Abstract
Enteroviruses can frequently target the human central nervous system to induce a variety of neurological diseases. Although enteroviruses are highly cytolytic, emerging evidence has shown that these viruses can establish persistent infections both in vivo and in vitro. Here, we investigated the susceptibility of three human brain cell lines, CCF-STTG1, T98G, and SK-N-SH, to infection with three enterovirus serotypes: coxsackievirus B3 (CVB3), enterovirus 71, and coxsackievirus A9. Persistent infection was observed in CVB3-infected CCF-STTG1 cells, as evidenced by prolonged detection of infectious virions, viral RNA, and viral antigens. Of note, infected CCF-STTG1 cells expressed the nonfunctional canonical viral receptors coxsackievirus-adenovirus receptor and decay-accelerating factor, while removal of cell surface chondroitin sulfate from CCF-STTG1 cells inhibited the replication of CVB3, suggesting that receptor usage was one of the major limiting factors in CVB3 persistence. In addition, CVB3 curtailed the induction of beta interferon in infected CCF-STTG1 cells, which likely contributed to the initiation of persistence. Furthermore, proinflammatory chemokines and cytokines, such as vascular cell adhesion molecule 1, interleukin-8 (IL-8), and IL-6, were upregulated in CVB3-infected CCF-STTG1 cells and human progenitor-derived astrocytes. Our data together demonstrate the potential of CCF-STTG1 cells to be a novel cell model for studying CVB3-central nervous system interactions, providing the basis toward a better understanding of CVB3-induced chronic neuropathogenesis.
INTRODUCTION
Enteroviruses (EVs) are small, nonenveloped, single-strand positive RNA viruses which belong to the Enterovirus genus of the Picornaviridae family. On the basis of the phylogenetic sequence analysis, the genus Enterovirus consists of 12 species (1, 2), including the notable members poliovirus (PV), coxsackievirus (CV), and enterovirus 71 (EV71). EV infections have a wide spectrum of clinical manifestations, ranging from mild febrile illness to potentially fatal multisystem disorders. Numerous pieces of clinical and epidemiologic evidence link EV infections to type 1 diabetes mellitus, myocarditis, and acute respiratory illness (3–5). In addition, EVs are the most frequent etiological agents of central nervous system (CNS) infections, which lead to aseptic meningitis and encephalitis, predominantly in the very young (6, 7).
In general, EVs are highly cytolytic to their host cells, mainly due to the rapid and pronounced shutdown of host transcriptional and translational machinery, resulting in substantial inhibition of host cell metabolism (8, 9). EV infections can also persist long after the initial infection in vivo as well as in vitro. Emerging evidence suggests that EV persistence is associated with a variety of chronic diseases, such as myalgic encephalomyelitis/chronic fatigue syndrome (10), dilated cardiomyopathy (11, 12), and postpolio syndrome (PPS) (13, 14), in clinical settings. Although a number of cell models of EV persistence, including human thymic epithelial cells (15, 16), microvascular endothelial cells (17), myocardial fibroblasts (18–20), glomerular and tubular kidney cells (21), as well as murine cardiac cells (22), have been described, the mechanisms responsible for EV persistence remain to be fully addressed. It has been proposed that selection of attenuated or replication-defective variants from viral quasispecies under host immunological pressure in vivo is likely to be the most pivotal factor. For instance, CVB mutants with mutations due to 5′-terminal deletions in the genome persist in host tissues, and RNAs of such variants can be stably detected from human cases of myocarditis (9, 23). It has been observed that CVB RNA can persist for dozens of months in skeletal muscle or the CNS through the formation of a stable double-stranded RNA complex, rather than through genetic alterations in the viral genome that generate replication-defective forms (24, 25). Persistent EV infections occurring in vitro seem to result from the coevolution of both host cells and viruses. While induced cellular responses, such as mutational alterations of the receptors, can inhibit virus replication and spread (26, 27), the emergence of viral variants with enhanced infectivity counteracts the host responses described above, leading to the establishment and maintenance of persistence (22, 28).
The relative inaccessibility of the CNS to the surveillance effectors of the immune system makes it particularly vulnerable to persistent virus infection (29). Indeed, various RNA and DNA viruses of different virus families, such as measles virus (MV), human immunodeficiency virus (HIV), and herpes simplex virus (HSV) (30), can persist in the human CNS. Virological evidence indicates that EVs may also persist in the human CNS. For instance, the detection of persistent viral RNA in brain tissue or cerebrospinal fluid implicates a close association of EVs with the late onset of neurological deterioration, exemplified by the development of PPS and amyotrophic lateral sclerosis (ALS) (14, 31, 32).
Despite the significance of EVs in human neurologic illnesses (e.g., aseptic meningitis, meningoencephalitis, and encephalitis), much remains to be elucidated about their neurotropism to different CNS cell types and their potential capacity for long-lasting infection establishment. In the present study, three established human brain cell lines, CCF-STTG1, T98G, and SK-N-SH, which retained numerous physiological properties (33, 34) were employed as in vitro models. It is disclosed herein that only CCF-STTG1 human astrocytoma cells support a persistent and productive coxsackievirus B3 (CVB3) infection, an outcome not previously demonstrated for CVB3 infection in human brain cells. Analysis of CCF-STTG1 cells persistently infected with CVB3 revealed that these cells (i) continued to release infectious virions up to 60 days postinfection (p.i.), (ii) did not express the functional canonical viral receptors coxsackievirus-adenovirus receptor (CAR) and decay-accelerating factor (DAF), and (iii) continued to secrete high levels of proinflammatory chemokines and cytokines. Our findings demonstrate the potential of CCF-STTG1 cells to be a novel cell model for studying the CVB3-CNS interaction and shed light on a new avenue for investigating CVB3-induced chronic neuropathogenesis.
MATERIALS AND METHODS
Virus source, inoculation, and titer determination.
CVB3 strain AH30 was isolated from a patient with encephalitis complications who was suspected of having enterovirus infection during an outbreak of hand, foot, and mouth disease (HFMD) in Anhui Province in central China. CVB3 strain AH30 was further purified by four sequential plaque purification assays on Vero (an African green monkey kidney cell line; ATCC CCL-81) cell monolayers and confirmed by immunofluorescence assay (IFA) with anti-CVB3 monoclonal antibody (MAb). Human coxsackievirus A9 (CVA9) strain Griggs (provided courtesy of the Institute of Biomedical Engineering, Chinese Academy of Medical Sciences, and Peking Union Medical College), CVB3 strain Nancy (provided courtesy of Z. Q. Yang from Wuhan University), and CVB3 strain AH30 were grown on 80 to 90% confluent monolayers of Vero cells. For EV71 propagation, rhabdomyosarcoma (RD; a human rhabdomyosarcoma cell line; ATCC CCL-136) cell monolayers were infected with EV71 strain BrCr and EV71 strain HN2 (provided courtesy of G. H. Chang from the Beijing Institute of Microbiology and Epidemiology). Virus stocks were prepared as described previously (35). Briefly, cell cultures displaying >90% cytopathicity were further disrupted by three sequential freeze-thaw cycles between −80°C and 37°C. The cellular debris was removed by centrifugation, and cell-free supernatants were filtrated through 0.22-μm-pore-size sterile membranes. Virus titers were determined as the 50% tissue culture infectious dose (TCID50) in Vero cells using the Reed-Muench formula.
Cell culture and NPC differentiation.
Vero, SK-N-SH (a human neuroblastoma cell line; ATCC HTB-11), and T98G (a human glioblastoma cell line; ATCC CRL-1690) cells were grown in minimal essential medium (MEM). CCF-STTG1 (a human astrocytoma cell line; ATCC CRL-1718) cells were cultured in RPMI 1640 medium. RD cells were grown in Dulbecco's modified Eagle's medium (DMEM)-MEM at a ratio of 1:1. All media were supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U ml−1 penicillin, and 100 mg ml−1 streptomycin (Life Technologies, NY). Human neural progenitor cells (NPCs) were obtained from human fetal brain tissue and characterized as previously described (36). For astrocyte differentiation, progenitor medium was replaced with DMEM–F-12 medium supplemented with 10% FBS, gentamicin (50 μg/ml), and amphotericin B (1.5 μg/ml). Cultures showing >95% positivity for glial fibrillary acidic protein (GFAP) after 21 days of differentiation were used (37).
Virus infection.
For virus infection experiments, cells (1 × 106 in 2 ml medium) were seeded into 6-well plates and incubated overnight at 37°C. Cells were washed once with phosphate-buffered saline (PBS) and infected with enteroviruses at the multiplicity of infection (MOI) indicated below in serum-free medium. Inoculated cell cultures were incubated at 37°C for 1 h, before being washed twice with PBS and replenished with complete medium. Mock infections were performed in parallel for each cell line. Enterovirus infection was monitored daily by evaluating the development of a cytopathic effect. At the indicated time points, culture supernatants and cell monolayers were harvested for further analysis.
Establishment and maintenance of persistently infected cell cultures.
To establish CVB3 persistent infection in CCF-STTG1 cells, cell cultures were infected with CVB3 strain Nancy at an MOI of 1. The cultures were maintained at 37°C and subcultured for 4 days p.i. The culture medium was changed every 4 days if the cell density was not sufficient for passage. Upon subculture, equal numbers of persistently infected cells and mock-infected cells were seeded in cell culture plates for comparison analyses. Cell culture samples were collected at the time points indicated below for viable cell counting (trypan blue staining). Samples of supernatants were harvested at various times for later determination of the TCID50 and further analyses.
LDH assay.
Virus-induced cytopathic effects were assessed as described previously (38, 39). Briefly, the culture supernatants from cell samples were analyzed for the release of the cytosolic enzyme lactate dehydrogenase (LDH), by using a commercial kit (cytotoxicity detection kit; Roche, Mannheim, Germany) according to the manufacturer's instructions. The cytotoxicity index was calculated as follows: [(LDH activity in the medium of virus-infected cells − background LDH activity)/(LDH activity in the medium of cells lysed with 1% Triton X-100 − background LDH activity)] × 100.
Cell viability assay.
To determine the impact of CVB3 persistent infection on CCF-STTG1 cell viability, equal numbers of persistently infected cells harvested at the time points p.i. indicated below and mock-infected cells were seeded in 96-well plates. Cell viability was measured using a Cell Counting Kit-8 (Beyotime, Nantong, China) according to the manufacturer's instructions. The relative viability of the infected cultures was calculated as a percentage versus the viability of uninfected parallel cultures.
Determination of growth characteristics of enteroviruses in human brain-derived cell lines.
Subconfluent monolayers of CCF-STTG1, T98G, and SK-N-SH cells grown in 24-well plates were inoculated with CVB3 Nancy, CVB3 AH30, EV71 BrCr, and CVA9 Griggs at an MOI of 1. Following incubation at 37°C for 1 h, the inoculum was removed and cell cultures were rinsed twice with PBS (pH 7.4) and subsequently incubated in complete medium for the times indicated below. Culture supernatants were then harvested and stored at −80°C. The infected CCF-STTG1 cells were scraped into the same volume of medium, and the preparation was frozen and thawed three times to release the intracellular virus. The virus titers in the culture supernatant (extracellular virus) and cell-associated fraction (intracellular virus) were determined by TCID50 assay.
