Abstract
Our previous studies on cutaneous beta human papillomavirus 38 (HPV38) E6 and E7 oncoproteins highlighted a novel activity of IκB kinase beta (IKKβ) in the nucleus of human keratinocytes, where it phosphorylates and stabilizes ΔNp73α, an antagonist of p53/p73 functions. Here, we further characterize the role of the IKKβ nuclear form. We show that IKKβ nuclear translocation and ΔNp73α accumulation are mediated mainly by HPV38 E7 oncoprotein. Chromatin immunoprecipitation (ChIP)/Re-ChIP experiments showed that ΔNp73α and IKKβ are part, together with two epigenetic enzymes DNA methyltransferase 1 (DNMT1) and the enhancer of zeste homolog 2 (EZH2), of a transcriptional regulatory complex that inhibits the expression of some p53-regulated genes, such as PIG3. Recruitment to the PIG3 promoter of EZH2 and DNMT1 resulted in trimethylation of histone 3 on lysine 27 and in DNA methylation, respectively, both events associated with gene expression silencing. Decreases in the intracellular levels of HPV38 E7 or ΔNp73α strongly affected the recruitment of the inhibitory transcriptional complex to the PIG3 promoter, with consequent restoration of p53-regulated gene expression. Finally, the ΔNp73α/IKKβ/DNMT1/EZH2 complex appears to bind a subset of p53-regulated promoters. In fact, the complex is efficiently recruited to several promoters of genes encoding proteins involved in DNA repair and apoptosis, whereas it does not influence the expression of the prosurvival factor Survivin. In summary, our data show that HPV38 via E7 protein promotes the formation of a multiprotein complex that negatively regulates the expression of several p53-regulated genes.
INTRODUCTION
A subgroup of cutaneous human papillomaviruses (HPV) that belongs to the genus beta of the HPV phylogenetic tree is suspected to be involved, together with UV radiation, in the development of nonmelanoma skin cancer (1, 2). Beta HPV types were originally isolated in patients suffering from a rare autosomal recessive cancer-prone genetic disorder, epidermodysplasia verruciformis (EV) and are consistently detected in nonmelanoma skin cancers from EV patients and immunocompromised and normal individuals (2). Many independent studies have demonstrated that the E6 and E7 proteins of several HPV types display transforming activities in in vitro and in vivo experimental models (3–12). We previously showed that E6 and E7 of beta HPV38 are able to efficiently immortalize primary human keratinocytes (4, 6). In addition, constitutive expression of HPV38 E6 and E7 in the skin of transgenic mice increases their susceptibility to chemical or UV-induced skin carcinogenesis (13).
The transforming activity of HPV38 is partly mediated by the accumulation of ΔNp73α (3, 6, 14, 15), an antagonist of p53/p73 functions involved in growth suppression and/or apoptosis (16–18). ΔNp73α cooperates with the oncogenic RasV12 in inducing transformation of primary mouse embryonic fibroblasts (MEFs), which acquire the property of inducing tumors upon injection into nude mice (19, 20). In addition, MEFs from ΔNp73α−/− mice expressing the adenoviral E1A oncoprotein, together with RasV12, are impaired in their ability to induce tumor formation when injected into athymic nude mice (21). Elevated ΔNp73α levels were found in some human cancers with poor prognosis (17, 22–24).
Similarly to HPV38, other viruses, such as cytomegalovirus and Epstein-Barr virus, were shown to be able to induce accumulation of ΔNp73α by different mechanisms (3, 14, 25, 26). We previously showed that expression of HPV38 E6 and E7 in keratinocytes or of Epstein-Barr virus LMP-1 in B cells promotes ΔNp73α transcriptional activation (3, 14). In contrast, cytomegalovirus does not alter ΔNp73α mRNA levels but promotes a rapid increase in ΔNp73α protein levels, most likely influencing its stability (26). We recently showed that HPV38, similarly to cytomegalovirus, is also able to increase ΔNp73α protein levels independent of its effect on gene expression (14). This event is mediated by the translocation of IκB kinase β (IKKβ) into the nucleus, where it binds and phosphorylates ΔNp73α at serine 422 (S422). Phosphorylation of S422 significantly increases the ΔNp73α half-life, resulting in transcriptional inhibition of p53-regulated genes, such as p21, PIG3, Fas, and Mdm2. In the present study, we evaluated the contribution of individual HPV38 oncoproteins in IKKβ nuclear translocation and ΔNp73α stabilization, and we further characterized the role of IKKβ and ΔNp73α in transcriptional regulation of p53-regulated genes.
