Abstract
The nucleoprotein (N) and phosphoprotein (P) interaction of nonsegmented negative-strand RNA viruses is essential for viral replication; this includes N0-P (N0, free of RNA) interaction and the interaction of N-RNA with P. The precise site(s) within N that mediates the N-P interaction and the detailed regulating mechanism, however, are less clear. Using a human parainfluenza virus type 3 (HPIV3) minigenome assay, we found that an N mutant (NL478A) did not support reporter gene expression. Using in vivo and in vitro coimmunoprecipitation, we found that NL478A maintains the ability to form NL478A0-P, to self-assemble, and to form NL478A-RNA but that NL478A-RNA does not interact with P. Using an immunofluorescence assay, we found that N-P interaction provides the minimal requirement for the formation of cytoplasmic inclusion bodies, which contain viral RNA, N, P, and polymerase in HPIV3-infected cells. NL478A was unable to form inclusion bodies when coexpressed with P, but the presence of N rescued the ability of NL478A to form inclusion bodies and the transcriptional function of NL478A, thereby suggesting that hetero-oligomers formed by N and NL478A are functional and competent to form inclusion bodies. Furthermore, we found that NL478A is also defective in virus growth. To our knowledge, we are the first to use a paramyxovirus to identify a precise amino acid within N that is critical for N-RNA and P interaction but not for N0-P interaction for the formation of inclusion bodies, which appear to be bona fide sites of RNA synthesis.
INTRODUCTION
Human parainfluenza virus type 3 (HPIV3) is a cytoplasmic, enveloped virus with a nonsegmented negative-strand (NNS) RNA genome that is classified in the Paramyxoviridae family, in the order Mononegavirales. It can cause severe respiratory tract diseases such as bronchiolitis, pneumonia, and croup in infants and young children (1). However, currently no valid antiviral therapy or vaccine is available. Thus, further exploration of its replication mechanism will be helpful in the development of novel therapeutic approaches. The RNA genome of HPIV3 consists of 15,462 nucleotides and is encapsidated by the nucleoprotein (N; 68 kDa) to form a helical nucleocapsid containing N-RNA that has the characteristic herringbone-like structure also observed in other Paramyxoviridae members (2–6). This N-RNA complex serves as a template to interact with the RNA-dependent RNA polymerase (RdRp) complex consisting of a large protein (L; 255 kDa) and a phosphoprotein (P; 90 kDa) cofactor; interaction between N-RNA and RdRp forms an active ribonucleoprotein (RNP) complex that is necessary for transcription and replication (2, 7) to generate six monocistronic mRNAs and an antigenome intermediate. P mRNA encodes a basic protein, designated C, via the translation of a +1 open reading frame of P mRNA, which is responsible for inhibiting viral RNA synthesis as well as counteracting the host interferon signaling pathway (8, 9). A synergic association between the L-P and N-RNA templates would therefore determine the ability of the RNA polymerase complex to transcribe or replicate.
Pairs of paramyxoviruses, such as HPIV3 and Sendai virus and canine distemper virus and measles virus, share about 50% nucleotide identity, despite the low level of sequence similarity among known paramyxovirus N genes by sequence comparisons (10–12). N consists of two major domains that are chemically opposite in nature: a highly conserved N-terminal core (about 80% of the sequence), which forms a globular body, and a hypervariable C-terminal tail (about 20% of the sequence), which extends from the N-terminal body (13). The N terminus contains all of the necessary components for N self-assembly and RNA binding to form N-RNA complex (14–16). Structural assays of the N-RNA complex of some NNS RNA viruses revealed that the RNA is sequestered between the N- and C-terminal lobes of the N-RNA complex (17, 18). The C terminus is mainly responsible for the binding of the N-RNA complex to P (3, 19–21). Thus, the C terminus is required for the binding of the N-RNA template to the RNA polymerase complex for viral RNA synthesis (22, 23). Studies of the nucleocapsid of Sendai virus showed that deletion of the C-terminal fragment abrogates the transcriptional activity of the nucleocapsid (24) and that the C terminus interacts with P in vitro (15). Furthermore, it is well known that in measles virus, a putative helical molecular recognition element within the C terminus of N can bind to the C terminus of P to undergo an induced folding (3, 25) for recruiting the polymerase complex to the N-RNA complex.
The P of NNS RNA virus is the best-characterized protein of the replicative complex according to structure analyses. Although no enzymatic activity has been detected in P, it acts as a cofactor of L, and L plays a catalytic function within the P-L RNA polymerase complex and has also been demonstrated to contain posttranscriptional modification activities such as capping, methylation, and polyadenylation of mRNAs. In addition, L alone is unable to bind to the N-RNA template, and P acts as a bridge for the binding of N-RNA template to the polymerase complex by forming the P-L complex (26, 27). P is also organized into two moieties that are functionally and structurally distinct: an N-terminal domain of P and a C-terminal domain of P. The N terminus of P chaperones N0 (free of RNA) and forms the N0-P complex to prevent the nonspecific aggregation of N (28) and regulate its assembly on the nascent genomic RNA during replication (29, 30). The main N0 binding site has been mapped to the extreme N terminus of P in some Paramyxovirinae (28, 31–34). The C terminus of P contains the regions responsible for the oligomerization of P and for the binding of P to L (35, 36), as well as the regions necessary for the binding of N-RNA template to P. The oligomeric nature of P of NNS RNA viruses is central to its various functions. For example, the binding of P to the N-RNA complex involves a simultaneous interaction between multiple C-terminal arms of the P oligomer and the exposed C terminus of assembled N (37). Previous studies demonstrated that P of HPIV3 also forms oligomers, and an oligomerization domain was mapped between amino acids 423 and 457 of P (38). In addition, studies by De et al. (39) indicated that the extreme N-terminal 40 amino acids of P of HPIV3 are required for the interaction of P with N to form N0-P, whereas the extreme C-terminal 20 amino acids of P are indispensable for the interaction of P with the N-RNA template. The most direct evidence for the extreme N terminus of P binding to N0 is from vesicular stomatitis virus (VSV), a prototype of NNS RNA viruses: N0 lacking the N-terminal 21 amino acids in complex with the N-terminal 60 amino acids of P was crystallized, and the formation of this complex blocked the intermolecular interaction of adjacent N molecules as well as access of RNA to the RNA binding region (40).
Because RNP is a highly structured RNA synthesis machine, the assembly of RNP is a critical step in the replication of NNS RNA viruses, and its function also requires that its individual components coordinate entirely and tightly. Although the interactions and roles of N, P, and L within RNP of Paramyxovirinae have been partially clarified, the detailed mechanisms that govern such functional coupling are largely unidentified. For example, it is well known that the functions of N of paramyxoviruses require it to have three essential properties. First, N needs to specifically bind to a single-strand RNA. Second, N has to polymerize in order to cover the entire length of the viral genome RNA. Third, the N-RNA template needs to bind to the P-L polymerase complex. However, what is not known is (i) how N0-P delivers N to viral RNA and how P accesses the N-RNA complex and (ii) the precise site(s) within N that contributes to N0 or N-RNA binding to P.
To address these unresolved questions, we explored the relationship between N-P interaction and RNA synthesis in HPIV3. To do so, we used in vivo coimmunoprecipitation and a minigenome assay to identify a critical residue, L478, within N that is required for N-RNA but not for N0 binding to P and to characterize its requirement for viral genome transcription. Via immunofluorescence analysis, we found that individually expressed N, P, L, and minigenome RNA were all recruited to cytoplasmic viral inclusion bodies in HPIV3-infected cells but exhibited a diffuse expression pattern in the cytoplasm of uninfected cells. We also demonstrated that the association of N with P provides the minimal requirements for inclusion body formation; the N mutant protein (NL478A) was unable to form inclusion bodies when coexpressed with P, but the presence of N rescued the inclusion body-forming ability and transcriptional function of NL478A.
MATERIALS AND METHODS
Cells and virus.
HeLa and LLC-MK2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 8% fetal bovine serum (FBS). BSR-T7 cells were maintained in DMEM containing 8% fetal bovine serum and 1 mg/ml G418. Recombinant vaccinia virus (vTF7-3) expressing the bacteriophage T7 RNA polymerase was propagated in HeLa cells, and HPIV3 (NIH 47885) was propagated in LLC-MK2 cells by inoculation at a multiplicity of infection of 0.1.
Plasmid constructs.
The plasmids pGEM4-N, pGEM4-P, pGEM4-L, HPIV3-MG (−), and pOCUS-HPIV3 encoding untagged N, P, L, HPIV3 minigenome, and HPIV3 genome, respectively, have been described previously. The pGEM4-N cDNA was used as the template for PCR amplification in the genetic manipulations to generate all constructs: cDNAs encoding wild-type N and mutants with a Myc tag at their C terminus were cloned in pcDNA3.0 (for vTF7-3-driven protein expression), cDNAs encoding wild-type N and NL478A with a Myc epitope tagged at their C terminus were also cloned in pCAGGS, cDNAs encoding wild-type N with a hemagglutinin (HA) tag at the C terminus were cloned in pcDNA3.0, and cDNA encoding wild-type N with a Flag tag at the C terminus was cloned in pCAGGS. The pGEM4-P cDNA was used as the template for PCR amplification. PCR products encoding wild-type P and P with the C-terminal 100 amino acids deleted (PΔC100) with an HA tag at their N terminus were cloned into pGADT7 (for vTF7-3-driven protein expression); PCR product encoding wild-type P with an HA tag at its N terminus was also cloned into pCAGGS. The plasmid encoding the wild-type L fused with enhanced green fluorescent protein (eGFP) was generated by PCR using pGEM4-L as the template, and PCR products were cloned into pEGFP-N1. The PCR-based approach also was applied to construct the HPIV3 minigenome plasmid HPIV3-MG-CAT carrying the chloramphenicol acetyltransferase (CAT) reporter gene used in the in vitro transcription. The PCR products of CAT were cloned into HPIV3-MG (−) to replace the luciferase gene. Plasmids carrying the HPIV3 genome containing point mutations within the N gene were constructed as follows. First, pOCUS-HPIV3 carrying the full-length genome of HPIV3 was digested with SalI and Eco91I, and the fragment of about 5.4 kb containing N, P, and M genes was purified and religated into pET-30a-c (+) to generate pET-30a-N-M. Point mutations in the N gene were introduced by overlapping PCR to generate desired mutations in pET-30a-N-M. Plasmid pET-30a-N-M containing desired mutations was used as the template for further amplification; the PCR products were ligated with pOCUS-HPIV3 digested by SalI and Eco91I to generate plasmids pOCUS-HPIV3-NL478A and pOCUS-HPIV3-NL478I. The details of the construction process of all the plasmids can be obtained upon request. All constructs were verified by sequencing.
