Abstract
The unique redox potential of iron is ideal for use as a cofactor in diverse biochemical reactions. Iron is therefore vital for the growth and proliferation of nearly all organisms, including pathogenic bacteria. Vertebrates sequester excess iron within proteins in order to alleviate toxicity and restrict the amount of free iron available for invading pathogens. Restricting the growth of infectious microorganisms by sequestering essential nutrients is referred to as nutritional immunity. In order to circumvent nutritional immunity bacterial pathogens have evolved elegant systems that allow for the acquisition of iron during infection. The Gram-positive extracellular pathogen Staphylococcus aureus is a commensal organism that can cause severe disease when it gains access to underlying tissues. Iron acquisition is required for S. aureus colonization and subsequent pathogenesis. Herein we review the strategies S. aureus employs to obtain iron through the production of siderophores and the consumption of host heme.
Introduction
Staphylococcus aureus are Gram-positive non-motile cocci, clinically distinguishable by a golden hue and the ability to clot vertebrate blood. S. aureus is often found as a part of the skin microflora and innocuously colonizes the nares of a third of the world’s population (42, 55). Upon breaching the epithelium this extracellular pathogen can cause severe ailments including bacteremia, pneumonia, osteomyelitis, endocarditis, and septic shock (40). Moreover, S. aureus develops resistance to antibiotics at a remarkable pace presenting a significant clinical challenge. Methicillin resistant S. aureus (MRSA) has recently become a serious problem in the clinical setting, highlighted by the fact that mortality due to MRSA infection has surpassed HIV-associated mortality in the United States (11). The decreasing efficacy of available antibiotics underscores the need to increase our understanding of the fundamental processes that promote S. aureus pathogenesis, as these processes could represent targets for novel therapeutics.
In the late 1800s Alexander Ogston discovered S. aureus cocci in pus isolated from an abscess (1), establishing the formation of tissue abscesses as a pathological hallmark of S. aureus infection. Within the abscess, S. aureus is confronted with a robust host immune response and an environment devoid of essential nutrients (13, 17). Nutrient iron is required for S. aureus growth and persistence within abscesses and hence must be acquired during infection (13, 71, 80, 82). Most vertebrate iron is utilized as a cofactor in biochemical reactions that occur intracellularly. This intracellular pool of iron is generally not available to extracellular pathogens such as S. aureus. Moreover, the amount of free iron found within the serum is negligible, as it is almost always complexed to high-affinity iron binding proteins. This process of iron sequestration by the host, also referred to as nutritional immunity, inhibits the growth of invading microorganisms (9, 84). In response to this severe iron limitation, S. aureus has evolved sophisticated strategies to obtain iron required to proliferate within vertebrates. This review seeks to provide a comprehensive analysis of the pathways S. aureus utilizes to obtain iron during infection.
Iron is sequestered within vertebrates
S. aureus is a commensal organism that can inflict life-threatening damage upon its host if it is able to gain access to underlying tissues. The ability of S. aureus to colonize nearly every major vertebrate organ underscores the considerable public health threat posed by this organism. S. aureus is the number one cause of heart and skin infections, the number one cause of soft tissue infections, the leading cause of hospital acquired infections, and a primary cause of bacterial pneumonia (7, 9, 28, 35, 47, 84). Each organ presents a unique challenge to colonization. Factors such as oxygen tension, organ-specific immune responses, and the availability of nutrients influence the outcome of staphylococcal pathogenesis.
Iron acquisition has been referred to as the “critical determinant” deciding the outcome of the host-pathogen interaction (84). Greater than 90% of the iron in mammals resides intracellularly and is therefore not a viable source of iron for extracellular pathogens unless it can be liberated from host cells (24). Extracellular iron is bound by high affinity iron-binding proteins such as transferrin, found in the serum, and lactoferrin, found in the lymph and mucosal secretions. These glycoproteins have a high affinity for free iron. S. aureus indirectly steals iron from lactoferrin or transferrin through the production of siderophores. Siderophores are secreted small molecules that have an extremely high affinity for iron and out-compete host iron-binding proteins. Siderophore-iron complexes are recognized by cognate receptors on the bacterial surface permitting the theft of iron from lactoferrin or transferrin.
In addition to being bound by proteins, iron is also complexed to the tetrapyrrole ring of heme. Heme represents 80% of iron within the host and is the preferred iron source of S. aureus (71). The most abundant hemoprotein within vertebrates is hemoglobin which binds four molecules of heme and is contained with circulating erythrocytes. In order to access this rich source of iron, S. aureus lyses erythrocytes through the secretion of hemolysins resulting in the liberation of hemoglobin. The host counters the displacement of hemoglobin through the action of the high-affinity hemoglobin binding protein haptoglobin. Haptoglobin is plentiful within the serum and becomes more abundant during inflammation (56). The hemoglobin-haptoglobin complex is one of the strongest noncovalent interactions reported in serum ensuring that the complex does not dissociate until the proteins and heme are recycled in the liver (37). Despite the strength of the hemoglobin-haptoglobin complex, S. aureus is capable of binding this complex and utilizing it as a source of iron (78).
Damage to erythrocytes also results in the release of free heme from liberated hemoglobin. To prevent bacterial access to this iron source, hemopexin binds heme with high affinity and traffics it to the liver where it is endocytosed by hepatocytes and cleared from the serum (77). This interaction favors the host as S. aureus cannot use the hemopexin-heme complex as a source of iron (78). In vitro studies have validated heme, hemoglobin, and haptoglobin-hemoglobin as sources of iron that support S. aureus proliferation. Scanning electron microscopy has established that erythrocytes are present with abscesses, confirming that S. aureus has abundant access to this critical nutrient source during tissue colonization (13). Iron homeostasis in the host is strictly regulated to ensure that iron is available for essential biochemical reactions while simultaneously preventing iron-associated toxicity and bacterial growth. Animal models have confirmed that disrupting host iron homeostasis increases the severity of S. aureus infections (31). Moreover, S. aureus infections in the healthcare setting are often associated with patients that have alterations in their iron status. Liver transplantation can result in iron overload, as the liver is the primary source of iron in the human body (24). The mortality rate in liver transplant patients that suffer from S. aureus bacteremia is over 80% (69). It is clear that disruptions in iron homeostasis are not only detrimental to the host, but also lead to increased S. aureus disease due to the promotion of bacterial replication during infection. The following sections provide a detailed overview of our current understanding of the mechanisms S. aureus employs to acquire iron during infection.