Immunofluorescence staining.
To detect CVB3 antigen in acutely infected cells, cell monolayers grown on glass slides were infected with CVB3 Nancy or CVB3 AH30 at an MOI of 1 for 24 h. Cells were fixed in 4% paraformaldehyde–PBS for 15 min at room temperature (RT), permeabilized with 0.2% Triton X-100–PBS for 10 min, and blocked in PBS with 3% bovine serum albumin (BSA) and 5% normal goat serum for 1 h at RT. Cells were subsequently incubated with the specific mouse anti-CVB3 MAb MAB948 (Chemicon International, CA) at RT for 45 min and washed three times with PBS, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Thermo Fisher Scientific Inc., IL) for 1 h at RT. After three washes with PBS, cell nuclei were counterstained with Hoechst 33258. For IFA to detect both the astrocyte maker GFAP and CVB3 antigen, cells harvested at the time points p.i. indicated below were seeded on glass slides. After 12 to 16 h, adherent cells were fixed and blocked as described above. The primary antibodies used were mouse anti-CVB3 MAb MAB948 and rabbit anti-GFAP polyclonal antibody (Boster, Wuhan, China). The slides were rinsed three times with PBS and incubated for 1 h with the secondary antibodies FITC-conjugated goat anti-mouse IgG and Texas Red-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific Inc.). After washes, cell nuclei were counterstained with Hoechst 33258. Cells were visualized under a PerkinElmer UltraVIEW VoX live-cell imaging system (PerkinElmer, CA), and data were obtained and processed using Volocity image analysis software.
IC assay.
An infectious center (IC) assay was used to determine the percentage of cultured CCF-STTG1 cells releasing infectious CVB3 as previously described (40). Briefly, single-cell suspensions were obtained by trypsinization, followed by incubation with CVB3-neutralizing antibody MAB948 at a titer of 1:6,000 for 1 h at RT. After three washes with PBS, 103 cells were seeded on HeLa cell monolayers. The cultures were incubated for 1 h at 37°C and then overlaid with 0.6% agar in complete medium. After 5 days, the monolayers were stained with 2% crystal violet and virus plaques were counted.
Strand-specific quantitative real-time PCR.
Primers and probe (Table 1) were synthesized by Invitrogen (Shanghai, China). To generate standard curves for strand-specific quantitative real-time PCR, CVB3-positive and -negative RNAs were transcribed in vitro from PCR products encompassing the CVB3 Nancy strain 5′ untranslated region (nucleotides 173 to 653). PCR products were synthesized from RNA extracted from a CVB3 stock solution using an EZNA viral RNA kit (Omega Bio-Tek, Inc., GA). Primer pairs Pos-Fwd/Pos-Rev and Neg-Fwd/Neg-Rev were used to obtain positive- and negative-strand amplicons, respectively, both of which contained a T7 promoter sequence. In vitro transcription and RNA isolation were performed using a MEGAscript kit (Life Technologies) according to the manufacturer's instructions. The concentration of RNA was determined by use of a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.), and the copy number of the transcript was calculated as previously described (41). cDNA synthesis of both polarities was performed with either the Tag-RP or Tag-FP primer endowed with a 5′ nonenterovirus Taq sequence to quantify the positive- or negative-strand viral RNA copies. The cDNAs used in the generation of the standard curve in the real-time PCR were stored at −20°C until use. Quantitative real-time PCR was carried out with a combination of Tag and FP primers and probe (for the positive strand) or Tag and RP primers and probe (for the negative strand) on DNA Engine Opticon real-time systems (Bio-Rad Laboratories, Inc., CA). Data acquisition and analysis were carried out with Opticon Monitor Software, version 3.1. The viral RNA copy numbers were determined from the threshold cycle values relative to those in standard curves and expressed as the number of viral copies per 1 μg cellular RNA.
Table 1.
Oligonucleotides used in this study
| Name | Nucleotide sequence (5′→3′)a | Genome positionb |
|---|---|---|
| Pos-Fwd | TAATACGACTCACTATAGGTCTGTTACCCCGGACTGAGT | 173–192 |
| Pos-Rev | GCTCTATTGGTCACCGGATG | 653–634 |
| Neg-Fwd | TAATACGACTCACTATAGGGCTCTATTGGTCACCGGATG | 653–634 |
| Neg-Rev | TCTGTTACCCCGGACTGAGT | 173–192 |
| Tag-FP | CGGTCATGGTGGCGAATAACAGACATGGTGCGAAGAGTCT | 407–427 |
| Tag-RP | CGGTCATGGTGGCGAATAAAGTAGTCGGTTCCGCTGCA | 547–529 |
| RP | AGTAGTCGGTTCCGCTGCA | 547–529 |
| FP | AGACATGGTGCGAAGAGTCT | 407–427 |
| Probe | FAM-TAGTCCTCCGGCCCCTGAATGC-TAMRA | 444–465 |
| Tag | CGGTCATGGTGGCGAATAA |
The T7 promoter sequence is shown in boldface. The nonenterovirus Taq sequence is indicated in italics. FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Relative to CVB3 strain Nancy.
CVB3-neutralizing antibody-mediated curing of persistent infection.
To determine whether CCF-STTG1 cells persistently infected with CVB3 could be cured, cell cultures (60 days p.i.) were maintained in medium containing either CVB3-neutralizing antibody MAB948 or normal mouse IgG control antibody at a titer of 1:6,000. Antibody was supplemented in medium over a period of 20 days. The cultures were then passaged in antibody-free medium. On the indicated days posttreatment, both the culture supernatant and the cell lysate were tested for infectious virus on Vero cell monolayers (24). Viral RNA was detected by a CVB3-specific nested RT-PCR as described previously (25).
Flow cytometry.
For CAR and DAF staining, cells were detached with trypsin-EDTA, followed by filtration through a 70-μm-pore-size cell strainer (BD Biosciences, NJ) to remove cell aggregates. After two washes with PBS containing 2% FBS, 105 cells were incubated with 0.5 μg anti-human DAF MAb (Biolegend, CA) or anti-CAR MAb clone RmcB (Millipore, MA) for 30 min at 4°C. Cells were subsequently washed twice with PBS containing 2% FBS and exposed to 0.5 μg of FITC goat anti-mouse IgG (minimal cross-reactivity) antibody (Biolegend, CA) for 30 min at 4°C in the dark. After two washes with PBS containing 2% FBS and 0.09% Na azide, cells were incubated with 1 μg/ml propidium iodide (Sigma, NY) for 10 min at 4°C to determine the proportion of living cells. Cells were then subjected to flow cytometry using a FACSDiva flow cytometer (Becton, Dickinson, NJ). A single cell population was identified by light scatter properties (forward scatter area [FSC-A] versus side scatter area [SSC-A]) on the P1 gate, followed by an FSC-A/propidium iodide area (PI-A) combined plot as the P2 gate to exclude dead cells. The P3 gate for FITC-A sorting was based on the fluorescence intensity of the isotype control [purified mouse IgG1(κ) isotype control antibody; Biolegend].
Antibody-mediated inhibition of CVB3 replication.
To determine the amount of antibodies for effective blocking to be used in the assay, permissive HeLa cells in 96-well plates (7.5 × 103 cells/well) were preincubated with anti-DAF or anti-CAR antibodies at concentrations ranging from 0.78 to 50 μg/ml of 2-fold serial dilutions or with a combination of both antibodies for 1 h at RT, followed by infection with CVB3 at an MOI of 1. Antibody blockade of CVB3-mediated cell lysis was monitored as previously described (42), and total inhibitory effects were observed with the combination of anti-DAF and anti-CAR antibodies at a concentration of 25 μg/ml. CCF-STTG1 cells were seeded into 96-well plates and incubated with anti-DAF antibody (25 μg/ml) or anti-CAR antibody (25 μg/ml), followed by infection with CVB3 Nancy or CVB3 AH30 at an MOI of 1. The presence of antibodies in the cell culture medium was maintained throughout the experiment thereafter. Viral titers were determined at the times indicated below to measure the efficiency of CVB3 replication in host cells.
Enzymatic removal of heparan sulfate (HS) and chondroitin sulfate (CS) from the surface of CCF-STTG1 cells.
Heparinase III (HSase; EC 4.2.2.8) and chondroitinase ABC (CSase; EC 4.2.2.4) (Sigma) were solubilized in resuspension buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, 4 mM CaCl2, and 0.01% bovine serum albumin (BSA). CCF-STTG1 cells were seeded in 96-well plates at 7.5 × 103 cells/well overnight, washed once with digestion buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 4 mM CaCl2, 0.1% BSA), and treated with heparinase III and chondroitinase ABC as previously described (43). Cells were then washed and inoculated with CVB3 at an MOI of 0.1 for 60 min in 50 μl of Opti-MEM I medium at 37°C in 5% CO2. The viral inoculum was then washed off, and the cells were incubated for the times indicated below. Thereafter, the presence of enzymes in the cell culture medium was maintained throughout the experiment. Viral titers at the times indicated below were determined to measure the efficiency of CVB3 replication in host cells.
CVB3 RNA transfection.
The DNA template for T7 RNA transcription was amplified by PCR using the full-length CVB3 cDNA as the template. The T7 promoter sequence and a poly(A) tail (44, 45) were added to the 5′ and 3′ ends of the DNA segment, respectively. In vitro transcription was carried out to generate viral genomic RNA using a MEGAscript kit. After lithium chloride (LiCl) precipitation, the RNA was quantified by spectrophotometry and stored at −80°C until use. CCF-STTG1 cells were transfected with viral genomic RNA using Lipofectin and Opti-MEM I (Life Technologies), as previously described (46). The presence of CVB3 in these cells and the corresponding supernatants was determined at the time points indicated below.
IFN-β or poly(I·C) treatment.
CCF-STTG1 cells (7.5 × 103 cells/well) were seeded into 96-well plates, followed by inoculation with CVB3 Nancy or CVB3 AH30 at an MOI of 1 for 6 h. Then, the cells were cultured with recombinant human beta interferon (IFN-β; 2,000 IU/ml; Millipore) or poly(I·C) (500 ng/ml; InvivoGen, CA) for an additional 36 h. Uninfected and CVB3-infected cells were set as negative and positive controls, respectively. Cells incubated with IFN-β or poly(I·C) alone were used to measure the direct cytotoxicity of each treatment. Titers of virus released with each treatment were determined at the time points indicated below, as described above. To investigate the long-term impact of IFN-β on CVB3 persistence, CCF-STTG1 cell cultures (60 days p.i.) were maintained in medium containing 20, 200, or 2,000 IU/ml of IFN-β at the times indicated below. Supernatants were collected, and virus production was titrated by the TCID50 assay.
ELISAs.