In vivo HPIV3 minigenome assay.
The in vivo minigenome assay was performed as described earlier (41) with minor modifications. HeLa cells in 12-well plates were grown to 90% confluence and infected with vTF7-3. One hour postinfection, the cells were transfected with 500 ng of plasmids encoding untagged N, Myc-tagged N (N-Myc), Myc-tagged N mutants, or HA-tagged N (N-HA); 250 ng of pGEM4-P or pGADT7-P encoding HA-tagged P (HA-P); 100 ng pGEM4-L; and 50 ng of plasmid carrying HPIV3 minigenome HPIV3-MG (−) carrying the luciferase reporter gene with Lipofectamine 2000 (Invitrogen). For investigating whether NL478A interferes with the RNA synthesis function of N, transfection was modified slightly. pGEM4-N (62.5 ng), pGEM4-P (250 ng), pGEM4-L (100 ng), and HPIV3-MG (−) (50 ng) were transfected with increasing amounts of pcDNA3.0-N-Myc or pcDNA3.0-NL478A-Myc (31.25 ng, 62.5 ng, 125 ng, and 250 ng). Total amounts of plasmid were always kept constant. Five hours later, the transfection medium was replaced by DMEM containing 4% FBS. After 24 h of transfection, cells were harvested and lysed in 150 μl of lysis buffer, from which 20-μl aliquots were used to determine luciferase activity according to the manufacturer's instructions (luciferase assay kit; Roche). The expression of N-Myc, Myc-tagged N mutant proteins, N-HA, and HA-P was detected via Western blotting with monoclonal anti-Myc antibody (Ab) sc-40 (Santa Cruz) and anti-HA antibody, respectively. All assays were repeated at least three times for accuracy.
In vivo coimmunoprecipitation.
HeLa cells in 6-well plates were infected with vTF7-3 at a multiplicity of infection of 3 for 1 h. Then, the cells were washed and transfected with the appropriate plasmids with Lipofectamine 2000. In order to examine the association of N mutant proteins and wild-type N, 1.5 μg of plasmid encoding Myc-tagged N mutants was coexpressed with 1 μg of plasmid encoding N-HA. To examine interactions between Myc-tagged N mutants and HA-P or P mutants, 1.5 μg of plasmid encoding Myc-tagged N mutants was coexpressed with 1 μg of plasmid encoding HA-P or HA-PΔC100. After 24 h of transfection, cells were harvested and lysed with 800 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% [wt/vol] Triton X-100, 1 mM EDTA [pH 8.0], 0.1% [vol/vol] SDS, and protease inhibitor cocktail) for 30 min. The soluble cytoplasmic extracts were collected by centrifugation for 30 min at 4°C and precleared by incubation with 20 μl protein G-Sepharose 4 Fast Flow medium for 1 h at 4°C with rotation. The precleared supernatants of lysates were incubated with polyclonal anti-Myc antibody sc-789 (Santa Cruz) or monoclonal anti-HA (Sigma) antibody for 4 h at 4°C with rotation. After brief centrifugation, supernatants were collected and mixed with protein G-Sepharose 4 Fast Flow medium and incubated overnight at 4°C with rotation. Beads were collected by centrifugation at 5,000 rpm for 2 min and extensively washed five times with washing buffer. The beads were then boiled for 10 min in SDS protein loading buffer, and the bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting using anti-Myc and anti-HA monoclonal antibodies.
N-RNA purification by CsCl gradient centrifugation.
HeLa cells cultured in 10-cm dishes were transfected with pCAGGS-N-Myc and pCAGGS-NL478A-Myc, respectively, with Lipofectamine 2000. After 48 h, cells were harvested and lysed in 600 μl of lysis buffer and then centrifuged at 13,000 × g at 4°C for 20 min. The cleared supernatants were loaded onto a discontinuous 25 to 40% (wt/wt) CsCl gradient and centrifuged for 1 h at 250,000 × g at 20°C. A visible band where N-RNA stays was collected and centrifuged at 200,000 × g at 4°C for 1 h to precipitate the contents. Then, the pellet was resuspended in 100 μl of phosphate-buffered saline (PBS) free of RNase, and half of the sample was analyzed by Western blotting. The other half was used for purification of RNA by phenol-chloroform extraction, and the RNA was analyzed in a 1% agarose gel.
In vitro coimmunoprecipitation.
N-RNA and NL478A-RNA were purified from HeLa cells by CsCl gradient centrifugation as described above, resuspended in 600 μl of lysis buffer, and divided into 3 equal aliquots. Each 200 μl of N-RNA or NL478A-RNA was mixed with 200 μl lysis buffer, HeLa cell lysates containing HA-P, or HA-PΔC100. Then, 400 μl of lysis buffer was added to each mixture to increase the volume to 800 μl for in vitro coimmunoprecipitation assay. The process of coimmunoprecipitation was performed as described for in vivo coimmunoprecipitation, and monoclonal anti-HA antibody was used for the immunoprecipitation.
Incorporation of BrUTP into minigenome RNA in vitro.
To decrease the size of the minigenome RNA transcribed in vitro, the CAT gene was inserted into minigenome to replace the luciferase reporter gene in HPIV3-MG (−) to construct plasmid HPIV3-MG-CAT carrying minigenome RNA containing CAT flanked with leader and trailer RNA of HPIV3. Because the plasmid HPIV3-MG-CAT does not have an appropriate restriction site downstream of the leader sequence, PCR products were chosen as the template for the transcription in vitro. Primer 1 (5′-AGATCTGTCGACAGGCCTGCAG-3′) and primer 2 (5′-ACCAAACAAGAGAAGAAACTTGTTCGG-3′) were used to amplify the fragment carrying minigenome RNA in the order 3′-leader-CAT-trailer-5′. One microgram of PCR product purified by phenol-chloroform extraction was used in transcription in vitro by using the Transcriptaid T7 high-yield transcription kit (Thermo; catalog no. K0441) in the presence of BrUTP. The mixtures of ATP, CTP, GTP, UTP, and BrUTP were added into the reaction system and incubated at 37°C for 2 h. The molar ratio of BrUTP to strand UTP is 1:3, and the final concentration of BrUTP is 2.5 mM. When the transcription was finished, 2 μl DNase I was added and incubated for 15 min at 37°C to remove the template DNA. Then, the reaction of DNase I was stopped by adding 2 μl 0.5 M EDTA (pH 8.0) and incubating the mixture at 65°C for 10 min. The RNA synthesized in vitro was purified using an RNeasy kit (Qiagen) and analyzed by agarose electrophoresis.
Confocal microscopy.
HeLa cells in 12-well plates were grown on coverslips and transfected with 0.25 μg of pCAGGS-N-Myc or pCAGGS-NL478A-Myc alone or together with 1 μg of pCAGGS-HA-P in the presence or absence of transfected minigenome RNA synthesized in vitro. At 24 h posttransfection, cells were washed three times with PBS, fixed with 0.4% paraformaldehyde, and then permeabilized with 0.2% Triton X-100 for 20 min at room temperature. After blocking by 3% bovine serum albumin in PBS for 30 min at room temperature, the cells were incubated with rabbit polyclonal anti-Myc antibody (Santa Cruz; 1:200), mouse monoclonal anti-HA (Sigma; 1:2,000), or mouse monoclonal bromodeoxyuridine (BrdU) (IIB5) antibody (Santa Cruz; 1:100) for 1 h at room temperature. Then, the cells were washed three times with 1% bovine serum albumin (BSA) in PBS and incubated with the goat anti-rabbit IgG rhodamine (Thermo; 1:100) or goat anti-mouse IgG fluorescein (Thermo; 1:200) secondary antibody for 1 h at room temperature. After being washed three times with 1% bovine serum albumin in PBS, the coverslips were turned over and put on a glass slide with a drop of Fluoroshield (Sigma) containing DAPI (4′,6-diamidino-2-phenylindole) for nuclear staining. Confocal images were collected by an Olympus confocal FV1000 microscope. To visualize the location of CAGGS-N-Myc, CAGGS-NL478A-Myc, CAGGS-HA-P, L-eGFP, or in vitro-transcribed minigenome RNA in HPIV3-infected cells, HeLa cells in 12-well plates grown on coverslips were grown to 40% confluence and infected with HPIV3 at a multiplicity of infection of 0.5 or mock infected. At 24 h postinfection, cells were transfected with 2 μg pCAGGS-N-Myc, pCAGGS-NL478A-Myc, pCAGGS-HA-P, pEGFP-N1-L-eGFP, and minigenome RNA, respectively. At 48 h postinfection, cells were fixed and processed as described above. To visualize the location of CAGGS-NL478A-Myc when it was coexpressed with CAGGS-N-Flag and CAGGS-HA-P, HeLa cells in 12-well plates grown on coverslips were transfected with 0.25 μg of pCAGGS-N-Flag, 0.25 μg of pCAGGS-NL478A-Myc, and 1 μg of pCAGGS-HA-P alone or in various combinations. After 24 h of transfection, cells were fixed and processed as described above. The mouse monoclonal anti-HA antibody was used for the visualization of HA-P, and the rabbit polyclonal anti-Myc antibody was used for the visualization of CAGGS-NL478A-Myc. The expression of CAGGS-N-Flag was detected via Western blotting with a monoclonal anti-Flag antibody.
Transfection and recovery of recombinant HPIV3.
BSR-T7 cells in 6-well plates, grown to 80% confluence, were transfected with pGEM4-N (400 ng), pGEM4-P (400 ng), pGEM4-L (200 ng), and pOCUS-HPIV3, pOCUS-HPIV3-NL478A, or pOCUS-HPIV3-NL478I (4 μg) with Lipofectamine 2000 (Invitrogen). After 5 h, the transfection medium was removed, and the cells were incubated with 2 ml of DMEM-4% FBS at 32°C for 7 days. Then, the cells were frozen, thawed, and scraped. The clarified supernatant was collected by centrifugation at 5,000 rpm at 4°C for 5 min and layered onto a fresh LLC-MK2 cell monolayer for amplification at 32°C for 4 days. Then, the LLC-MK2 cells were frozen, thawed, and scraped; the clarified supernatant was harvested; and titers were determined. Single recovered recombinant HPIV3 was isolated by being picked up as agar plugs during the titer determination. The agar plugs were placed in 500 μl of Opti-MEM at 4°C overnight, and 250 μl was used to infect fresh LLC-MK2 cell monolayers for amplification of the plaque isolates.
