Abstract
Although serious human diseases have been correlated with human herpesvirus 6A (HHV-6A) and HHV-6B, the lack of animal models has prevented studies which would more definitively link these viral infections to disease. HHV-6A and HHV-6B have recently been classified as two distinct viruses, and in this study we focused specifically on developing an in vivo model for HHV-6A. Here we show that Rag2−/−γc−/− mice humanized with cord blood-derived human hematopoietic stem cells produce human T cells that express the major HHV-6A receptor, CD46. Both cell-associated and cell-free viral transmission of HHV-6A into the peritoneal cavity resulted in detectable viral DNA in at least one of the samples (blood, bone marrow, etc.) analyzed from nearly all engrafted mice. Organs and cells positive for HHV-6A DNA were the plasma and cellular blood fractions, bone marrow, lymph node, and thymic samples; control mice had undetectable viral DNA. We also noted viral pathogenic effects on certain T cell populations. Specific thymocyte populations, including CD3− CD4+ CD8− and CD3+ CD4− cells, were significantly modified in humanized mice infected by cell-associated transmission. In addition, we detected significantly increased proportions of CD4+ CD8+ cells in the blood of animals infected by cell-free transmission. These findings provide additional evidence that HHV-6A may play a role in human immunodeficiencies. These results indicate that humanized mice can be used to study HHV-6A in vivo infection and replication as well as aspects of viral pathogenesis.
INTRODUCTION
Human herpesvirus 6 (HHV-6) is a member of the betaherpesvirus subfamily and was identified in 1986 (1). Recently this virus has been reclassified as two distinct variants, HHV-6A and HHV-6B, based upon differences in tropism, disease, and epidemiology. These two variants have an overall nucleotide identity of 90% (2, 3), and serological assays to differentiate the variants are in development (4). The main cellular receptor for HHV-6A is CD46, which is expressed on all nucleated cells (5). CD134 has recently been identified as a cellular receptor for HHV-6B (6). While HHV-6B infection is ubiquitous in humans and is known to cause roseola infantum (7), the prevalence of HHV-6A and its role in human disease are poorly understood. HHV-6 has been implicated in diseases that include multiple sclerosis (8–10), encephalitis, graft-versus-host disease (11, 12), other clinical complications of solid-organ transplants and hematopoietic stem-cell transplants (13, 14), drug-induced hypersensitivity syndrome (15, 16), malignancies, myocarditis, and cardiomyopathy (17, 18). HHV-6A has an impact upon human T cell populations (19) and can enhance human immunodeficiency virus type 1 (HIV-1) replication (20). HHV-6A infects helper T cells, as does HIV-1, and HHV-6A has been suggested as a potential cofactor in AIDS progression (18, 20–22).
A variety of animal models have been explored for HHV-6 studies but with limited success. Early reports indicated that HHV-6A was able to replicate in T cells isolated from chimpanzees (23) and pigtailed macaques (24). More recent reports have shown that nonhuman primate (NHP) models exhibit signs of disease following infection with HHV-6. Leibovitch et al. recently showed that common marmosets can be infected with HHV-6A and that infection is accompanied by neurological symptoms (25). Lusso et al. demonstrated that HHV-6A replicates in vivo in pigtailed macaques and that coinfection of macaques with HHV-6A and simian immunodeficiency virus (SIV) resulted in faster depletion of CD4+ T cells (22). The requirement for specialized facilities and the expenses involved in NHP research have been detrimental to further studies.
A small-animal model of HHV-6 infection would allow for further investigation of viral pathogenesis without the costs and facilities required for NHP research. The viral target cells in humanized mice are human immune cells; hence, viral infection in humanized mice may be more reflective of human infection than infection in NHP due to differences in human and NHP genetics. Additionally, humanized mice can be infected with HIV-1, as opposed to the genetically distinct SIV isolates used in NHP models. Humanized mice infected with HIV-1 manifest symptoms of AIDS (26) for studies of HHV-6A as a cofactor in AIDS progression.
Here we report on the use of a newer generation of humanized mice to study HHV-6A replication and pathogenesis in vivo. Rag2−/−γc−/− mice (RAG-hu mice) are engrafted with human hematopoietic stem cells (HSCs) and undergo multilineage hematopoiesis to produce a variety of human blood cell types which are dispersed throughout the lymphoid and nonlymphoid organs. These mice (and other similar HSC-humanized mouse models) have been shown to support replication and viral pathogenesis after challenge with the herpesviruses Epstein-Barr virus (EBV) (27–30), Kaposi's sarcoma-associated herpesvirus (KSHV) (31, 32), and human cytomegalovirus (hCMV) (33). Here RAG-hu mice were challenged with recombinant HHV-6A expressing green fluorescent protein (GFP) via either cell-free or cell-associated transmission. Our findings show that viral DNA is detectable in blood and lymphoid organs for up to 8 weeks after infection. Viral DNA was detected in the plasma, blood cells, thymus, lymph nodes, and bone marrow; although no single mouse tested positive for viral DNA in all of these compartments, 11 of 12 mice were positive in at least one. Specific thymocyte populations were found to be significantly modified in animals infected for longer periods (and via cell-associated transmission), while animals infected via cell-free transmission showed a significant increase in CD4+ CD8+ cells in blood. These findings suggest that humanized mice represent a new in vivo model to study HHV-6A replication and immunopathogenesis.
MATERIALS AND METHODS
Cells.
Human cord blood samples were obtained with permission from the University of Colorado Cord Blood Bank. The Institutional Review Board does not require a protocol for human cord blood because samples are shipped without patient identifiers. HSCs were purified from human cord blood based upon the CD34 marker, using an EasySep human cord blood CD34-positive selection kit (StemCell Technologies). Cells were cultured for 2 days in Iscove's modified Dulbecco's medium (IMDM; Invitrogen) supplemented with 10% fetal calf serum (FCS) and 10 ng/ml each of human interleukin-3 (IL-3), IL-6, and stem cell factor (SCF) (R&D Systems).
Virus propagation.
Bacterial artificial chromosome (BAC)-derived HHV-6A, strain U1102, was previously engineered to express GFP (34). BAC-derived HHV-6A DNA was isolated from overnight Escherichia coli cultures grown at 32°C in LB containing chloramphenicol (15 μg/ml) and purified using NucleoBond PC 100 columns (Clontech) per the manufacturer's protocols. HHV-6A BAC DNA (5 μg) and 1 μg of the human cytomegalovirus pp71-expressing plasmid pCGN1-pp71 (35) were transfected into 5 × 106 Jjhan cells with transfection reagent “V,” utilizing a Nucleofector instrument (Lonza AG) per the manufacturer's protocols. After transfection, the cells were maintained in 3 ml RPMI medium containing 8% fetal bovine serum (Sigma) and supplemented with 100 U/ml each of penicillin and streptomycin. After 5 to 7 days, the medium was changed and supplemented with 20 ng/ml tetradecanoyl phorbol acetate (TPA; Sigma) and 3 mM sodium butyrate (Sigma) for 24 h. Cells were washed 3 times with phosphate-buffered saline (PBS) to remove the TPA and sodium butyrate and cocultured with an equal number of HSB-2 cells that were prestimulated for 24 h with 2 pg/ml IL-2 (Sigma) and 5 ng/ml phytohemagglutinin (PHA; Sigma). Fresh prestimulated HSB2 cells were added every 4 to 6 days to allow accumulation of the virus by cell-to-cell spread.