The concentrations of IFN-β, vascular cell adhesion molecule 1 (VCAM-1), interleukin-6 (IL-6), IL-8, and monocyte chemotactic protein 1 (MCP-1) in the supernatants of virus-infected cells were measured using commercialized enzyme-linked immunosorbent assay (ELISA) kits (PBL Biomedical Laboratories, NJ, or Boster). Briefly, cells were infected with CVB3, EV71, or CVA9 as described above. IFN-β production in poly(I·C)-treated cells was measured as a positive control. The specificity of virus-induced chemokine or cytokine release was ascertained using a mock heat-inactivated virus suspension. For kinetic assays, supernatant samples of the cell culture were collected at the time points indicated below and processed as recommended by the manufacturer. The absorbance at 450 nm was recorded for each well. The target protein levels were determined by plotting the optical densities (ODs) using a 4-parameter fit for the standard curve (Gen5 data analysis software; BioTek).
Nucleotide sequence accession number.
The nucleotide acid sequence of CVB3 AH30 was deposited in the GenBank database (accession number KC481610).
RESULTS
Susceptibility of human brain-derived cell lines to enterovirus infection.
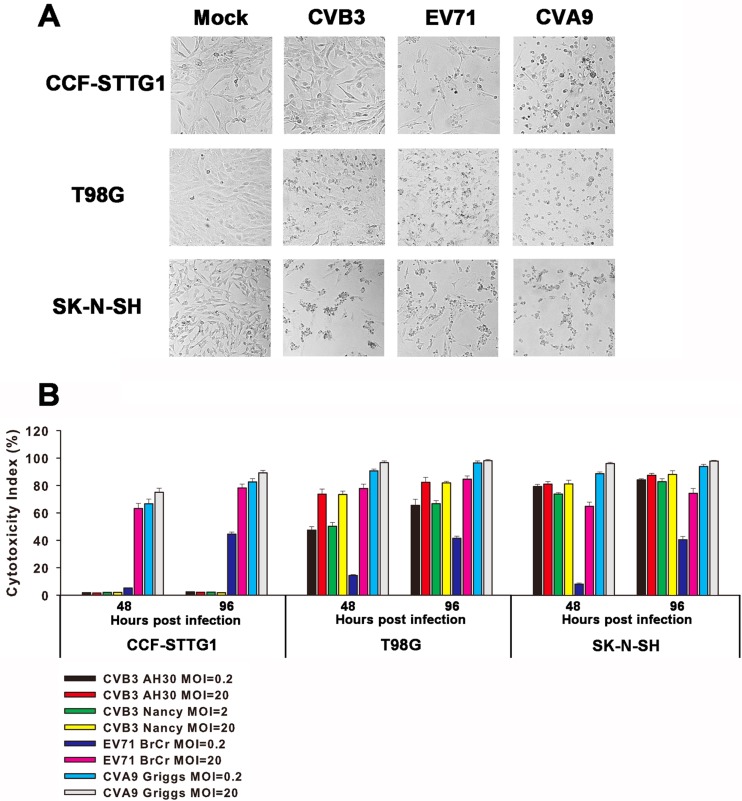
To investigate the susceptibility of human brain-derived cell lines to enteroviruses in vitro, SK-N-SH, CCF-STTG1, and T98G cells were infected with CVB3, EV71, and CVA9. At an MOI of 5, visible cytopathic effects, such as cell rounding and detachment, were observed at 48 h p.i. in all three cell lines infected with EV71 or CVA9. As for CVB3 infection, typical cytopathic effects were observed for SK-N-SH and T98G cells as early as 48 h p.i., but little, if any, cytopathicity was observed for CCF-STTG1 cells up to 7 days p.i. (Fig. 1A).
Fig 1.
Effects of enterovirus acute infection on human brain-derived cell lines. (A) Effects of enteroviruses on cell morphology. CCF-STTG1, T98G, and SK-N-SH cells were infected with CVB3, EV71, and CVA9 at an MOI of 5 for 48 h. Photographs were taken under a phase-contrast microscope. The results of one representative experiment out of three are shown. Magnification, ×100. (B) Enterovirus infection-induced cytotoxicity. Cell lines were inoculated with enteroviruses at an MOI of 0.2 or 20 for the indicated times, and the culture supernatants were assayed for the release of the cytosolic enzyme LDH to measure cytotoxicity. The results are presented as the means ± standard deviations of triplicate measurements.
To quantify the cell damage induced by enterovirus infection, culture supernatants were analyzed for the release of the cytosolic enzyme LDH to measure cytotoxicity. As shown in Fig. 1B, EV71 or CVA9 infection led to a significant increase in cytotoxicity in all three cell lines. CVB3 infection also remarkably induced cell damage in SK-N-SH or T98G cells but did not trigger significant cytotoxicity in CCF-STTG1 cell monolayers. In addition, two CVB3 strains of different origins showed almost identical results, indicating that the impact of CVB3 infection on CCF-STTG1 cells was not strain specific.
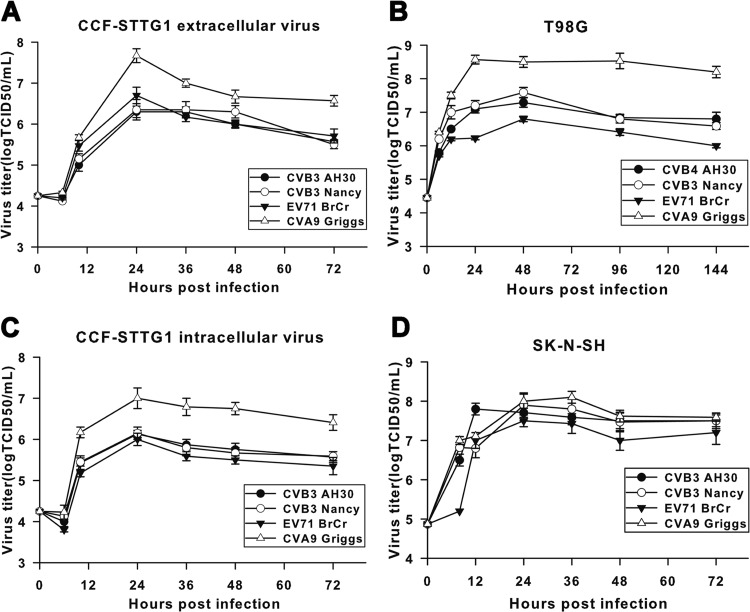
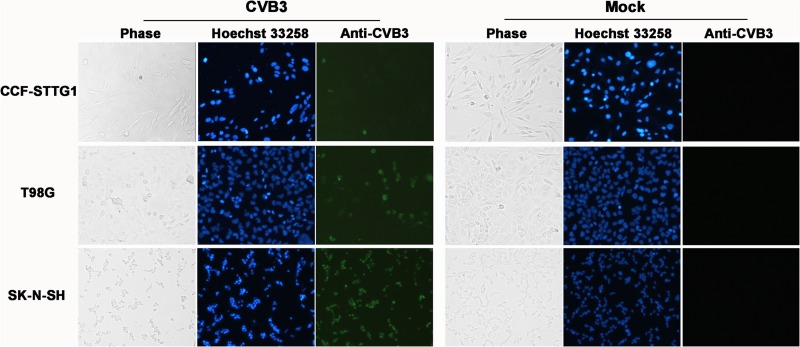
We next sought to determine the kinetics of enterovirus replication in the three cell lines described above. All three enteroviruses tested replicated efficiently in CCF-STTG1 (Fig. 2A), T98G (Fig. 2B), and SK-N-SH (Fig. 2D) cells, as indicated by a rapid increase of virus titers at early stages of infection. The highest titers were detected after 24 to 48 h p.i., followed by the plateau effect, with a slow decrease for the remainder of the infection. Despite the quite different susceptibilities of CCF-STTG1 cells to EV71 and CVB3 (see above), the infectious virus production kinetics of both extracellular (Fig. 2A) and intracellular (Fig. 2C) CVB3 was not markedly different from that of EV71, and virus titers at various times were of approximately the same order of magnitude. In addition, the ratios of maximum intracellular to extracellular virus for CCF-STTG1 cells were approximately the same for the three enteroviruses tested (data not shown), suggesting that virion synthesis and liberation were not defective for CVB3 in CCF-STTG1 cells. However, compared with SK-N-SH and T98G cells, a relatively lower level of increase in virus abundance was detected CCF-STTG1 cell cultures. This suggests that CVB3 exhibited a host cell-specific, lower replication rate in CCF-STTG1 cells than SK-N-SH and T98G cells. Moreover, as indicated in Fig. 3, only a small proportion of CCF-STTG1 cells (about 5%) was found to be viral antigen positive at 24 h postinoculation by IFA. On the contrary, both SK-N-SH and T98G cells were infected at a higher rate with the same MOI of 1 (Fig. 3), and this high rate of infection presumably contributes to high yields of infectious virus progeny and predisposition to acute cytocidal effects. Taken together, these data demonstrate that CCF-STTG1 human astrocytoma cells support CVB3 replication but do not display significant cytolytic changes usually associated with enterovirus infection in vitro.
Fig 2.
Replication kinetics of enteroviruses in human brain-derived cell lines. Subconfluent monolayers of CCF-STTG1, T98G, and SK-N-SH cells were infected with enteroviruses at an MOI of 1 in 24-well plates. At various times after infection, the virus titers in the culture supernatant (extracellular virus) for all three cell lines tested and the cell-associated fraction (intracellular virus) for CCF-STTG1 cells were determined by the TCID50 assay. The results are presented as the means ± standard deviations obtained from three independent experiments.
Fig 3.
Detection of CVB3 antigens by IFA in human brain-derived cell lines. Cells were infected with CVB3 Nancy or CVB3 AH30 at an MOI of 1 for 24 h. CVB3 antigen (green) was detected by IFA using the specific mouse anti-CVB3 MAb MAB948, followed by FITC-conjugated goat anti-mouse secondary antibody. Nuclei were counterstained with Hoechst 33258 (blue). Magnification, ×100. The results of one representative experiment out of three are shown.
Persistent CVB3 infection of CCF-STTG1 cells.
