Abstract
Yep-phi is a T7-related bacteriophage specific to Yersinia pestis, and it is routinely used in the identification of Y. pestis in China. Yep-phi infects Y. pestis grown at both 20°C and 37°C. It is inactive in other Yersinia species irrespective of the growth temperature. Based on phage adsorption, phage plaque formation, affinity chromatography, and Western blot assays, the outer membrane proteins of Y. pestis Ail and OmpF were identified to be involved, in addition to the rough lipopolysaccharide, in the adsorption of Yep-phi. The phage tail fiber protein specifically interacts with Ail and OmpF proteins, and residues 518N, 519N, and 523S of the phage tail fiber protein are essential for the interaction with OmpF, whereas residues 518N, 519N, 522C, and 523S are essential for the interaction with Ail. This is the first report to demonstrate that membrane-bound proteins are involved in the adsorption of a T7-related bacteriophage. The observations highlight the importance of the tail fiber protein in the evolution and function of various complex phage systems and provide insights into phage-bacterium interactions.
INTRODUCTION
Yersinia pestis has caused three major pandemics of plague during human history. It is one of several virulent microorganisms known to have changed human civilization pathways and has attracted great attention due to its bioterrorism potential (1–3). Naturally occurring and genetically engineered antibiotic-resistant Y. pestis strains increase our concerns about the threat of its misuse as a biological weapon or bioterrorism agent. For example, the isolation of a multidrug-resistant (MDR) Y. pestis strain (IP275) in 1995 caused considerable alarm in the public health and biodefense communities (4). Thus, there is an urgent need for improved treatments and detection methods. Phage therapy has been proposed since their discovery in the early 20th century (5). However, the advent of antibiotics led to this therapeutic approach being largely abandoned (5, 6). Nevertheless, renewed interest in phage therapy has grown over the past decade due to the rapid emergence of antibiotic resistance in bacterial pathogens (7, 8), with several phages shown to be effective in treating various bacterial infections (5, 9).
Five Y. pestis phages have been fully sequenced, including L-413C (GenBank accession number NC004745) (10), PhiA1122 (AY247822) (11), Yep-phi (HQ333270) (12), Berlin (AM183667) (12), and Yepe2 (EU734170) (12). L-413C is a temperate phage belonging to the Myoviridae family (10). The other four phages are classified as members of the Podoviridae family, with genomes that are colinear with those of the Escherichia coli phages T7 and T3, as well as the Y. enterocolitica phage PhiYeO3-12 (12, 13). Whole-genome phylogenies have indicated that the phage Yep-phi is most closely related to the Berlin and Yepe2 phages; thus, Berlin, Yepe2, and Yep-phi are classified in the Yep-phi subgroup, while PhiA1122 belongs to the other subgroup (12).
Yep-phi is a T7-related bacteriophage that is routinely used as a diagnostic agent for the identification of Y. pestis in China. It is thought that the third pandemic of plague originated from China (14), and there are several epidemic foci of Y. pestis in China (12). The phage forms clear plaques on all isolates of Y. pestis in China but is inactive on other Yersinia species irrespective of the growth temperature. Based on the spectra of lytic activity, Yep-phi is much more specific for Y. pestis than the other diagnostic phage, PhiA1122, which is used by the Centers for Disease Control and Prevention of the United States for the identification of Y. pestis. Y. pseudotuberculosis is also infected by PhiA1122 when grown at 37°C (15). The PhiA1122 receptor is demonstrated to be the common lipopolysaccharide (LPS) core of Y. pestis and Y. pseudotuberculosis. The receptor used by phage Yep-phi for adsorption has not yet been identified (15).
The interaction of a phage with the bacterial surface (adsorption) is the first step in the infection process (16). This step involves the recognition of, and binding to, one or more cell envelope constituents, which leads to the ejection of the phage DNA from the capsid (17). The implementation of efficient phage therapies to target pathogenic bacteria requires detailed knowledge of the bacterial receptors recognized by the phages (18). Phages tend to use structures that are exposed on the outer membrane of the host bacteria as receptors, since they are easily accessible (8). Porins, affinity transporters, and LPS are the most common outer membrane components acting as receptors for tailed phages that infect Gram-negative bacteria (19, 20). Outer membrane proteins (OMPs), known to act as phage receptors, all share a channel-forming β-barrel region organized by a variable number of antiparallel β-strands. Such proteins can function as monomeric (FhuA) or trimeric (OmpA, OmpC, and LamB) proteins (19, 21). LPS is the major component of the outer membrane of Gram-negative bacteria and is a surface structure encountering the surrounding environment. The LPS of Y. pestis is O antigen deficient and restricted to the core and lipid A moieties (22). Much of the available detailed information about the receptors of Y. pestis phages focuses on the LPS (15, 23). The LPS of Y. pestis has been studied extensively in terms of structure, activity, composition, modification, virulence, and assembly (15, 22–24). The receptor was identified as the LPS of Y. pestis for eight bacteriophages, including L-413C, P2 vir1, PhiJA1, PhiA1122, Pokrovskaya, T7Yp, Y, and PST (23). The phages were demonstrated to employ different components of the LPS core as a specific receptor (23).