S. aureus responds to iron starvation through the activity of the transcriptional regulator Fur
S. aureus responds to the iron-restricted environment of the host by dramatically altering its protein expression profile. This change in protein expression is mediated by the iron-dependent ferric uptake regulator (fur) (29, 80). In the presence of iron, Fur binds a consensus DNA sequence known as the fur box, found upstream of fur-regulated genes. When iron becomes limiting, Fur releases from the DNA, alleviating Fur-mediated transcriptional repression (5). This results in transcriptional repression of the Fur regulon when S. aureus is iron replete and de-repression when S. aureus is iron starved. Hence, S. aureus fur-deficient strains exhibit a gene expression profile that mirrors that of iron starved organisms. In vivo imaging of S. aureus infected mice has demonstrated that Fur-regulated genes are expressed in heart and kidneys abscesses, suggesting that staphylococci are starved for iron in these organs (60, 63). A comparison of cytoplasmic protein profiles between wild type S. aureus and an isogenic fur mutant found twenty staphylococcal proteins that are more abundant in the absence of Fur, suggesting that these proteins are negatively regulated by Fur (29). In addition to iron acquisition systems, this analysis revealed an increase in glycolytic and fermentative enzymes, indicating that S. aureus modulates its metabolism in order to adapt to the iron-starved environment of the host. Under iron-limiting conditions S. aureus increases fermentative metabolism, resulting in the production of lactate. Secretion of lactate lowers the pH of the microenvironment and the affinity of transferrin for iron (29). These data demonstrate that S. aureus alters the host environment in a way that promotes the release of iron from host proteins presumably increasing iron availability.
Fur regulates the expression of staphylococcal virulence factors that are involved in attachment to host cells, biofilm formation, and manipulation of host wound healing (3, 12, 36, 38). Fur also regulates the expression of secreted cytolytic and immunomodulatory toxins, which play a profound role in decreasing the host immune response to favor bacterial survival (54). In fur mutants the cytolytic toxins are more abundantly expressed while immunomodulatory toxins are decreased compared to wild type cells. This altered exoprotein profile leads to an increase in susceptibility to neutrophil killing and consequently, reduced virulence in a murine pneumonia model of infection (79). These findings highlight the importance of Fur to staphylococcal pathogenesis.
S. aureus siderophore production
Extracellular iron complexed to host proteins, such as transferrin and lactoferrin, are targets for the iron scavenging activity of siderophores. Siderophores are small molecules that are secreted by bacteria and have an exceptionally high affinity for iron. S. aureus produces two distinct siderophores named staphyloferrin A and staphyloferrin B that share many properties (Fig. 1A). Bacterial siderophore biosynthesis proceeds through two different pathways, the non-ribosomal peptide synthetase pathway and the non-ribosomal peptide synthetase independent pathway (NIS). Both staphyloferrin A and staphyloferrin B are synthesized via the NIS pathway and are part of the carboxylate family of siderophores. The genes involved in the biosynthesis and import of both staphyloferrin A and B are regulated by Fur and are therefore maximally expressed in iron-limiting environments (29, 45). These siderophores have been synthesized in vitro and the chemically synthesized siderophores restore the iron scavenging defects of mutants unable to produce the respective siderophore. The presence of a third S. aureus siderophore named aureochelin has been suggested but this molecule has not been characterized (19). Supernatants isolated from mutants unable to synthesize both staphyloferrin A and staphyloferrin B are unable to support the growth of wild type S. aureus in an iron-depleted environment (6). This finding highlights the importance of staphyloferrin A and B to S. aureus iron acquisition and calls into question the relevance and/or existence of aureochelin. In vitro studies have demonstrated that siderophore-mediated iron acquisition is the dominant mechanism by which S. aureus steals iron from transferrin, suggesting that S. aureus does not produce a transferrin receptor involved in iron acquisition (58).
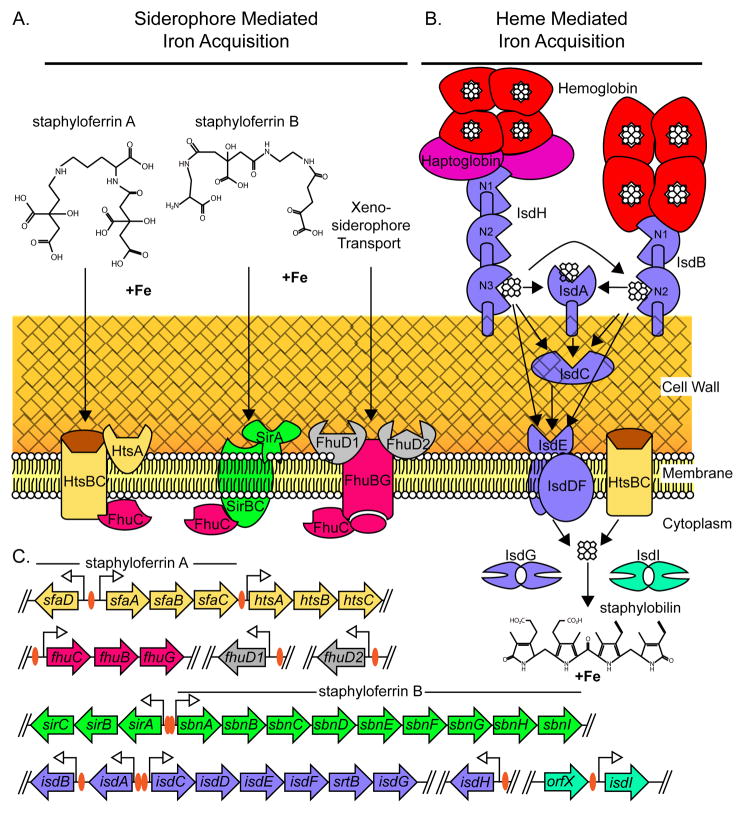
Figure 1. A model of the Staphylococcus aureus iron acquisition pathways.
A. S. aureus produces two siderophores, staphyloferrin A and staphyloferrin B. Staphyloferrin A import is mediated by the HtsA lipoprotein and HtsBC permease. The SirA lipoprotein is the receptor for staphyloferrin B and the SirBC permease mediates the translocation of staphyloferrin B across the membrane. S. aureus imports xenosiderophores produced by other bacteria through the binding activity of FhuD1 and FhuD2 receptor lipoproteins and the FhuBG permease. The energy needed for siderophore uptake is provided by the FhuC ATPase. B. Heme acquisition is mediated by the Isd system. IsdH binds hemoglobin-haptoglobin and IsdB binds hemoglobin. Heme is passed through the NEAT domains of IsdH (N1-N3) IsdB (N1-N2), IsdA, and IsdC. Heme can also be passed from IsdH or IsdB directly to the IsdE heme-receptor lipoprotein. Heme transport across the membrane occurs through either the IsdDF or HtsBC permeases. Once in the cytoplasm heme is a substrate for the heme-degrading enzymes IsdG and IsdI. S. aureus degradation of heme leads to the release of iron and the production of staphylobilin. C. Genetic loci involved in S. aureus iron acquisition pathways. The sfa operon encodes the genes required for staphyloferrin A biosynthesis, while the genes within the sbn operon encode the staphyloferrin B synthesis genes. Promoter regions containing a consensus fur box are indicated with an orange oval. Genes are not drawn to scale.