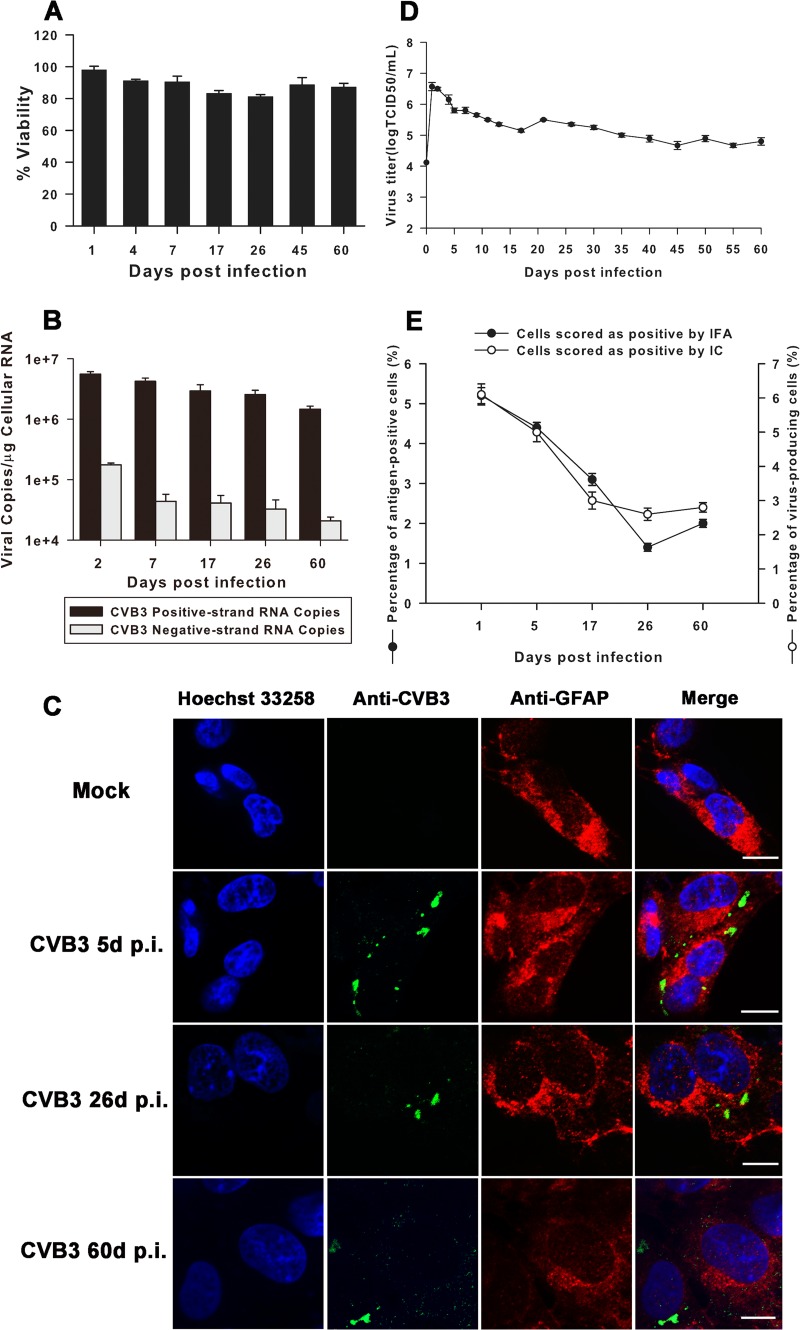
After confirmation of productive and noncytopathic CVB3 infection in CCF-STTG1 cells, we focused our investigation on determining whether CVB3 could establish a long-standing persistent infection in this cell line. CCF-STTG1 cells were infected with CVB3 Nancy at an MOI of 1, and cell morphology was monitored daily for 60 days under an inverted microscope. CCF-STTG1 cells kept growing as compact and adherent monolayers for up to 60 days p.i. with no significant cytopathic effect (data not shown). The viabilities of CVB3-infected CCF-STTG1 cells presented at steady-state levels ranging from approximately 81% to 97% over all time points p.i., comparable to those of the corresponding mock-infected controls (Fig. 4A). These data indicate that the vast majority of CVB3-infected CCF-STTG1 cells escape from cell lysis and remain intact and viable up to 60 days p.i.
Fig 4.
Human astrocytoma cell line CCF-STTG1 supports persistent CVB3 infection. (A) Impact of CVB3 persistent infection on viability of CCF-STTG1 cells. Equal numbers of cells persistently infected for the indicated times p.i. and mock-infected cells were seeded in 96-well plates. Relative cell viability was measured with a Cell Counting Kit-8 using the corresponding uninfected cells (mock) as controls (for which viability was set equal to 100%). The results are presented as the means ± standard deviations of triplicate measurements. (B) Persistence of both positive- and negative-strand CVB3 RNA in CCF-STTG1 cells. The levels of viral RNA replication dynamics of both positive and negative polarity were determined in persistently infected CCF-STTG1 cells by strand-specific real-time PCR. The results are presented as the means ± standard deviations of triplicate measurements. (C) Detection of viral antigen (green) and GFAP (red) in CCF-STTG1 cells persistently infected with CVB3 by IFA. Cell nuclei (blue) were counterstained with Hoechst 33258. Bars, 10 μm. The results of one representative experiment out of three are shown. (D) CCF-STTG1 cells persistently infected with CVB3 showed continuous detectable levels of infectious virus progeny ranging from 104.8 to 106.6 TCID50s/ml. Means and standard deviations of three independent experiments are shown. (E) The percentage of CVB3-infected CCF-STTG1 cells was determined by both IFA and IC assay at the indicated times p.i. The results are presented as the means ± standard deviations of triplicate measurements.
Next, the copy numbers of both positive- and negative-strand viral RNA present during acute and persistent infection phases were determined using strand-specific quantitative real-time PCR. The assay sensitivity ranged from 102 to 109 copies. Using the standard curves, the viral copy number/μg of cellular RNA was calculated for each sample. Overall, the kinetic profiles of positive- and negative-strand viral RNA proved to be similar. As shown in Fig. 4B, the largest amounts of both polarities were observed at day 2 p.i., with 5.57 × 106 ± 0.55 × 106 and 1.76 × 105 ± 0.13 ×105 copies per μg cellular RNA detected, respectively. Thereafter, the copy numbers of both strands gradually decreased over the following four time points, and by day 60 p.i. the levels of positive- and negative-strand RNA were approximately 3- and 8-fold lower, respectively, than they were at day 2 p.i. Restricted replication of CVB3 RNA is likely to be a hallmark of establishing long-standing infection, and perturbation of virus RNA synthesis probably contributes to the conversion of CVB3 infection from acute to persistent in CCF-STTG1 cells.
CVB3-infected CCF-STTG1 cells were costained with antibodies against CVB3 and the astrocyte-specific marker GFAP at various days p.i. Both green signals (indicating the presence of CVB3 antigen) and red signals (indicating the presence of GFAP) were recognized in the cytoplasm, confirming that persistent CVB3 infection did not alter the astrocyte lineage of CCF-STTG1 cells (Fig. 4C). In addition to the long-term presence of CVB3 antigen and viral RNA, infectious particles were also detected in CVB3-infected CCF-STTG1 cells, as determined by titration on Vero cells after establishing virus persistence (Fig. 4D). The highest virus titer (106.6 TCID50s/ml) was found at day 1 p.i., and virus production decreased thereafter but remained at relatively constant levels with moderate fluctuations up to 60 days p.i., suggesting that CVB3 persistently replicated in CCF-STTG1 cells. As shown in Fig. 4E, the proportions of CVB3 antigen-positive CCF-STTG1 cells were quite stable: about 5% at day 1 p.i. in the acute phase of infection and 1% to 3% further in the persistent phase thereafter. Similar results were obtained by the infectious center (IC) assay. About 6% of CCF-STTG1 cells scored positive at day 1 p.i., and the percentage of virus-producing cells remained at a level of about 3% in the persistent phase. Taken together, these data indicate that CVB3 can readily establish persistent infection in CCF-STTG1 cells.
To evaluate whether persistently infected CCF-STTG1 cells could be cured, anti-CVB3 neutralizing antibody was added to cultures at the beginning of 60 days p.i. By the time that the cells were cultured for 8 days, the cell-free supernatant and cell lysate were negative for both infectious virus and viral RNA. Continued culturing in the presence of anti-CVB3 antibody resulted in the cure of viral persistence (Table 2). We did not detect virus or viral RNA after completion of antibody treatment (data not shown).
Table 2.
CCF-STTG1 cells are cured of persistent infection following anti-CVB3 treatment
| Treatment | Detection of CPEa/viral RNA on the following days posttreatment: |
||||
|---|---|---|---|---|---|
| 4 | 8 | 12 | 16 | 20 | |
| Anti-CVB3 antibody | +/+ | −/− | −/− | −/− | −/− |
| Mouse IgG antibody | +/+ | +/+ | +/+ | +/+ | +/+ |
CPE, cytopathic effect.
CAR and DAF are not involved in CVB3 infection of CCF-STTG1 cells.
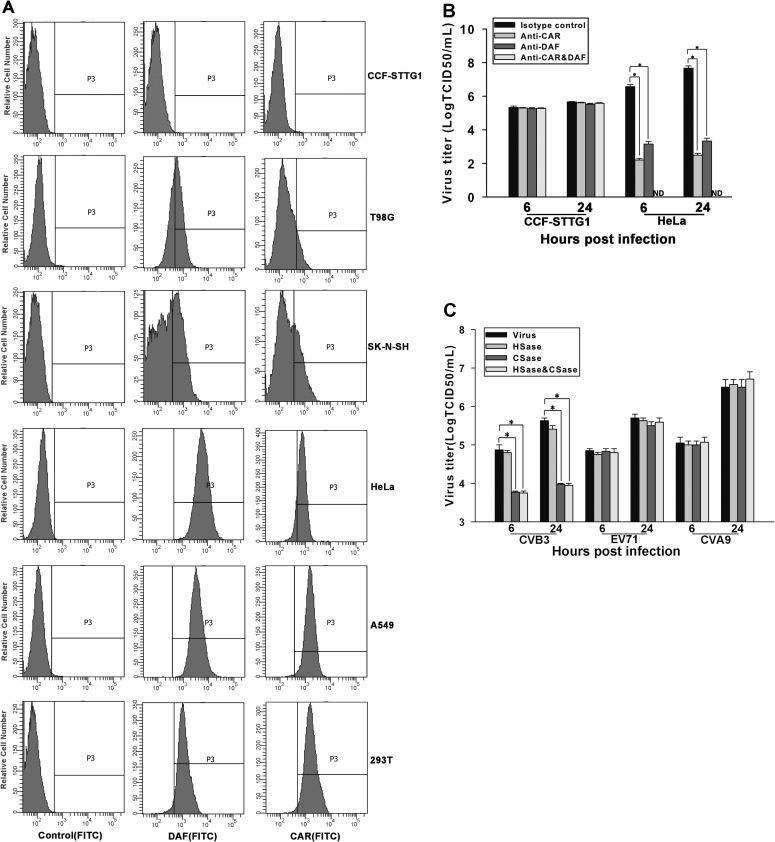
The restricted expression of virus receptor on the cell surface has been shown to be an important cellular determinant of viral persistence in cultured cells (22, 47). Therefore, we intended to explore whether persistent infection of CVB3 in CCF-STTG1 cells was due to a low abundance of the CVB3 receptors CAR and DAF. The expression levels of CAR and DAF in six cell lines, CCF-STTG1, T98G, SK-N-SH, HeLa, A549, and 293T, were measured by flow cytometry analysis. Overall, CCF-STTG1, T98G, and SK-N-SH cells expressed remarkably lower levels of both CAR and DAF than HeLa, A549, and 293T cells (Fig. 5A). It is worth noting that the CAR and DAF expression levels in CCF-STTG1 cells, with regard to both the percentage of positive cells and the relative receptor density, were close to those of control cells. Therefore, it is reasonable to conclude that CCF-STTG1 cells do not express CAR and DAF or express very low levels of these two receptors below the detection limit of flow cytometry analysis.
Fig 5.