RESULTS
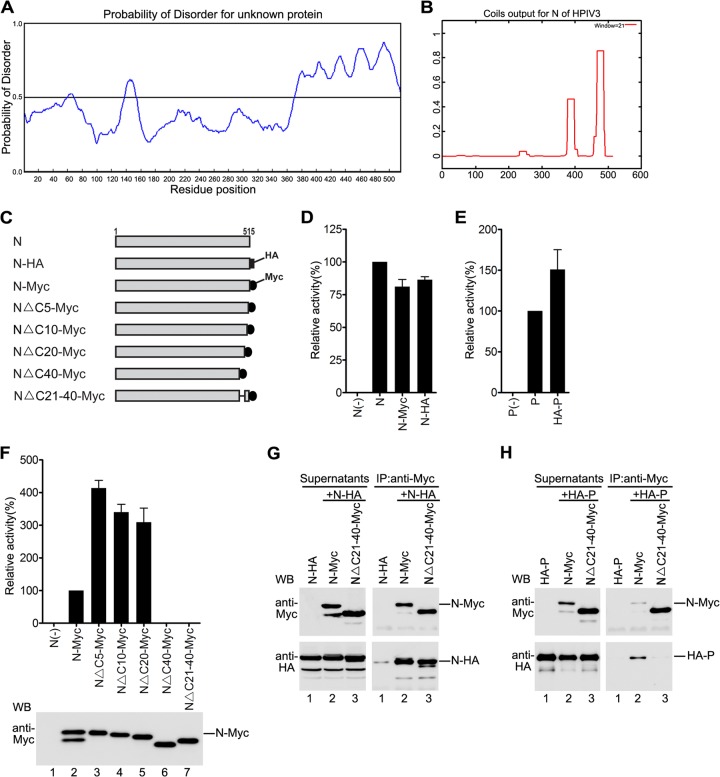
Amino acids 21 to 40 from the C terminus of N are required for the function of N and the interaction of N with P.
Recent studies of some members of the Paramyxoviridae family such as Nipah, Hendra, and measles viruses indicated that interactions between N and P are regulated by an intrinsically disordered region (IDR) and that a disorder-to-order transition determines the binding of N and P (42). Thus, we sought to determine whether N of HPIV3 also contains an IDR and, if so, whether that IDR also mediates the interaction of N with P. To do so, PONDR prediction used for analysis of the IDR of measles virus N (42) was carried out to predict the structural disorder of N, and we found that the C terminus of N contains a long IDR (amino acids 350 to 515) and that the 40 amino acids of the extreme C terminus of N are especially highly disordered (Fig. 1A). In addition, a coiled-coil motif within the protein usually participates in a plethora of protein-protein interactions and recognitions (43). Thus, we also analyzed the sequence of N of HPIV3 for the presence of such a motif using the COILS program (43). COILS analysis revealed the presence of a high-probability coiled-coil motif within the extreme C terminus of N (amino acids 475 to 500) (Fig. 1B). Thus, the extreme C terminus of 40 amino acids of N might be involved in regulating the interaction of N with P and might be required for the function of N. To test our hypothesis, we constructed plasmids encoding four progressively truncated C-terminal mutants of N with Myc tag (Fig. 1C) for studying interactions with P and functions of mutants. To facilitate our study, we first generated two plasmids encoding wild-type N with Myc or HA tag (Fig. 1C) or one plasmid encoding wild-type P with HA tag and sought to determine whether N-HA/Myc or HA-P was as functional as untagged wild-type N or P in analysis of the synthesis of a well-developed HPIV3 minigenome RNA that contains a luciferase reporter gene flanked by trailer and leader sequences of the HPIV3 genome and expresses the luciferase reporter gene when coexpressed with three support proteins of N, P, and L (41); thus, plasmids encoding untagged P, L, and N or plasmids encoding N-HA/Myc or HA-P as described in Fig. 1D and E, respectively, were transfected into HeLa cells expressing the T7 RNA polymerase through vTF7-3 infection. The results showed that N-HA/Myc or HA-P supported the luciferase expression similarly to untagged N and P, suggesting that the functions of N-HA/Myc and HA-P are similar to those of untagged N and P, respectively.
Fig 1.
NΔC21-40 maintains N-N interaction but is defective in N-P interaction. (A) PONDR prediction of structural disorder within HPIV3 N. The disorder prediction value for a given residue is plotted against the residue number. The significance threshold, above which residues are considered to be disordered, set to 0.5, is shown. (B) The COILS program was used to determine the potential coiled-coil motif of N of HPIV3. The graph displays the probability of coiled-coil formation as a function of residue number with a 21-amino-acid scan window. (C) Schematic representation of full-length HPIV3 N and mutants used. (D and E) Minigenome RNA synthesis analysis of tagged N and P. vTF7-3-infected HeLa cells were transfected with plasmids as described in Materials and Methods. The relative luciferase expression levels of cells transfected with plasmids encoding untagged N (D) or P (E) were defined as 100%. Expression of N-Myc/HA or HA-P was detected via Western blotting with anti-Myc antibody and anti-HA antibody. (F) Minigenome RNA synthesis activity of the deletion mutant of N. Cells were transfected and processed as described for panel D. Expression of N-Myc and mutants was detected via Western blotting with anti-Myc antibody. (G) Interactions of the deletion mutants of N and N-HA. HeLa cells were cotransfected with plasmids as described in Materials and Methods. At 24 h posttransfection, the cells were harvested and subjected to coimmunoprecipitation assay. The immunoprecipitation was performed using anti-Myc polyclonal antibody. Lysates were detected via Western blotting using anti-Myc and anti-HA monoclonal antibodies and are indicated on the left side. The immune complexes were detected with anti-Myc and anti-HA monoclonal antibodies and are indicated on the right side. (H) Interactions between the N deletion mutants and HA-P. Cells were transfected and processed as in panel G, except that N-HA was replaced with HA-P.
Next, the RNA synthesis activities of four Myc-tagged C-terminally truncated mutants of N were also determined in a minigenome RNA synthesis assay. As shown in Fig. 1F, mutants NΔC5-Myc, NΔC10-Myc, and NΔC20-Myc supported much higher luciferase expression (3- to 4-fold) than did N-Myc, but luciferase expression was not detected when NΔC40-Myc was used (upper panel). To further confirm that amino acids 21 to 40 from the C terminus of N are indeed indispensable for the function of N, we evaluated the RNA synthesis activity of another mutant, NΔC21-40-Myc. Similar to NΔC40-Myc, NΔC21-40-Myc was unable to support luciferase expression (Fig. 1F, upper panel), although all the mutants have comparable expression levels (Fig. 1F, bottom panel), suggesting that amino acids 21 to 40 from the C terminus are critical for the function of N.
Because oligomerization of N and interaction of N with P are indispensable for the function of N, next we sought to determine whether NΔC21-40-Myc could associate with N and P. Plasmids encoding N-HA were transfected either individually or jointly with plasmids encoding N-Myc or NΔC21-40-Myc into HeLa cells as shown in Fig. 1G. At 24 h posttransfection, whole-cell lysates were pelleted and supernatants were subjected to immunoprecipitation with an anti-Myc polyclonal antibody. N-HA could be coimmunoprecipitated similarly by N-Myc or NΔC21-40-Myc (Fig. 1G, bottom right panel, lanes 2 and 3), but N-HA alone could not be coimmunoprecipitated in the absence of N-Myc or NΔC21-40-Myc (Fig. 1G, bottom right panel, lane 1). These results strongly suggest that amino acids 21 to 40 from the C terminus are not required for oligomerization of N. However, when HA-P was coexpressed with N-Myc or NΔC21-40-Myc and the interactions were evaluated via a similar coimmunoprecipitation assay, HA-P could be coimmunoprecipitated by N-Myc but not by NΔC21-40-Myc (Fig. 1H, bottom right panel, lanes 2 and 3), demonstrating that amino acids 21 to 40 from the C terminus of N are indispensable for interaction of N with P. Of note, we always observed two bands for expressed N-Myc or N-HA in Western blot analysis using monoclonal anti-Myc or anti-HA antibodies: the intense upper band represents the authentic size of N-Myc or N-HA and the faint lower band probably is the degraded product of N (Fig. 1F, bottom panel, lane 2; Fig. 1G, left panel, lanes 1 and 2).
NL478A maintains NL478A-N interaction but is defective in NL478A-P interaction.