On the other hand, little attention has been devoted to the interactions between phages and Y. pestis OMPs. In the current report, we analyzed the host factors attached by Yep-phi. Although most of the Y. pestis phages require only a single bacterial receptor (LPS) (23), Yep-phi infection requires Y. pestis OMPs Ail and OmpF as well as the LPS receptor. Furthermore, amino acid residues critical for the specific binding of the tail fiber protein gene product (gp) 17 to Ail and OmpF were identified. To the best of our knowledge, the current study is the first to report T7-related phages employing OMPs in the adsorption process, thereby providing novel insights into the interactions between the host and these phages.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
Y. pestis and phage cultures were incubated at 26°C, whereas Y. pseudotuberculosis and E. coli cultures were incubated at 37°C (Table 1). The wild-type (WT) Y. pestis biovar microtus strain 201, which is avirulent to humans but highly lethal to mice (25), was used. Brain heart infusion and Luria-Bertani (LB) media were used for bacterial liquid cultures, and soft-agar medium included an additional 0.4% (wt/vol) agar.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics |
|---|---|
| Y. pestis strain | |
| 201 (wild type) | Avirulent to humans, highly lethal to mice |
| 201pKD46 | 201 harboring pKD46 vector, Ampr |
| 201ΔwaaA(YPO0055) | ΔwaaA, derived from strain 201pKD46, Kmr cassette replacement of waaA gene |
| 201ΔlamB(YPO3711) | ΔlamB, derived from strain 201pKD46, Kmr cassette replacement of lamB gene |
| 201ΔompF(YPO1411) | ΔompF, derived from strain 201pKD46, Kmr cassette replacement of ompF gene |
| 201ΔompA(YPO1435) | ΔompA, derived from strain 201pKD46, Kmr cassette replacement of ompA gene |
| 201Δail(YPO2905) | Δail, derived from strain 201pKD46, Kmr cassette replacement of ail gene |
| 201C-lamB | Complemented ΔlamB strains, transformed with a pACYC184 vector with cloned lamB |
| 201C-ompF | Complemented ΔompF strains, transformed with a pACYC184 vector with cloned ompF |
| 201C-ompA | Complemented ΔompA strains, transformed with a pACYC184 vector with cloned ompA |
| 201C-ail | Complemented Δail strains, transformed with a pACYC184 vector with cloned ail |
| 201C-waaA | Complemented ΔwaaA strains, transformed with a pACYC184 vector with cloned waaA |
| 201ΔailΔompA | Δail ΔompA, derived from strain 201ΔompA, Cmr cassette replacement of ail gene |
| 201ΔlamBΔail | ΔlamB Δail, derived from strain 201Δail, Cmr cassette replacement of lamB gene |
| 201ΔompFΔail | ΔompF Δail, derived from strain 201Δail, Cmr cassette replacement of ompF gene |
| 201ΔompFΔompA | ΔompF ΔompA, derived from strain 201ΔompA, Cmr cassette replacement of ompF gene |
| 201ΔompFΔlamB | ΔompF ΔlamB, derived from strain 201ΔlamB, Cmr cassette replacement of ompF gene |
| 201ΔompAΔlamB | ΔompA ΔlamB, derived from strain 201ΔlamB, Cmr cassette replacement of ompA gene |
| 201ΔompFΔailΔompA | ΔompF Δail ΔompA, derived from strain 201ΔailΔompA, Gmr cassette replacement of ompFgene |
| 201ΔailΔompAΔlamB | Δail ΔompA ΔlamB, derived from strain 201ΔompAΔlamB, Gmr cassette replacement of ail gene |
| 201ΔompFΔompAΔlamB | ΔompF ΔompA ΔlamB, derived from strain 201ΔompAΔlamB, Gmr-cassette replacement of ompF gene |
| 201ΔailΔompFΔwaaA | Δail ΔompF ΔwaaA, derived from strain 201ΔompFΔail, Gmr cassette replacement of waaA gene |
| Y. pseudotuberculosis | Pa3606 Human, Japan Yp 1b |
| Plasmids | |
| pKD46 | Red recombinase plasmid; Ampr |
| pET28a::gp17 | Expression vector with a PCR-amplified gp17 gene in the NheI/HindIII site; Kmr |
| pET28a::lamB | Expression vector with a PCR-amplified lamB gene in the NheI/HindIII site; Kmr |
| pET32a::ompF | Expression vector with a PCR-amplified ompF gene in the BamHI/SalI site; Ampr |
| pET32a::ompA | Expression vector with a PCR-amplified ompA gene in the BamHI/SalI site; Ampr |
| pET32a::ail | Expression vector with a PCR-amplified ail gene in the BamHI/SalI site; Ampr |
| pACYC184::waaA | Cloning vector with a PCR fragment covering a region from 500-bp fragment upstream to 300-bp fragment downstream of the waaA gene in the BamHI/HindIII site;Cmr |
| pACYC184::lamB | Cloning vector with a PCR fragment covering a region from 500-bp fragment upstream to 300-bp fragment downstream of the lamB gene in the BamHI/HindIII site; Cmr |
| pACYC184::ompF | Cloning vector with a PCR fragment covering a region from 500-bp fragment upstream to 300-bp fragment downstream of the ompF gene in the BamHI/HindIII site; Cmr |
| pACYC184::ompA | Cloning vector with a PCR fragment covering a region from 500-bp fragment upstream to 300-bp fragment downstream of the ompA gene in the BamHI/HindIII site; Cmr |
| pACYC184::ail | Cloning vector with a PCR fragment covering a region from 500-bp fragment upstream to 300-bp fragment downstream of the ail gene in the BamHI/HindIII site; Cmr |
Phage adsorption assays.
Approximately 8 × 105 PFU of Yep-phi in 100 μl was mixed with a 500-μl sample of bacteria (A600 = 1.2). Y. pseudotuberculosis was cultured overnight, and Y. pestis was cultured for 48 h. The suspension was incubated at room temperature for 5 min and centrifuged at 16,000 × g for 3 min, after which the phage titer remaining in the supernatant (i.e., residual PFU%), was determined. LB was used as a nonadsorbing control in each assay, and the phage titer in the control supernatant was set to 100%. Each assay was performed in duplicate and repeated twice (15).
Periodate and proteinase K treatments.
To test the effects of proteinase K treatment on Yep-phi adsorption, the protocol described here was used (15). Y. pestis cultures (2 ml) were treated with proteinase K (0.2 mg/ml; Promega) at 37°C for 3 h and then washed with 2 ml of LB. The phage adsorption assay was performed as described above. A control without proteinase K was used to ensure that the possible effect was not due to the incubation at 37°C.
To test the effects of periodate, Y. pestis culture (1.5 ml) was centrifuged at 16,000 × g for 1 min, and the bacterial pellet was suspended in 1.5 ml sodium acetate (50 mM, pH 5.2), or sodium acetate containing 100 mM IO4−, to determine whether periodate can destroy the phage receptor (15). The cells were incubated for 2 h (protected from light), centrifuged according to the above-mentioned process, washed with 1.5 ml of LB, and then centrifuged again and suspended in LB. The A600 of the bacterial suspension was adjusted to 1, and the phage adsorption assay was carried out.
Identification of the bacterial targets of phage-encoded tail fiber protein gp17.
Tail fiber protein gp17 was expressed as His6 fusions and purified from E. coli BL21(λDE3) (26). Recombinant protein was covalently coupled to Affi-Gel resin (Bio-Rad) using HNG buffer (20 mM HEPES-KOH, pH 7.5, 150 mM sodium chloride, 10% [vol/vol] glycerol). The unreacted sites were blocked with 10 mM ethanolamine, with a pH of 7.5, at 4°C for 30 min. Lysates were prepared from exponential-phase Y. pestis cells, resuspended in 20 ml of HNG buffer (1 mM EDTA, 1% [vol/vol] Triton X-100, 1 mM [each] dithiothreitol and phenylmethylsulfonyl fluoride), followed by sonication. Cell debris was removed by centrifugation for 0.5 h at 12,000 rpm. Affinity chromatography was performed by mixing 100 μl of resin with 1 ml of Y. pestis lysate at 4°C overnight. The resin was washed extensively with HNG buffer with increasing concentrations (0.1 M to 1 M) of NaCl. Bound proteins were eluted with 1% (wt/vol) SDS, resolved by SDS-PAGE, and stained with Coomassie brilliant blue. Specific bands were excised, and proteins were digested with trypsin prior to being subjected to matrix-assisted laser desorption ionization–time of flight mass spectrometry, in conjunction with postsource decay and collision-induced decay for the determination of protein identity (27, 28).
Preparation and verification of mutants.
The genes that can encode the possible receptors of Yep-phi from Y. pestis were subjected to single, double, or triple deletion, by replacing the target gene(s) with kanamycin (single), chloramphenicol (double), and/or gentamicin (triple) resistance cassettes (29). Mutant genotypes were confirmed by PCR. A PCR-generated DNA fragment containing the target gene-coding region, with its promoter-proximal region (∼500 bp upstream from the coding sequence) and transcriptional terminator (∼300 bp downstream), was cloned into the pACYC184 vector (30) and then verified by DNA sequencing. The recombinant plasmid was subsequently introduced into the mutants, yielding the respective complemented mutant strains. All the DNA sequences mentioned in this study were derived from the genomic data of strain CO92. The characteristics of the strains and the plasmids are presented in Table 1. All the primers used in this study, designed using Primer Premier 5.0 software, are listed in Table 2.