Staphyloferrin A
The importance of staphyloferrin A to S. aureus iron acquisition was demonstrated by the discovery that mutants unable to produce staphyloferrin B sustain limited growth in serum (6). A bioinformatic screen looking for homologues of known NIS synthetases in S. aureus identified the genes that encode for staphyloferrin A production, the sfnABCD operon (6, 18). Genetic and biochemical approaches subsequently confirmed the assignment of sfnABCD as encoding for staphyloferrin A synthesis (6, 18). Inactivation of sfnABCD does not attenuate growth in serum, but inactivation of sfnABCD in a mutant unable to produce staphyloferrin B severely impairs S. aureus growth in serum (6). Biochemical experiments revealed that staphyloferrin A has a mass of 479 Daltons and is composed of a molecule of D–ornithine that links together two molecules of citrate via amide bonds (41). Mixing citrate, D-ornithine, ATP, SnfB and SnfD is sufficient for the in vitro synthesis of staphyloferrin A (18). Consistent with the structure of staphyloferrin A, addition of D-ornithine to S. aureus growth media results in the increased production of staphyloferrin A (51).
ABC transporters specific for siderophores are often found in close proximity to the genes responsible for siderophore production. Genes encoding the ABC transporter HtsABC are found adjacent to the sfn operon; however HtsBC has been reported to transport heme (Fig. 1A). In a series of elegant genetic experiments, Beasley et al. determined that the HtsABC transporter is also required for staphyloferrin A import (Fig. 1A)(6). These results suggest that HtsABC might be a promiscuous system involved in the transport of multiple iron sources.
The crystal structure of HtsA, the lipoprotein receptor of staphyloferrin A, supports its involvement in siderophore transport (6, 34). This structure represents the first siderophore receptor to be structurally characterized from a Gram-positive bacterium. The conformational change that highlights HtsA staphyloferrin A binding leads to an unprecedented ligand entrapment (34). Given the structural features of HtsA-staphyloferrin A binding it is unlikely that heme is a ligand for HtsA. This suggests that an alternative lipoprotein acts as a receptor for heme transport through HtsBC although this supposition remains to be experimentally verified.
Staphyloferrin B
The structure of staphyloferrin B is distinct from that of staphyloferrin A (Fig. 1A). Staphyloferrin B is an alpha-hydroxycarboxylate siderophore composed of L-2,3-diaminopropionic acid (Dap), 1,2-diaminoethane (Dae), and α-ketoglutaric acid with a molecular mass of 448 Daltons. The synthesis of staphyloferrin B is encoded by the sbnABCDEFGHI operon (14). In vitro synthesis of staphyloferrin B requires only the NIS synthetases SbnC, SnbE, SbnF and the decarboxylase SbnH. These proteins synthesize staphyloferrin B when mixed with ATP, magnesium, Dap, citrate, Dae and α-ketoglutaric acid (14). Inactivation of sbnE leads to a decrease in bacterial load in the kidneys of systemically infected mice, underscoring the importance of staphyloferrin B-mediated iron acquisition to pathogenesis (20). Staphyloferrin B import is mediated by the staphylococcal iron regulated transporter (sirABC) (21). SirBC is predicted to be the membrane permease that supports the transport of staphyloferrin B into the cytoplasm. SirA is the lipoprotein receptor component of the staphyloferrin B import system and SirA undergoes a conformational change leading to the enclosure of staphyloferrin B in the binding pocket, in a manner analogous to HtsA (33). The fact that staphyloferrin A and B bind their cognate lipoprotein receptors with dissociation constants in the low nanomolar range underscores the exquisite evolution of this organism to obtain iron from its host during infection.
As staphyloferrin A and B are likely the only siderophores produced by S. aureus it is possible that these molecules perform unique roles during infection. Staphyloferrin A and B may scavenge iron from different host proteins or reach maximal iron-binding capacity at different sites of colonization. Alternatively, the metabolic pathways that supply the precursors for the biosynthesis of each siderophore may be favored at specific sites of colonization. Future work will refine the role of each siderophore during infection.
Transport of siderophores across the staphylococcal membrane
Transport of molecules across the membrane requires energy and therefore ABC transporters are typically associated with an ATPase. One unusual feature of sirABC and htsABC is that neither operon encodes a putative ATPase (72). The ferric hydroxymate uptake operon fhuCBG encodes a putative ATPase named fhuC (also referred to as fhuA). FhuC is the ATPase necessary for transport of staphyloferrin A and B (6, 72). This finding was established by the observation that growth of an fhuCBG mutant is inhibited in an iron-deplete environment and cannot be restored by the addition of supernatants containing either staphyloferrin A or B. Growth of this mutant can be complemented by fhuC provided in trans and supplementation with supernatant containing either staphyloferrin A or B (6). Therefore, inhibiting FhuC could have debilitating effects on the ability of S. aureus to procure iron during infection, establishing FhuC as a promising therapeutic target.
Despite the fact that S. aureus has not been demonstrated to produce hydroxymate-type siderophores, this organism has the capacity to utilize these siderophores as a source of iron. The ability to scavenge hydroxymate siderophores produced by other bacteria, also known as xenosiderophores, may allow S. aureus to establish residence amongst the microbiome. The uptake of xenosiderophores by S. aureus is also dependent upon fhuCBG (65). The lipoprotein receptors FhuD1 and FhuD2 are required for fhuCBG-mediated xenosiderophore uptake (Fig. 1A). The two genes encoding FhuD1 and FhuD2 (fhuD1 and fhuD2) are located within distinct loci in the S. aureus genome (Fig. 1C). Unlike SirA and HtsA, FhuD1 and FhuD2 undergo only a modest conformational change upon siderophore binding (66, 67). This feature of the lipoprotein receptors probably reduces the affinity for any one siderophore, but facilitates a broad spectrum binding ability for many xenosiderophores. S. aureus can therefore out-compete other bacteria by stealing xenosiderophores while retaining staphyloferrin A and B-mediated iron uptake. This feature likely contributes to the observation that S. aureus is a significant clinical problem for patients that have been administered siderophore-based therapy to treat iron-overload disease. For example, S. aureus is the leading cause of bacterial infection in thalassemic patients that have been treated with the siderophore desferrioxamine to reduce iron overload after a blood transfusion (8, 62). Since FhuD1 and FhuD2 increase the variety of iron sources that can be utilized by S. aureus, inhibiting these receptors could reduce the competitive advantage of S. aureus in bacterial communities and decrease the degree to which this pathogen is found as part of the normal flora. Considering that most hospital-acquired staphylococcal infections originate from the patients normal flora, this decolonization strategy would have significant clinical impact (83).
Heme uptake in staphylococci
Heme represents the primary reservoir of iron within vertebrates and S. aureus can fulfill its iron requirement by obtaining iron from heme (24). In fact, S. aureus preferentially imports and utilizes heme-iron when grown in the presence of both transferrin and heme or hemoglobin, suggesting that heme-iron is the preferred source of iron during infection (71, 78). Hemoglobin is the most abundant hemoprotein in vertebrates making this protein an attractive source of iron for S. aureus. The ability to lyse erythrocytes through the secretion of hemolysins promotes the release of hemoglobin from this intracellular reservoir. In this regard, S. aureus can utilize intact erythrocytes as sole iron source in vitro (78). These experiments suggest that S. aureus encodes systems dedicated to the acquisition of heme-iron bound to erythrocyte hemoglobin.