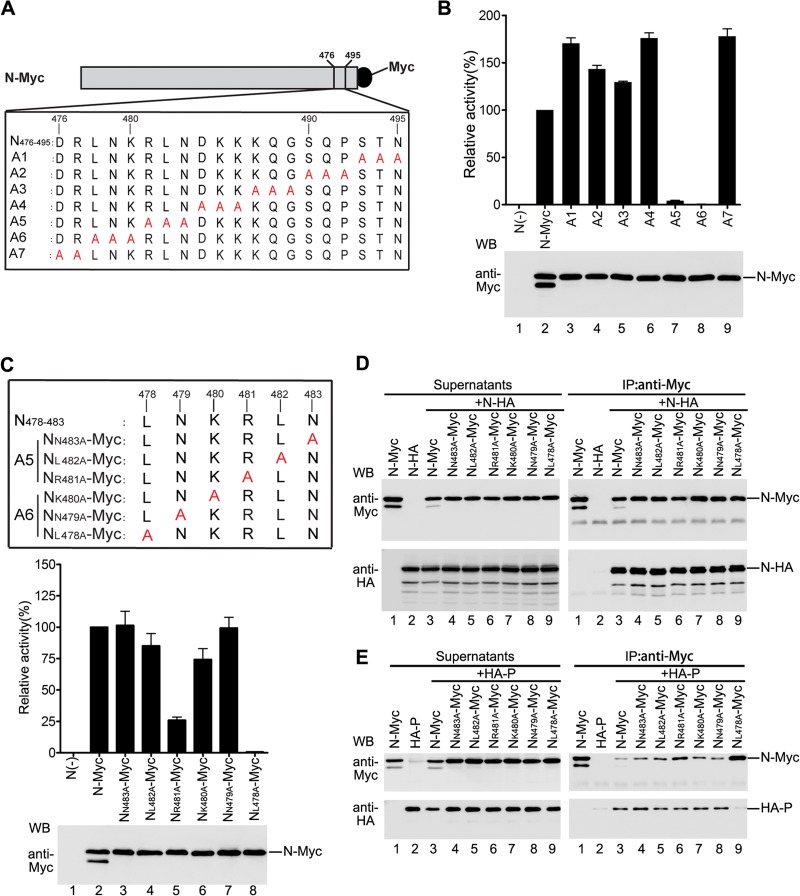
Having established that amino acids 21 to 40 from the C terminus are indispensable for the RNA synthesis function of N and the interaction of N with P, we sought to identify the precise site(s) in this region contributing to the functions of N and the interaction of N with P by employing alanine-scanning mutagenesis. First, we generated seven plasmids encoding N mutants with Myc tag via mutating triple (A1 to A6) or double (A7) amino acids in this region to alanines (Fig. 2A). To determine whether the individual alanine-scanning mutants could support luciferase expression in a minigenome RNA synthesis assay, we evaluated the luciferase expression of HeLa cells expressing HA-P, L, minigenome, and N-Myc or individual N mutants (A1 to A7). As shown in Fig. 2B, all the N mutants, with the exception of one mutant (A6), supported luciferase expression (upper panel). The relative luciferase activity of five mutants (A1 to A4 and A7) was higher than that observed with N-Myc, and the relative luciferase activity of one mutant (A5) was lower (13%) than that observed with N-Myc (upper panel). All the mutants were expressed similarly to N-Myc in transfected cells (Fig. 2B, bottom panel). Therefore, amino acids within A5 and A6 might be important for the RNA synthesis function of N.
Fig 2.
Amino acid L478 within N is critical for the function of N and N-P interaction. (A) Myc-tagged alanine-scanning mutants of N used in this study. Triple amino acids or double amino acids within amino acids 476 to 495 of N were replaced with alanine (shown in red). The names of the mutants are shown on the left. (B) Minigenome RNA synthesis activities of the mutants. Plasmids encoding P, L, minigenome, and N-Myc or mutants were transfected into vTF7-3-infected HeLa cells. Relative luciferase expression levels were measured, and the expression of N-Myc and the mutants was detected via Western blotting as described for Fig. 1F. (C) Minigenome RNA synthesis activities of the alanine-scanning mutants with a single-site mutation within A5 and A6. Amino acids in the region comprising positions 478 to 483 (LNKRLN) of N were replaced with alanine (shown in red). The names of the mutants are shown on the left. Their minigenome RNA synthesis activities were measured as described for Fig. 1F. (D) Interaction of mutants used in panel C with N-HA. Plasmid encoding N-HA was transfected singly or cotransfected with plasmids encoding N-Myc or mutants. Immunoprecipitation was performed with anti-Myc polyclonal antibody. The expression of the proteins was detected by Western blotting using anti-Myc and anti-HA monoclonal antibodies (left). The immune complexes were detected with anti-Myc and anti-HA monoclonal antibodies (right). (E) Interactions between mutants used in panel C and P. Cells were transfected and processed as in panel D, except that the N-HA was replaced by HA-P.
Next, we sought to narrow the scope of amino acids that are important for the RNA synthesis function of N; thus, we also generated six plasmids encoding N mutants with Myc tag containing unique alanine mutations within A5 and A6 (Fig. 2C, upper panel). We repeated the above-mentioned experiment using these individual mutants; all the mutants have expression levels similar to that of N-Myc (Fig. 2C, bottom panel); clearly, with the exception of mutant NL478A-Myc, four mutants, NN483A-Myc, NL482A-Myc, NK480A-Myc, and NN479A-Myc, supported luciferase expression levels comparable to that supported by N-Myc, and mutant NR481A-Myc supported lower luciferase expression (25%) than did N-Myc (Fig. 2C, middle panel). Therefore, amino acid L478 within N is essential for the RNA synthesis function of N. Then, we sought to determine whether NL478A-Myc would generate an effect on oligomerization of N and N-P interaction similar to that generated by NΔC21-40-Myc. Coimmunoprecipitation experiments similar to those described for Fig. 1G and H were performed. The results showed that N-HA was specifically coimmunoprecipitated by NL478A-Myc or other mutants as efficiently as by N-Myc (Fig. 2D, right panel), indicating that NL478A-Myc had no adverse effect on NL478A-N interaction for forming hetero-oligomeric N. Subsequently, in a coimmunoprecipitation assay to examine interaction of N or mutants with P, NL478A-Myc failed to coimmunoprecipitate HA-P even though much higher quantities of NL478A-Myc were immunoprecipitated with anti-Myc antibody (Fig. 2E, bottom right panel, lane 9). However, other mutants interact with HA-P as efficiently as does N-Myc (Fig. 2E, bottom right panel, lanes 3 to 8). Thus, our results suggest that NL478A-Myc is defective in NL478A-P interaction.
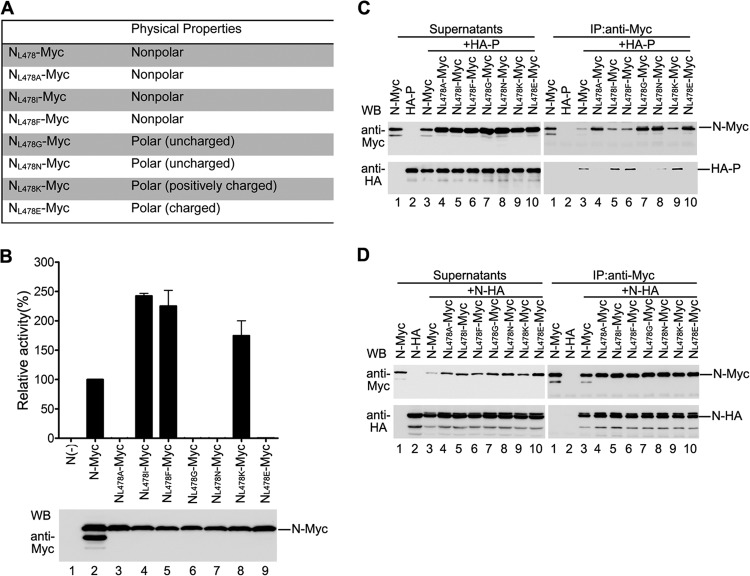
Because our results showed that L478 is a critical amino acid for N-P interaction and the RNA synthesis function of N, we sought to characterize how the amino acid in this site determines N-P interaction and RNA synthesis function of N. Thus, we created six N mutants in which L478 was mutated to different polar amino acids (G, N, K, and E), a nonpolar amino acid (I), or an aromatic residue (F) (Fig. 3A). The RNA synthesis function of these mutants was examined, and we found that NL478I-Myc, NL478F-Myc, and NL478K-Myc supported higher luciferase expression (1.5- to 2.5-fold) than that supported by N-Myc (Fig. 3B, upper panel, lanes 4, 5, and 8), whereas mutants NL478G-Myc, NL478N-Myc, and NL478E-Myc did not support any luciferase expression (Fig. 3B, upper panel, lanes 6, 7, and 9) even if they were expressed at similar levels as N-Myc (Fig. 3B, bottom panel). Furthermore, consistent with the RNA synthesis function of mutants, NL478I-Myc, NL478F-Myc, and NL478K-Myc interacted well with HA-P (Fig. 3C, right panel, lanes 5, 6, and 9), but the NL478G-Myc, NL478N-Myc, and NL478E-Myc mutants failed to interact with HA-P (Fig. 3C, right panel, lanes 7, 8, and 10). However, the capacity of all the mutants for interaction with N-HA was similar to that of N-Myc (Fig. 3D). Taken together, these results suggest that the nonpolar hydrophobic or polar positively charged nature of the residue in this site is critical for the RNA synthesis function of N and N-P interaction.
Fig 3.
Effect of amino acid nature in site L478 on the function of N, N-N interaction, and N-P interaction. (A) Amino acid L478 was mutated to other amino acids with different physical properties. (B) Minigenome RNA synthesis activities of L478 mutants. Plasmids encoding P, L, minigenome, and N-Myc or L478 mutants were transfected into vTF7-3-infected HeLa cells. Relative luciferase expression was measured, and the expression of N-Myc and L478 mutants was detected via Western blotting as described for Fig. 1F. (C) Interaction between L478 mutants and HA-P. HeLa cells were transfected with plasmids encoding N-Myc or mutants with HA-P as described in Materials and Methods. Coimmunoprecipitation was performed as described for Fig. 1G. (D) Interaction between L478 mutants and N-HA. Cells were transfected and processed as in panel C, except that HA-P was replaced with N-HA.
Of note, when detecting interaction of N or mutants with P, we repeatedly observed that higher quantities of N-Myc were immunoprecipitated by anti-Myc antibody when N-Myc was expressed alone than when coexpressed with HA-P (Fig. 2E and 3C, right and upper panel, lanes 1 and 3), suggesting that N-P interaction results in less efficient binding of anti-Myc antibody to Myc tag; therefore, less N-Myc was immunoprecipitated by anti-Myc antibody. It is possible that either N-P interaction directly blocks binding of anti-Myc antibody to Myc tag or N-P interaction induces the conformational change of N which covers the Myc tag.