Table 2.
Oligonucleotides used in this work
| Target | Primer (forward/reverse, 5′–3′)a |
|---|---|
| Protein expression | |
| lamB | CTAGCTAGCCATGGTTATGCGCGT/CCCAAGCTTTTACCACCAGGCTTCG |
| ail | GGGATCCAGTATTTCTATTGGATATGC/GGTCGACTTAACCCACTTTCACATCATCG |
| ompF | GGGATCCTACGGTAAAGTTGACGC/GGTCGACTTAGGACTGAACATAACCCAG |
| ompA | GGGATCCGGTTTAGTATGGCGTGC/GGTCGACTTAAGCCTGTGGCTGAGTC |
| gp17 | CTAGCTAGCATGGCTAACAAAATTTC/CCCAAGCTTTCAGTTTCGAACTAAG |
| gp17(1–469) | CTAGCTAGCATGGCTAACAAAATTTC/CCCAAGCTTTCAATCGACCATCTTG |
| gp17(1–119) | CTAGCTAGCATGGCTAACAAAATTTC/CCCAAGCTTTCAAGTATCCGCAG |
| gp17(470–569) | CTAGCTAGCGGACTACTCACTACCG/CCCAAGCTTTCAGTTTCGAACTAAG |
| Construction of mutants | |
| Kanamycin resistance cassette | |
| waaA | ATGAATAACCACGGCTCAATGTGGTGCTCCGCCCTCCTAGGTGTAGGCTGGAGCTGCTTC/TTAGTGGCTCCGTTGTGGCAGATAAGGCTCCAACAAATGTCATATGAATATCCTCCTTA |
| lamB | ATGATTACTCTGCGCAAGTTACCAATAGCACTGGCTGTTGGTGTAGGCTGGAGCTGCTTC/TTACCACCAGGCTTCGAATTGCGCACCGAAGGTGACTTCACATATGAATATCCTCCTTA |
| ail | ATGGTTTTTATGAATAAGACATTACTGGTCTCTTCTTTAAGTGTAGGCTGGAGCTGCTTC/TTAGAACCGGTAACCCGCGCCAAGCATCCAAGTACCCACTCATATGAATATCCTCCTTA |
| ompF | ATGAGGGTAATAATAATGATGAAGCGCAATATTCTTGCAGGTGTAGGCTGGAGCTGCTTC/TTAGAACTGGTAAACCAAGCCAACAGCAACAACGTTGTCGCATATGAATATCCTCCTTA |
| ompA | TTGCCTATTTGGATGATAATGAGGCGTAAAATGAAAAAGAGTGTAGGCTGGAGCTGCTTC/TTAAGCCTGTGGCTGAGTCACAACTTCTTTGTAGCCTTTCCATATGAATATCCTCCTTA |
| Chloromycetin resistance cassette | |
| lamB | ATGATTACTCTGCGCAAGTTACCAATAGCACTGGCTGTTGAGATTGCAGCATTACACGTC/TTACCACCAGGCTTCGAATTGCGCACCGAAGGTGACTTCATGTAACGCACTGAGAAGC |
| ail | ATGGTTTTTATGAATAAGACATTACTGGTCTCTTCTTTAAAGATTGCAGCATTACACGTC/TTAGAACCGGTAACCCGCGCCAAGCATCCAAGTACCCACTTGTAACGCACTGAGAAGC |
| ompF | ATGAGGGTAATAATAATGATGAAGCGCAATATTCTTGCAGAGATTGCAGCATTACACGTC/TTAGAACTGGTAAACCAAGCCAACAGCAACAACGTTGTCGTGTAACGCACTGAGAAGC |
| ompA | TTGCCTATTTGGATGATAATGAGGCGTAAAATGAAAAAGAAGATTGCAGCATTACACGTC/TTAAGCCTGTGGCTGAGTCACAACTTCTTTGTAGCCTTTCTGTAACGCACTGAGAAGC |
| Gentamicin resistance cassette | |
| lamB | ATGATTACTCTGCGCAAGTTACCAATAGCACTGGCTGTTGTTAGGTGGCGGTACTTGGG/TTACCACCAGGCTTCGAATTGCGCACCGAAGGTGACTTCACGTCAGGTGGCACTTTTCG |
| ail | ATGGTTTTTATGAATAAGACATTACTGGTCTCTTCTTTAATTAGGTGGCGGTACTTGGG/TTAGAACCGGTAACCCGCGCCAAGCATCCAAGTACCCACTCGTCAGGTGGCACTTTTCG |
| ompF | ATGAGGGTAATAATAATGATGAAGCGCAATATTCTTGCAGTTAGGTGGCGGTACTTGGG/TTAGAACTGGTAAACCAAGCCAACAGCAACAACGTTGTCGCGTCAGGTGGCACTTTTCG |
| Verification of the mutants | |
| waaA | CTGCGGCAGGTTACAAGAAG/ATGTGTGCTAGTCGCAATCC |
| lamB | TACTCAAGAAGGTTACTCG/GTTGGCATAAGTTGCGAAG |
| ail | ACTGGGGAGTGATAGGTTC/TTACTTCTACTCTCTGATTG |
| ompF | CGGCAACGCAACCCGTCTG/CTTCAGAACGGTCATTTTTG |
| ompA | GGGTGGTTTAGTATGGCGTG/TCCTGACCAAAGCGGTAAG |
| Complementation of mutants | |
| lamB | CGCGGATCCGCCCAGAACATC/CCCAAGCTTGTACCAGACAGAGTG |
| ompF | CGCGGATCCCCGTTGCGGCGATGGATG/CCCAAGCTTTTTTCAAACATGTCAGAG |
| ail | CGCGGATCCCCATTGAGCGTGTGAAAC/CCCAAGCTTCAACTTTACCGTCTCAAC |
| ompA | CGCGGATCCTACAGTGTTGTTCTGGG/CCCAAGCTTTACTGAACCCCC |
| waaA | CGCGGATCCGTGCAAAAGCCGTAGTC/CCCAAGCTTCGATTTTAAGCTCTGGG |
Underlining indicates endonuclease restriction sites.
Phage plaque formation assays.
The lysis activity of each phage was examined using spot tests on Y. pestis strains (9, 31). A series of exponential-phase cultures were prepared for each mutant strain. One hundred-microliter cultures were concentrated by centrifugation, dissolved in 10 ml LB, and then mixed with 3 ml of liquefied soft-agar medium. The mixture was then vortexed and poured onto individual LB plates to produce double-layer agar plates. When the medium solidified, the serial dilutions of parental phage stocks (3 μl) were spotted onto the surface of the plates. The plates were inverted, incubated at 26°C overnight, and then examined and photographed.
Anti-OMP antibodies and immunoblot analysis.