S. aureus heme and hemoglobin receptors
Gram-positive bacteria are shielded from the environment by a thick cell wall, composed of the disaccharide N-acetylmuramic acid-(β1-4)-N-acetylglucosamine (MurNAc-GlcNAc) and the cell wall tetrapeptide (L-Ala-D-isoGln-L-Lys-D-Ala). Chains of MurNAc-GlcNAc are bound to the cell wall peptide via an amide bond between MurNAc and the L-Ala. In S. aureus, the glycan chains and the cell wall tetrapeptide are cross-linked via a pentaglycine crossbridge producing a continuous layer of peptidoglycan (30) (23). This network of peptidoglycan protects the cytoplasmic membrane from environmental insult and is the region of the bacterium that directly interfaces with the host. Sortase A (SrtA) is a transpeptidase that covalently anchors proteins to the cell wall of S. aureus (48–50). SrtA substrates are characterized by a hydrophobic domain, followed by an LPXTG motif and a charged tail located at the C-terminus of the protein. Surface localized proteins are linked to the cell wall via SrtA-mediated cleavage of the LPXTG domain between the threonine and glycine, and a subsequent transpeptidation between the modified LPXTG domain and the glycine peptide of an immature peptidoglycan subunit (for review of S. aureus SrtA see ref. 53). S. aureus srtA mutants are severely attenuated in a murine abscess model of infection (14, 49).
The thickness of the cell wall presents a considerable barrier to the import of nutrients such as heme-iron. Heme was the first molecule for which a mechanism of import through the Gram-positive cell wall was elucidated. Much of what is currently known about heme import was initiated by a genome wide search for proteins homologous to SrtA. This search uncovered a second sortase encoded within S. aureus named sortase B (SrtB). SrtB is also a transpeptidase; however this enzyme anchors proteins that contain an NPTQN motif in place of LPXTG. The genomic region surrounding srtB contains several features suggestive of a heme utilization system that has since been named the iron regulated surface determinant (Isd) system. The S. aureus Isd system is encoded within five operons: isdA, isdB, isdCDEFsrtBisdG, isdH, orfXisdI (Fig. 1C). A consensus fur box is located upstream of each of these operons and therefore all of these genes are iron-regulated (49, 63). IsdA, IsdB, and IsdH each contain an LPXTG motif and hence are covalently anchored to the cell wall by SrtA. IsdC contains an NPQTN motif and is anchored to the cell wall by SrtB. IsdDEF is a membrane localized ABC transporter complex, suggesting a role in transporting iron-containing molecules across the cytoplasmic membrane (48). IsdA, IsdB, IsdC, IsdD, IsdE, IsdG, and IsdI bind heme (48, 70). Heme uptake into S. aureus requires SrtA, SrtB, IsdA, and IsdE (48). The current model for Isd-mediated heme import proposes that IsdA, IsdB, and IsdH are surface-exposed hemoprotein receptors that pass heme to IsdC. IsdC then transports heme through the cell wall to the membrane localized IsdDEF ABC transport system (Fig. 1B).
The transfer of heme between the Isd heme receptors to IsdE has recently been described in vitro. IsdA, IsdB, IsdH, and IsdC contain NEAr iron Transporter (NEAT) domains. NEAT domains are conserved stretches of 125 amino acids found within proteins that neighbor putative iron transporters (2). Each NEAT domain binds one heme molecule within a groove that coordinates binding to the heme-iron via a tyrosine residue (32, 68). IsdA and IsdC each contain one NEAT domain, while IsdB contains two NEAT domains. Heme is transferred unidirectionally from NEAT domain 2 of IsdB to the NEAT domains of either IsdA or IsdC. Heme is also transferred in a unidirectional manner from the NEAT domain of IsdA to the NEAT domain of IsdC (46). Transfer of heme through the cell wall-localized Isd proteins IsdA, IsdB, or IsdC to the membrane-localized heme receptor IsdE was confirmed in vitro using magnetic circular dichroism (MCD) and electrospray ionization mass spectrometry (ESI-MS). The NEAT domain of IsdC can transfer heme to IsdE. Heme is not transferred between IsdA and IsdE (52). These in vitro studies provide a structural basis for heme-binding and transport through the cell wall that can be used to develop competitive inhibitors that disrupt this process.
In vivo heme is complexed to proteins. Hemoglobin represents an abundant reservoir of heme that is targeted by the S. aureus hemoglobin receptor IsdB (78). The haptoglobin-hemoglobin complex is also a viable source of iron for S. aureus and this complex is bound by IsdH (25, 27, 78) (Fig 1. B). IsdH contains three NEAT domains (25). Structural analysis of the IsdH NEAT domains revealed that NEAT domain 1 binds both hemoglobin and haptoglobin. NEAT domain 2 binds hemoglobin with nanomolar affinities and NEAT domain 3 weakly binds heme. Heme can be transferred in a bidirectional fashion from NEAT domain 3 of IsdH to NEAT domain 2 of IsdB, and in a unidirectional fashion to the NEAT domains of IsdA, IsdC, and IsdE (52, 86).These results suggest a model whereby IsdH strips heme from the haptoglobin-hemoglobin complex through the sequential activity of the NEAT domains and transfers heme to IsdB, IsdA, IsdC or IsdE (Fig. 1B) (26, 59).
The in vitro experiments defining heme transfer via the NEAT domains of IsdH, IsdB, and IsdA implies that these proteins interact in such a way that allows for the transfer of heme between the proteins in vivo. Indeed IsdA and IsdB are colocalized at the cell surface in an iron-dependent fashion (60). Mild iron starvation results in the visualization of discreet IsdA and IsdB foci, while more severe iron starvation leads to the nearly circumferential distribution of both proteins. Recent findings suggest that subcellular protein localization in Gram-positive cocci can be dictated by a YSIRK/GS motif encoded within the N-terminus of the protein (10, 22). The signal sequence of IsdB contains a YSIRK/GS motif while IsdA does not. Visualization of newly synthesized IsdB using gold-labeled IsdB antibodies and electron miscopy revealed that IsdB is found specifically localized to the sites of cell division. IsdA localization is more ubiquitous but a significant portion of IsdA is also found at the site of cell division (60). Despite the more uniform nature of IsdA localization, its contact with IsdB has been detected using immuno-coprecipitation highlighting the strength of this interaction. Hemoglobin binding to the cell surface also colocalizes with IsdB at discrete iron-regulated foci. In keeping with the role of IsdB and IsdH as hemoprotein receptors, inactivation of isdB or IsdH reduces hemoglobin binding to S. aureus. In fact, mutating isdH or isdA in an isdB background severely abrogates the hemoglobin binding activity of the cell. These results highlight the cooperative nature of hemoglobin binding between these proteins and provide support for the in vitro observations that heme can be transferred between IsdH, IsdB, and IsdA.