CAR and DAF are not involved in CVB3 infection of CCF-STTG1 cells. (A) CAR and DAF expression in various cell lines. The levels of expression of CAR and DAF on the cell surface were analyzed using flow cytometry. (Left) Fluorescence with isotype antibody as a control; (middle and right) expression of CAR and DAF, respectively. Representative histograms are shown. (B) CVB3 replication in CCF-STTG1 cells in the presence of anti-CAR or anti-DAF antibodies. CCF-STTG1 and HeLa cells were incubated with anti-CAR antibody or anti-DAF antibody, or with both, followed by inoculation with CVB3. Virus titers in culture supernatants were determined at the indicated times p.i. The means of three independent experiments are shown, and error bars represent the standard deviations. ND, not detectable. (C) Inhibition of CVB3 replication in CCF-STTG1 cells by treatment with CSase. CCF-STTG1 cells were treated with chondroitinase ABC or with heparinase III, or with both, prior to inoculation of CVB3, EV71, or CVA9. Virus titers were measured as described for panel B. Bars represent the means ± the standard deviations of three independent experiments. Asterisks, significant differences between groups (P < 0.01, determined by Student's t test).
By antibody-blocking studies, we next sought to determine whether CAR and/or DAF is required for CVB3 infection of CCF-STTG1 cells. CVB3 replicated efficiently in CCF-STTG1 cells pretreated with anti-CAR or anti-DAF antibodies alone or in combination. However, the replication of CVB3 was completely blocked with the combination of both antibodies or was significantly reduced with the presence of each antibody in HeLa cells (Fig. 5B). These results indicate that CVB3 infection of CCF-STTG1 cells is mediated by interactions between the virus and cell surface molecules other than CAR and DAF.
To investigate the possible role of cell surface glycosaminoglycans (GAGs) in CVB3 infection of CCF-STTG1 cells, HS and CS was enzymatically cleaved with heparinase (HSase) and chondroitinase (CSase), respectively. HSase treatment induced a less than 2-fold reduction in virus titers over all time points of infection, whereas CSase treatment or combination HSase and CSase treatment caused a sharp decrease in virus titers by 15- to 20-fold at 24 h p.i. However, neither CSase nor HSase significantly inhibited or enhanced EV71 and CVA9 replication in CCF-STTG1 cells (Fig. 5C) or CVB3 infection in HeLa cells (data not shown). These results implicate that CS is important for CVB3 infection of CCF-STTG1 cells and that alternative receptor usage probably confers a low rate of infection for CVB3, which is one of the limiting factors for establishing virus persistence.
Transfection of CVB3 genomic RNA results in significant cell death in CCF-STTG1 cell monolayers.
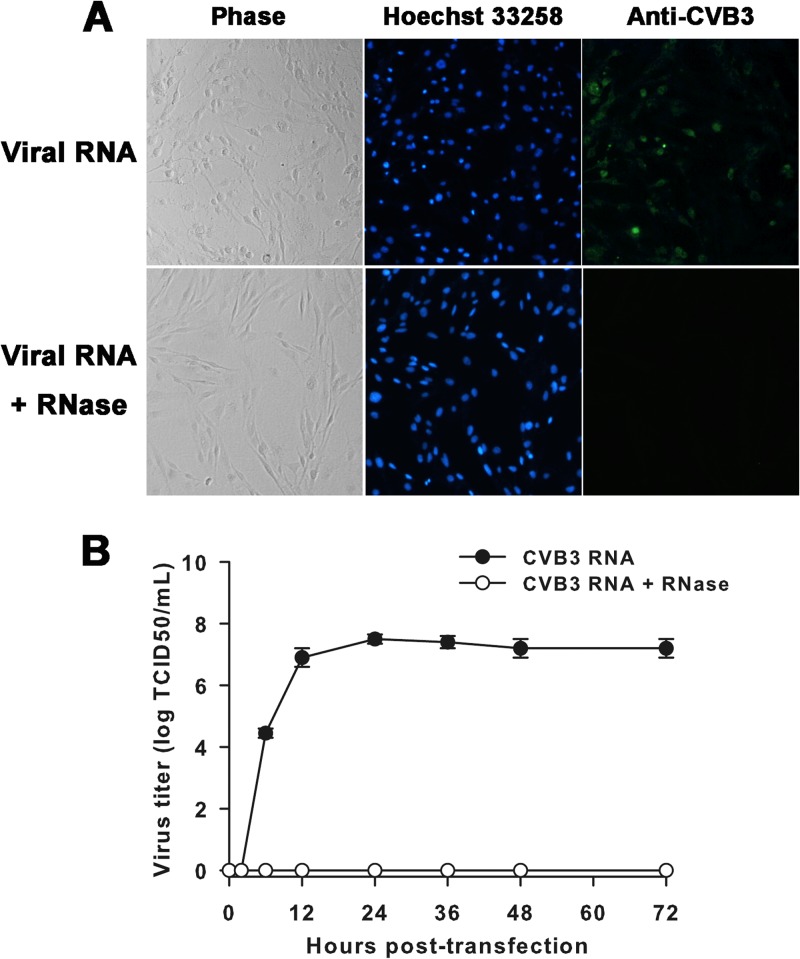
To test the hypothesis that abated entry of CVB3 in CCF-STTG1 cells was a critical element for regulating persistence, we examined the impact of virus infection on cells by direct genomic RNA transfection, bypassing the surface receptor-mediated restriction. Following transfection of CCF-STTG1 cells, high percentages of cells (40%) were found to be CVB3 antigen positive by IFA, and an extensive cytopathic effect was observed in the cultures on day 1 (Fig. 6A). Significant levels of virus replication were also evident, with infectious virus detected in the culture medium as early as 6 h after transfection, and then the levels increased, reaching a maximum of 107.5 TCID50s/ml by 24 h posttransfection. In contrast, the infectivity of synthetic RNA was lost upon treatment with RNase, and no infectious virus progeny could be identified at any time point (Fig. 6B).
Fig 6.
Cell morphology change, viral antigen detection, and virus yield following RNA transfection. (A) CCF-STTG1 cells were transfected with CVB3 genomic RNA or RNA upon treatment with RNase. An extensive cytopathic effect was observed in the cultures on day 1 following RNA transfection, and high percentages of cells (40%) were shown to be CVB3 antigen positive (green) by IFA. Nuclei were counterstained with Hoechst 33258 (blue). Magnification, ×100. The results of one representative experiment out of three are shown. (B) At the indicated times posttransfection, samples of supernatant were harvested and virus titers were determined. The means and standard deviations of three independent experiments are shown.
Next, we determined whether the virus progeny produced by RNA transfection could induce a significant cytopathic effect in naive CCF-STTG1 cells. Though maximal yields of about 106 TCID50s/ml of infectious particles were detected in the supernatant, no significant cytopathic effect was observed within 7 days p.i. Of note, about 5% of cells were found to be viral antigen positive (data not shown). Taken together, these data implicate that the initiation of CVB3 persistence is somewhat dependent on the inherent early blockage of virus entry by naive CCF-STTG1 cells.
CVB3 infection blunts IFN-β release in CCF-STTG1 cells.
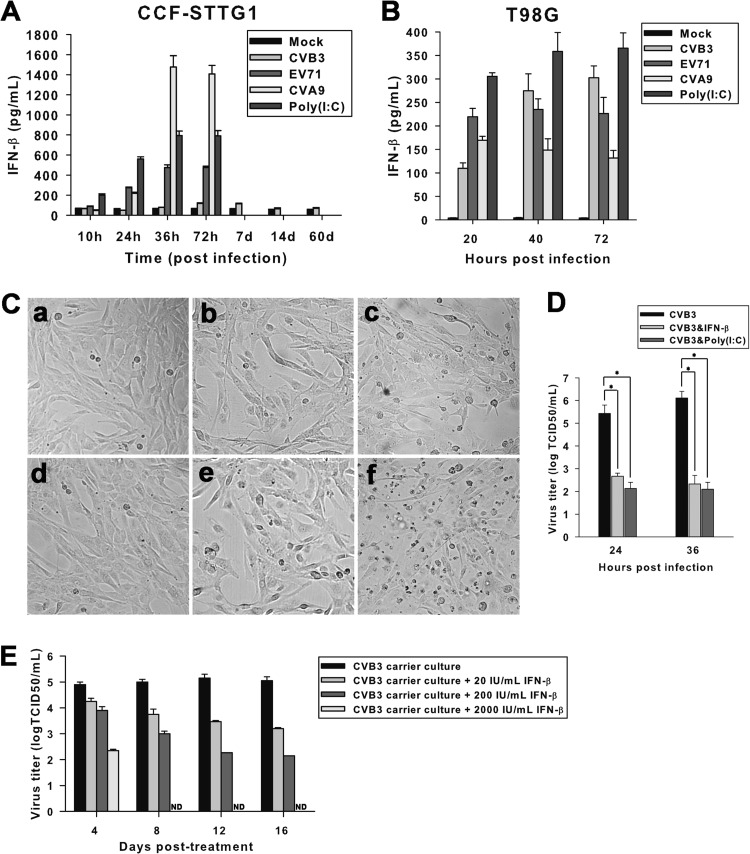
Production of innate antiviral mediators by host cells may contribute to the initiation of CVB persistence (48). To investigate the possible role of IFN in persistent CVB3 infection of CCF-STTG1 cells, the expression kinetics of IFN-β was determined by ELISA. Superinduction with the double-stranded RNA mimic poly(I·C) demonstrated that both CCF-STTG1 and T98G cells were capable of expressing IFN-β. Both EV71 and CVA9 infections effectively induced IFN-β protein production in CCF-STTG1 cells. However, little or no IFN-β induction was observed in culture supernatants of CVB3-infected CCF-STTG1 cells either early after infection or during CVB3 persistence (Fig. 7A). In contrast, almost equal efficiencies of IFN-β induction were observed in CVB3-infected T98G cells and EV71- or CVA9-infected control cells, suggesting that the CVB3-induced blunted expression of IFN-β in CCF-STTG1 cells is likely to be a cell type-specific response (Fig. 7B).
Fig 7.
CVB3 curtails host IFN-β induction in CCF-STTG1 cells. CCF-STTG1 (A) and T98G (B) cells were infected with CVB3, EV71, or CVA9 for the indicated times. The expression kinetics of IFN-β in cell culture supernatants were measured by ELISA. IFN-β production in poly(I·C)-treated cells was measured as a positive control. (C) Exogenous addition of IFN-β or poly(I·C) induces cytopathicity in CCF-STTG1 cells acutely infected with CVB3. CCF-STTG1 cells inoculated with CVB3 Nancy or CVB3 AH30 for 6 h were cultured with IFN-β (e) or poly(I·C) (f) for an additional 36 h. Uninfected (a) and CVB3-infected (d) cells were set as negative and positive controls, respectively. Cells incubated with only IFN-β (b) or poly(I·C) (c) were used to measure the direct cytotoxicity of each treatment. Photographs were taken under a phase-contrast microscope. Magnification, ×100. The results of one representative experiment out of three are shown. (D) Effect of IFN-β or poly(I·C) treatment on virus yields of CCF-STTG1 cells acutely infected with CVB3. CVB3-infected cells were treated with IFN-β or poly(I·C), and 24 or 36 h later, samples of medium were retrieved and viral production was determined by the TCID50 assay. Asterisks, significant differences between groups (P < 0.01, determined by Student's t test). (E) CVB3 titers in persistently infected CCF-STTG1 cells treated with recombinant human IFN-β. Bars represent the means ± standard deviations of three independent experiments. ND, not detectable.