To isolate virus, we pelleted the cultures by low-speed centrifugation and reserved the supernatants. Infected cells were resuspended in 10 ml of medium and sonicated to release virus from infected cells. The medium was then cleared of cellular debris, and the supernatant was added to the reserved medium. Virus was then purified by ultracentrifugation through a 20% sorbitol cushion in a SW 28 rotor for 90 min at 53,000 × g. The resulting pellet was resuspended in medium supplemented with 1.5% bovine serum albumin (BSA), and aliquots were stored at −80°C following snap-freezing in liquid nitrogen.
Determination of virus titers.
Titers of HHV-6A were calculated using standard 50% tissue culture infective dose (TCID50) assays (36). Briefly, Jjhan cells were plated onto a 96-well plate at 1 × 105 cells per well and incubated overnight at 37°C. Aliquots of HHV-6A were thawed at 37°C and briefly sonicated. The stock was serially diluted in 10-fold increments and used to inoculate Jjhan cells. The cultures were incubated in a 37°C incubator with 5% CO2 for 10 to 14 days. GFP-positive (GFP+) wells were scored to determine the titers of the stock.
Animals.
BALB/c-Rag2−/−γc−/− mice were humanized by engraftment with CD34+ human HSCs purified from human umbilical cord blood as described previously (37). Mice were maintained in a specific-pathogen-free room at the Brigham Young University Central Animal Care Facility. These studies have been reviewed and approved by the Institutional Animal Use and Care Committee (protocol 120101). Briefly, 1- to 5-day-old mice were conditioned by irradiation with 350 rads and then injected intrahepatically with 2 × 105 to 5 × 105 human CD34+ cells. Mice were screened for human cell engraftment at 8 weeks postengraftment. Peripheral blood was collected by tail bleed and stained with antibodies specific to either human or mouse CD45. Fluorescence-activated cell sorter (FACS) analysis was performed to determine percent peripheral blood engraftment of human cells (38, 39).
Preparation of carrier cells for viral transmission to humanized mice.
Fresh CD34-depleted cord blood mononuclear cells (1 × 106) were cultured in basal medium (RPMI 1640 plus 10% FCS plus 1× penicillin-streptomycin) and stimulated with PHA (20 μg/ml) for 48 h followed by IL-2 (100 units/ml) for 10 days. We chose to use these cells because (i) they are routinely used for HHV-6A infections and (ii) they are readily available in our lab. DNA extraction and quantitative PCR (Q-PCR) were performed on a portion of the cells to verify lack of endogenous HHV-6A. Cells (1.7 × 106) were infected with 3.3 × 105 infectious units (i.u.) of a recombinant strain of HHV-6A expressing GFP under the CMV immediate early (IE) promoter and in the U1102 strain background (34) (hereafter referred to as HHV-6A) plus 5 μg/ml Polybrene (Sigma), and mock-infected cells were resuspended in basal medium with Polybrene. Samples were infected for 2 h and shaken every 30 min. The final multiplicity of infection (MOI) was 0.02. After incubation, cells were resuspended in 3 ml basal medium and plated on a 6-well plate. IL-2 was added, and cells were incubated for 48 h. FACS analysis was performed on infected and mock-infected cells to detect and quantify HHV-6A-infected cells prior to mouse infection. GFP was used to identify infected cells, and samples were stained with anti-human CD3, CD4, and CD8 antibodies (see below) to characterize infected cell types.
HHV-6A transmission to humanized mice.
In the cell-associated viral-transmission study, 1 × 105 cells (of which ∼20% were GFP+) in 100 μl serum-free RPMI 1640 were injected intraperitoneally (i.p.) into mice. Uninfected mice were injected similarly with uninfected cells in the same medium as that for infected mice. In the cell-free viral-transmission study, cell-free HHV-6A was thawed and immediately diluted in RPMI 1640 (no serum or antibiotics). One hundred microliters of cell-free virus (4.3 × 105 i.u./mouse) was injected i.p. into mice.
Measurement of blood viral load.
Blood was collected by tail bleed for 6 weeks. Seventy microliters of whole blood was collected per time point, and the blood was usually centrifuged to separate cellular and plasma fractions. DNA was then extracted with a QIAamp DNA blood minikit (Qiagen). Q-PCR was performed using an Applied Biosystems StepOne machine to detect and quantify the presence of viral genomes using a published assay (40). Serial dilutions (10-fold) of a plasmid containing the target HHV-6A sequence were used as copy number standards; the sensitivity of the assay was previously reported to be 10 DNA copies (40). The limit of detection of the assay was 1,000 normalized copies in plasma/ml and 400 copies in bone marrow, lymph node, thymus, and spleen (see below).
Organ collection and measurement of viral load in organs.
In the cell-associated study, mice were sacrificed at time points ranging from 6.5 to 9.5 weeks postinfection (p.i.) (Table 1), and lymphoid organs (thymus, bone marrow, lymph nodes, and spleen) were collected. Bone marrow was extracted from both femurs. The thymus was divided in half for Q-PCR or FACS analysis in one infected and one mock-infected animal. Subsequently, single-cell suspensions were made and divided in half in order to perform both FACS and Q-PCR analyses. For Q-PCR, DNA was extracted using a QIAamp DNA blood minikit and analyzed by Q-PCR as described above. Similar methods were used for organ collection in the cell-free viral-transmission study except that mice were sacrificed at 1 week p.i.
Table 1.
Characteristics of humanized mice used for HHV-6A infectionsa
| Mouse no. | Injected with HHV-6A | Age (mo) | Engraftment (%) | Irradiated | Sacrificed (wk) |
|---|---|---|---|---|---|
| 33 | +, CA | 2 | 0 | − | 9.5 |
| 34 | +, CA | 2 | 0 | − | 6.5 |
| 35 | +, CA | 2 | 0 | − | 9.5 |
| 36 | +, CA | 2 | 0 | − | 9.5 |
| 685 | +, CA | 3 | 0 | + | 9.5 |
| 686 | +, CA | 3 | 0 | + | 9.5 |
| 687 | +, CA | 3 | 0 | + | 9.5 |
| 688 | +, CA | 3 | 0 | + | 6.5 |
| 708 | +, CA | 5 | 29 | + | 8 |
| 3089 | +, CA | 4 | 58 | + | 8 |
| 3090 | +, CA | 4 | 56 | + | 8 |
| 3092 | +, CA | 4 | 71 | + | 6.5 |
| 3099 | +, CA | 5 | 36 | + | 8 |
| 3100 | +, CA | 5 | 49 | + | 8 |
| 698 | −, M | 3 | 41 | + | 8 |
| 3095 | −, M | 4 | 42 | + | 6.5 |
| 3096 | −, M | 4 | 32 | + | 8 |
| 3098 | −, M | 4 | 39 | + | 8 |
| 759 | −, U | 6 | 18 | + | NA |
| 767 | −, U | 6 | 43 | + | NA |
| 769 | −, U | 6 | 68 | + | NA |
| 37 | +, CF | 2 | 0 | − | 1 |
| 38 | +, CF | 2 | 0 | − | 1 |
| 39 | +, CF | 2 | 0 | − | 1 |
| 40 | +, CF | 2 | 0 | − | 1 |
| 711 | +, CF | 7 | 38 | + | 1 |
| 715 | +, CF | 7 | 75 | + | 1 |
| 755 | +, CF | 4 | 46 | + | 1 |
| 756 | +, CF | 4 | 36 | + | 1 |
| 778 | +, CF | 5 | 48 | + | 1 |
| 781 | +, CF | 5 | 35 | + | 1 |
CA, cell-associated virus; M, mock-infected cells; U, uninfected; CF, cell-free virus; NA, not applicable.