Heme transport into the cytoplasm
Compared to our understanding of the transport of heme through the cell wall, much less is known about the molecular details involved in transporting heme across the membrane. The current model of IsdDEF function is that the IsdE lipoprotein receives heme from IsdC. IsdE then passes heme to IsdF, the ABC permease, which transports heme through the membrane using energy provided by the ATP hydrolyzing activity of IsdD. This model is supported by experiments showing IsdE and IsdD bind heme and apo-IsdE receives heme from the NEAT domain of IsdC (32, 48, 52). However, inactivation of isdDEF impairs, but does not abolish, S. aureus growth on heme as the sole iron source. This implies that an additional membrane transporter(s) imports heme-iron. A homology search for alternative ABC-type iron transporters revealed a previously uncharacterized fur-regulated transport system that was named hts (heme transport system). The htsABC system, like isdDEF, comprises a membrane-associated lipoprotein (HtsA) and two putative ABC transporters (HtsB and HtsC). HtsB and HtsC have significant homology to HmuU, the heme transport permease of Corynebacterium diphtheriae. Inactivation of htsB and htsC reduces the ability of S. aureus to import heme-iron, which results in a decrease in bacterial burden in the liver and kidneys of mice infected with these mutants (71). The binding pocket of HtsA is a large basic patch of positively-charged amino acids, making it difficult to envision that HtsA binds heme, a hydrophobic molecule (34). A plausible explanation for these two apparently conflicting findings is that HtsBC could interact with IsdE to transport heme and with HtsA to transport staphyloferrin A. HtsABC does not encode an ATPase, and FhuC was been implicated as the ATPase involved in siderophore transport [65]. The protein that provides the energy needed for HtsBC to transport heme across the membrane is unknown. Inactivation of isdE together with htsA leads to a significant reduction in virulence demonstrating the importance of these transporters to S. aureus survival in the host (47).
Release of iron from heme
In order to utilize heme as a source of iron, bacterial pathogens must have mechanisms of opening the macrocyclic conjunction of heme to release the iron atom. S. aureus encodes two cytoplasmic heme-degrading proteins within the Isd system named IsdG and IsdI. isdG is co-transcribed along with the other genes in the isdCDEFsrtBisdG operon (70). isdI is an intrachromosomal paralogue of isdG that is also Fur-regulated and is encoded in a bicistronic operon with an open reading frame of unknown function (70). The amino acid sequences of IsdG and IsdI share 64% identity and the crystal structures of IsdG and IsdI can be superimposed with a root mean square deviation of 1.0 Å, underscoring the significant similarity between these proteins (85). IsdG and IsdI were the first identified members of a family of heme degrading enzymes that bears their name, the IsdG-family of heme oxygenases.
The degradation of heme is a reaction that occurs in both eukaryotes and bacteria. Before the discovery of the IsdG-family of heme oxygenases, all characterized heme degradation reactions resulted in the production of biliverdin, carbon monoxide, and iron. In vertebrates, biliverdin is then further reduced to the potent antioxidant bilirubin by biliverdin reductase (75). Three major features distinguish the IsdG-family of heme oxygenases from the classical heme oxygenase family. First, IsdG and IsdI are structurally distinct from the classical heme oxygenases (85). Second, the binding of heme to IsdG or IsdI results in the severe distortion of the planarity of heme to the point where it appears ruffled (44). This represents the highest degree of heme distortion for any known heme-binding protein and plays a significant role in the third distinctive feature of IsdG-family members: IsdG and IsdI degrade heme to a novel chromophore. Heme degradation catalyzed by the IsdG-family of heme oxygenases results in the production of a mixture of β- and σ-isomers of oxo-bilirubin, molecules collectively referred to as staphylobilin (Fig. 1B) (64). The heme degradation reactions occur in vitro in the presence of an electron donor, such as NADPH-cytochrome P450 (70). The electron donor used to catalyze IsdG-family-mediated heme degradation in vivo is unknown. isdGI mutants are attenuated in the murine model of systemic infection implicating heme degradation and staphylobilin formation as being important for pathogenesis (63).
The physiological significance of encoding two seemingly identical enzymes has been the subject of considerable research and has revealed subtle differences in the regulation of IsdG and IsdI. Expression of both isdG and isdI is transcriptionally regulated by Fur (63). IsdG stability is also dependent on the presence of heme such that IsdG is degraded in the absence of heme while IsdI is not (63). These results suggest that S. aureus can tailor the amount of heme degradation in order to fulfill the nutrient iron requirement of this organism. Targeting IsdG-family enzymes has significant clinical potential due to the requirement for these enzymes during colonization and the absence of this enzyme family within humans.
Despite the fact that the primary function of heme uptake systems is to satisfy the nutrient iron need of S. aureus, appreciable heme is brought into the cell during iron replete conditions. In this case, isdG and isdI are not expressed and heme is instead distributed intact to the bacterial membrane (71). It is likely that this exogenously acquired heme is destined for proteins involved in respiration since heme is an essential cofactor for proteins involved in the transfer of electrons. Consistent with this model, the respiratory pathways of some Gram positive pathogens rely entirely on the ability of the bacteria to scavenge heme. Taken together, these results imply that S. aureus differentially utilizes heme depending upon its metabolic needs. In conditions of iron starvation, exogenously acquired heme is degraded to release free iron. In contrast, when iron is abundant, heme is acquired and utilized intact to populate cytochromes of the electron transport chain. This reduces the requirement for endogenous heme synthesis and in effect decreases the metabolic burden on the bacterium. How iron levels within the host impact the subcellular fate of heme within S. aureus remains to be uncovered.
The response of S. aureus to heme toxicity
Heme is used as a cofactor in many biological systems due the redox potential of the encircled iron atom. However, this redox active molecule can also be toxic at high levels. This creates a paradox for microorganisms that utilize heme as a source of iron. S. aureus resolves this paradox by adapting to heme toxicity, as evidenced by its resistance to lethal levels of heme when first exposed to sub-lethal concentrations of heme (81). A mechanistic explanation for this adaptation was provided by the observation that S. aureus exposed to sub-lethal concentrations of heme up-regulates an ABC transport system 45-fold compared to untreated controls (29). Based on the heme responsiveness of this system, it has been named the heme-regulated transporter (hrtAB) (Fig. 2). The ATPase of this system (hrtA) is encoded in a bicistronic operon with the predicted permease (hrtB), and both are required for adaptation to heme-mediated toxicity. The ATPase activity of HrtA is influenced by various physiochemical conditions including: the concentration of ATP, temperature, pH, and metal cofactors. Mutants of hrtA that are unable to hydrolyze ATP are unable to adapt to heme toxicity. These findings mechanistically link the ATPase activity of HrtA with the alleviation of heme toxicity (73).
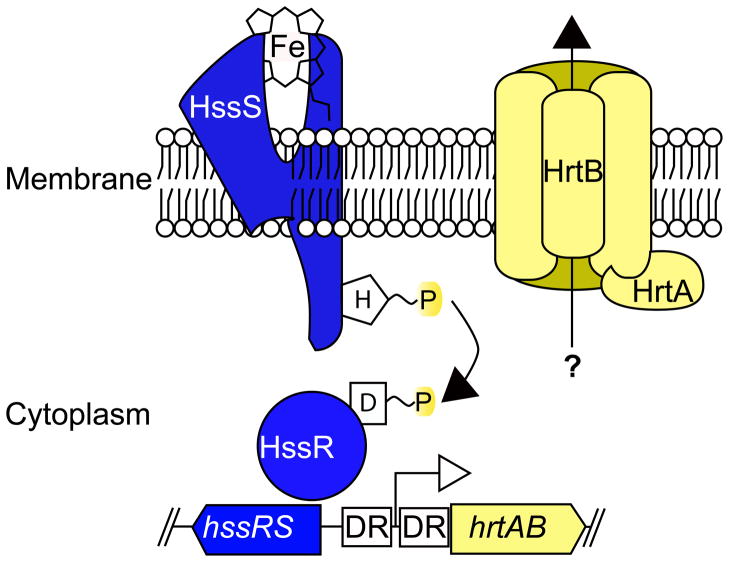
Figure 2. Sensing and alleviation of heme-associated toxicity.