The OMPs were expressed as His6 fusions and purified from E. coli BL21(λDE3) cells (26). For polyclonal antibody production, New Zealand White rabbits were subcutaneously injected with 1 mg of purified Ail or OmpF proteins mixed in 0.5 ml of phosphate-buffered saline (PBS) and emulsified with an equal volume of complete Freund's adjuvant (CFA) for a total of three immunizations. Three days after the final boost, all rabbits were exsanguinated by heart puncture under general anesthesia. Sera were separated from blood cells by centrifugation at 12,000 × rpm for 15 min at 4°C. This crude serum was purified using saturated ammonium sulfate precipitation and stored at −80°C. For immunoblot analysis, SDS-PAGE and Western blotting were performed according to standard protocols (32). The captured proteins were subjected to electrophoresis by 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The PVDF membranes were blocked in PBST (PBS and 0.2% Tween 20) containing 5% skimmed milk for 2 h and subsequently incubated with our polyclonal antibody for 1 h at room temperature or overnight at 4°C. The membranes were washed with PBST 3 times and probed by goat anti-rabbit secondary antibody, conjugated with horseradish peroxidase (Roche, USA), for 1 h at room temperature. After washing the membranes 3 times with PBST, the immunoblots were developed with the enhanced chemiluminescence system SuperSignal West Pico Chemiluminescent Substrate (Pierce, USA).
RESULTS
Destruction of phage receptors by both periodate and proteinase K.
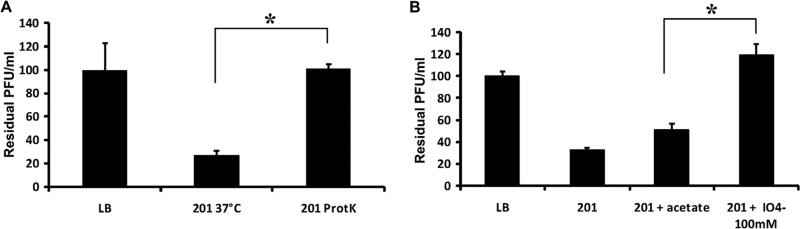
The surface of Gram-negative bacteria, exposed to the environment, consists mainly of proteins and polysaccharide structures (24). It is important to test whether the degradation of cell surface proteins, or LPS, can destroy the Yep-phi receptor, as the bacteriophage could exploit both LPS and OMPs as receptors (15). Bacteria were treated with either proteinase K (for destroying surface proteins) or periodate (for destroying surface carbohydrates), prior to the phage adsorption assay, in order to determine the possible nature of the phage receptor (15). Yep-phi exhibited high infection efficiency when mixed with Y. pestis cells (Fig. 1); 5 min after mixing, most phages were removed from the suspension by binding to Y. pestis cells. When the phage was incubated with proteinase K or periodate-treated Y. pestis, the majority of phages remained in the supernatant. The significant increase of free phage particles indicated that the virions were unable to gain efficient adsorption on the treated bacteria. Therefore, the Y. pestis receptors (which were recognized and bound by Yep-phi) likely contain both protein and carbohydrate structures. In this respect, they differ notably from the features of PhiA1122 and T7 (11, 15, 19).
Fig 1.
Effects of the different treatments of bacteria on Yep-phi adsorption, which are shown as residual PFU percentages. (A) Effect of proteinase K treatment on the adsorption of Yep-phi to Yersinia pestis strain 201. (B) Effect of periodate treatment on the adsorption of Yep-phi to Y. pestis strain 201. The control (LB and “201+acetate”), untreated strain (strain 201), and treatment (“201 ProtK” for proteinase K treatment and “201+IO4−” for periodate treatment) groups were tested for adsorptions as indicated in the x axes. Error bars denote statistical variations. Significance was determined by one Student t test for comparison between the treated group and the WT group. *, P < 0.05.
Alignment of RBP tail fiber protein gp17.
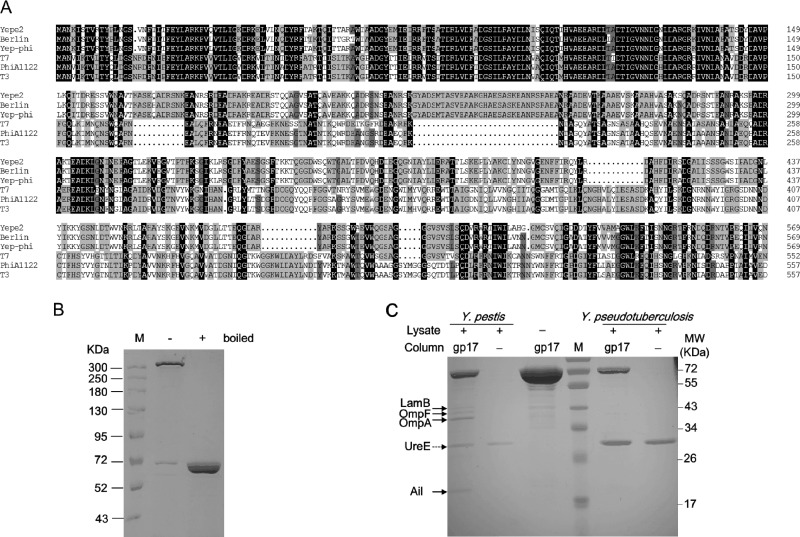
For T7-related phages, the primary determinant of the host receptor is the tail fiber protein gp17 (11). The six T7 tail fibers, which are the homotrimers of gp17, are responsible for the first specific, albeit reversible attachment to E. coli LPS (11). As a T7-related phage, Yep-phi has a unique host specificity that can be attributed to its tail fiber gp17 protein. It has been reported that the receptor binding protein (RBP) of phages in the same species has a conserved N-terminal moiety due to its involvement in interactions with other tail proteins, whereas the C-terminal portion diverges in the region that was proposed to recognize the host (33). This pattern of sequence conservation is also observed in the alignment of gp17 sequences of Y. pestis phages (Yep-phi, PhiA1122, Yepe2, and Berlin) and E. coli phages (T7, T3) (Fig. 2A). The alignment shows a highly conserved N-terminal moiety (amino acids 1 to 149), which belongs to domain family PHA00430 (34), indicating that they have the same mechanism of attaching the fiber to the phage as numerous T7-related phages. Aside from this region, the other sequence of the Yep-phi subgroup diversified from that of the other three (T7, T3, and PhiA1122). It is known that T7, T3, and PhiA1122 tail fibers adsorb to the host surface LPS (11, 15), so we speculate that the Yep-phi subgroup adsorbs to different receptors.
Fig 2.
(A) Alignment of tail fiber proteins. Identical residues are shaded in black, residues sharing >75% homology are shaded in deep gray, and those sharing >50% homology are shaded in light gray. Gaps (indicated by dotted lines) were introduced into the sequences to maximize the alignments. Numbering is based on the N-terminal methionine. Multiple sequence alignment was analyzed and edited with DNAman software (Lynnonon Biosoft, Quebec, Canada). The names of phages are indicated on the left. (B) SDS-PAGE gel showing the SDS-resistant multimer formed by gp17. Symbols “+” and “−” above the gel denote the purified protein boiled previously and unboiled, respectively. (C) Identification of cell wall-anchored protein targets of gp17 by affinity chromatography. The protein bands found to interact specifically with gp17 are indicated by black arrows. UreE protein binding to the nickel column is denoted by a dotted arrow.