FACS analysis.
Anti-human CD45 (eBioscience) and anti-mouse CD45 (eBioscience) were used for screening mice preinfection to determine percent engraftment. Anti-human CD3 (BioLegend), anti-human CD4 (eBioscience), anti-human CD8 (eBioscience), and anti-human CD46 (eBioscience) were used in FACS analyses of regular tail bleeds and on the harvested organs. Samples were run on a BD FACSCanto flow cytometer and analyzed with Summit version 4.3 software.
RESULTS
RAG-hu mice produce cells that express the HHV-6A receptor.
RAG-hu mice were engrafted with human CD34+ hematopoietic stem cells isolated from cord blood as described previously (37) and as outlined in Materials and Methods. Mice were screened for human cell engraftment at 8 weeks postreconstitution by FACS analysis of peripheral blood for the panleukocyte markers human CD45 (hCD45) and mouse CD45 (mCD45).
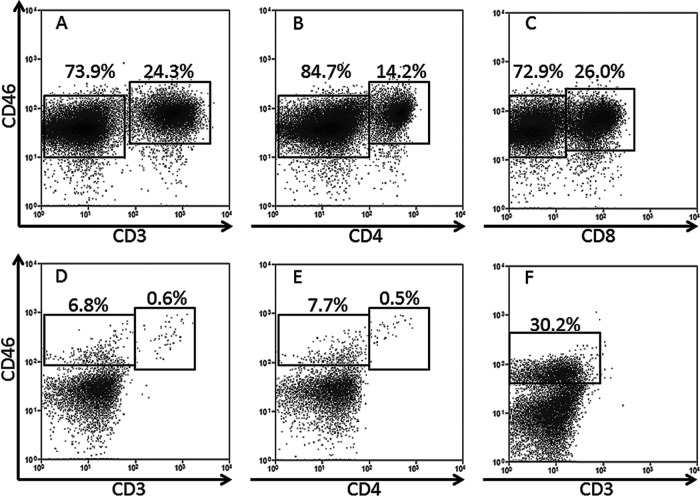
CD46 is a known receptor involved in HHV-6A entry (5) and serves as an inhibitor of complement-mediated cell lysis. CD46 is thought to be expressed in all nucleated human cells but in mice is expressed only in testis (41), which may explain murine resistance to HHV-6A infection. Thus, we stained cells from RAG-hu mice for the presence of CD46 in order to determine if this animal model might be useful for HHV-6A research. We found that RAG-hu mice produce human CD46+ cells in the blood, thymus, and bone marrow (Fig. 1). CD3+ T cells were CD46+ as well as some CD3− cells that were not characterized further.
Fig 1.
Humanized mice harbor human cells that express the HHV-6A receptor. (A to C) RAG-hu thymocytes are CD46+, including CD3+ T cells and the two major subsets of T cells (A), CD4+ helper T cells (B), and CD8+ cytotoxic T cells (C). (D and E) RAG-hu blood also contains CD46+ cells, some of which are also CD3+ (D) and CD4+ (E). (F) The bone marrow also harbors CD46+ cells, but only a minimal number of T cells were detected in this organ.
Infection of RAG-hu mice with HHV-6A.
Initial attempts at HHV-6A infection used cell-associated virus because HHV-6 is known to be a highly cell-associated virus (42) and because a recent study using the related hCMV in humanized mice was unable to achieve infection with cell-free virus but was successful using infected fibroblasts as carrier cells (33). We also attempted cell-free viral transmission with a high-titer stock of HHV-6A to determine if this mechanism would also be viable for inoculation of humanized mice. Detection of cell-free transmission provides additional evidence for the permissiveness of in vivo infection because in cell-associated transmission the input virus may subsequently be detected whether transmission to the graft takes place or not.
In the cell-associated transmission study, mice were divided into 5 groups: (i) nonhumanized Rag2−/−γc−/− mice, never irradiated, inoculated with infected cells, (ii) 0%-engrafted Rag2−/−γc−/− mice, irradiated, inoculated with infected cells, (iii) >30%-engrafted Rag2−/−γc−/− mice inoculated with infected cells, (iv) >30%-engrafted Rag2−/−γc−/− mice inoculated with uninfected cells, and (v) engrafted Rag2−/−γc−/− mice, not inoculated, uninfected (Table 1). Animals with >30% peripheral blood engraftment [defined as (hCD45+ cells)/(hCD45+ cells + mCD45+ cells)] were used in order to ensure that the human immune system was sufficient to support viral infection. Group 1 served as a control for determining if HHV-6A could infect and/or persist in nonhumanized immunocompromised mice. Group 2 served as a control for determining if HHV-6A could infect mice that had been sublethally irradiated (thus becoming further immunocompromised) and reconstituted but had undetectable engraftment. Group 3 was the experimental group to determine if HHV-6A could infect engrafted RAG-hu mice. Groups 4 and 5 served as uninfected controls. Cells used for viral transmission were not donor matched with cell samples used to engraft, similar to a previous study where successful hCMV transmission was accomplished with allogeneic human fibroblasts in a related humanized-mouse model (33). In the cell-free transmission study, mice were divided into 2 groups: (i) nonhumanized, never irradiated, inoculated with cell-free virus and (ii) >30% engrafted, inoculated with cell-free virus. Engraftment of mice in the >30% groups in both studies ranged from 30 to 75% (Table 1).
All RAG-hu mice were tested for the presence of HHV-6A DNA by Q-PCR prior to experimental infection. This verification was necessary because a low percentage of human cord blood samples (which are used to initially engraft the humanized mice) are contaminated with HHV-6A (43). All mice tested negative for HHV-6A DNA in whole-blood samples analyzed prior to cell-associated HHV-6A inoculation as did PHA-plus-IL-2-stimulated cord blood cells prior to HHV-6A infection (used as carrier cells for transmission).
Generation of infected cells for use in cell-associated transmission study.
PHA- and IL-2-stimulated CD34-depleted cord blood mononuclear cells were infected with HHV-6A or uninfected for 2 h as described previously in Materials and Methods. Cells were cultured for 48 h p.i. (with green cells present upon visual inspection by fluorescence microscopy at 20 h p.i. in infected samples; data not shown). We observed an increase in cell size in the infected group compared to cell size in the uninfected group, which is a common cytopathic effect of HHV-6A (data not shown). We also observed a CD3low CD4+ subgroup in the infected sample (Fig. 2D) that was not present in the uninfected sample (Fig. 2C); downregulation of CD3 is also common upon HHV-6A infection of T cells (44). Approximately 20% of lymphocytes were GFP+ in the infected sample (Fig. 2B) immediately prior to injection into RAG-hu mice, and there was a low background of GFP expression in the uninfected sample (Fig. 2A).
Fig 2.
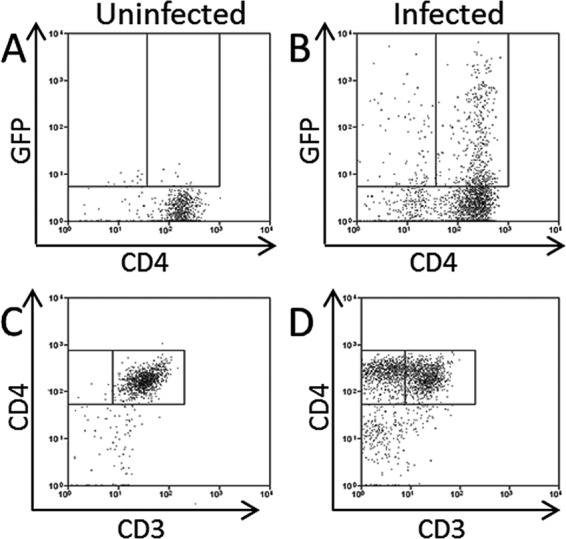
HHV-6A-infected carrier cells used in cell-associated transmission to humanized mice. (A to D) Flow cytometry of uninfected (A and C) and HHV-6A-infected (B and D) cells prior to injection into RAG-hu mice. (A and B) Detection of GFP expression. (C and D) Analysis of CD3 and CD4 expression.