HssS senses exposure to heme through an unknown mechanism. This results in the autophosphorylation of HssS which is followed by a phosphorelay between histidine 279 of HssS and aspartate 52 of HssR. Phosphorylated HssR binds to a direct repeat sequence (DR) found within the hrtAB promoter region resulting in the expression of those genes. The HrtA ATPase and the HrtB permease form an ABC-type transport system that alleviates heme-mediated toxicity through an unknown mechanism.
While the ATPase activity of HrtA has been characterized, the permease activity of HrtB has yet to be confirmed. Moreover, the substrate of HrtAB and the mechanism by which HrtAB detoxifies heme are unknown. The current model predicts that HrtAB acts as an efflux pump that expels a toxic metabolite that accumulates as a result of heme exposure. HrtB expression in the absence of HrtA results in a decrease in the integrity of the membrane and an altered protein secretion profile (4). This phenomenon has a dramatic effect on the virulence of S. aureus (discussed below).
The up-regulation of HrtA in response to heme is dependent on a two-component system known as the heme sensor system (hssRS) (Fig. 2). Bacterial two-component systems are typically comprised of a response regulator and a histidine kinase. These systems are critical for the proper physiological response to environmental cues. In the case of HssRS, the environmental cue is heme toxicity and the physiological response is the up-regulation of hrtAB. Heme exposure activates the histidine kinase, HssS, resulting in autophosphorylation. HssS then transphophorylates HssR, the response regulator component of the system. Phosphorylated HssR binds to a direct repeat sequence within the hrtAB promoter in order to induce transcription of hrtAB (74) (Fig. 2). Expression of hrtAB is dependent on HssRS and inactivation of either hssS or hssR results in an inability to adapt to heme toxicity. Despite the fact that heme triggers HssRS-dependent hrtAB activation, the ligand that induces the initial autophosphorylation of HssS is unknown. Identification of the ligand for HssRS will provide much needed information regarding the mechanisms by which heme promotes cellular toxicity, a finding which will have broad biological implications.
The role of heme acquisition during infection
Considering the requirement for iron during infection, the Isd system plays a major role during pathogenesis. In addition to binding heme, IsdA is capable of binding a wide variety of host proteins, promotes adherence to cells in tissue culture, and acts in concert with IsdB to promote resistance to neutrophil killing (16, 57). Because IsdA binds multiple host proteins, the specific contribution of IsdA-mediated heme-binding to infection has been difficult to assess. Inactivation of isdA leads to decreased ability to colonize human skin and a decrease in bacterial load in murine kidneys five days post systemic infection (14, 15). These results suggest that IsdA is important for the colonization and persistence of S. aureus in these two host environments. IsdA is expressed in the hearts and livers of infected animals, but isdA mutants display wild type levels of colonization in both of these organs. Heme transport via IsdA may be overshadowed in this infection model because in vitro data have demonstrated the NEAT domains of IsdH and IsdB can bind heme and transfer it directly to IsdC or IsdE (52). Similar to the isdA mutant, inactivation of isdC leads to only a modest decrease in bacterial loads in infected kidneys after five days of infection (14). In light of the in vitro evidence demonstrating that heme can be passed directly from IsdH and IsdB to the heme lipoprotein receptor IsdE it remains possible that heme transport across the cell wall could occur via this route in vivo (Fig. 1B).
IsdB appears to be the lynchpin of heme acquisition in vivo. S. aureus inactivated for isdB display a log decrease in the number of bacteria recovered from the spleen and kidneys following systemic infection of mice (60). IsdB expression is prominent in the heart of infected animals and colonization of the heart by the isdB mutant is reduced by two orders of magnitude (60). This latter result suggests that hemoglobin recognition is particularly important to cardiac colonization. Inactivation of isdH alone does not affect bacterial burden in this model implicating IsdB as the critical hemoprotein receptor during systemic infection. The importance of IsdB to S. aureus infection combined with the surface exposed localization of the protein establishes IsdB as a potential vaccine candidate. Mice immunized with purified IsdB exhibit greater survival after subsequent challenge with S. aureus than mock-immunized mice (43). Full protection is observed when the IsdB vaccine includes the cell wall anchored proteins IsdA, SdrD, and SdrE (76). Moreover, antibodies against IsdA and IsdB rescue animals from lethal staphylococcal challenge and inhibit abscess formation (39). Future studies will focus on determining the efficacy of this vaccination strategy in human subjects.
Animal models of infection are limited due to the fact that virulence factors expressed by human pathogens have evolved to bind human-specific molecules. Advances in the genetic manipulation of mice have made it possible to engineer “humanized” mice that express human versions of proteins that are targeted by bacterial virulence factors. This technology has been used to investigate the interaction between S. aureus and human hemoglobin. There is a considerable amount of divergence between the surface exposed amino acids of human and murine hemoglobin. These amino acids likely represent the binding platform for the S. aureus hemoglobin receptor IsdB. This is supported by the observation that the affinity of IsdB for human hemoglobin is significantly higher than the affinity for mouse hemoglobin. Moreover, human hemoglobin is more readily utilized as an iron source by S. aureus in culture. In keeping with this, the burden of S. aureus is increased by an order of magnitude in the hearts and livers of mice expressing a human hemoglobin transgene following systemic infection (61). This increase in virulence is dependent on IsdB, as S. aureus mutants inactivated for isdB proliferate to similar levels in both wild type mice and mice expressing human hemoglobin. These results establish the human hemoglobin-expressing mice as a humanized mouse model that more accurately represents the nutrient environment encountered by S. aureus during infection. This infection model will be valuable for determining the contribution of hemoglobin-iron acquisition to colonization of a variety of infection sites, and in the search for inhibitors specific for the human-hemoglobin-IsdB interaction.
The last step to obtaining iron from heme is mediated by the heme oxygenases IsdG and IsdI. These two enzymes are differentially regulated at the posttranslational level through proteolytic degradation of IsdG in the absence of heme (63). A clue to the physiological importance of this differential regulation comes from the organic-specific colonization profile of isdG and isdI mutants. Inactivation of isdG alone or in combination with isdI results in a decrease in bacterial burden in both the heart and kidneys of infected animals. Inactivation of isdI leads to a decrease in bacterial burden only in the heart (63). The finding that both IsdI and IsdG are independently required for full virulence suggests that these enzymes have distinct roles during vertebrate colonization. In addition, the requirement for each of these enzymes during systemic infection establishes the heme degradation machinery as a target for therapeutic intervention.