Identification of cell wall-anchored protein targets of tail fiber protein gp17.
Considering that the initial binding of phage to bacteria is mediated by the interaction of its six gp17 tail fibers with bacterial surface receptors, gp17 protein of Yep-phi was expressed and purified as a His6 fusion (11, 34). Consistent with a previous report of T7 (34), we found that the purified recombinant gp17 protein of Yep-phi existed mainly as a multimer in solution. The multimer fraction was SDS resistant unless previously boiled (Fig. 2B). The bacterial targets of phage tail fiber protein gp17 were identified by affinity chromatography of Y. pestis lysates using a gp17-coupled Affi-Gel resin column (Fig. 2C). The protein bands were recovered specifically from a Y. pestis lysate compared with other controls, including Y. pseudotuberculosis lysate. Trypsinolysis and mass spectrometry revealed that the captured polypeptides were the Y. pestis OMPs LamB, OmpF, OmpA, and Ail (Table 3). The common band in all lanes was UreE protein, which was nonspecifically bound to the nickel column.
Table 3.
Functional features of the proteins on polyacrylamide gels
| Protein | MWa | Locus tag |
Product | Function in phage receptor(s): | |
|---|---|---|---|---|---|
| CO92 | 201 | ||||
| LamB | 46,783 | YPO3711 | YP_3073 | Maltoporin | λ |
| OmpF | 40,362 | YPO1411 | YP_1182 | Outer membrane porin protein F | K20, RB69, SV76, RB51, T2 |
| OmpA | 39,193 | YPO1435 | YP_1326 | Outer membrane protein A | K3, M1, Ox2, TuII, RB32, PST |
| Ail | 20,153 | YPO2905 | YP_2550 | Attachment invasion locus protein | NDb |
| UreEc | 25,943 | YPO2668 | YP_2469 | Urease accessory protein ureE | ND |
MW, molecular weight.
ND, not determined.
UreE protein exists both in Y. pestis and Y. pseudotuberculosis and is able to bind the nickel column.
LamB is found in numerous species of Gram-negative bacteria and is an abundant integral outer membrane porin responsible for maltose uptake. LamB also serves as the receptor for bacteriophage lambda (35). OmpA protein plays a structural role in the integrity of the bacterial cell surface (36). It is proposed to interact specifically with the peptidoglycan layer (36) and serves as a receptor for several T-even phages infecting E. coli (31). OmpF is often regarded as a classical porin, which is present in large quantities, and is also required as a receptor by several phages of E. coli (31, 36, 37). Ail protein is essential for Y. pestis to resist complement-mediated killing, and Ail-mediated resistance to human serum depends on the LPS core (38, 39). However, the function of Ail in phage receptors has yet to be fully explained. It is worth mentioning that we employed the single nucleotide polymorphism (SNP) method to scan the sequence polymorphisms of the proteins in the available 133 sequenced Y. pestis strains (40). The results indicated that the ail gene displayed the highest variability. Information for four SNP loci, i.e., a T-to-G change at position 136 encoded by ail [ail136(T-G)], ail238 (T-G), ail240 (C-A), and ail253 (G-T), were acquired in the relatively small 549-bp coding region. The mutation rate was unusually higher than that of the other genes. Ail was predicted to consist of eight transmembrane β-sheets and four cell surface-exposed loops (41, 42). In the current work, the resulting amino acid alteration Ail46 (Y-D) was located at loop 1, whereas Ail80 (V-F/L) and Ail85 (Y-D) were located at loop 2. The results are consistent with the notion that the external loops of the OMPs, between the transmembrane β-strands, undergo rapid mutational alterations as they interact with the elements of the external world, including phages (36, 43).
Phage propagation on the various mutant host strains.
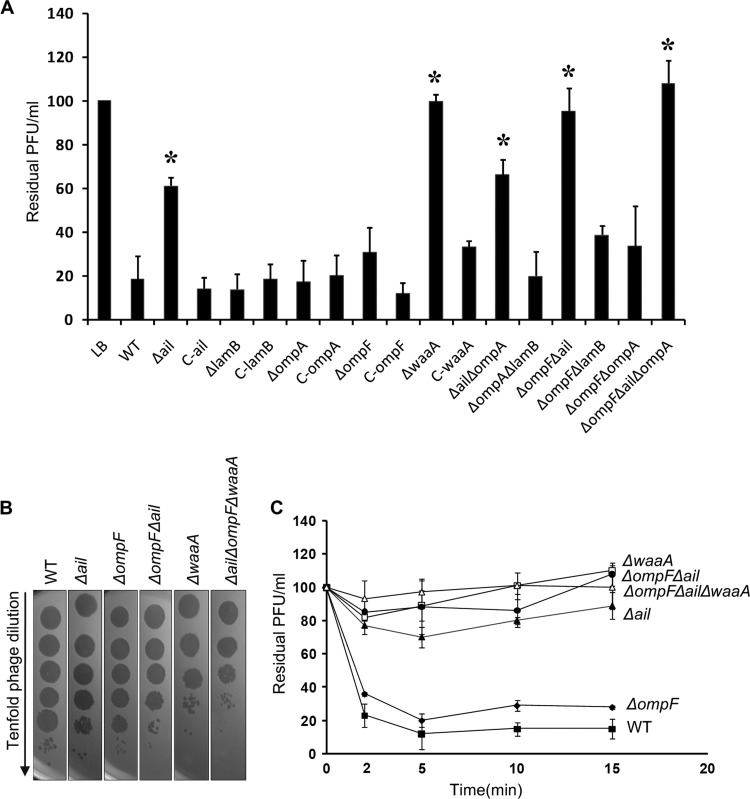
Single-, double-, or triple-gene inactivation was accomplished in Y. pestis using PCR products with kanamycin, chloramphenicol, and/or gentamicin resistance cassettes, in order to test the respective roles of the four proteins in Yep-phi infection (Table 1). The waaA gene of the LPS core biosynthetic pathway was eliminated to confirm the role of LPS core in the phage receptor, as previously shown (15). Each single gene mutant was complemented in trans by the corresponding wild-type gene to avoid the phenotype caused by a polar effect, or a spontaneous mutation, elsewhere in the genome (Table 1).
The results show that the waaA mutant lost most of its phage-binding activity during the adsorption assay (Fig. 3A), thereby confirming that the rough LPS is a critical part of the receptor (15). This result is in agreement with the notion that the closely related phages T7 and T3 utilize the LPS core as their receptors (16, 19). ΔlamB and ΔompA mutants had phage adsorption rates comparable to that of the WT strain, while the ΔompF mutant showed a small defect, and the defect increased with Δail. The ΔompF Δail double mutant was accompanied by a decrease at a level comparable to that of the waaA mutant. The results indicated that the Y. pestis LPS core was a major target of the phage, but Ail and OmpF were also involved in the phage infection. We speculate that the loss of the ail and ompF products may result in defective phage receptors.
Fig 3.