HHV-6A DNA detection in blood and lymphoid organs.
All animals were bled regularly, and DNA was extracted for Q-PCR analysis of the viral genome from different blood fractions (plasma, blood cell, or whole blood, as indicated in Tables 2 and 3). In addition, animals were sacrificed at various time points (Table 1) in order to examine various organs for viral genome detection and/or to analyze cellular populations for GFP expression and for depletion or enrichment of specific T cell populations. Viral DNA was detected by Q-PCR in nearly all animals at at least one time point in the >30%-engrafted, HHV-6A-inoculated groups (both cell-free transmission and cell-associated transmission) (Tables 2, 3, and 4).
Table 2.
Viral DNA detected in blood and lymphoid organs 1 week after cell-free infectiona
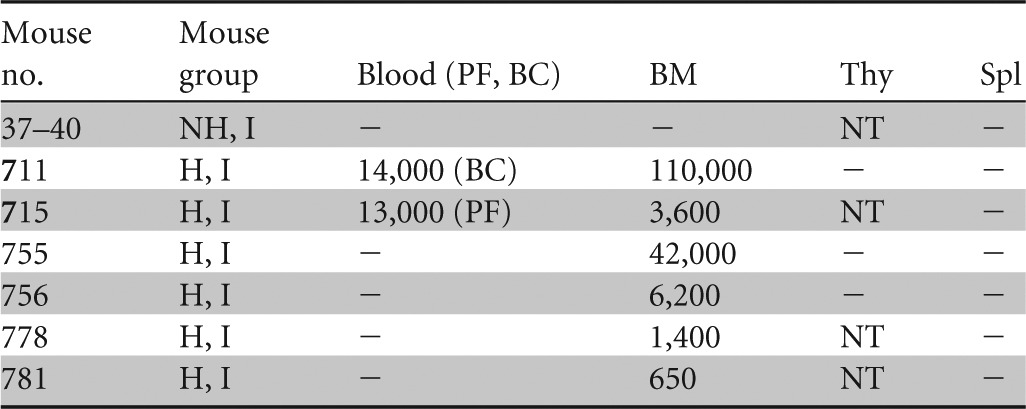
NH, nonhumanized; H, humanized; I, infected; PF, plasma fraction; BC, blood cell fraction; BM, bone marrow; Thy, thymus; Spl, spleen; −, below limit of detection; NT, not tested. Results for plasma and blood cell fractions are reported in DNA copies/ml, for BM in DNA copies per femur, for Thy in DNA copies per half thymus, and for Spl in DNA copies per one-third spleen. Blood fractions analyzed are indicated in the column headings.
Table 3.
Viral DNA detected in blood after cell-associated infectiona
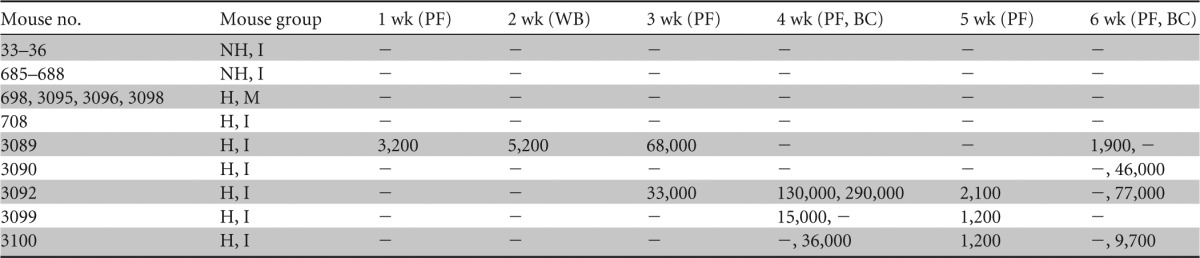
NH, nonhumanized; H, humanized; I, infected; M, mock-infected; PF, plasma fraction; BC, blood cell fraction; WB, whole blood; −, below limit of detection. Results for plasma and blood cell fractions are reported in DNA copies/ml. Not all samples were analyzed by Q-PCR each week. Blood fractions analyzed are indicated in the column headings.
Table 4.
Viral DNA detected in lymphoid organs from mice sacrificed 6 1/2 to 9 1/2 weeks after cell-associated infection
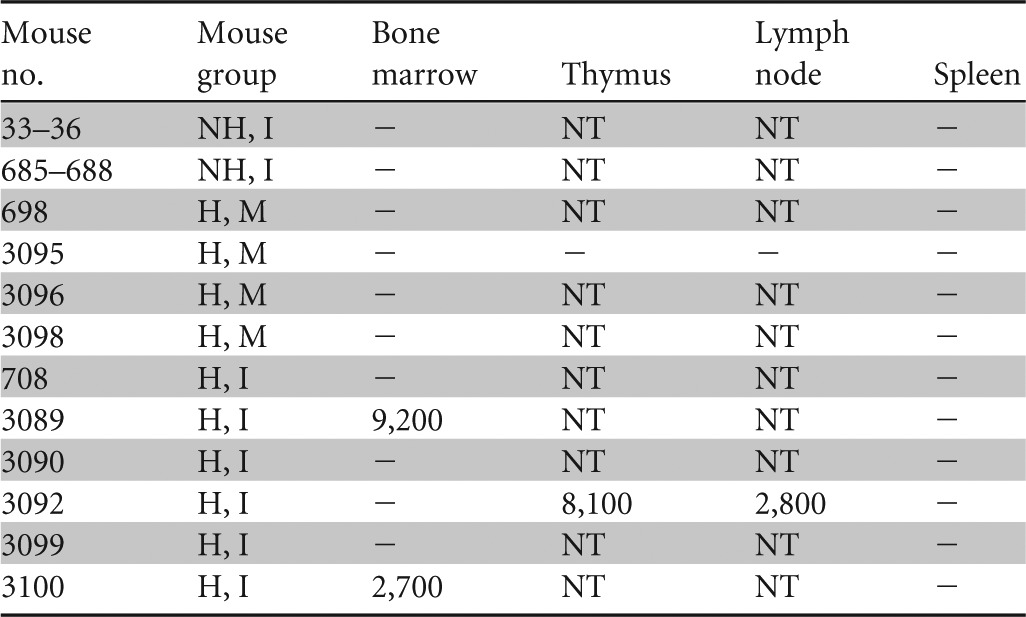
NH, nonhumanized; H, humanized; I, infected; M, mock-infected; −, below limit of detection; NT, not tested. Results for bone marrow are reported in DNA copies per femur, for thymus in DNA copies per half thymus, for lymph node in DNA copies per node, and for spleen in DNA copies per one-third spleen.
In the cell-free transmission study, viral DNA was detected in the bone marrow of all 6 >30%-engrafted, HHV-6A-inoculated mice, while no viral DNA was detectable in the nonhumanized mice (n = 4) (Table 2). Two of the >30%-engrafted, HHV-6A-inoculated mice had detectable viral DNA present in the blood, whereas no viral DNA was detected in the blood of infected, unengrafted mice. No viral DNA was detected in analyzed thymic (n = 3) or splenic (n = 6) tissues from RAG-hu mice infected by cell-free transmission. All samples in the cell-free transmission study were analyzed at the time of sacrifice (1 week postinfection).