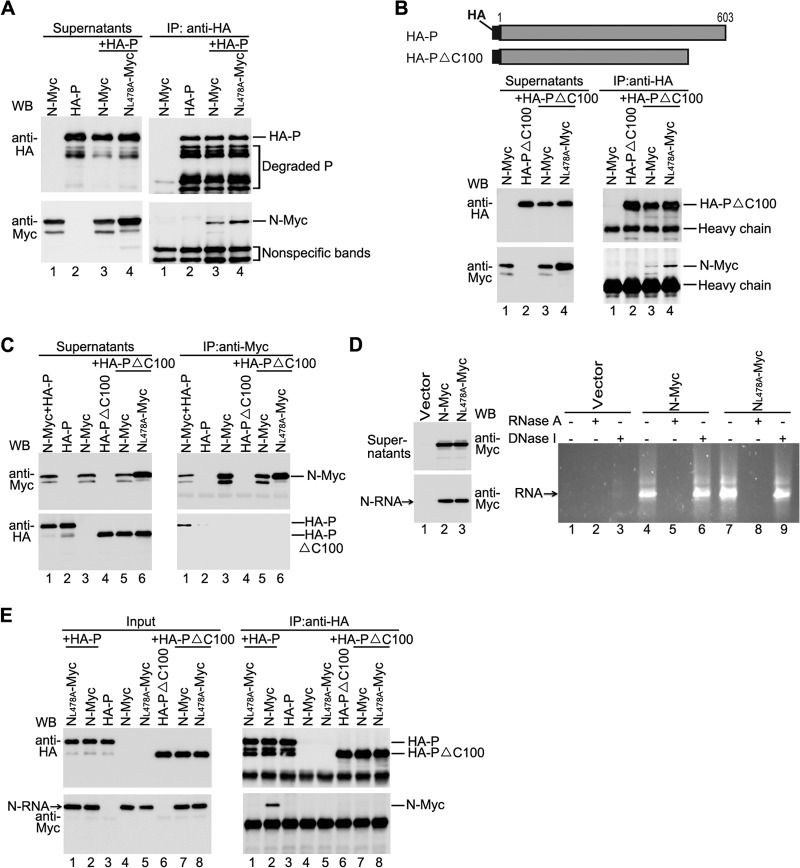
NL478A is defective in NL478A-RNA interaction but not in NL478A0-P interaction.
In all the above-mentioned assays for coimmunoprecipitation, the supernatants of whole-cell lysates expressing N consist of two types of N: N0 and N-RNA complex, both of which can interact with P. Abolishment of either N0-P or N-RNA-P interaction will definitely result in the defect of the RNA synthesis function of N. Based on the facts that NL478A-Myc fails to coimmunoprecipitate P using anti-Myc antibody and that N-P interaction results in less efficient binding of anti-Myc antibody to N-Myc, we sought to know whether NL478A-Myc is defective in one of two types of interaction or both. For this purpose, we performed a converse coimmunoprecipitation experiment using anti-HA antibody as described in Fig. 4A. Comparable N-Myc and NL478A-Myc were specifically coimmunoprecipitated by HA-P using anti-HA antibody, suggesting that NL478A-Myc still maintains one of two types of interaction, either NL478A0-P or NL478A-RNA-P, and this interaction blocks anti-Myc antibody binding to Myc tag. It has been shown that the C terminus of P is indispensable for the binding of N-RNA to P (39); thus, we generated a P mutant with deletion of the C-terminal 100 amino acids (HA-PΔC100) which interacts with N0 but not N-RNA. N-Myc or NL478A-Myc was coexpressed with HA-PΔC100, and coimmunoprecipitation results clearly showed that N-Myc and NL478A-Myc were coimmunoprecipitated similarly by HA-PΔC100 using anti-HA antibody, but N-Myc alone could not be (Fig. 4B, bottom right panel, lanes 1, 3, and 4), indicating that NL478A-Myc maintains N0-P interaction. Furthermore, we performed a converse coimmunoprecipitation experiment with the same protein expression combinations with anti-Myc antibody. Neither N-Myc nor NL478A-Myc was able to coimmunoprecipitate HA-PΔC100, even though N-Myc and NL478A-Myc were well immunoprecipitated (Fig. 4C, right panel, lanes 5 and 6). Taken together, the results indicate that NL478A-Myc indeed maintains NL478A0-P interaction and this interaction blocks the binding of anti-Myc antibody to Myc tag.
Fig 4.
NL478A maintains NL478A0-P interaction but is defective in NL478A-RNA-P. (A) NL478A-Myc interacted with P when immunoprecipitation was performed using anti-HA monoclonal antibody. Plasmids encoding N-Myc or NL478A-Myc and HA-P were cotransfected into HeLa cells. At 24 h posttransfection, the cells were harvested and subjected to immunoprecipitation using anti-HA monoclonal antibody. The expression of N-Myc, NL478A-Myc, and HA-P was detected via Western blot analysis using anti-Myc and anti-HA monoclonal antibodies (left), and the immune complexes were detected with anti-Myc and anti-HA monoclonal antibodies (right). (B) NL478A-Myc interacted with HA-PΔC100 when immunoprecipitation was performed using anti-HA monoclonal antibody. A schematic diagram of HA-PΔC100 is shown in the upper panel, and the interaction of NL478A-Myc and HA-PΔC100 is shown in the lower panel. Protein expression (left) and immunoprecipitation (right) were detected as described for panel A. (C) Neither N-Myc nor NL478A-Myc interacted with HA-PΔC100 when immunoprecipitation was performed using anti-Myc polyclonal antibody. Plasmids encoding HA-P or HA-PΔC100 were cotransfected with plasmids encoding N-Myc or NL478A-Myc. Immunoprecipitation was performed using anti-Myc polyclonal antibody. Protein expression (left) and immune complexes (right) were detected using anti-Myc and anti-HA monoclonal antibodies. (D) N-RNA purification via CsCl gradient centrifugation. HeLa cells were transfected with empty plasmid or plasmids encoding N-Myc and NL478A-Myc individually. At 48 h posttransfection, the cells were lysed, and the supernatants were subjected to CsCl gradient centrifugation as described in Materials and Methods. The lysates and purified N-RNA were detected via Western blot analysis (left). The RNAs (lanes 1, 4, and 7) or RNAs treated with RNase A (lanes 2, 5, and 8) or DNase I (lanes 3, 6, and 9) were analyzed in a 1% agarose gel. (E) Interactions between HA-PΔC100 and purified N-RNA in vitro. The same amounts of purified N-RNA or NL478A-RNA were mixed with cell lysates expressing HA-P or HA-PΔC100, respectively, and processed as described in Materials and Methods. Immunoprecipitation was performed using anti-HA monoclonal Ab. The input of HA-PΔC100, HA-P, NL478A-RNA, and N-RNA (left) and the immune complexes (right) was detected with anti-Myc and anti-HA monoclonal antibodies.
Next, we sought to confirm that the RNA binding ability of NL478A-Myc was similar to that of N-Myc and that NL478A-Myc is indeed defective in interaction of NL478A-RNA with P. N-Myc and NL478A-Myc were expressed individually; supernatants from the whole-cell lysates (Fig. 4D, left upper panel, lanes 2 and 3) were added onto a discontinuous CsCl gradient for further ultracentrifugation as described in Materials and Methods. A visible band where the N-RNA complex was located was extracted, pelleted, and resuspended in PBS as purified N-RNA complex for Western blot analysis (Fig. 4D, left bottom panel, lanes 2 and 3) and nucleic acid extraction. As shown in Fig. 4D (right panel), the nucleic acids extracted from the pellet ran as a smear with a bright main band in a 1% agarose gel only when N-Myc or NL478A-Myc but not vector was expressed (lanes 1, 4, and 7), and they were not digested by DNase I treatment (lanes 6 and 9) but were digested by RNase A treatment (lanes 5 and 8), thereby demonstrating that N-Myc and NL478A-Myc bind to RNAs at similar levels. Next, purified N-RNA or NL478A-RNA was mixed with cell lysates expressing HA-P or HA-PΔC100 in vitro, respectively, as shown in Fig. 4E (left panel), and in vitro coimmunoprecipitation was carried out using anti-HA antibody. As shown in Fig. 4E (right panel), purified N-RNA could be coimmunoprecipitated by HA-P but NL478A-RNA could not be, even though similar amounts of N-RNA and NL478A-RNA were used (lanes 1 and 2), suggesting that NL478A-RNA is indeed defective in interaction with HA-P. In addition, neither N-RNA nor NL478A-RNA was able to be coimmunoprecipitated by HA-PΔC100 (right panel, lanes 7 and 8), further suggesting that HA-PΔC100 indeed interacts with N0 but does not interact with the N-RNA complex (Fig. 4B, right panel, lanes 3 and 4; Fig. 4E, right panel, lanes 7 and 8). Taking the data together (Fig. 2E and 4A, B, C, and E) clearly demonstrates that N0-P interaction or conformational change of N induced by N0-P interaction blocks the anti-Myc antibody binding to Myc tag, and NL478A-Myc maintains NL478A0-P interaction and RNA binding ability but forms an aberrant NL478A-RNA complex that is defective in interaction with P.
Coexpression of N, but not NL478A, with P supports cytoplasmic inclusion body formation.
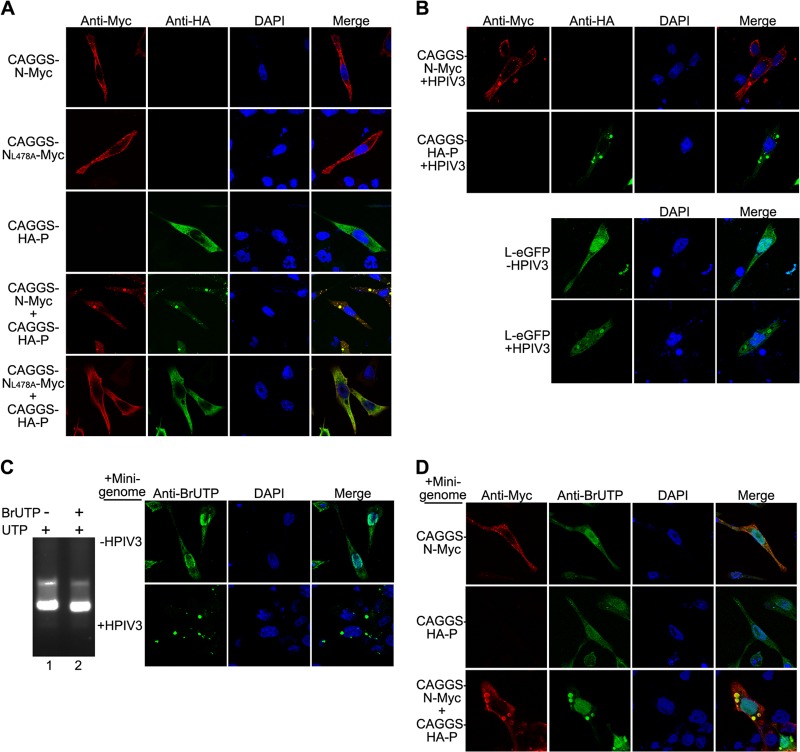
To further evaluate the localization of NL478A when coexpressed with P, we also constructed three plasmids encoding Myc-tagged N (CAGGS-N-Myc), Myc-tagged NL478A (CAGGS-NL478A-Myc), and HA-tagged P (CAGGS-HA-P) for immunofluorescence assay. CAGGS-N-Myc, CAGGS-NL478A-Myc, and CAGGS-HA-P were expressed individually or coexpressed with plasmid encoding CAGGS-HA-P in HeLa cells, as shown in Fig. 5A. An immunofluorescence assay was performed as described in Materials and Methods. Expression of CAGGS-N-Myc or CAGGS-NL478A-Myc alone displayed a diffuse pattern with some tiny particles throughout the cell (Fig. 5A, first and second rows). When CAGGS-HA-P was expressed alone in cells, it was evenly distributed throughout the cells (Fig. 5A, third row). In contrast, we observed that when coexpressed, CAGGS-N-Myc and CAGGS-HA-P clearly formed discrete, punctate, cytoplasmic inclusion-like structures that were heterogeneous in size (Fig. 5A, fourth row from top). However, no inclusion-like structures were observed when CAGGS-NL478A-Myc and CAGGS-HA-P were coexpressed (Fig. 5A, bottom row).