MATERIALS AND METHODS
Plasmid constructs.
The genes of interest were expressed in the following expression vectors: pBabe (27), pLXSN (Clontech, Le Pont Claix, France), pRetroSuper (Screeninc, Amsterdam, Netherlands), and pcDNA3.1 (Invitrogen). The constructs pRetroSuper-HPV38 E6E7, pBabe-puro-Flag-DN-IKKβ (dominant-negative kinase-dead IKKβ), pBabe-puro-ΔN-IκBα (lacking the first 36 N-terminal amino acids), pLXSN-HPV38 E6E7, pRetroSuper-HPV38 E6E7, pcDNA3-HA-ΔNp73α, pcDNA3-Flag-IKKβ, and pBabe-hygro-CRE were previously described (6, 14). The construct pBabe-puro-HPV38 E6E7LoxP/LoxP was generated by cloning HPV38 E6E7 open reading frames into the pBabe-LoxP-puro vector. p53RE-mutated PIG3 promoter was generated with the QuikChange Lightning site-directed mutagenesis kit (Qiagen) using pGL3-wild-type PIG3 promoter vector as a template and the following oligonucleotides: sense, 5′-GCAGCACCCAGATTACCCACCTATACTCAAGATGGGCGGG-3′; and antisense, 5′-CCCGCCCATCTTGAGTATAGGTGGGTAATCTGGGTGCTGC-3′.
Cell culture and treatment.
Human foreskin keratinocytes (HFK) were grown together with NIH 3T3 feeder layers in FAD medium containing 3 parts Ham F-12, 1 part Dulbecco modified Eagle medium, 5% fetal calf serum, insulin (5 μg/ml), epidermal growth factor (10 ng/ml), cholera toxin (8.4 ng/ml), adenine (24 μg/ml), and hydrocortisone (0.4 μg/ml). Feeder layers were prepared by mitomycin C-induced cell cycle arrest of NIH 3T3 cells.
HFK stably expressing HPV38 E6 or E7 were generated by retroviral transduction. Cells transduced with the empty retroviral vector (pLXSN) were used as negative control. Due to the limited life span of HFK expressing HPV38 E6 or E7, the experiments shown in Fig. 4B were performed with cells expressing human telomerase reverse transcriptase (hTERT), together with each of the viral proteins. For DNA methylation analysis, HFK expressing HPV38 E7 and E6 genes (38E6E7 HFK) were treated with 10 μM 5-aza-2′-deoxycytidine (5-Aza; Sigma-Aldrich) or dimethyl sulfoxide (DMSO) for 4 days, with drug replacement every 24 h. At the end of the treatment, cells were collected for DNA methylation and reverse transcription-quantitative PCR (RT-qPCR) analysis. For transient transfection, 38E6E7 HFK (105 cells/well) were seeded in a 24-well plate and transfected using JetPrime reagent (Polyplus) according to the manufacturer's protocol.
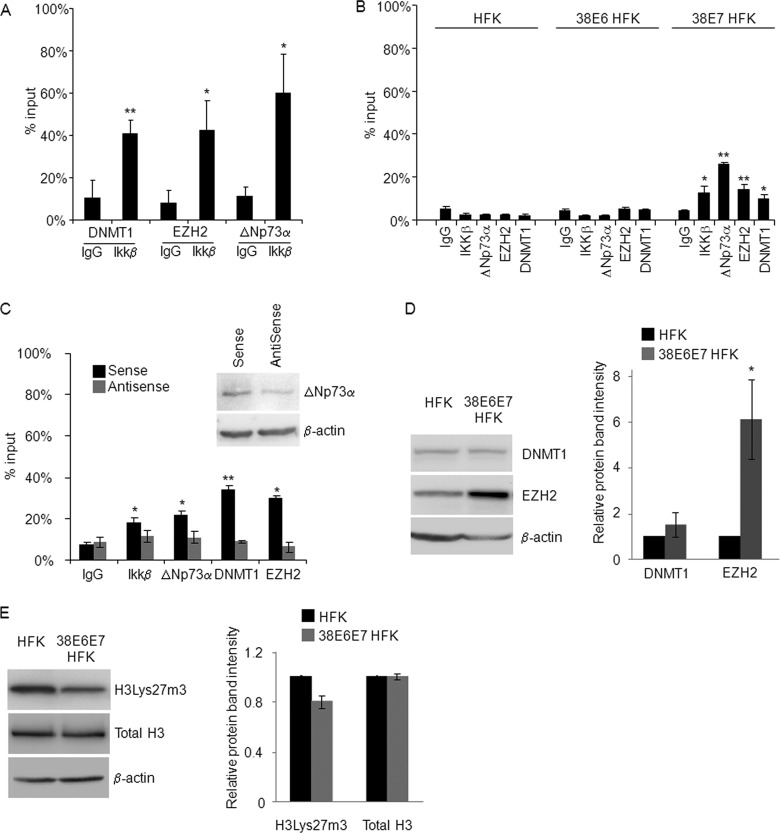
Fig 4.