In the cell-associated transmission study, HHV-6A DNA was detected by Q-PCR in plasma and cellular fractions of blood (Table 3) and in the bone marrow, lymph node, and thymus but not in spleen (Table 4). In the >30%-engrafted, HHV-6A-inoculated group, viral DNA was detected in the plasma fraction in 4 of 6 mice, in the blood cell fraction in 3 of 6 mice tested, and in 1 of 6 whole-blood samples tested. In addition, 2 of 6 mice in this group had detectable viral DNA in the bone marrow, and the thymic and lymph node samples from the lone mouse tested from this group had detectable viral DNA (Table 4). We noted that viral DNA was detected mostly in the plasma from weeks 1 to 5 and that plasma viral load decreased in copy number after week 4 and in frequency of detection after week 5. Four of 18 samples collected from engrafted, humanized infected mice during the first 3 weeks of the cell-associated transmission experiment were positive for viral DNA, while 15 of 38 samples tested from week 4 onward were positive and had generally higher levels of viral DNA. In total, 5 of 6 mice in the >30%-engrafted, HHV-6A-inoculated group had viral DNA present in blood or organs. Mouse 708 had undetectable viral DNA in blood and lymphoid organs, but this animal was inadvertently inoculated with about half of the volume of infected carrier cells into the subcutaneous space and the other half into the intended intraperitoneal cavity. No viral DNA was detected in any of the three control groups in the cell-associated transmission study.
Detection of HHV-6A-infected cells in vivo via GFP expression.
We attempted to detect GFP+ cells as an additional way to verify successful infection in both the cell-associated transmission and cell-free transmission studies. However, no GFP+ cells were detected in the cell-associated study in blood samples collected weeks 1 and 3 postinfection and analyzed by flow cytometry. Additionally, no GFP+ cells were detected when lymphoid organs were collected at the time of sacrifice. A single mesenteric lymph node sample (mouse 715) from the cell-free transmission study was found to harbor GFP+ cells when animals were sacrificed and analyzed at 1 week postinfection (data not shown). CD4+ cells (0.04%) in the lymph node were GFP+, while 0.28% of CD8+ cells were GFP+. Of the CD4+ GFP+ cells, most were CD3− (95%). Of the GFP+ cells detected in this sample, 4% were CD3+, 55% were CD4+, and 7% were CD8+.
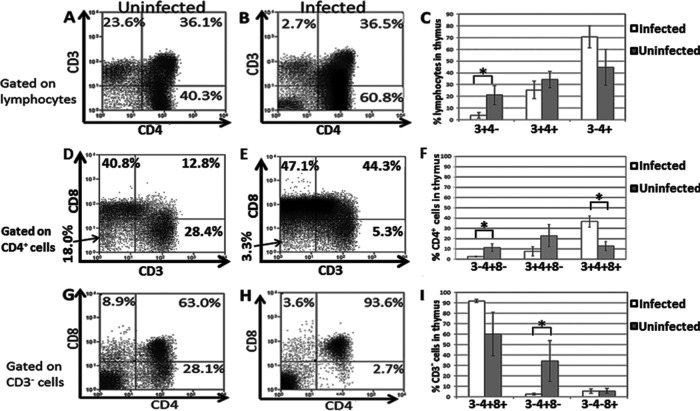
Thymocyte populations are significantly changed in HHV-6A-infected RAG-hu mice.
Previous work with humanized mice (SCID-hu thy/liv model) infected with HHV-6A or HHV-6B indicated that these viruses are capable of modifying thymic populations after direct viral inoculation into the thymic graft (19). We thus analyzed thymic populations taken from RAG-hu mice infected by either cell-associated or cell-free transmission. Animals infected by cell-free transmission had undetectable levels of viral DNA in the thymus at 1 week postinfection (3 of 3 tested) (Table 2), and their thymocyte populations were similar to those of uninfected animals (data not shown). Thus, we focused our thymocyte analysis on animals infected by the cell-associated transmission route, noting that these animals were also infected for a longer duration. RAG-hu mice infected by cell-associated transmission did exhibit significant shifts in thymic populations (Fig. 3). We noted a significant decrease (P = 0.05) in CD3 expression on CD4+ thymocytes, with means of 3.9% and 17.9% of thymocytes that were CD3+ CD4− in infected (n = 4) and uninfected (n = 6) groups, respectively. This is similar to a previous report indicating that HHV-6 can downregulate CD3 expression (45). We also detected a significant loss (P = 0.04) of intrathymic T progenitor cells (CD3− CD4+ CD8−), with means of 2.5% for infected mice (n = 3) and 11.3% for uninfected mice (n = 5) when gating on the CD4+ population. When analyzing this same population on a CD3− gate, we again found a significant depletion (P = 0.03), with means of 2.7% for infected mice (n = 3) and 32.2% for uninfected mice (n = 5). We also detected a significant (P = 0.02) increase in the number of CD3+ CD4+ CD8+ thymocytes, with means of 36.6% for infected mice (n = 3) and 12.8% for uninfected mice (n = 5). The CD3− CD4+ CD8+ population appeared to increase in infected animals, but the difference was not significant (P = 0.08), and the CD3+ CD4+ CD8− population appeared to decrease in infected animals, but the difference was not significant (P = 0.25).
Fig 3.
Depletion of specific thymocyte populations in HHV-6A-infected mice. RAG-hu mice infected with HHV-6A by cell-associated viral transmission showed modulation of specific thymocyte populations. (A to C) The CD3+ CD4− population was depleted, while the CD3− CD4+ population was increased in infected animals. Mouse 3095 (uninfected) is shown in panel A and mouse 3099 (infected) is shown in panel B. Samples were gated on a lymphocyte gate. n = 4 infected; n = 6 uninfected. (D to F) The CD3− CD4+ CD8− and CD3+ CD4+ CD8− populations were depleted and the CD3+ CD4+ CD8+ population was increased when analysis was gated upon CD4+ cells. Mouse 3098 (uninfected) is shown in panel D and mouse 3099 (infected) is shown in panel E. n = 3 infected; n = 5 uninfected. (G to I) The CD3− CD4+ CD8+ population expanded and the CD3− CD4+ CD8− population was reduced when analysis was gated on the CD3− population. Mouse 3098 (uninfected) is shown in panel G and mouse 3089 (infected) is shown in panel H. n = 3 infected; n = 5 uninfected. When data analysis was performed, the two single-positive and the double-positive cell populations were normalized to 100%. Mouse 708 was excluded from these analyses because no viral DNA was detected in that mouse at any time point and we concluded that the mouse likely was not successfully infected. Standard errors are indicated; Student's t test was used for statistical analysis. *, P ≤ 0.05.
Detection of CD4+ CD8+ T cells in HHV-6A-infected RAG-hu blood.
Previous studies have indicated that HHV-6A infection can induce CD4 expression on primary human CD8 T cells in vitro (46). Thus, we analyzed samples by FACS for the presence of CD4+ CD8+ T cells. We detected significantly increased ratios (P = 0.04) of CD3+ CD4+ CD8+ cells in the blood of RAG-hu mice infected by the cell-free transmission route (Fig. 4). This population represented a mean of 8.8% of CD3+ T cells, while in uninfected humanized mice these cells were 3.1% of all T cells. CD4+ CD8+ cells are normally rare in human blood, with one report showing an average of 2.91% of CD4+ CD8+ cells in normal human blood (n = 10) (47), which is similar to our results for uninfected humanized mouse blood. Other blood T cell populations, including CD3+ CD4− CD8+, CD3+ CD4+ CD8−, CD3− CD4+ CD8+, CD3− CD4+ CD8−, and CD3− CD4− CD8+ populations, were not significantly altered in infected samples.