(A) Yep-phi adsorption to Y. pestis outer membrane protein and lipopolysaccharide mutants, shown as residual PFU percentages. The control and strains used for adsorptions are presented below the columns (complete strain names and descriptions can be found in Table 1). Error bars denote ranges. Significance was determined by one Student t test for comparison between the mutant group and the WT group; *, P < 0.05. (B) Tenfold dilution of lysates of Yep-phi applied to bacterial lawns of wild-type and mutant Y. pestis strains. (C) Adsorption kinetics of Yep-phi to Y. pestis LPS and OMP mutants, shown as residual PFU percentages. Standard errors are indicated by vertical lines.
It is likely that there are at least three molecules participating in the adsorption process; therefore, we constructed the Δail ΔompF ΔwaaA mutant. To test whether the mutant could express antiphage activity, the plaquing efficiency of Yep-phi on a series of mutants was measured (Fig. 3B). Significant differences were observed in the ability of the phage to lyse the mutants. Additionally, Yep-phi formed plaques very robustly on the Δail ΔompF ΔwaaA mutant compared to the WT strain (Fig. 3B). The results suggest that under the same culture conditions, the antiphage activity is the highest on the Δail ΔompF ΔwaaA mutant, followed by the ΔwaaA and the ΔompF Δail mutants. None of the defined mutants completely prevented the phage attachment or altered the ability of the phage to lyse the mutant bacteria, hinting at the complexity of the adsorption process that has yet to be uncovered. However, the mutation appeared to alter a subsequent step, which was measured by a reduction in plaque-forming efficiency (Fig. 3B). The adsorption kinetics assay (Fig. 3C) further demonstrated that the phage receptors were affected in the Δail, ΔompF Δail, and ΔwaaA strains. It is clear that LPS is the major contributor to the phage fibers' adsorption. It seems likely that the OmpF protein did not affect the adsorption alone but may be able to cooperate with Ail to make a contribution to the attachment. Fibers “walk” across the cell surface to find the right receptor for the tail (44), but binding of fibers to bacterial receptors is weak. Hypothetically, the two OMPs could cooperate with LPS to assist the fibers in binding to the correct sites as soon as possible. That is why, when the two OMPs are not available, the phage can still infect the cell, but with a lower efficiency. Effective adsorption needs cooperative binding among fibers (44). Perhaps the six fibers bind different sites, with some of the fibers binding OMPs, and some binding LPS. Normally, the fibers bind to both sites, making the binding stable and fast, which results in higher infection efficiency. However, when either OMPs or LPS are deficient, the fibers can bind only one site. This causes the binding to be weak, ultimately reducing infection efficiency.
To the best of our knowledge, the OMPs of Gram-negative bacteria have not exhibited receptor activity for T7-related phages. In this study, we found that a T7-related phage, Yep-phi, required not only LPS but also Ail and OmpF for its efficient infection. Deficiencies of the three molecules exhibited a remarkable decrease in adsorption efficiency, although it was not enough to cause complete resistance against phage infection. The failure to inhibit the infection implies that other factors remain to be discovered in order to gain a better understanding of the phage system.
Specific binding with the C-terminal region (510 amino acids [aa] to 529 aa) of gp17.
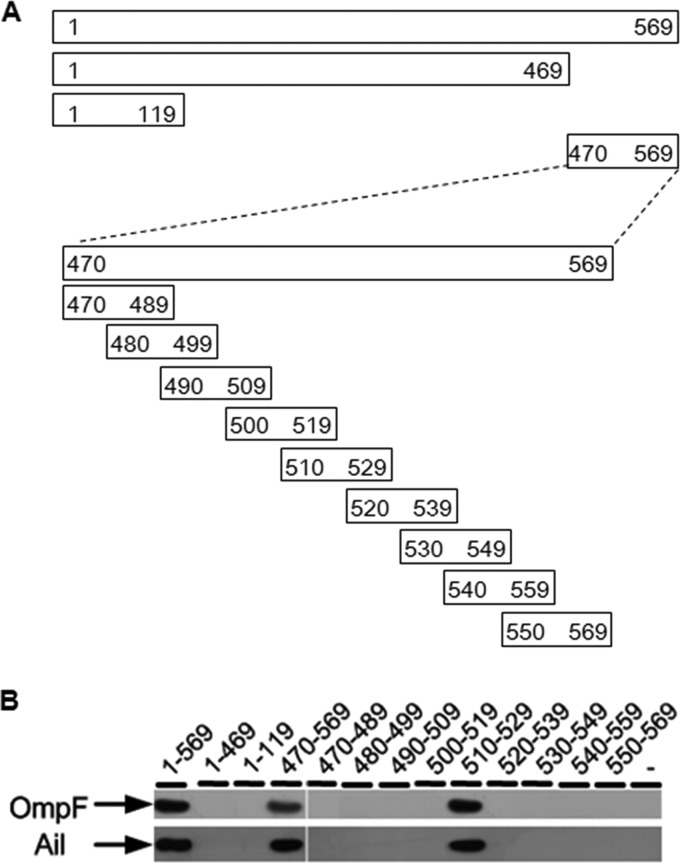
To analyze the locations of the OMPs' binding region, three fragments spanning different regions of gp17 protein were expressed as His6 fusions and purified from E. coli BL21(λDE3) (Fig. 4A, upper part). The purified fusions were used as baits to capture OmpF and Ail proteins from the lysates of Δail ΔompA ΔlamB and ΔompF ΔompA ΔlamB mutants, respectively. The triple gene mutants were used to eliminate the interplay between the OMPs, and the captured proteins were examined by immunoblotting, using rabbit anti-OmpF or anti-Ail polyclonal antibodies. The result showed that fragment gp17470–569 was responsible for the interaction with Ail and OmpF, while no interactions were observed with fragments gp171–469 and gp171–119 (Fig. 4B). Fragment gp17470–569 contains the tip domain of the tail fiber protein. The tip domain plays an important role in determining the host range for the T7 phage. The structure of the tip domain has been resolved and is composed of an eight-stranded β-sandwich (BCDEFGHI) and four loops (i.e., BC, DE, FG, and HI loops) at the top, which are thought to interact with the receptors (34). We performed alignment of the tip domains of the Yep-phi subgroup, T7, PhiA1122, and T3 gp17 proteins. The alignment showed conserved hydrophobic positions, indicating a common fold (Fig. 5A). Of special interest is the mosaic nature of the T7 tip domain; the BC loop displayed significant similarity with the Yep-phi subgroup, whereas the DE, FG, and HI loops had the highest similarity to T3 and PhiA1122.
Fig 4.
(A) Positions of the fragments on the gp17 protein. The thick line represents the fragments, and the numbers on the two ends of each fragment denote the starting and ending amino acid positions. (B) OmpF and Ail detected by Western blotting using either rabbit anti-OmpF or anti-Ail polyclonal antibody after affinity chromatography.
Fig 5.