Fig 5.
Interaction of N and P provides the minimal requirement for inclusion body formation. (A) N but not NL478A coexpressed with P forms inclusion body-like structures. HeLa cells were transfected with plasmids encoding CAGGS-N-Myc or CAGGS-NL478A-Myc individually or coexpressed with plasmid encoding CAGGS-NL478A-Myc-HA-P. At 24 h posttransfection, cells were fixed and stained with antibodies and visualized via confocal microscopy as described in Materials and Methods. Nuclear staining was obtained by treatment with DAPI. (B) Visualization of CAGGS-N-Myc, CAGGS-HA-P, and L-eGFP in HPIV3-infected cells. Cells were infected with HPIV3 at a multiplicity of infection of 0.5 or mock infected. At 24 h postinfection, cells were transfected with plasmids encoding CAGGS-N-Myc, CAGGS-HA-P, and L-eGFP. At 48 h postinfection, cells were fixed and stained with antibodies and visualized via confocal microscopy. (C) Transcribed minigenome RNA localizes to inclusion bodies in HPIV3-infected cells. BrUTP was incorporated into minigenome RNA transcribed in vitro as described in Materials and Methods. Minigenome RNA-incorporated BrUTP was then analyzed in a 1% agarose gel (left). HeLa cells were infected with HPIV3 at a multiplicity of infection of 0.5 or mock infected. At 24 h postinfection, minigenome RNA synthesized in vitro was transfected into the HeLa cells. At 48 h postinfection, cells were harvested and processed as in panel A and examined via confocal microscopy. (D) Transcribed minigenome RNA localizes to inclusion bodies formed in the presence of N and P. HeLa cells were transfected with plasmids encoding CAGGS-N-Myc and transcribed minigenome RNA in vitro. At 24 h posttransfection, cells were processed as in panel A and examined via confocal microscopy.
Previous studies showed that infection by some NNS RNA viruses can induce the formation of punctate structures of concentrated viral proteins termed viral inclusion bodies (44, 45) which contain viral RNA, N, P, and L and appear to be bona fide sites of RNA synthesis (46–49). We sought to further demonstrate that structures formed by CAGGS-N-Myc and CAGGS-HA-P are indeed inclusion bodies; thus, CAGGS-N-Myc, CAGGS-HA-P, and eGFP-tagged-L (L-eGFP) were individually expressed in HPIV3-infected cells. In each case, CAGGS-N-Myc, CAGGS-HA-P, or L-eGFP was also recruited to punctate, cytoplasmic viral inclusion bodies that were structurally similar to those that we observed when CAGGS-N-Myc and CAGGS-HA-P were coexpressed (Fig. 5B); however, when expressed alone in the absence of HPIV3 infection, CAGGS-N-Myc, CAGGS-HA-P, and L-eGFP were all localized throughout the cell (Fig. 5A, fourth row from top; Fig. 5B, bottom panel, top row). To determine whether these inclusion bodies contain viral RNA, we first synthesized the minigenome RNAs in an in vitro transcription reaction driven by T7 RNA polymerase in the presence of 5-BrUTP, and the minigenome RNAs that incorporated 5-BrUTP migrated with a slightly higher mobility than that generated from UTP only on an agarose gel (Fig. 5C, left panel). Then, 5-BrUTP-labeled minigenome RNAs were transfected into HPIV3-infected cells. Clearly, minigenome RNAs were completely distributed to typical inclusion bodies, but minigenome RNAs were distributed only throughout the cells in the absence of HPIV3 infection (Fig. 5C, right panel). We also sought to determine whether the interaction of CAGGS-N-Myc and CAGGS-HA-P efficiently recruits minigenome RNAs to inclusion bodies; thus, 5-BrUTP-labeled minigenome RNAs were transfected into cells expressing CAGGS-N-Myc, CAGGS-HA-P, or both CAGGS-N-Myc and CAGGS-HA-P. As shown in Fig. 5D, coexpressed CAGGS-N-Myc and CAGGS-HA-P could redistribute minigenome RNAs into inclusion bodies, but individually expressed CAGGS-N-Myc or CAGGS-HA-P could not. Taken together, these data show that coexpression of N with P efficiently supports cytoplasmic inclusion body formation, but coexpression of NL478A with P does not, and inclusion bodies formed in HPIV3-infected cells contain all the components required for transcription and replication.
N rescues the function of NL478A via the interaction of NL478A and N.
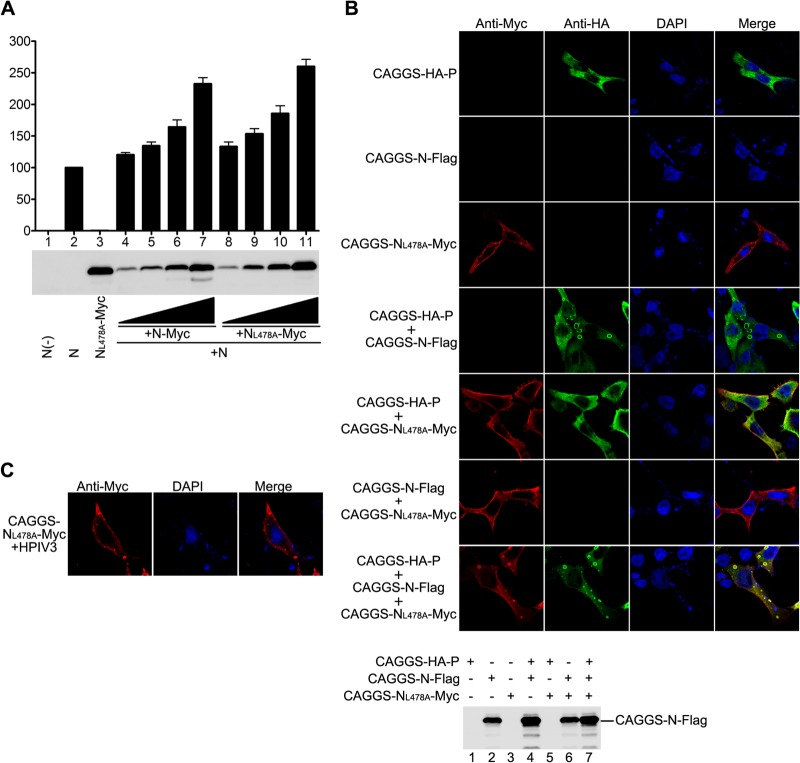
The observation that NL478A-Myc is defective in RNA synthesis function but maintains interaction with N prompted us to investigate whether NL478A-Myc has a dominant negative effect on the RNA synthesis function of N in a minigenome assay. A low concentration of untagged N was used so that it would be sufficient for a basal level of luciferase expression. We then evaluated the roles of NL478A-Myc by cotransfecting increasing amounts of plasmids expressing N-Myc or NL478A-Myc in the presence of constant plasmids expressing low concentrations of untagged N. As shown in Fig. 6A, a basal level of luciferase expression was detected and counted as 100% by using a low concentration of untagged N (upper panel, lane 2), but an optimal concentration of NL478A-Myc was unable to support any luciferase expression (upper panel, lane 3), as previously described (Fig. 2C, middle panel, lane 8). Surprisingly, expression of increasing concentrations of NL478A-Myc or N-Myc gradually increased the luciferase expression with comparable levels in the presence of constant untagged N (upper panel, lanes 4 to 11), suggesting that the hetero-oligomers consisting of N and NL478A are active in RNA synthesis. To ascertain whether the effect observed is indeed caused by increased protein expression, we also performed Western blot analysis using anti-Myc antibody for expression of NL478A-Myc and N-Myc. As shown in Fig. 6A (bottom panel), increased luciferase expression correlated with increased expression of NL478A-Myc and N-Myc (lanes 4 to 11). Next, we sought to know whether N would also rescue NL478A for inclusion body formation in the presence of P. Thus, we constructed a plasmid encoding Flag-tagged N (N-Flag) and performed triple transfection with plasmids encoding N-Flag, CAGGS-NL478A-Myc, and CAGGS-HA-P; correspondingly, single or two-plasmid combination transfections were also used as controls. Of note, we only labeled CAGGS-NL478A-Myc using anti-Myc antibody or CAGGS-HA-P using anti-HA antibody for immunofluorescence analysis, and the expression of N-Flag was detected by Western blotting using anti-Flag antibody (Fig. 6B, bottom panel). As shown in Fig. 6B, coexpression of CAGGS-NL478A-Myc and CAGGS-HA-P induced the formation of inclusion bodies in the presence of N-Flag (Fig. 6B, top panel, bottom row). In contrast, coexpression of CAGGS-HA-P with CAGGS-NL478A-Myc or CAGGS-NL478A-Myc with N-Flag failed to induce the formation of inclusion bodies (Fig. 6B, top panel, fifth and sixth rows from top). Similarly, CAGGS-NL478A-Myc also formed inclusion bodies in HPIV3-infected cells (Fig. 6C). Taken together, these data suggest that inclusion bodies may be active RNA synthesis sites and that hetero-oligomers consisting of N and NL478A can interact with P for inclusion body formation and are functional in RNA synthesis.