Fig 4.
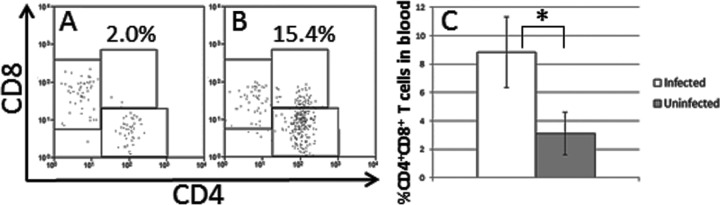
Detection of CD4+ CD8+ T cells in blood of HHV-6A-infected mice. (A to C) Levels of CD4+ CD8+ T cells in blood were quantified by flow cytometry. (A) Uninfected mouse. (B) HHV-6A-infected mouse 715 (cell-free transmission and highest amount of CD4+ CD8+ cells). All samples were first gated on CD3. (C) Mean levels of CD4+ CD8+ cells were quantified in cell-free-infected mice (n = 6) and uninfected mice (n = 4). Standard errors are indicated; Student's t test was used for statistical analysis. *, P ≤ 0.05.
DISCUSSION
Here we have shown that RAG-hu mice are susceptible to infection with HHV-6A by either cell-associated or cell-free transmission. Viral DNA was detected in blood (cellular and plasma fractions), bone marrow, lymph node, and thymic tissues (Tables 2, 3, and 4), although no single mouse tested positive for viral DNA in all of these compartments, as mentioned previously. Following cell-associated transmission, viral DNA was detectable for up to 8 weeks postinfection, indicating a persistent infection. No viral DNA was detected in any of the three control groups in the cell-associated study, indicating that viral transmission from the infected carrier cells to the originally engrafted cells and subsequent replication were successful because no viral DNA was detectable in either blood or lymphoid organs in control mice without an HSC graft. Irradiated but nonengrafted animals were included as a control because of the higher level of murine immunodeficiency of irradiated mice, and they also had undetectable viral DNA after transmission. Some animals in the cell-associated transmission study had detectable viral DNA at early time points and not later, and some animals had no detectable DNA at early time points but detectable DNA at later time points. We attribute this to a relatively high limit of detection in the assay, because only small blood samples can be obtained from mice. We noted that plasma viral DNA levels peaked at 3 to 4 weeks postinfection, but levels decreased to near the level of detection in plasma by 5 weeks and only a single animal had detectable viral DNA in the plasma at 6 weeks. Viral DNA was still detected in the cellular fraction of blood at 6 weeks in three animals, while plasma viral DNA was found in a single mouse at that time point, potentially indicating a shift from lytic infection (extracellular DNA) to latency (intracellular DNA).
Cell-associated transmission was attempted because a similar previous experiment with hCMV was successful only with this method (33). However, cell-free transmission was successful in all 6 animals in our study. We also noted a greater tendency to detect viral DNA in the bone marrow of animals infected by cell-free transmission, but it is not clear if that finding is due to a different mode of transmission or to different sacrificial time points (1 week for cell-free transmission and 6.5 to 9.5 weeks for cell-associated transmission). We also detected a significant increase in CD4+ CD8+ cells in the blood of mice infected by cell-free transmission (Fig. 4). This was possibly due to CD4 upregulation in CD8 T cells, which was previously shown in vitro in HHV-6A-infected cells (46). When we correlated Q-PCR results for blood cells and plasma with detection of these CD4+ CD8+ cells, there was not a clear trend because one animal (715) had high proportions of these cells and detectable viral DNA in plasma, while other animals also had high proportions of the cells but undetectable viral DNA in either blood fraction. Another animal (711) had low proportions of the cells with only intracellular blood viral DNA. The frequent detection of this effect combined with the relatively rare detection of viral DNA in either blood fraction indicates that these cells may be uninfected by HHV-6A. These cells largely maintained CD3 expression, which also indicates a lack of infection. It is possible that HHV-6A infection promotes the release of these cells from the thymus because CD4+ CD8+ cells are rare outside of the thymus. However, we failed to detect viral DNA in the thymus of 3 animals tested, so if infection promotes a release of these cells from the thymus then it must occur from a distal site. There is evidence that the presence of CD4+ CD8+ cells in human blood is upregulated following viral infection, including after infection with persistent viruses such as the herpesvirus EBV (47).
We attempted to use GFP expression from a recombinant virus to further demonstrate successful infection. However, the only animal with detectable GFP+ cells was mouse 715 from the cell-free transmission group, and those cells were from the mesenteric lymph node. FACS analysis of GFP+ cells indicated that they were mostly CD3− CD4+, which is in accordance with our in vitro results shown in Fig. 2D and previously published data showing a tropism for CD4+ T cells and a downregulation of CD3 after infection (45, 48). We later determined that the GFP cassette in this virus is driven by the CMV IE promoter (Y. Mori, personal communication). Since the cell-associated mice were sacrificed at 6.5 to 9.5 weeks p.i., it is possible that the virus was in a latent state at the time of organ collection; this hypothesis is supported by the shift from extracellular to intracellular DNA seen in blood. The activity of the CMV IE promoter in the context of a latent HHV-6A infection is currently unknown, and if that promoter is inactive during latency it may explain a lack of GFP+ cells in any of the mice infected by cell-associated transmission.
We made the interesting observation that several thymocyte populations were altered in HHV-6A-infected animals versus those in uninfected animals (Fig. 3). These observations further support that successful viral transmission occurred, because similar findings have been reported for in vitro studies and for another humanized-mouse study. In those studies, CD3 depletion occurred only in infected (not in bystander) cells (19, 46, 48). Thymocyte depletion was detected only in animals infected by the cell-associated pathway, but these animals were also infected for a longer period. It is possible that virus had not trafficked to the thymus in mice infected by cell-free transmission, a finding supported by our Q-PCR data, where 0 of 3 of these thymic samples harbored viral DNA (Table 2). We noted significant depletion of the CD3+ CD4− and CD3− CD4+ CD8− populations in HHV-6A-infected animals. We also noted a significant increase in the CD3+ CD4+ CD8+ subset, with a marginally significant increase in the CD3− CD4+ CD8+ population. The significant loss of CD3− CD4+ CD8− thymocytes was similarly reported by Gobbi et al. when HHV-6A was directly inoculated into the thymic organoid of SCID-hu thy/liv mice (19). In contrast to that report, our results show a significant increase in the CD3+ CD4+ CD8+ subset. These discrepancies may be explained by the use of different virus isolates, with strain GS used in that report and a recombinant isolate based upon strain U1102 used here. In addition, we have used a newer generation of humanized mice with a wider scope of human cell types and a much broader distribution in the mouse, and we inoculated at a site distant from the thymus. Several of these thymocyte populations that were modified by infection in vivo can be explained by a tropism and cytopathogenicity of the virus for CD4+ T cells. Additionally, the tendency of the virus to downregulate CD3 and/or to upregulate CD4 expression can also explain shifting populations (e.g., CD3+ CD4− and CD3+ CD4+ CD8− populations expected to decrease and CD3− CD4+ CD8+ population expected to increase). The CD3+ CD4+ CD8− population was previously shown to be more infectible with HHV-6A than other thymocyte populations (19). We have proposed that in the cell-associated study the virus was predominantly latent at the time points at which the thymic samples were collected. We are not aware of any studies documenting CD3 downregulation or CD4 upregulation in latently infected primary cells, so it is currently not clear if lytic replication is required for these effects upon host cell gene expression.