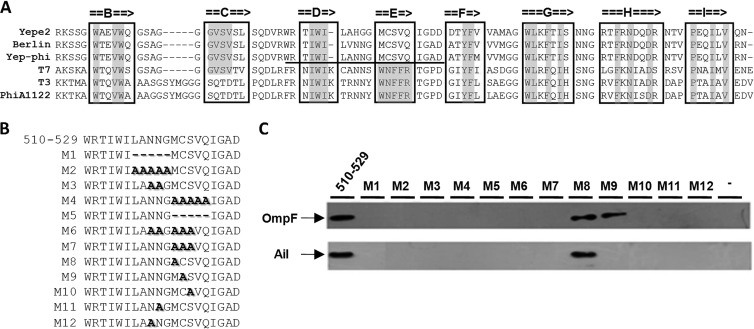
(A) Alignment of the sequence of the tip domain of T7 gp17 with its homologs from E. coli phage T3 and Y. pestis phages Yep-phi, Berlin, Yepe2, and PhiA1122. Beta-strands are denoted by arrows. The residues of beta-strands that are identical with each other are marked with gray. The domain necessary for the phage Yep-phi and Y. pestis interaction is underlined. (B) Locations of deletions and alanine replacements used in this study are indicated by M1 to M12. (C) OmpF and Ail were detected by Western blotting using either rabbit anti-OmpF or anti-Ail polyclonal antibody after affinity chromatography.
To decipher the minimal portion of the tip domain required to bind the two OMPs, fragment gp17470–569 was divided into nine N-terminally 6×His-tagged peptides (the lower part of Fig. 4A). Immunoblotting was performed to test the interaction, and the results showed that one of the peptides (510 aa to 529 aa), with only 20 amino acids, was sufficient for the interaction with the OMPs, which is located at the DE loop of the tip domain. We thus speculate that the DE loop may provide a suitable location for specific binding. We noticed that β-strand E of the Yep-phi subgroup consisted of unique hydrophobic residues that were distantly diverged from PhiA1122, T7, and T3. The dissimilarities existing between them may account for different receptor specificities.
In order to localize the residue(s) necessary for the binding activity, 12 deletion/replacement mutations (designated M1 to M12, respectively [Fig. 5B]), located at the polarity residues throughout the β-strand E region, as well as the loop region, were constructed. Mutational analysis suggested that both the DE loop and the β-strand E regions were involved in the binding. Mutations at residues 518N, 519N, or 523S were completely deficient in terms of binding to OmpF, whereas residues 518N, 519N, 522C, or 523S were found to be essential for the interaction with Ail (Fig. 5C).
DISCUSSION
Phages bind to unique host-specific structures, allowing them to recognize a suitable host in a mixed bacterial population (18). LPS is currently recognized as the receptor for T7 and T3 phages (34). Four T7-related phages have been sequenced in prior studies of Y. pestis-specific phages (11, 12). However, only the receptor of PhiA1122 has been identified (15). The other three phages, represented by Yep-phi (used in a phage lysis assay in China for the identification of Y. pestis isolates), have significantly different characteristics, either at the genome or at the phenotype level, from those of PhiA1122. To date, the molecular interactions between Yep-phi and Y. pestis have not been fully understood.
In this study, the host components that interact with Yep-phi were found to be the rough LPS and two OMPs (Ail and OmpF). First, the different specificities of the infection on Y. pseudotuberculosis indicate that Yep-phi and PhiA1122 adsorb different surface structures. Second, both periodate and proteinase K treatments of Y. pestis inhibited phage binding. Third, the spontaneous Yep-phi resistance mutants of Y. pestis could not be isolated, indicating that the phage had more complex behavior. Fourth, affinity chromatography revealed that tail fiber protein of Yep-phi is likely responsible for binding to four OMPs of Y. pestis, among which two (Ail and OmpF) were identified to be involved in the adsorption process. Fifth, an LPS mutant and a series of OMP mutants were accompanied by a decrease of phage adsorption efficiency.
Furthermore, our data indicate that residues 510 to 529 of tail fiber protein gp17 are sufficient to allow binding to Ail and OmpF in vitro. Residues 518N, 519N, and 523S were determined to be essential for the interaction with OmpF, while residues 518N, 519N, 522C, and 523S were found to be essential for the interaction with Ail.
The results reveal novel phage-host interactions and the infection mechanisms of Y. pestis. Three kinds of host factors were specifically identified in this study (LPS, Ail, and OmpF). Although the phage can still lyse the Δail ΔompF ΔwaaA strain, it is with significantly decreased infection activity. One possible reason it could still lyse the strain is that there might be additional membrane components cooperatively determining the phage infection efficiency. The method employed in this study for isolating OMP receptors may not have been suitable for isolating proteins of low concentration. Another plausible explanation is that this interaction is presumably followed by a secondary, irreversible attachment of the tail to an unknown receptor (34). Because the irreversible binding triggers the release of naked genome from the virion, followed by the transit of viral DNA to the host cell cytoplasm (20), tail fiber protein gp17 may just keep the phage near the bacterial surface to make productive tail interactions with its receptor more likely (34). Although other factors may also contribute to the adaptation of a phage to a certain host, at least three distinct binding sites are required for the phage to proliferate efficiently on its host. The data presented in this study, together with the available knowledge, allow the proposition of a comprehensive model explaining the events leading to genome ejection from the phage particle.
LPS is required for the structural integrity of numerous OMPs and has significant effects on the cell surface protein composition (10, 16, 45). Close LPS-protein interactions are required for protein support, proper folding, and biological activity (10). For example, a drastic decrease of the concentration of OmpF has been observed in E. coli LPS mutants (45). Ail-mediated resistance to human serum depends on the LPS core (39), and changes in the LPS structure may influence the Ail conformation and specific activity (38). However, whether the receptor functions of Ail and OmpF are related to the LPS core remains unknown.
OMPs Ail and OmpF of Y. pestis have never been characterized as phage receptors. The current study reports that the host specificity of Yep-phi is mediated, at least in part, through the Ail and OmpF molecules. Affinity chromatography assay showed four OMPs responsible for interacting with the phage tail fiber protein, but the central roles of LamB and OmpA in phage adsorption have yet to be fully described. They are possibly related with the locations of the phage on the cell surface or for a later step of phage DNA injection.
Since Y. pestis is classified as a potential biowarfare or bioterror agent, phage therapy may be considered as an approach to counter such a threat (46, 47). A major concern regarding the use of phages in the treatment of infectious diseases still remains the emergence of phage-resistant mutants (23). The mutation of the attachment site is the most frequent route to phage resistance of the host (8). The phage Yep-phi infection has more-complex recognition specificity, requiring at least three host molecules. Various attachment sites can reduce the impact of phage resistance and overcome this problem. Thus, Yep-phi would be highly amenable for use in plague treatments.
Overall, our results represent the first ever demonstration of membrane-bound proteins of Y. pestis involved in a T7-related phage adsorption. This work is also the first to identify the residues of the phage tail fiber protein essential for the interaction with OMPs of Gram-negative bacteria. These observations highlight the importance of the tail fiber protein in the evolution and function of various complex phage systems. This information may also contribute to the application of bacteriophages in various settings, such as bacteria typing or phage therapy, through the rational design of mutants binding different receptors or the engineering of artificial, chimeric phage fibers.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 31200137).
We also thank Jin Wang, Lili Zhang, and Xiangxiu Qin for their help in the experiments.