To get a glance into the role of CVB3-induced attenuation of IFN-β release in persistent infection, CCF-STTG1 cells inoculated with CVB3 Nancy or CVB3 AH30 were cocultured with IFN-β or poly(I·C). Similar to the observations described above, CVB3 inoculation alone induced no obvious alteration in cell morphology (Fig. 7Cd) compared to that for the mock-infected control (Fig. 7Ca). Treatment of CCF-STTG1 cells with IFN-β (Fig. 7Cb) or poly(I·C) (Fig. 7Cc) alone elicited a modest impact on cell morphology, causing sporadic cell death. However, CVB3 inoculation plus treatment with IFN-β (Fig. 7Ce) or poly(I·C) (Fig. 7Cf) augmented the effect of cell death, with cells exhibiting signs of clustered cell shrinkage and detachment. To extend our analysis, virus titers were measured in CVB3-infected and IFN-β- or poly(I·C)-treated CCF-STTG1 cells at acute phase. As shown in Fig. 7D, application of IFN-β or poly(I·C) to the CVB3-infected cell culture resulted in a substantial reduction in the production of infectious virus progeny. The long-term effect of exogenous IFN-β treatment on CCF-STTG1 cell cultures persistently infected with CVB3 (60 days p.i.) was determined (Fig. 7E). Compared to untreated control CVB3-infected carrier cells, a consistently dose-dependent reduction of virus titers in culture supernatants was observed with treatment with IFN-β. Moreover, the 2,000-IU/ml IFN-β regimen was able to eliminate infectious virus as early as 8 days posttreatment, and cell lysates were negative for viral RNA thereafter (data not shown). The observations presented above demonstrate that curtailing of IFN-β release by CVB3 contributes to both the initiation and maintenance of persistent infection in CCF-STTG1 cells.
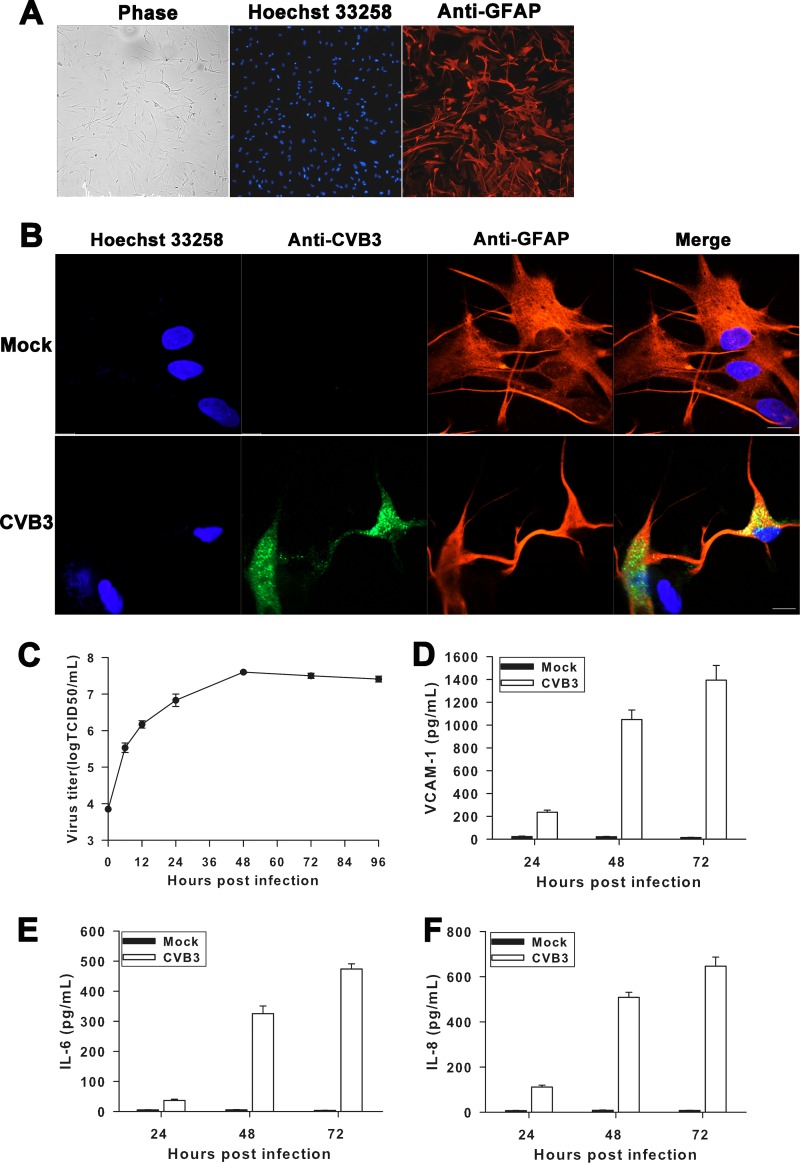
Persistent CVB3 infection modulates secretion of proinflammatory chemokines and cytokines in CCF-STTG1 cells.
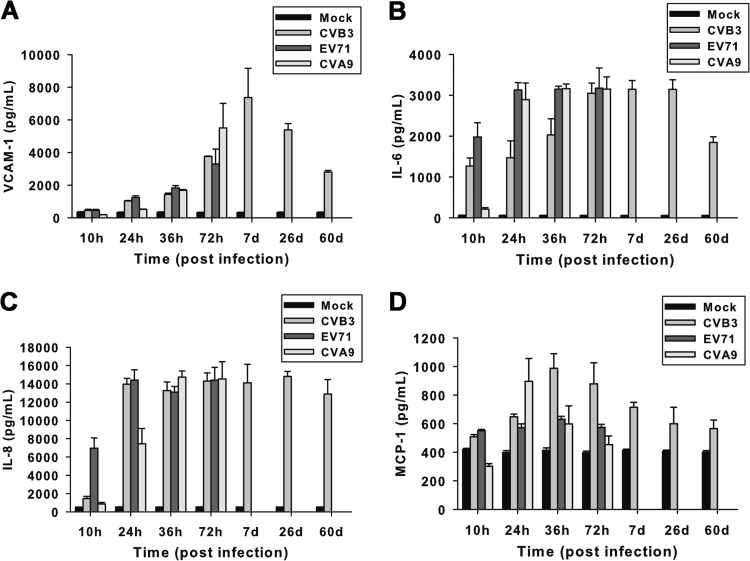
The levels of VCAM-1, IL-6, IL-8, and MCP-1 in the supernatants of virus-infected cell cultures were measured in this study to elucidate their potential roles as mediators of cytotoxic effects, maintenance of infection, and disease progression. CCF-STTG1 cells were infected with CVB3, EV71, or CVA9 for various times at an MOI of 1. The release kinetics of VCAM-1, IL-6, IL-8, and MCP-1 (Fig. 8A to D) in cell culture supernatants were measured by ELISA. CVB3 infection induced an increased production of VCAM-1, IL-6, and IL-8 up to 72 h p.i., as did lytic infection with EV71 or CVA9. Prolonged high levels of VCAM-1, IL-6, and IL-8 proteins were also detected in culture supernatants of CCF-STTG1 cells persistently infected with CVB3 up to 60 days p.i. Moreover, a modestly increased level of MCP-1 relative to that in mock-infected cultures was detected in CVB3-infected CCF-STTG1 cells at 36 h p.i., and thereafter, the production of MCP-1 decreased progressively.
Fig 8.
Chronic production of chemokines and cytokines in CCF-STTG1 cells persistently infected with CVB3. CCF-STTG1 cells were infected with CVB3, EV71, or CVA9 at an MOI of 1 for the indicated times. The release kinetics of VCAM-1 (A), IL-6 (B), IL-8 (C), and MCP-1 (D) in cell culture supernatants were measured by ELISA. Bars represent the means ± standard deviations of three independent experiments.
CVB3 infection induces secretion of chemokine and cytokine proteins in human progenitor-derived astrocytes.
Further experiments were conducted using human progenitor-derived astrocytes (HPDAs) as a model to examine whether CVB3 infects primary astrocytes and induces chemokine and cytokine expression. By 21 days after serum induction, astrocytic differentiation of progenitors resulted in staining for the astrocytic marker GFAP by more than 95% of the astrocytes, which showed a large, flattened, and pleomorphic morphology (Fig. 9A), as described previously (37, 49). The ability of CVB3 to infect HPDAs was assayed at an MOI of 0.01, and susceptibility was demonstrated by viral antigen expression. By 24 h after exposure to CVB3, approximately 30% of HPDAs showed signs of infection, indicating high CVB3 susceptibility. Anti-CVB3 antigen reactivity was clearly localized in the cytoplasm of cells which were intensely stained for GFAP, indicating that the infected cells retained their astrocytic phenotype (Fig. 9B). The kinetics of CVB3 replication in HPDAs is illustrated in Fig. 9C. HPDAs infected with CVB3 showed an increase in infectious progeny virus, with a peak in virus titer of 107.6 TCID50s/ml at day 2 p.i. and then with a plateau at between approximately 107.4 and 107.5 TCID50s/ml for the remaining observation period (day 4). To test whether CVB3 infection would lead to the synthesis and secretion of proinflammatory chemokines and cytokines, HPDAs were infected with CVB3 and secretion of VCAM-1, IL-6, and IL-8 was detected at various times p.i. using ELISA. As indicated in Fig. 9D to F, all three proteins tested were efficiently synthesized and secreted into the culture supernatants of CVB3-infected HPDAs, compared with the findings for mock-infected cultures. The pattern of induction was virtually identical to that seen in CVB3-infected CCF-STTG1 cells, reinforcing the potential role of CVB3 infection in modulating neuroinflammatory responses.
Fig 9.
Induction of chemokines and cytokines in CVB3-infected human progenitor-derived astrocytes. (A) Human neural progenitor cells differentiated into astrocytes and immunostained with antibodies against GFAP (red), with nuclei stained with Hoechst 33258 (blue). Magnification, ×100. The results of one representative experiment out of three are shown. (B) Detection of CVB3 antigen in HPDAs by double IFA for CVB3 antigen (green) and GFAP (red) on HPDAs at 24 h p.i. with CVB3. Nuclei were counterstained with Hoechst 33258 (blue). Bars, 20 μm. (C) Production of infectious CVB3 by HPDAs. HPDAs were infected with CVB3 at an MOI of 0.01, and the production of infectious virus particles was tested by the TCID50 assay at the indicated times p.i. Bars represent the means ± standard deviations of three independent experiments. (D to F) Expression of chemokine and cytokine proteins by CVB3-infected HPDAs. HPDAs were infected with CVB3 at an MOI of 0.01, and supernatants were collected and analyzed for the expression of VCAM-1, IL-6, and IL-8 by ELISA. Bars represent the means ± the standard deviations of three independent experiments.
DISCUSSION
Neurologic illnesses, including those occurring in the context of HFMD outbreaks, predominate among the clinical manifestations of EV71 and CVA9 infections (50). In addition, CVB3-associated aseptic meningitis has also become an emerging concern of public health in recent years (51). Using in vitro cell culture systems, we investigated the potential tropisms of CVB3, EV71, and CVA9 in human brain-derived cell lines of three different origins. Our results demonstrated that SK-N-SH, CCF-STTG1, and T98G cells were permissive to infection by CVB3, CVA9, and EV71, indicating the broad host range of these three enteroviruses. However, these viruses showed distinct efficiencies of replication which correlated with the severity of cytopathicity, virus one-step growth curves, and their impacts on cellular viability.