We and others have previously shown that RAG-hu mice are also highly susceptible to HIV-1 infection (26, 38, 49, 50). Our current findings indicate higher proportions of CD4+ cells in HHV-6A-infected animals, similar to those shown by Lusso et al. in vitro, where they showed that HHV-6A-infected CD8+ T cells began to express CD4 and were able to replicate HIV-1 (46). If HHV-6A is able to convert CD8+ T cells to become infectible by HIV-1 in vivo, then those cells may be depleted by HIV-1 and/or by HHV-6A. Downregulation of CD3, as our results herein have indicated, is expected to cause immunosuppression because CD3 serves as the signaling subunit of the T cell receptor. Hence, T cells could engage the T cell receptor but not be able to respond effectively. Either of these two effects would support the hypothesis that HHV-6A is a cofactor in AIDS progression (21, 22). Our future directions include plans to perform coinfection studies of HHV-6A and HIV-1 in humanized mice in order to determine if there is a synergistic effect between the two viruses in the progression to AIDS as well as to determine if RAG-hu mice can be infected with HHV-6B.
ACKNOWLEDGMENTS
We are grateful to Yasuko Mori (National Institute of Biomedical Innovation, Osaka, Japan) for giving permission to use a recombinant strain of HHV-6A produced in her laboratory. BALB/c-Rag2−/−γc−/− mice were generously provided by Ramesh Akkina at Colorado State University.
This work was funded by a Pilot Grant award from the HHV-6 Foundation to B.K.B. This work was partially supported by National Institutes of Health grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) to E.A.M. A.T. was supported by the Brigham Young University Cancer Research Center and the HHV-6 Foundation.
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo RC. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601 [DOI] [PubMed] [Google Scholar]
- 2.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emery VC, Clark DA. 2007. Chapter 49. HHV-6A, 6B, and 7: persistence in the population, epidemiology and transmission. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 4.Burbelo PD, Bayat A, Wagner J, Nutman TB, Baraniuk JN, Iadarola MJ. 2012. No serological evidence for a role of HHV-6 infection in chronic fatigue syndrome. Am. J. Transl. Res. 4:443–451 [PMC free article] [PubMed] [Google Scholar]
- 5.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827 [DOI] [PubMed] [Google Scholar]
- 6.Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, Yamanishi K, Mori Y. 2013. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc. Natl. Acad. Sci. U. S. A. 110:9096–9099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067 [DOI] [PubMed] [Google Scholar]
- 8.Virtanen JO, Farkkila M, Multanen J, Uotila L, Jaaskelainen AJ, Vaheri A, Koskiniemi M. 2007. Evidence for human herpesvirus 6 variant A antibodies in multiple sclerosis: diagnostic and therapeutic implications. J. Neurovirol. 13:347–352 [DOI] [PubMed] [Google Scholar]
- 9.Sola P, Merelli E, Marasca R, Poggi M, Luppi M, Montorsi M, Torelli G. 1993. Human herpesvirus 6 and multiple sclerosis: survey of anti-HHV-6 antibodies by immunofluorescence analysis and of viral sequences by polymerase chain reaction. J. Neurol. Neurosurg. Psychiatry 56:917–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behzad-Behbahani A, Mikaeili MH, Entezam M, Mojiri A, Pour GY, Arasteh MM, Rahsaz M, Banihashemi M, Khadang B, Moaddeb A, Nematollahi Z, Azarpira N. 2011. Human herpesvirus-6 viral load and antibody titer in serum samples of patients with multiple sclerosis. J. Microbiol. Immunol. Infect. 44:247–251 [DOI] [PubMed] [Google Scholar]
- 11.Appleton AL, Sviland L, Peiris JS, Taylor CE, Wilkes J, Green MA, Pearson AD, Kelly PJ, Malcolm AJ, Proctor SJ. 1995. Human herpes virus-6 infection in marrow graft recipients: role in pathogenesis of graft-versus-host disease. Newcastle upon Tyne Bone Marrow Transport Group. Bone Marrow Transpl. 16:777–782 [PubMed] [Google Scholar]
- 12.Agut H. 2011. Deciphering the clinical impact of acute human herpesvirus 6 (HHV-6) infections. J. Clin. Virol. 52:164–171 [DOI] [PubMed] [Google Scholar]
- 13.Le J, Gantt S, AST Infectious Diseases Community of Practice 2013. Human herpesvirus 6, 7 and 8 in solid organ transplantation. Am. J. Transplant. 13(Suppl 4):128–137 [DOI] [PubMed] [Google Scholar]
- 14.Razonable RR, Zerr DM, AST Infectious Diseases Community of Practice 2009. HHV-6, HHV-7 and HHV-8 in solid organ transplant recipients. Am. J. Transplant. 9(Suppl 4):S97–S100 [DOI] [PubMed] [Google Scholar]
- 15.Saraya T, Mikoshiba M, Kamiyama H, Yoshizumi M, Tsuchida S, Tsukagoshi H, Ishioka T, Terada M, Tanabe E, Tomioka C, Ishii H, Kimura H, Kozawa K, Shiohara T, Takizawa H, Goto H. 2013. Evidence for reactivation of human herpesvirus 6 in generalized lymphadenopathy in a patient with drug-induced hypersensitivity syndrome. J. Clin. Microbiol. 51:1979–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tohyama M, Hashimoto K, Yasukawa M, Kimura H, Horikawa T, Nakajima K, Urano Y, Matsumoto K, Iijima M, Shear NH. 2007. Association of human herpesvirus 6 reactivation with the flaring and severity of drug-induced hypersensitivity syndrome. Br. J. Dermatol. 157:934–940 [DOI] [PubMed] [Google Scholar]
- 17.Yao K, Crawford JR, Komaroff AL, Ablashi DV, Jacobson S. 2010. Review part 2: human herpesvirus-6 in central nervous system diseases. J. Med. Virol. 82:1669–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ablashi DV, Devin CL, Yoshikawa T, Lautenschlager I, Luppi M, Kühl U, Komaroff AL. 2010. Review part 3: human herpesvirus-6 in multiple non-neurological diseases. J. Med. Virol. 82:1903–1910 [DOI] [PubMed] [Google Scholar]
- 19.Gobbi A, Stoddart CA, Malnati MS, Locatelli G, Santoro F, Abbey NW, Bare C, Linquist-Stepps V, Moreno MB, Herndier BG, Lusso P, McCune JM. 1999. Human herpesvirus 6 (HHV-6) causes severe thymocyte depletion in SCID-hu Thy/Liv mice. J. Exp. Med. 189:1953–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emery VC, Atkins MC, Bowen EF, Clark DA, Johnson MA, Kidd IM, McLaughlin JE, Phillips AN, Strappe PM, Griffiths PD. 1999. Interactions between beta-herpesviruses and human immunodeficiency virus in vivo: evidence for increased human immunodeficiency viral load in the presence of human herpesvirus 6. J. Med. Virol. 57:278–282 [DOI] [PubMed] [Google Scholar]
- 21.Lusso P, Gallo RC. 1995. Human herpesvirus 6 in AIDS. Immunol. Today 16:67–71 [DOI] [PubMed] [Google Scholar]