Fig 6.
N and NL478A form functional hetero-oligomers. (A) N-Myc rescues the RNA synthesis activity of NL478A-Myc. HeLa cells infected with vTF7-3 were transfected with plasmids encoding untagged N (62.5 ng), P (250 ng), L (100 ng), and minigenome (50 ng) with increasing amounts of plasmids encoding N-Myc or NL478A-Myc. Luciferase activity and protein expression were detected as described for Fig. 1F. (B) N rescues the inclusion body formation of NL478A in the presence of P. HeLa cells were transfected with plasmids encoding N-Flag, CAGGS-NL478A-Myc, and CAGGS-HA-P individually or in various combinations. At 24 h posttransfection, cells were fixed and stained with anti-Myc and anti-HA antibodies and examined via confocal microscopy. The expression of N-Flag was detected via Western blotting with anti-Flag monoclonal antibody. (C) NL478A localizes to inclusion bodies in HPIV3-infected cells. HeLa cells, grown on coverslips, were infected with HPIV3 at a multiplicity of infection of 0.5 or mock infected. At 24 h postinfection, cells were transfected with 2 μg of plasmid encoding CAGGS-NL478A-Myc. At 48 h postinfection, cells were processed as described for panel B and visualized via confocal microscopy.
NL478A is defective in virus growth.
To further determine the role of the NL478A in the virus replication cycle, we sought to know whether recombinant HPIV3 expressing NL478A can be rescued by reverse genetics. Thus, HPIV3 genome cDNA in plasmid pOCUS-HPIV3 was used as the template to generate a new plasmid, pOCUS-HPIV3-NL478A, in which the N coding region was replaced with NL478A. Because NL478I-Myc supported the highest RNA synthesis activity in the minigenome assay (Fig. 3B, lane 4), as a control, we also generated another plasmid, pOCUS-HPIV3-NL478I, in which the N coding region was replaced with NL478A. Next, plasmids pOCUS-HPIV3, pOCUS-HPIV3-NL478A, and pOCUS-HPIV3-NL478I were then transfected along with three support plasmids (41) to recover viruses as described in Materials and Methods. Recombinant viruses encoding wild-type or mutant N were designated HPIV3, HPIV3-NL478A, or HPIV3-NL478I. The results show that HPIV3 and HPIV3-NL478I all could be easily rescued from several independent transfection experiments. However, HPIV3-NL478A could not be recovered even after several independent rescue assays and also trying various amounts of supporting plasmid combinations for transfection. Taken together, our results suggest that NL478A is unable to support virus growth.
DISCUSSION
In the present study, we extensively examined the interaction of N with P and RNA synthesis function of N mutants. We can draw several conclusions from our findings. (i) NL478A maintains the NL478A0-P interaction and possesses the same ability as N for binding to RNA; however, NL478A forms an aberrant N-RNA complex that is unable to be bound by P and is defective for its template function (Fig. 4). (ii) The interaction of N with P provides the minimal requirement for the formation of inclusion bodies, which contain all the components of RNP in HPIV3-infected cells, and NL478A is defective in inclusion body formation when coexpressed with P (Fig. 5). (iii) Hetero-oligomers containing N and NL478A can form inclusion bodies in the presence of P and are also functional in RNA synthesis (Fig. 6).
Although the C terminus of N of several paramyxoviruses contains an IDR and serves as the anchor point of P for binding the polymerase complex (22, 23, 42, 50), so far, structural and molecular information on parainfluenza virus N is scarce. We sought to determine whether N of HPIV3 also contains an IDR and whether that IDR regulates its interaction with P. Indeed, the C-terminal 40 amino acids of N are highly disordered, suggesting its possible involvement in the interaction of N with P and RNA synthesis function (Fig. 1A). By in vivo minigenome RNA synthesis and coimmunoprecipitation assay, we found that amino acids 21 to 40 from the C terminus of N are indispensable for the RNA synthesis function of N and for interaction of N with P (Fig. 1F and H). However, to our surprise, we observed that N with deletion of the C-terminal 5, 10, and 20 amino acids significantly increases RNA synthesis activity of N (Fig. 1F). It is possible that the interaction between the polymerase complex and the N-RNA template has to be relatively weak to allow the polymerase to cartwheel on the N-RNA template during RNA synthesis; thus, for HPIV3, full-length N may result in an excessively strong association between P and N-RNA, which may not be beneficial for transcription and replication. Similar results were obtained from minireplicon studies using measles virus N constructs with C-terminal truncations, in which progressive deletions of up to amino acid 501 resulted in increases in minireplicon reporter gene expression (21). It has been shown that the major inducible heat shock protein (Hsp70) destabilizes the tight N-RNA-P complex by interacting with the C terminus of N in measles virus, resulting in efficient induction of transcription and replication (21, 50). We also could not exclude another possibility that the C-terminal 20 amino acids of N may contain the binding site(s) of a cellular factor(s) which negatively regulates RNA synthesis; deletion of the binding site(s) will also eliminate the inhibitory effect of the cellular factor(s) on RNA synthesis.
Loss of RNA synthesis function of NL478A-Myc caused by defects in NL478A-P interaction could derive from two possibilities: (i) abolishment of NL478A0-P interaction, which is critical for viral genome RNA encapsidation, and (ii) abolishment of the interaction of NL478A-RNA with P, which is the prerequisite for the RNA polymerase complex to gain access to the N-RNA template. Via a series of in vivo and in vitro coimmunoprecipitation assays, we found that NL478A-Myc maintains the NL478A0-P interaction and has the same ability as N for encapsidating RNA (Fig. 4). Similar studies have suggested that the hypervariable C-terminal tail of the Sendai virus N is required for template function but not for RNA encapsidation (22). A recent study revealed that the deletion of 128 amino acids from the C terminus of N of Nipah virus does not impair the formation of the N herringbone-like nucleocapsid structure (51), indicating that the C terminus of N is not required for N-RNA complex formation and self-assembly. Furthermore, one of the binding sites of P was mapped to the 29 amino acids of the C terminus of N of Nipah virus (52). In contrast, deletion or mutations in the extreme C terminus of VSV N completely abolished its ability to encapsidate RNA in an in vitro assembly experiment and in vivo VSV infectious cDNA clone analysis (53, 54). These findings might reflect the different properties of N of paramyxoviruses and rhabdoviruses.
We provided the following evidence to confirm that N0-P interaction or conformational change of N induced by N0-P interaction blocks anti-Myc antibody binding to Myc tag and that NL478A-Myc maintains NL478A0-P but is defective in NL478A-RNA-P interaction: (i) much higher quantities of N-Myc were immunoprecipitated by anti-Myc antibody when N-Myc was expressed alone than coexpressed with HA-P (Fig. 2E, right and upper panels, lanes 2 and 3); (ii) NL478A-Myc is unable to coimmunoprecipitate HA-P using anti-Myc antibody (Fig. 2E, right and bottom panels, lanes 9), but HA-P can efficiently coimmunoprecipitate NL478A-Myc using anti-Myc antibody (Fig. 4A, right and bottom panels, lanes 4); (iii) HA-PΔC100, which interacts with N0 but not with N-RNA, can coimmunoprecipitate NL478A-Myc using anti-HA antibody (Fig. 4B, right and bottom panels, lanes 4), but NL478A-Myc is unable to coimmunoprecipitate HA-PΔC100 using anti-Myc antibody (Fig. 4C, right and bottom panels, lanes 6); (iv) HA-P interacts with purified N-RNA complex but not with purified NL478A-RNA complex (Fig. 4E, right and bottom panels, lanes 1 and 2); (v) HA-PΔC100 can coimmunoprecipitate neither purified N-RNA complex nor purified NL478A-RNA complex using anti-HA antibody (Fig. 4E, right and bottom panels, lanes 7 and 8).
The fact that interaction of N with P provides the minimal requirement for the formation of inclusion bodies, which contain all the components for RNA synthesis, and the observations that NL478A is defective in inclusion body formation and that N rescues inclusion body formation and the RNA synthesis function of NL478A indicate that inclusion bodies might be involved in RNA synthesis (Fig. 5 and 6). However, we also could not exclude the possibility that the effects could be separate. It is of interest that NL478A does not form inclusion bodies even though it maintains NL478A0-P interaction. This indicates that the minimum requirement for the formation of inclusion bodies is interaction of RNA-N with P, not N0 with P. So far, studies of inclusion bodies of NNS RNA viruses have been extensively reported. Derdowski et al. (55) showed that in a recently discovered paramyxovirus, human metapneumovirus, N and P proteins provide the minimal viral requirements for cytoplasmic inclusion body formation in the absence of viral infection. Respiratory syncytial virus forms cytoplasmic inclusion bodies that contain viral RNA, N, and P and the viral polymerase complex and are thought to be sites of nucleocapsid accumulation and viral RNA synthesis (46, 47). Furthermore, recent studies of rabies virus showed that inclusion bodies appear to be bona fide sites of RNA synthesis (48, 49), and although VSV does not require a specialized site for primary RNA transcription, protein expression redirects VSV RNA synthesis to cytoplasmic inclusion bodies (56, 57). Also, studies of parainfluenza virus type 5 showed that viral genomes are targeted into cytoplasmic inclusion bodies that contain the viral replication machinery (58).
It is very interesting to find that when a small amount of untagged N is coexpressed with gradually increased NL478A-Myc in the minigenome RNA synthesis assay, the hetero-oligomeric complex containing NL478A-Myc and wild-type N did not inhibit RNA synthesis in vivo but gradually increased the reporter gene luciferase expression (Fig. 6A), suggesting that the hetero-oligomer containing wild-type N and NL478A is active in RNA synthesis and also indicating that the presence of a small amount of wild-type N in a hetero-oligomer is sufficient for the formation of functional N-RNA template. Since oligomerization of N is a continuous process, it is possible that recruitment of small amounts of the wild-type N to excess of NL478A efficiently corrects the structure of template in final hetero-oligomers.
In summary, we identified and characterized a critical amino acid (L478) in N involved in N-P interaction and RNA synthesis. We provided evidence that NL478A is defective in NL478A-RNA-P interaction but maintains the NL478A0-P interaction. We also demonstrated that N-P interaction provides the minimal requirement for the formation of inclusion bodies and NL478A is defective in inclusion body formation but that wild-type N can rescue defection of inclusion body formation and RNA synthesis of NL478A. To our knowledge, our results provide the first detailed experimental characterization of the interaction between N and P in a paramyxovirus and designate the N-P interaction as a valuable target for rational antiviral approaches.