To date, limited data are available concerning the susceptibility of human CNS cell lines to CVA9 infection. Our results showed that, compared to CVB3 and EV71, CVA9 replicated more aggressively and quickly in all three tested cell lines. In addition, CVA9 infection was the most destructive to infected cells. Although they produced lower levels of infectious virus progeny than CVA9, EV71 strains BrCr and HN2 (data not shown) were also capable of inducing acute cytopathic effects in all three brain-derived cell lines, which was consistent with the findings of previous studies (52, 53). Infection of T98G or SK-N-SH cells by CVB3 was productive and cytopathic, suggesting the fully permissive nature of these two cell lines during acute cytolytic virus infection. These observations support the notion that common EV infections are able to trigger direct cell death and the loss of function within the CNS (54–56). In contrast, cells of the human astrocytoma cell line CCF-STTG1 seem to be refractory to CVB3-induced cell death, though they support productive virus replication. Thus, a clear difference in the susceptibility of CCF-STTG1 cells to different EVs exists, and this difference is presumably due to the distinct patterns of virus-host interactions.
Our study established an in vitro cell model for persistent productive CVB3 infection in CCF-STTG1 cells. The prolonged presence of positive- and negative-strand viral RNA in cultures, as well as the detection of both infectious particles and CVB3 antigens, collectively demonstrates that continuous virus replication occurs in persistently infected cell cultures. During the course of persistent infection, only a small proportion of cells scored positive. Furthermore, CCF-STTG1 cell cultures persistently infected with CVB3 could be readily cured by the addition of virus-specific antibodies. Thus, our model of persistent CVB3 infection in CCF-STTG1 cells was probably established through the mechanism of a carrier-state culture involving CVB3 replication among a minority of cells at any given time, in line with previous reports of CVB3 persistence in human vascular endothelial cells (57), a lymphoid cell line (58), myocardial fibroblasts (18, 20), and a murine cardiac myocyte cell line (22).
Of great interest, persistent CVB3 infection of CCF-STTG1 cells appeared to involve an entry pathway distinct from that of acutely infected cells, such as HeLa cells. CCF-STTG1 cells did not express detectable CAR and DAF on the cell surface. In addition, there was no inhibition of CVB3 infection of CCF-STTG1 cells by MAbs against the CAR and DAF molecules. Notably, CAR knockdown resulted in decreased acute CVB3 infection and virus-induced death of cardiac cells in vitro (59, 60). Furthermore, inhibited expression of CAR was found in a murine cardiac myocyte cell line persistently infected with CVB3 (22). The present study also hinted that CVB3 infection was much less efficient in CCF-STTG1 cells expressing restricted levels of CAR and DAF. Such cell-dependent resistance appeared to be at the level of entry, because transfection of abundant viral genomic RNA into CCF-STTG1 cells resulted in significant cell death and virus replication. Therefore, it is speculated that host cell resistance due to a limited receptor distribution likely contributes to the establishment of CVB3 persistence in CCF-STTG1 cells.
Cell surface glycosaminoglycan (GAG) chain components, including heparan sulfate (HS) and/or chondroitin sulfate (CS), have been reported to serve as initial receptors for various viruses (61–68). The expression of HS and CS in astrocytes of the central nervous system both in vivo and in vitro (69) raises the possibility that HS and/or CS in astrocytic cells may function in viral binding during an infection. The present work disclosed that removal of cell surface CS significantly reduced the ability of CVB3 to infect CCF-STTG1 cells, whereas enzymatic removal of HS did not affect CVB3 infection. These results suggest that cell surface CS rather than HS plays an important role in CVB3 infection of CCF-STTG1 cells. It should be noted that removal of CS did not completely abolish CVB3 infection, implying that there might be additional receptor molecules involved in CVB3 entry in this system.
In addition to the modulation of virus receptors, several other factors, such as the production of antiviral mediators, can also have a great influence on the initiation and maintenance of CVB persistence (21, 48). IFN-β exerts its antiviral function in two ways: one is to elicit an antiviral state in uninfected cells and the other is to selectively trigger apoptotic cell death in infected cells (70, 71). The current study showed that very limited IFN-β secretion was detected in CCF-STTG1 cells persistently infected with CVB3, indicating that CVB3 persistence attenuated inducible IFN-β release in CCF-STTG1 cells. Moreover, exogenous addition of IFN-β led to the elimination of infection in CCF-STTG1 cells persistently infected with CVB3. Therefore, CVB3-induced curtailing of IFN-β release likely facilitates the establishment of persistent CVB3 infection in CCF-STTG1 cells, while IFN-β treatment could be an effective means of eradicating CVB3 from persistently infected CCF-STTG1 cells.
It was also disclosed in this study that CVB3 infection stimulated the production of proinflammatory chemokines and cytokines, such as VCAM-1, IL-6, and IL-8, in both CCF-STTG1 cells and HPDAs. Induced production of chemokines may promote the chemoattraction of a variety of inflammatory cells and blood-brain barrier (BBB) dysfunction (72). Chemokines and cytokines may also be directly toxic to neuronal and glial cells (73). Because astrocytes are the most abundant cell type in the CNS (74), CVB3 replication accompanied by the induced production of chemokines and cytokines in astrocytes, if it occurs in vivo, is detrimental to the largely non-self-renewing brain tissues. Notably, EV71 or CVA9 infection also stimulated the production of VCAM-1, IL-6, and IL-8 in CCF-STTG1 cells, suggesting that the induced production of chemokines and cytokines is probably common during EV infection of astrocytic cells. Further work is needed to determine whether our findings in primary cells and cell lines translate to appropriate animal models. Such work was beyond the scope of this study.
In conclusion, our results show that CCF-STTG1 human astrocytoma cells can support a persistent CVB3 infection in vitro. Inherent early blockage of virus entry by naive CCF-STTG1 cells may account for the unique outcome of persistent CVB3 infection in this cell line. Through it curtails IFN-β expression, CVB3 may modulate antiviral responses to maintain the balance of virus-host interactions and thus facilitate the establishment and maintenance of persistence. The remarkable properties of infected astrocytic cells to produce chemokines and cytokines may have important implications for the pathogenesis of CVB3-induced neuroinflammatory diseases.
ACKNOWLEDGMENTS
We thank Minhua Luo and Gengfu Xiao for providing human brain-derived cell lines and HPDAs. We also thank Huipeng Chen for providing the full-length CVB3 cDNA.
This work was supported by 973 Project grant no. 2011CB504902 from the Ministry of Science and Technology of China and National Natural Science Foundation of China grant no. 81071351.
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Nasri D, Bouslama L, Pillet S, Bourlet T, Aouni M, Pozzetto B. 2007. Basic rationale, current methods and future directions for molecular typing of human enterovirus. Expert Rev. Mol. Diagn. 7:419–434 [DOI] [PubMed] [Google Scholar]
- 2.Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2012. Picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA [Google Scholar]
- 3.Hober D, Sauter P. 2010. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat. Rev. Endocrinol. 6:279–289 [DOI] [PubMed] [Google Scholar]
- 4.Baboonian C, Treasure T. 1997. Meta-analysis of the association of enteroviruses with human heart disease. Heart 78:539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacques J, Moret H, Minette D, Leveque N, Jovenin N, Deslee G, Lebargy F, Motte J, Andreoletti L. 2008. Epidemiological, molecular, and clinical features of enterovirus respiratory infections in French children between 1999 and 2005. J. Clin. Microbiol. 46:206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michos AG, Syriopoulou VP, Hadjichristodoulou C, Daikos GL, Lagona E, Douridas P, Mostrou G, Theodoridou M. 2007. Aseptic meningitis in children: analysis of 506 cases. PLoS One 2:e674. 10.1371/journal.pone.0000674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. 2011. Enterovirus infections of the central nervous system. Virology 411:288–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitton JL, Cornell CT, Feuer R. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765–776 [DOI] [PubMed] [Google Scholar]
- 9.Kim K-S, Tracy S, Tapprich W, Bailey J, Lee C-K, Kim K, Barry W, Chapman N. 2005. 5′-terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 79:7024–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chia J, Chia A, Voeller M, Lee T, Chang R. 2010. Acute enterovirus infection followed by myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and viral persistence. J. Clin. Pathol. 63:165–168 [DOI] [PubMed] [Google Scholar]
- 11.Chapman NM, Kim KS. 2008. Persistent coxsackievirus infection: enterovirus persistence in chronic myocarditis and dilated cardiomyopathy. Curr. Top. Microbiol. 323:275–292 [DOI] [PubMed] [Google Scholar]
- 12.Muir P, Nicholson F, Tilzey AJ, Signy M, English TA, Banatvala JE. 1989. Chronic relapsing pericarditis and dilated cardiomyopathy: serological evidence of persistent enterovirus infection. Lancet i:804–807 [DOI] [PubMed] [Google Scholar]
- 13.Bridgens R, Sturman S, Davidson C, Expert BPFs. 2010. Post-polio syndrome—polio's legacy. Clin. Med. 10:213–214 [DOI] [PubMed] [Google Scholar]
- 14.Leparc-Goffart I, Julien J, Fuchs F, Janatova I, Aymard M, Kopecka H. 1996. Evidence of presence of poliovirus genomic sequences in cerebrospinal fluid from patients with postpolio syndrome. J. Clin. Microbiol. 34:2023–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaidane H, Caloone D, Lobert PE, Sane F, Dardenne O, Naquet P, Gharbi J, Aouni M, Geenen V, Hober D. 2012. Persistent infection of thymic epithelial cells with coxsackievirus B4 results in decreased expression of type 2 insulin-like growth factor. J. Virol. 86:11151–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brilot F, Chehadeh W, Charlet-Renard C, Martens H, Geenen V, Hober D. 2002. Persistent infection of human thymic epithelial cells by coxsackievirus B4. J. Virol. 76:5260–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanone MM, Favaro E, Conaldi PG, Greening J, Bottelli A, Perin PC, Klein NJ, Peakman M, Camussi G. 2003. Persistent infection of human microvascular endothelial cells by coxsackie B viruses induces increased expression of adhesion molecules. J. Immunol. 171:438–446 [DOI] [PubMed] [Google Scholar]
- 18.Heim A, Canu A, Kirschner P, Simon T, Mall G, Hofschneider PH, Kandolf R. 1992. Synergistic interaction of interferon-beta and interferon-gamma in coxsackievirus B3-infected carrier cultures of human myocardial fibroblasts. J. Infect. Dis. 166:958–965 [DOI] [PubMed] [Google Scholar]
- 19.Heim A, Grumbach I, Pring-Akerblom P, Stille-Siegener M, Muller G, Kandolf R, Figulla HR. 1997. Inhibition of coxsackievirus B3 carrier state infection of cultured human myocardial fibroblasts by ribavirin and human natural interferon-alpha. Antiviral Res. 34:101–111 [DOI] [PubMed] [Google Scholar]