Heme toxicity and infection
S. aureus strains inactivated for individual components of the Isd system are unable to satisfy their nutrient iron need and are therefore attenuated for growth within vertebrates. Conversely, an inability to cope with heme stress through inactivation of hssR and hrtA results in a hypervirulent phenotype. In a murine abscess model of infection hssR or hrtA mutants exhibit more overt symptoms of infection and the number of abscesses in the liver is elevated compared to mice infected with wild type S. aureus (81). The hypervirulence of ΔhrtA is specific to the liver and is due to increased expression of immunomodulatory proteins in these mutant strains [98]. This results in an impairment in neutrophil migration and activation which promotes bacterial survival. The increased secretion of immunomodulatory proteins that leads to the hypervirulent phenotype is due to a loss of membrane integrity in the hrtA mutant (4). The mechanism leading to the hypervirulence of the hssR mutant has yet to be elucidated. The ΔhrtA phenotype highlights the delicate balance of the host-pathogen interaction and raises questions regarding the mechanism by which S. aureus senses and responds to membrane damage induced by the host immune response. The observation that orthologues of hssRS and hrtAB are conserved in many Gram-positive pathogens that come in contact with vertebrate blood provides further support that the ability to sense heme-dependent toxicity is important for pathogenesis.
Concluding remarks
Nutritional immunity protects the host from invading microorganisms by sequestering iron and other essential nutrients. To combat this immune strategy S. aureus utilizes a multifaceted approach that targets the most abundant iron reservoirs within the host. The efficiency with which S. aureus is able to overcome nutritional immunity is clinically relevant given the significant public health threat posed by this pathogen. The successful treatment of staphylococcal infections with antibiotics is proving to be difficult due to the prevalence of antibiotic resistant strains. Therefore, it is necessary to explore alternative strategies to combat this pathogen that is becoming resistant to all clinically relevant antibiotics.
The importance of iron acquisition systems to S. aureus virulence suggests these systems are viable targets for therapeutic intervention. S. aureus siderophores steal iron from host proteins while the Isd system consumes host heme and releases iron. Both pathways are expressed within the iron-limited host environment. The functional redundancy built into staphylococcal iron acquisition systems guarantees the pathogen obtains enough iron to successfully colonize a variety of diverse niches within the host. Colonization is decreased but not abolished when either pathway is inactivated, suggesting that both siderophore-iron and heme-iron are critical to pathogenesis. The finding that S. aureus grown in iron-replete conditions transports exogenous heme to the membrane intact implies that S. aureus has alternative uses for heme. Clearly there is much to be learned regarding the nutrient uptake and processing pathways that allow this pathogen to proliferate during infection.
Supplementary Material
Acknowledgments
We thank members of the Skaar laboratory for critically evaluating this manuscript. Work in the Skaar laboratory is supported by NIH grants AI0169233, AI073843 and AI057157. EPS is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. NDH is supported by the National Institute of Health Ruth L. Kirschstein fellowship F32-AI091244-01.
Literature Cited
- 1.Classics in infectious diseases. “On abscesses”. Alexander Ogston (1844–1929) Rev Infect Dis. 1984;6:122–8. [PubMed] [Google Scholar]
- 2.Andrade MA, Ciccarelli FD, Perez-Iratxeta C, Bork P. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002;3:RESEARCH0047. doi: 10.1186/gb-2002-3-9-research0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanasopoulos AN, Economopoulou M, Orlova VV, Sobke A, Schneider D, et al. The extracellular adherence protein (Eap) of Staphylococcus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood. 2006;107:2720–7. doi: 10.1182/blood-2005-08-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attia AS, Benson MA, Stauff DL, Torres VJ, Skaar EP. Membrane damage elicits an immunomodulatory program in Staphylococcus aureus. PLoS Pathog. 2010;6:e1000802. doi: 10.1371/journal.ppat.1000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–29. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 6.Beasley FC, Vines ED, Grigg JC, Zheng Q, Liu S, et al. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol. 2009;72:947–63. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- 7.Benito N, Miro JM, de Lazzari E, Cabell CH, del Rio A, et al. Health care-associated native valve endocarditis: importance of non-nosocomial acquisition. Ann Intern Med. 2009;150:586–94. doi: 10.7326/0003-4819-150-9-200905050-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brock JH, Ng J. The Effect of Desferrioxamine on the Growth of Staphylococcus-Aureus, Yersinia-Enterocolitica and Streptococcus-Faecalis in Human-Serum - Uptake of Desferrioxamine-Bound Iron. Fems Microbiology Letters. 1983;20:439–42. [Google Scholar]
- 9.Bullen JJ. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–38. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–6. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 11.CDC. HIV/AIDS Surveillance Report. 2007. p. 17. [Google Scholar]
- 12.Chavakis T, Hussain M, Kanse SM, Peters G, Bretzel RG, et al. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med. 2002;8:687–93. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 13.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. Faseb J. 2009;23:3393–404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol Microbiol. 2009;74:594–608. doi: 10.1111/j.1365-2958.2009.06880.x. [DOI] [PubMed] [Google Scholar]
- 15.Clarke SR, Mohamed R, Bian L, Routh AF, Kokai-Kun JF, et al. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe. 2007;1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Clarke SR, Wiltshire MD, Foster SJ. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol Microbiol. 2004;51:1509–19. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- 17.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–5. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 18.Cotton JL, Tao J, Balibar CJ. Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry. 2009;48:1025–35. doi: 10.1021/bi801844c. [DOI] [PubMed] [Google Scholar]
- 19.Courcol RJ, Trivier D, Bissinger MC, Martin GR, Brown MR. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect Immun. 1997;65:1944–8. doi: 10.1128/iai.65.5.1944-1948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect Immun. 2004;72:29–37. doi: 10.1128/IAI.72.1.29-37.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dale SE, Sebulsky MT, Heinrichs DE. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J Bacteriol. 2004;186:8356–62. doi: 10.1128/JB.186.24.8356-8362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. Embo J. 2008;27:2656–68. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dmitriev BA, Toukach FV, Holst O, Rietschel ET, Ehlers S. Tertiary structure of Staphylococcus aureus cell wall murein. J Bacteriol. 2004;186:7141–8. doi: 10.1128/JB.186.21.7141-7148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drabkin D. Metabolism of the Hemin Chromoproteins. Physiological Reviews. 1951;31:345–431. doi: 10.1152/physrev.1951.31.4.345. [DOI] [PubMed] [Google Scholar]
- 25.Dryla A, Gelbmann D, Von Gabain A, Nagy E. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol. 2003;49:37–53. doi: 10.1046/j.1365-2958.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 26.Dryla A, Hoffmann B, Gelbmann D, Giefing C, Hanner M, et al. High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded beta-barrel fold. J Bacteriol. 2007;189:254–64. doi: 10.1128/JB.01366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etz H, Minh DB, Henics T, Dryla A, Winkler B, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci U S A. 2002;99:6573–8. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fowler VG, Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, et al. Staphylococcus aureus endocarditis: a consequence of medical progress. Jama. 2005;293:3012–21. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 29.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus Redirects Central Metabolism to Increase Iron Availability. PLoS Pathog. 2006:2. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghuysen JM, Strominger JL. Structure of the Cell Wall of Staphylococcus Aureus, Strain Copenhagen. I. Preparation of Fragments by Enzymatic Hydrolysis. Biochemistry. 1963;2:1110–9. doi: 10.1021/bi00905a035. [DOI] [PubMed] [Google Scholar]