ACKNOWLEDGMENTS
We acknowledge the generosity of Amiya Banerjee (Cleveland Clinic) in providing plasmids pGEM4-N, pGEM4-P, pGEM4-L, HPIV3-MG (−), and pOCUS-HPIV3 for reverse genetics study. We gratefully acknowledge Markeda Wade for professionally editing the manuscript.
This work was supported by a grant from the China Natural Science Foundation (grant 81271816) and Major State Basic Research Development Program (973 Program) (2012CB518906).
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Collins PL, Chanock RM, McIntosh K. 1996. Parainfluenza viruses, p 1205–1241 In Fields BN, Knipe DM, Howley PM. (ed), Fields virology, 3rd ed. Lippincott-Raven Publications, Philadelphia, PA [Google Scholar]
- 2.Banerjee AK, Barik S, De BP. 1991. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol. Ther. 51:47–70 [DOI] [PubMed] [Google Scholar]
- 3.Longhi S, Receveur-Bréchot V, Karlin D, Johansson K, Darbon H, Bhella D, Yeo R, Finet S, Canard B. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638–18648 [DOI] [PubMed] [Google Scholar]
- 4.Schoehn G, Mavrakis M, Albertini A, Wade R, Hoenger A, Ruigrok RW. 2004. The 12 A structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 339:301–312 [DOI] [PubMed] [Google Scholar]
- 5.Bhella D, Ralph A, Murphy LB, Yeo RP. 2002. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83:1831–1839 [DOI] [PubMed] [Google Scholar]
- 6.Bhella D, Ralph A, Yeo RP. 2004. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340:319–331 [DOI] [PubMed] [Google Scholar]
- 7.Banerjee AK. 1987. The transcription complex of vesicular stomatitis virus. Cell 48:363–364 [DOI] [PubMed] [Google Scholar]
- 8.Luk D, Sánchez A, Banerjee AK. 1986. Messenger RNA encoding the phosphoprotein (P) gene of human parainfluenza virus 3 is bicistronic. Virology 153:318–325 [DOI] [PubMed] [Google Scholar]
- 9.Galinski MS. 1991. Paramyxoviridae: transcription and replication. Adv. Virus Res. 39:129–162 [DOI] [PubMed] [Google Scholar]
- 10.Rozenblatt S, Eizenberg O, Ben-Levy R, Lavie V, Bellini WJ. 1985. Sequence homology within the morbilliviruses. J. Virol. 53:684–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinski MS, Mink MA, Lambert DM, Wechsler SL, Pons MW. 1986. Molecular cloning and sequence analysis of the human parainfluenza 3 virus RNA encoding the nucleocapsid protein. Virology 149:139–151 [DOI] [PubMed] [Google Scholar]
- 12.Jambou RC, Elango N, Venkatesan S, Collins PL. 1986. Complete sequence of the major nucleocapsid protein gene of human parainfluenza type 3 virus: comparison with other negative strand viruses. J. Gen. Virol. 67:2543–2548 [DOI] [PubMed] [Google Scholar]
- 13.Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 14.Morgan EM, Re GG, Kingsbury DW. 1984. Complete sequence of the Sendai virus NP gene from a cloned insert. Virology 135:279–287 [DOI] [PubMed] [Google Scholar]
- 15.Buchholz CJ, Retzler C, Homann HE, Neubert WJ. 1994. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204:770–776 [DOI] [PubMed] [Google Scholar]
- 16.Myers TM, Pieters A, Moyer SA. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322–335 [DOI] [PubMed] [Google Scholar]
- 17.Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363 [DOI] [PubMed] [Google Scholar]
- 18.Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360 [DOI] [PubMed] [Google Scholar]
- 19.Harty RN, Palese P. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863–2867 [DOI] [PubMed] [Google Scholar]
- 20.Liston P, Batal R, DiFlumeri C, Briedis DJ. 1997. Protein interaction domains of the measles virus nucleocapsid protein (NP). Arch. Virol. 142:305–321 [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Glendening C, Linke H, Parks CL, Brooks C, Udem SA, Oglesbee M. 2002. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76:8737–8746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. 1993. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67:4358–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston RL, Baase WA, Gay LS. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630–8640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heggeness MH, Scheid A, Choppin PW. 1981. The relationship of conformational changes in the Sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology 114:555–562 [DOI] [PubMed] [Google Scholar]
- 25.Bourhis JM, Johansson K, Receveur-Bréchot V, Oldfield CJ, Dunker KA, Canard B, Longhi S. 2004. The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res. 99:157–167 [DOI] [PubMed] [Google Scholar]
- 26.Horikami SM, Moyer SA. 1995. Alternative amino acids at a single site in the Sendai virus L protein produce multiple defects in RNA synthesis in vitro. Virology 211:577–582 [DOI] [PubMed] [Google Scholar]
- 27.Mellon MG, Emerson SU. 1978. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J. Virol. 27:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curran J, Marq JB, Kolakofsky D. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard M, Wertz G. 1989. Vesicular stomatitis virus RNA replication: a role for the NS protein. J. Gen. Virol. 70:2683–2694 [DOI] [PubMed] [Google Scholar]
- 30.Chen M, Ogino T, Banerjee AK. 2007. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J. Virol. 81:13478–13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishio M, Tsurudome M, Kawano M, Watanabe N, Ohgimoto S, Ito M, Komada H, Ito Y. 1996. Interaction between nucleocapsid protein (NP) and phosphoprotein (P) of human parainfluenza virus type 2: one of the two NP binding sites on P is essential for granule formation. J. Gen. Virol. 77:2457–2463 [DOI] [PubMed] [Google Scholar]
- 32.Precious B, Young DF, Bermingham A, Fearns R, Ryan M, Randall RE. 1995. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J. Virol. 69:8001–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaji D, Shaila MS. 1999. Domains of Rinderpest virus phosphoprotein involved in interaction with itself and the nucleocapsid protein. Virology 258:415–424 [DOI] [PubMed] [Google Scholar]
- 34.Tober C, Seufert M, Schneider H, Billeter MA, Johnston IC, Niewiesk S, ter Meulen V, Schneider-Schaulies S. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liston P, DiFlumeri C, Briedis DJ. 1995. Protein interactions entered into by the measles virus P, V, and C proteins. Virus Res. 38:241–259 [DOI] [PubMed] [Google Scholar]
- 36.Smallwood S, Ryan KW, Moyer SA. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154–163 [DOI] [PubMed] [Google Scholar]
- 37.Curran J. 1998. A role for the Sendai virus P protein trimer in RNA synthesis. J. Virol. 72:4274–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary SK, Malur AG, Huo Y, De BP, Banerjee AK. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373–382 [DOI] [PubMed] [Google Scholar]
- 39.De BP, Hoffman MA, Choudhary S, Huntley CC, Banerjee AK. 2000. Role of NH(2)- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J. Virol. 74:5886–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leyrat C, Yabukarski F, Tarbouriech N, Ribeiro EA, Jr, Jensen MR, Blackledge M, Ruigrok RW, Jamin M. 2011. Structure of the vesicular stomatitis virus N(0)-P complex. PLoS Pathog. 7:e1002248. 10.1371/journal.ppat.1002248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman MA, Banerjee AK. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201–211 [DOI] [PubMed] [Google Scholar]
- 42.Habchi J, Longhi S. 2012. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol. Biosyst. 8:69–81 [DOI] [PubMed] [Google Scholar]
- 43.Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82–88 [DOI] [PubMed] [Google Scholar]
- 44.Compans RW, Holmes KV, Dales S, Choppin PW. 1966. An electron microscopic study of moderate and virulent virus-cell interactions of the parainfluenza virus SV5. Virology 30:411–426 [DOI] [PubMed] [Google Scholar]
- 45.Nakai M, Imagawa DT. 1969. Electron microscopy of measles virus replication. J. Virol. 3:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243–247 [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Barreno B, Delgado T, Melero JA. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, Blondel D. 2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J. Virol. 83:7948–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ménager P, Roux P, Mégret F, Bourgeois JP, Le Sourd AM, Danckaert A, Lafage M, Préhaud C, Lafon M. 2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri bodies. PLoS Pathog. 5:e1000315. 10.1371/journal.ppat.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Bourhis JM, Longhi S, Carsillo T, Buccellato M, Morin B, Canard B, Oglesbee M. 2005. Hsp72 recognizes a P binding motif in the measles virus N protein C terminus. Virology 337:162–174 [DOI] [PubMed] [Google Scholar]
- 51.Ong ST, Yusoff K, Kho CL, Abdullah JO, Tan WS. 2009. Mutagenesis of the nucleocapsid protein of Nipah virus involved in capsid assembly. J. Gen. Virol. 90:392–397 [DOI] [PubMed] [Google Scholar]
- 52.Chan YP, Koh CL, Lam SK, Wang LF. 2004. Mapping of domains responsible for nucleocapsid protein-phosphoprotein interaction of Henipaviruses. J. Gen. Virol. 85:1675–1684 [DOI] [PubMed] [Google Scholar]
- 53.Das T, Chakrabarti BK, Chattopadhyay D, Banerjee AK. 1999. Carboxy-terminal five amino acids of the nucleocapsid protein of vesicular stomatitis virus are required for encapsidation and replication of genome RNA. Virology 259:219–227 [DOI] [PubMed] [Google Scholar]
- 54.Heinrich BS, Morin B, Rahmeh AA, Whelan SP. 2012. Structural properties of the C terminus of vesicular stomatitis virus N protein dictate N-RNA complex assembly, encapsidation, and RNA synthesis. J. Virol. 86:8720–8729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derdowski A, Peters TR, Glover N, Qian R, Utley TJ, Burnett A, Williams JV, Spearman P, Crowe JE., Jr 2008. Human metapneumovirus nucleoprotein and phosphoprotein interact and provide the minimal requirements for inclusion body formation. J. Gen. Virol. 89:2698–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das SC, Nayak D, Zhou Y, Pattnaik AK. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 80:6368–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog. 6:e1000958. 10.1371/journal.ppat.1000958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlos TS, Young DF, Schneider M, Simas JP, Randall RE. 2009. Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J. Gen. Virol. 90:2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]