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Bossi P, Bricaire F. 2003. The plague, possible bioterrorist act. Presse Med. 32:804–807 (In French.) [PubMed] [Google Scholar]
- 2.Zhou D, Han Y, Yang R. 2006. Molecular and physiological insights into plague transmission, virulence and etiology. Microbes Infect. 8:273–284 [DOI] [PubMed] [Google Scholar]
- 3.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527 [DOI] [PubMed] [Google Scholar]
- 4.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. 10.1371/journal.pone.0000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morello E, Saussereau E, Maura D, Huerre M, Touqui L, Debarbieux L. 2011. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: first steps towards treatment and prevention. PLoS One 6:e16963. 10.1371/journal.pone.0016963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagens S, Habel A, von Ahsen U, von Gabain A, Blasi U. 2004. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 48:3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 14:524–531 [DOI] [PubMed] [Google Scholar]
- 8.Shin H, Lee JH, Kim H, Choi Y, Heu S, Ryu S. 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS One 7:e43392. 10.1371/journal.pone.0043392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul VD, Sundarrajan S, Rajagopalan SS, Hariharan S, Kempashanaiah N, Padmanabhan S, Sriram B, Ramachandran J. 2011. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol. 11:195. 10.1186/1471-2180-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia E, Chain P, Elliott JM, Bobrov AG, Motin VL, Kirillina O, Lao V, Calendar R, Filippov AA. 2008. Molecular characterization of L-413C, a P2-related plague diagnostic bacteriophage. Virology 372:85–96 [DOI] [PubMed] [Google Scholar]
- 11.Garcia E, Elliott JM, Ramanculov E, Chain PS, Chu MC, Molineux IJ. 2003. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 185:5248–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Wu W, Qi Z, Cui Y, Yan Y, Guo Z, Wang Z, Wang H, Deng H, Xue Y, Chen W, Wang X, Yang R. 2011. The complete genome sequence and proteomics of Yersinia pestis phage Yep-phi. J. Gen. Virol. 92:216–221 [DOI] [PubMed] [Google Scholar]
- 13.Pajunen M, Kiljunen S, Skurnik M. 2000. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiljunen S, Datta N, Dentovskaya SV, Anisimov AP, Knirel YA, Bengoechea JA, Holst O, Skurnik M. 2011. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. J. Bacteriol. 193:4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruger DH, Schroeder C. 1981. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol. Rev. 45:9–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sao-Jose C, Baptista C, Santos MA. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186:8337–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen MC, van Alphen LB, Fodor C, Crowley SM, Christensen BB, Szymanski CM, Brondsted L. 2012. Phase variable expression of capsular polysaccharide modifications allows Campylobacter jejuni to avoid bacteriophage infection in chickens. Front. Cell. Infect. Microbiol. 2:11. 10.3389/fcimb.2012.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindberg AA. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241 [DOI] [PubMed] [Google Scholar]
- 20.Sao-Jose C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. 2006. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281:11464–11470 [DOI] [PubMed] [Google Scholar]
- 21.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267:512–514 [DOI] [PubMed] [Google Scholar]
- 22.Dentovskaya SV, Anisimov AP, Kondakova AN, Lindner B, Bystrova OV, Svetoch TE, Shaikhutdinova RZ, Ivanov SA, Bakhteeva IV, Titareva GM, Knirel AY. 2011. Functional characterization and biological significance of Yersinia pestis lipopolysaccharide biosynthesis genes. Biochemistry (Mosc.) 76:808–822 [DOI] [PubMed] [Google Scholar]
- 23.Filippov AA, Sergueev KV, He Y, Huang XZ, Gnade BT, Mueller AJ, Fernandez-Prada CM, Nikolich MP. 2011. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS One 6:e25486. 10.1371/journal.pone.0025486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skurnik M. 2012. Yersinia surface structures and bacteriophages. Adv. Exp. Med. Biol. 954:293–301 [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Qiu Y, Gao H, Guo Z, Han Y, Song Y, Du Z, Wang X, Zhou D, Yang R. 2009. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9:128. 10.1186/1471-2180-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Gao H, Qin L, Li B, Han Y, Guo Z, Song Y, Zhai J, Du Z, Wang X, Zhou D, Yang R. 2008. Identification and characterization of PhoP regulon members in Yersinia pestis biovar Microtus. BMC Genomics 9:143. 10.1186/1471-2164-9-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22:185–191 [DOI] [PubMed] [Google Scholar]
- 28.Parker CE, Warren MR, Mocanu V.2010. Mass spectrometry for proteomics, p 71–92 In Alzate O. (ed), Neuroproteomics. CRC Press, Boca Raton, FL: [PubMed] [Google Scholar]
- 29.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose RE. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. 10.1093/nar/16.1.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM. 2011. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage's host specificity. Genome Biol. Evol. 3:674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minskaia E, Nicholson J, Ryan MD. 2013. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 13:67. 10.1186/1472-6750-13-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veesler D, Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Doval C, van Raaij MJ. 2012. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl. Acad. Sci. U. S. A. 109:9390–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs KA, Isaac DD, Xu J, Hendrix RW, Silhavy TJ, Theriot JA. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53:1771–1783 [DOI] [PubMed] [Google Scholar]
- 36.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239–253 [DOI] [PubMed] [Google Scholar]
- 37.Bekhit A, Fukamachi T, Saito H, Kobayashi H. 2011. The role of OmpC and OmpF in acidic resistance in Escherichia coli. Biol. Pharm. Bull. 34:330–334 [DOI] [PubMed] [Google Scholar]
- 38.Kolodziejek AM, Schnider DR, Rohde HN, Wojtowicz AJ, Bohach GA, Minnich SA, Hovde CJ. 2010. Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect. Immun. 78:5233–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartra SS, Styer KL, O'Bryant DM, Nilles ML, Hinnebusch BJ, Aballay A, Plano GV. 2008. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect. Immun. 76:612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, Zhang Y, Zheng H, Qin N, Xiao X, Wu M, Wang X, Zhou D, Qi Z, Du Z, Wu H, Yang X, Cao H, Wang H, Wang J, Yao S, Rakin A, Falush D, Balloux F, Achtman M, Song Y, Yang R. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 110:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller VL, Beer KB, Heusipp G, Young BM, Wachtel MR. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 41:1053–1062 [DOI] [PubMed] [Google Scholar]
- 42.Yamashita S, Lukacik P, Barnard TJ, Noinaj N, Felek S, Tsang TM, Krukonis ES, Hinnebusch BJ, Buchanan SK. 2011. Structural insights into Ail-mediated adhesion in Yersinia pestis. Structure 19:1672–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 339:576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ried G, Hindennach I, Henning U. 1990. Role of lipopolysaccharide in assembly of Escherichia coli outer membrane proteins OmpA, OmpC, and OmpF. J. Bacteriol. 172:6048–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwudke D, Ergin A, Michael K, Volkmar S, Appel B, Knabner D, Konietzny A, Strauch E. 2008. Broad-host-range Yersinia phage PY100: genome sequence, proteome analysis of virions, and DNA packaging strategy. J. Bacteriol. 190:332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anisimov AP, Amoako KK. 2006. Treatment of plague: promising alternatives to antibiotics. J. Med. Microbiol. 55:1461–1475 [DOI] [PubMed] [Google Scholar]