- 22.Lusso P, Crowley RW, Malnati MS, Di Serio C, Ponzoni M, Biancotto A, Markham PD, Gallo RC. 2007. Human herpesvirus 6A accelerates AIDS progression in macaques. Proc. Natl. Acad. Sci. U. S. A. 104:5067–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusso P, Markham PD, DeRocco SE, Gallo RC. 1990. In vitro susceptibility of T lymphocytes from chimpanzees (Pan troglodytes) to human herpesvirus 6 (HHV-6): a potential animal model to study the interaction between HHV-6 and human immunodeficiency virus type 1 in vivo. J. Virol. 64:2751–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lusso P, Secchiero P, Crowley RW. 1994. In vitro susceptibility of Macaca nemestrina to human herpesvirus 6: a potential animal model of coinfection with primate immunodeficiency viruses. AIDS Res. Hum. Retroviruses 10:181–187 [DOI] [PubMed] [Google Scholar]
- 25.Leibovitch E, Wohler JE, Cummings Macri SM, Motanic K, Harberts E, Gaitán MI, Maggi P, Ellis M, Westmoreland S, Silva A, Reich DS, Jacobson S. 2013. Novel marmoset (Callithrix jacchus) model of human herpesvirus 6A and 6B infections: immunologic, virologic and radiologic characterization. PLoS Pathog. 9:e1003138. 10.1371/journal.ppat.1003138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berges BK, Rowan MR. 2011. The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment. Retrovirology 8:65. 10.1186/1742-4690-8-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocco M, Bellan C, Tussiwand R, Corti D, Traggiai E, Lazzi S, Mannucci S, Bronz L, Palummo N, Ginanneschi C, Tosi P, Lanzavecchia A, Manz MG, Leoncini L. 2008. CD34+ cord blood cell-transplanted Rag2−/−γc−/− mice as a model for Epstein-Barr virus infection. Am. J. Pathol. 173:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma SD, Hegde S, Young KH, Sullivan R, Rajesh D, Zhou Y, Jankowska-Gan E, Burlingham WJ, Sun X, Gulley ML, Tang W, Gumperz JE, Kenney SC. 2011. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 85:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strowig T, Gurer C, Ploss A, Liu YF, Arrey F, Sashihara J, Koo G, Rice CM, Young JW, Chadburn A, Cohen JI, Münz C. 2009. Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J. Exp. Med. 206:1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yajima M, Imadome K, Nakagawa A, Watanabe S, Terashima K, Nakamura H, Ito M, Shimizu N, Honda M, Yamamoto N, Fujiwara S. 2008. A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses. J. Infect. Dis. 198:673–682 [DOI] [PubMed] [Google Scholar]
- 31.Wu W, Vieira J, Fiore N, Banerjee P, Sieburg M, Rochford R, Harrington WJ, Feuer G. 2006. KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo. Blood 108:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons CH, Adang LA, Overdevest J, O'Connor CM, Taylor JRJ, Camerini D, Kedes DH. 2006. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J. Clin. Invest. 116:1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith MS, Goldman DC, Bailey AS, Pfaffle DL, Kreklywich CN, Spencer DB, Othieno FA, Streblow DN, Garcia JV, Fleming WH, Nelson JA. 2010. Granulocyte-colony stimulating factor reactivates human cytomegalovirus in a latently infected humanized mouse model. Cell Host Microbe 8:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H, Kawabata A, Yoshida M, Oyaizu H, Maeki T, Yamanishi K, Mori Y. 2010. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology 407:360–367 [DOI] [PubMed] [Google Scholar]
- 35.Kalejta RF, Bechtel JT, Shenk T. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 37.Traggiai E, Chicha L, Mazzucchelli L, Bronz L, Piffaretti JC, Lanzavecchia A, Manz MG. 2004. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science 304:104–107 [DOI] [PubMed] [Google Scholar]
- 38.Berges BK, Wheat WH, Palmer BE, Connick E, Akkina R. 2006. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−γc−/− (RAG-hu) mouse model. Retrovirology 3:76. 10.1186/1742-4690-3-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akkina R, Berges BK, Palmer BE, Remling L, Neff CP, Kuruvilla J, Connick E, Folkvord J, Gagliardi K, Kassu A, Akkina SR. 2011. Humanized Rag1−/−γc−/− mice support multilineage hematopoiesis and are susceptible to HIV-1 infection via systemic and vaginal routes. PLoS One 6:e20169. 10.1371/journal.pone.0020169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautheret-Dejean A, Manichanh C, Thien-Ah-Koon F, Fillet AM, Mangeney N, Vidaud M, Dhedin N, Vernant JP, Agut H. 2002. Development of a real-time polymerase chain reaction assay for the diagnosis of human herpesvirus-6 infection and application to bone marrow transplant patients. J. Virol. Methods 100:27–35 [DOI] [PubMed] [Google Scholar]
- 41.Tsujimura A, Shida K, Kitamura M, Nomura M, Takeda J, Tanaka H, Matsumoto M, Matsumiya K, Okuyama A, Nishimune Y, Okabe M, Seya T. 1998. Molecular cloning of a murine homologue of membrane cofactor protein (CD46): preferential expression in testicular germ cells. Biochem. J. 330:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhen Z, Bradel-Tretheway B, Sumagin S, Bidlack JM, Dewhurst S. 2005. The human herpesvirus 6 G protein-coupled receptor homolog U51 positively regulates virus replication and enhances cell-cell fusion in vitro. J. Virol. 79:11914–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams O, Krempe C, Kögler G, Wernet P, Scheid A. 1998. Congenital infections with human herpesvirus 6. J. Infect. Dis. 178:544–546 [DOI] [PubMed] [Google Scholar]
- 44.Braun DK, Dominguez G, Pellett PE. 1997. Human herpesvirus 6. Clin. Microbiol. Rev. 10:521–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lusso P, Malnati M, De Maria A, Balotta C, DeRocco SE, Markham PD, Gallo RC. 1991. Productive infection of CD4+ and CD8+ mature human T cell populations and clones by human herpesvirus 6. Transcriptional down-regulation of CD3. J. Immun. 147:685–691 [PubMed] [Google Scholar]
- 46.Lusso P, De Maria A, Malnati M, Lori F, DeRocco SE, Baseler M, Gallo RC. 1991. Induction of CD4 and susceptibility to HIV-1 infection in human CD8+ T lymphocytes by human herpesvirus 6. Nature 349:533–535 [DOI] [PubMed] [Google Scholar]
- 47.Nascimbeni M, Shin EC, Chiriboga L, Kleiner DE, Rehermann B. 2004. Peripheral CD4+CD8+ T cells are differentiated effector memory cells with antiviral functions. Blood 104:478–486 [DOI] [PubMed] [Google Scholar]
- 48.Grivel JC, Santoro F, Chen S, Fagá G, Malnati MS, Ito Y, Margolis L, Lusso P. 2003. Pathogenic effects of human herpesvirus 6 in human lymphoid tissue ex vivo. J. Virol. 77:8280–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baenziger S, Tussiwand R, Schlaepfer E, Mazzucchelli L, Heikenwalder M, Kurrer MO, Behnke S, Frey J, Oxenius A, Joller H, Aguzzi A, Manz MG, Speck RF. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−γc−/− mice. Proc. Natl. Acad. Sci. U. S. A. 103:15951–15956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Kovalev GI, Su L. 2007. HIV-1 infection and pathogenesis in a novel humanized mouse model. Blood 109:2978–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]