence of the segmentation process in arthropods. Dev. Dyn. 2007;236:1379–1391. doi: 10.1002/dvdy.21157. [DOI] [PubMed] [Google Scholar]
- Damen W.G.M., Janssen R., Prpic N.M. Pair rule gene orthologs in spider segmentation. Evol. Dev. 2005;7:618–628. doi: 10.1111/j.1525-142X.2005.05065.x. [DOI] [PubMed] [Google Scholar]
- Damen W.G.M., Weller M., Tautz D. Expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proc. Natl. Acad. Sci. USA. 2000;97:4515–4519. doi: 10.1073/pnas.97.9.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis G.K., Jaramillo C.A., Patel N.H. Pax group III genes and the evolution of insect pair-rule patterning. Development. 2001;128:3445–3458. doi: 10.1242/dev.128.18.3445. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibner, C., 2010. Segment formation in the centipede Strigamia maritima with emphasis on segment polarity genes and environmental factors (doctoral thesis). National University of Ireland Galway, Galway.
- Farzana L., Brown S.J. Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev. Genes Evol. 2008;218:181–192. doi: 10.1007/s00427-008-0207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. Intersecting signalling and transcriptional pathways in Drosophila heart specification. Semin. Cell Dev. Biol. 1999;10:61–71. doi: 10.1006/scdb.1998.0279. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Handel K., Basal A., Fan X., Roth S. Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Dev. Genes Evol. 2005;215:13–31. doi: 10.1007/s00427-004-0446-9. [DOI] [PubMed] [Google Scholar]
- Hannibal R.L., Price A.L., Patel N.H. The functional relationship between ectodermal and mesodermal segmentation in the crustacean, Parhyale hawaiensis. Dev. Biol. 2012;361:427–438. doi: 10.1016/j.ydbio.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Hodcroft, E., 2012. TreeCollapserCL 〈http://emmahodcroft.com/TreeCollapseCL3.html〉
- Ingham P., Gergen P. Interactions between the pair-rule genes runt, hairy, even-skipped and fushi-tarazu and the establishment of periodic pattern in the Drosophila embryo. Development. 1988;104:51–60. [Google Scholar]
- Ingham P.W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Jaeger J., Surkova S., Blagov M., Janssens H., Kosman D., Kozlov K.N., Manu, Myasnikova E., Vanario-Alonso C.E., Samsonova M., Sharp D.H., Reinitz J. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- Janssen R., Budd G.E., Prpic N.-M., Damen W.G. Expression of myriapod pair rule gene orthologs. EvoDevo. 2011;2:5. doi: 10.1186/2041-9139-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle C., Johnstone J., Jowett T., Arthur H., Arthur W. The pattern of segment formation, as revealed by engrailed expression, in a centipede with a variable number of segments. Evol. Dev. 2003;5:198–207. doi: 10.1046/j.1525-142x.2003.03027.x. [DOI] [PubMed] [Google Scholar]
- Lawrence P.A., Johnston P., Macdonald P., Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi-tarazu and even-skipped genes. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Lee H.H., Frasch M. Survey of forkhead domain encoding genes in the Drosophila genome: classification and embryonic expression patterns. Dev. Dyn. 2004;229:357–366. doi: 10.1002/dvdy.10443. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Mazet F., Yu J.K., Liberles D.A., Holland L.Z., Shimeld S.M. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene. 2003;316:79–89. doi: 10.1016/s0378-1119(03)00741-8. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Pankratz M.J., Jaeckle H. Blastoderm segmentation. In: Bate M., Martinez Arias A., editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; USA: 1993. pp. 467–516. [Google Scholar]
- Patel N.H., Condron B.G., Zinn K. Pair-rule expression patterns of even-skipped are found in both short-germ and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- Peel A.D., Chipman A.D., Akam M. Arthropod segmentation: beyond the Drosophila paradigm. Nat. Rev. Genet. 2005;6:905–916. doi: 10.1038/nrg1724. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., 2009. FigTree 〈http://tree.bio.ed.ac.uk/software/figtree/〉.
- Riechmann V., Irion U., Wilson R., Grosskortenhaus R., Leptin M. Control of cell fates and segmentation in the Drosophila mesoderm. Development. 1997;124:2915–2922. doi: 10.1242/dev.124.15.2915. [DOI] [PubMed] [Google Scholar]
- Romano L.A., Wray G.A. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Sanchez R., Serra F., Tarraga J., Medina I., Carbonell J., Pulido L., de Maria A., Capella-Gutierrez S., Huerta-Cepas J., Gabaldon T., Dopazo J., Dopazo H. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin A.F., Peel A.D., Averof M. A segmentation clock with two-segment periodicity in insects. Science. 2012;336:338–341. doi: 10.1126/science.1218256. [DOI] [PubMed] [Google Scholar]
- Schroeder M.D., Greer C., Gaul U. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development. 2011;138:3067–3078. doi: 10.1242/dev.062141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.D., Pearce M., Fak J., Fan H.Q., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:1396–1410. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.C., Sher D., Eisenstein M., Shigesada K., Reitzel A.M., Marlow H., Levanon D., Groner Y., Finnerty J.R., Gat U. The evolutionary origin of the Runx/CBFbeta transcription factors—studies of the most basal metazoans. BMC Evol. Biol. 2008;8 doi: 10.1186/1471-2148-8-228. Article number 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swantek D., Gergen J.P. Ftz modulates Runt-dependent activation and repression of segment-polarity gene transcription. Development. 2004;131:2281–2290. doi: 10.1242/dev.01109. [DOI] [PubMed] [Google Scholar]
- True J.R., Haag E.S. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Walrad P.B., Hang S., Gergen J.P. Hairless is a cofactor for Runt-dependent transcriptional regulation. Mol. Biol. Cell. 2011;22:1364–1374. doi: 10.1091/mbc.E10-06-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.J., Dearden P.K. Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera) Plos One. 2012:7. doi: 10.1371/journal.pone.0046490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1