- 31.Gladstone GP, Walton E. The effect of iron and haematin on the killing of staphylococci by rabbit polymorphs. Br J Exp Pathol. 1971;52:452–64. [PMC free article] [PubMed] [Google Scholar]
- 32.Grigg J, Vermeiren C, Heinrichs D, Murphy M. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–49. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 33.Grigg JC, Cheung J, Heinrichs DE, Murphy ME. Specificity of Staphyloferrin B recognition by the SirA receptor from Staphylococcus aureus. J Biol Chem. 2010;285:34579–88. doi: 10.1074/jbc.M110.172924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grigg JC, Cooper JD, Cheung J, Heinrichs DE, Murphy ME. The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J Biol Chem. 2010;285:11162–71. doi: 10.1074/jbc.M109.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho KM, Robinson JO. Risk factors and outcomes of methicillin-resistant Staphylococcus aureus bacteraemia in critically ill patients: a case control study. Anaesth Intensive Care. 2009;37:457–63. doi: 10.1177/0310057X0903700320. [DOI] [PubMed] [Google Scholar]
- 36.Hussain M, Becker K, von Eiff C, Schrenzel J, Peters G, Herrmann M. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J Bacteriol. 2001;183:6778–86. doi: 10.1128/JB.183.23.6778-6786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang PK, Greer J. Interaction between hemoglobin subunits in the hemoglobin. haptoglobin complex. J Biol Chem. 1980;255:3038–41. [PubMed] [Google Scholar]
- 38.Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76:1756–65. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28:6382–92. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 41.Konetschny-Rapp S, Jung G, Meiwes J, Zahner H. Staphyloferrin A: a structurally new siderophore from staphylococci. Eur J Biochem. 1990;191:65–74. doi: 10.1111/j.1432-1033.1990.tb19094.x. [DOI] [PubMed] [Google Scholar]
- 42.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193:172–9. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 43.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun. 2006;74:2215–23. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee WC, Reniere ML, Skaar EP, Murphy ME. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–63. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsay JA, Riley TV. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect Immun. 1994;62:2309–14. doi: 10.1128/iai.62.6.2309-2314.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M, Tanaka WN, Zhu H, Xie G, Dooley DM, Lei B. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J Biol Chem. 2008;283:6668–76. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mason WJ, Skaar EP. Assessing the contribution of heme-iron acquisition to Staphylococcus aureus pneumonia using computed tomography. PLoS One. 2009;4:e6668. doi: 10.1371/journal.pone.0006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazmanian S, Skaar E, Gaspar A, Humayun M, Gornicki P, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–09. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 49.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97:5510–5. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–3. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 51.Meiwes J, Fiedler HP, Haag H, Zahner H, Konetschny-Rapp S, Jung G. Isolation and characterization of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol Lett. 1990;55:201–5. doi: 10.1111/j.1574-6968.1990.tb13863.x. [DOI] [PubMed] [Google Scholar]
- 52.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, Stillman MJ. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283:28125–36. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Noble WC, Valkenburg HA, Wolters CH. Carriage of Staphylococcus aureus in random samples of a normal population. J Hyg (Lond) 1967;65:567–73. doi: 10.1017/s002217240004609x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliviero S, Morrone G, Cortese R. The human haptoglobin gene: transcriptional regulation during development and acute phase induction. Embo J. 1987;6:1905–12. doi: 10.1002/j.1460-2075.1987.tb02450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180:500–9. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 58.Park RY, Sun HY, Choi MH, Bai YH, Shin SH. Staphylococcus aureus siderophore-mediated iron-acquisition system plays a dominant and essential role in the utilization of transferrin-bound iron. J Microbiol. 2005;43:183–90. [PubMed] [Google Scholar]
- 59.Pilpa RM, Robson SA, Villareal VA, Wong ML, Phillips M, Clubb RT. Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J Biol Chem. 2009;284:1166–76. doi: 10.1074/jbc.M806007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect Immun. 2009;77:2624–34. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pishchany G, McCoy AL, Torres VJ, Krause JC, Crowe JE, Jr, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–50. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahav G, Volach V, Shapiro M, Rund D, Rachmilewitz EA, Goldfarb A. Severe infections in thalassaemic patients: prevalence and predisposing factors. Br J Haematol. 2006;133:667–74. doi: 10.1111/j.1365-2141.2006.06082.x. [DOI] [PubMed] [Google Scholar]
- 63.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–15. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–38. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebulsky MT, Hohnstein D, Hunter MD, Heinrichs DE. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J Bacteriol. 2000;182:4394–400. doi: 10.1128/jb.182.16.4394-4400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sebulsky MT, Shilton BH, Speziali CD, Heinrichs DE. The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J Biol Chem. 2003 doi: 10.1074/jbc.M305073200. [DOI] [PubMed] [Google Scholar]
- 67.Sebulsky MT, Speziali CD, Shilton BH, Edgell DR, Heinrichs DE. FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: a case of gene duplication and lateral transfer. J Biol Chem. 2004;279:53152–9. doi: 10.1074/jbc.M409793200. [DOI] [PubMed] [Google Scholar]
- 68.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal Structure of the Heme-IsdC Complex, the Central Conduit of the Isd Iron/Heme Uptake System in Staphylococcus aureus. J Biol Chem. 2007;282:10625–31. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 69.Singh N, Sun HY. Iron overload and unique susceptibility of liver transplant recipients to disseminated disease due to opportunistic pathogens. Liver Transpl. 2008;14:1249–55. doi: 10.1002/lt.21587. [DOI] [PubMed] [Google Scholar]
- 70.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–43. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 71.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–8. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 72.Speziali CD, Dale SE, Henderson JA, Vines ED, Heinrichs DE. Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J Bacteriol. 2006;188:2048–55. doi: 10.1128/JB.188.6.2048-2055.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stauff DL, Bagaley D, Torres VJ, Joyce R, Anderson KL, et al. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol. 2008;190:3588–96. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stauff DL, Torres VJ, Skaar EP. Signaling and DNA-binding Activities of the Staphylococcus aureus HssR-HssS Two-component System Required for Heme Sensing. J Biol Chem. 2007;282:26111–21. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
- 75.Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–6. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 76.Stranger-Jones YK, Bae T, Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2006;103:16942–7. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 78.Torres V, Pishchany G, Humayun M, Schneewind O, Skaar E. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–29. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, et al. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010 doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–9. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, et al. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host & Microbe. 2007;1:109–19. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trivier D, Courcol RJ. Iron depletion and virulence in Staphylococcus aureus. FEMS Microbiol Lett. 1996;141:117–27. doi: 10.1111/j.1574-6968.1996.tb08373.x. [DOI] [PubMed] [Google Scholar]
- 83.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–6. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 84.Weinberg ED. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–6. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu H, Xie G, Liu M, Olson JS, Fabian M, et al. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J Biol Chem. 2008;283:18450–60. doi: 10.1074/jbc.M801466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.