- 20.Heim A, Brehm C, Stille-Siegener M, Muller G, Hake S, Kandolf R, Figulla HR. 1995. Cultured human myocardial fibroblasts of pediatric origin: natural human interferon-alpha is more effective than recombinant interferon-alpha 2a in carrier-state coxsackievirus B3 replication. J. Mol. Cell. Cardiol. 27:2199–2208 [DOI] [PubMed] [Google Scholar]
- 21.Conaldi PG, Biancone L, Bottelli A, De Martino A, Camussi G, Toniolo A. 1997. Distinct pathogenic effects of group B coxsackieviruses on human glomerular and tubular kidney cells. J. Virol. 71:9180–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinkert S, Klingel K, Lindig V, Dorner A, Zeichhardt H, Spiller OB, Fechner H. 2011. Virus-host coevolution in a persistently coxsackievirus B3-infected cardiomyocyte cell line. J. Virol. 85:13409–13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman NM, Kim K-S, Drescher KM, Oka K, Tracy S. 2008. 5′ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology 375:480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam PE, Messner RP. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feuer R, Ruller CM, An N, Tabor-Godwin JM, Rhoades RE, Maciejewski S, Pagarigan RR, Cornell CT, Crocker SJ, Kiosses WB. 2009. Viral persistence and chronic immunopathology in the adult central nervous system following coxsackievirus infection during the neonatal period. J. Virol. 83:9356–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gosselin A-S, Simonin Y, Guivel-Benhassine F, Rincheval V, Vayssiere J-L, Mignotte B, Colbere-Garapin F, Couderc T, Blondel B. 2003. Poliovirus-induced apoptosis is reduced in cells expressing a mutant CD155 selected during persistent poliovirus infection in neuroblastoma cells. J. Virol. 77:790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavio N, Couderc T, Girard S, Sgro JY, Blondel B, Colbere-Garapin F. 2000. Expression of mutated poliovirus receptors in human neuroblastoma cells persistently infected with poliovirus. Virology 274:331–342 [DOI] [PubMed] [Google Scholar]
- 28.Gibson JP, Righthand VF. 1985. Persistence of echovirus 6 in cloned human cells. J. Virol. 54:219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin DE. 2003. Immune responses to RNA-virus infections of the CNS. Nat. Rev. Immunol. 3:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergmann CC, Lane TE, Stohlman SA. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muir P, Nicholson F, Sharief MK, Thompson EJ, Cairns NJ, Lantos P, Spencer GT, Kaminski HJ, Banatvala JE. 1995. Evidence for persistent enterovirus infection of the central nervous system in patients with previous paralytic poliomyelitis. Ann. N. Y. Acad. Sci. 753:219–232 [DOI] [PubMed] [Google Scholar]
- 32.Berger MM, Kopp N, Vital C, Redl B, Aymard M, Lina B. 2000. Detection and cellular localization of enterovirus RNA sequences in spinal cord of patients with ALS. Neurology 54:20–25 [DOI] [PubMed] [Google Scholar]
- 33.Stacey G, Viviani B. 2001. Cell culture models for neurotoxicology. Cell Biol. Toxicol. 17:319–334 [DOI] [PubMed] [Google Scholar]
- 34.Stein GH. 1979. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J. Cell. Physiol. 99:43–54 [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Zhang X, Zhou G, Xiang X, Ji X, Zheng Z, He Z, Wang H. 2012. Simultaneous determination of human enterovirus 71 and coxsackievirus B3 by dual-color quantum dots and homogeneous immunoassay. Anal. Chem. 84:3200–3207 [DOI] [PubMed] [Google Scholar]
- 36.Luo MH, Hannemann H, Kulkarni AS, Schwartz PH, O'Dowd JM, Fortunato EA. 2010. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J. Virol. 84:3528–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo MH, Schwartz PH, Fortunato EA. 2008. Neonatal neural progenitor cells and their neuronal and glial cell derivatives are fully permissive for human cytomegalovirus infection. J. Virol. 82:9994–10007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arndt U, Wennemuth G, Barth P, Nain M, Al-Abed Y, Meinhardt A, Gemsa D, Bacher M. 2002. Release of macrophage migration inhibitory factor and CXCL8/interleukin-8 from lung epithelial cells rendered necrotic by influenza A virus infection. J. Virol. 76:9298–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C-J, Lin H-R, Liao C-L, Lin Y-L. 2008. Cholesterol effectively blocks entry of flavivirus. J. Virol. 82:6470–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber S, Haisch C, Lodge P. 1990. Functional diversity in vascular endothelial cells: role in coxsackievirus tropism. J. Virol. 64:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bessaud M, Autret A, Jegouic S, Balanant J, Joffret ML, Delpeyroux F. 2008. Development of a TaqMan RT-PCR assay for the detection and quantification of negatively stranded RNA of human enteroviruses: evidence for false-priming and improvement by tagged RT-PCR. J. Virol. Methods 153:182–189 [DOI] [PubMed] [Google Scholar]
- 42.Shafren DR, Williams DT, Barry RD. 1997. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 71:9844–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schowalter RM, Pastrana DV, Buck CB. 2011. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 7:e1002161. 10.1371/journal.ppat.1002161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klump WM, Bergmann I, Muller BC, Ameis D, Kandolf R. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melchers WJ, Hoenderop JG, Bruins Slot HJ, Pleij CW, Pilipenko EV, Agol VI, Galama JM. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harkins S, Cornell CT, Whitton JL. 2005. Analysis of translational initiation in coxsackievirus B3 suggests an alternative explanation for the high frequency of R+4 in the eukaryotic consensus motif. J. Virol. 79:987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawicki SG, Lu JH, Holmes KV. 1995. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 69:5535–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chehadeh W, Kerr-Conte J, Pattou F, Alm G, Lefebvre J, Wattre P, Hober D. 2000. Persistent infection of human pancreatic islets by coxsackievirus B is associated with alpha interferon synthesis in beta cells. J. Virol. 74:10153–10164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferenczy MW, Johnson KR, Marshall LJ, Monaco MC, Major EO. 2013. Differentiation of human fetal multipotential neural progenitor cells to astrocytes reveals susceptibility factors for JC virus. J. Virol. 87:6221–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA. 2006. Enterovirus surveillance—United States, 1970-2005. MMWR Surveill Summ. 55(SS-08):1–20 [PubMed] [Google Scholar]
- 51.Tao Z, Song Y, Li Y, Liu Y, Jiang P, Lin X, Liu G, Song L, Wang H, Xu A. 2012. Coxsackievirus B3, Shandong Province, China, 1990-2010. Emerg. Infect. Dis. 18:1865–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YF, Chou CT, Lei HY, Liu CC, Wang SM, Yan JJ, Su IJ, Wang JR, Yeh TM, Chen SH, Yu CK. 2004. A mouse-adapted enterovirus 71 strain causes neurological disease in mice after oral infection. J. Virol. 78:7916–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen YY, Chang TY, Chen ST, Li C, Liu HS. 2003. Comparative study of enterovirus 71 infection of human cell lines. J. Med. Virol. 70:109–118 [DOI] [PubMed] [Google Scholar]
- 54.Girard S, Couderc T, Destombes J, Thiesson D, Delpeyroux F, Blondel B. 1999. Poliovirus induces apoptosis in the mouse central nervous system. J. Virol. 73:6066–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabor-Godwin JM, Ruller CM, Bagalso N, An N, Pagarigan RR, Harkins S, Gilbert PE, Kiosses WB, Gude NA, Cornell CT, Doran KS, Sussman MA, Whitton JL, Feuer R. 2010. A novel population of myeloid cells responding to coxsackievirus infection assists in the dissemination of virus within the neonatal CNS. J. Neurosci. 30:8676–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feuer R, Mena I, Pagarigan RR, Harkins S, Hassett DE, Whitton JL. 2003. Coxsackievirus B3 and the neonatal CNS: the roles of stem cells, developing neurons, and apoptosis in infection, viral dissemination, and disease. Am. J. Pathol. 163:1379–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conaldi PG, Serra C, Mossa A, Falcone V, Basolo F, Camussi G, Dolei A, Toniolo A. 1997. Persistent infection of human vascular endothelial cells by group B coxsackieviruses. J. Infect. Dis. 175:693–696 [DOI] [PubMed] [Google Scholar]
- 58.Matteucci D, Paglianti M, Giangregorio AM, Capobianchi MR, Dianzani F, Bendinelli M. 1985. Group B coxsackieviruses readily establish persistent infections in human lymphoid cell lines. J. Virol. 56:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werk D, Schubert S, Lindig V, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J. 2005. Developing an effective RNA interference strategy against a plus-strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus-adenovirus receptor. Biol. Chem. 386:857–863 [DOI] [PubMed] [Google Scholar]
- 60.Fechner H, Pinkert S, Wang X, Sipo I, Suckau L, Kurreck J, Dorner A, Sollerbrant K, Zeichhardt H, Grunert HP, Vetter R, Schultheiss HP, Poller W. 2007. Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor. Gene Ther. 14:960–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zautner AE, Korner U, Henke A, Badorff C, Schmidtke M. 2003. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection. J. Virol. 77:10071–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan CW, Poh CL, Sam IC, Chan YF. 2013. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J. Virol. 87:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Boer SM, Kortekaas J, de Haan CA, Rottier PJ, Moormann RJ, Bosch BJ. 2012. Heparan sulfate facilitates Rift Valley fever virus entry into the cell. J. Virol. 86:13767–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLeish NJ, Williams CH, Kaloudas D, Roivainen MM, Stanway G. 2012. Symmetry-related clustering of positive charges is a common mechanism for heparan sulfate binding in enteroviruses. J. Virol. 86:11163–11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zautner AE, Jahn B, Hammerschmidt E, Wutzler P, Schmidtke M. 2006. N- and 6-O-sulfated heparan sulfates mediate internalization of coxsackievirus B3 variant PD into CHO-K1 cells. J. Virol. 80:6629–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodfellow IG, Sioofy AB, Powell RM, Evans DJ. 2001. Echoviruses bind heparan sulfate at the cell surface. J. Virol. 75:4918–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ. 2006. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J. Virol. 80:3487–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim E, Okumura M, Sawa H, Miyazaki T, Fujikura D, Yamada S, Sugahara K, Sasaki M, Kimura T. 2011. Paradoxical effects of chondroitin sulfate-E on Japanese encephalitis viral infection. Biochem. Biophys. Res. Commun. 409:717–722 [DOI] [PubMed] [Google Scholar]
- 69.Dow KE, Wang W. 1998. Cell biology of astrocyte proteoglycans. Cell. Mol. Life Sci. 54:567–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka N, Sato M, Lamphier MS, Nozawa H, Oda E, Noguchi S, Schreiber RD, Tsujimoto Y, Taniguchi T. 1998. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3:29–37 [DOI] [PubMed] [Google Scholar]
- 71.Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J. Virol. 74:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nair A, Frederick TJ, Miller SD. 2008. Astrocytes in multiple sclerosis: a product of their environment. Cell. Mol. Life Sci. 65:2702–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosking MP, Lane TE. 2010. The role of chemokines during viral infection of the CNS. PLoS Pathog. 6:e1000937. 10.1371/journal.ppat.1000937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Narasipura SD, Henderson LJ, Fu SW, Chen L, Kashanchi F, Al-Harthi L. 2012. Role of beta-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J. Virol. 86:1911–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]