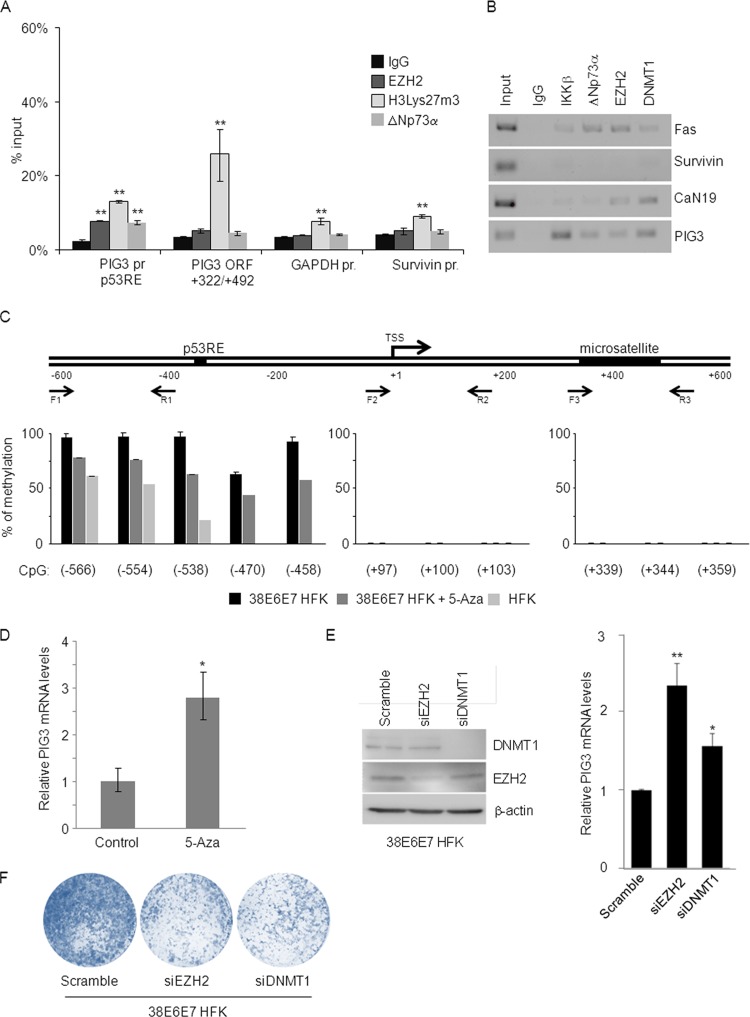
Endogenous EZH2, DNMT1, IKKβ, and ΔNp73α cobind on the PIG3 promoter. (A) 38E6E7 HFK were harvested and formaldehyde fixed. After sonication, the chromatin was subjected to ChIP assay using DNMT1, EZH2, or ΔNp73α antibodies. The eluted DNA was processed for Re-ChIP with anti-IKKβ antibody or nonspecific IgG control. Input and coimmunoprecipitation DNA were analyzed by qPCR for the p53RE region of the PIG3 promoter. The results are the average of two independent experiments performed in duplicate. * and **, P < 0.05 and P < 0.01, respectively, versus the corresponding IgG control. (B) HFK, 38E6 HFK, and 38E7 HFK were subjected to ChIP assay using antibodies for the indicated proteins or nonspecific IgG control. Input and eluted DNA were analyzed by qPCR for the p53RE region of the PIG3 promoter. The results were generated as indicated for Fig. 2A and are the average of two independent experiments performed in duplicate. * and **, P < 0.05 and P < 0.01, respectively, versus the corresponding IgG control. (C) Sense and ΔNp73α-AntiSense oligonucleotides were transfected into 38E6E7 HFK. The cells were collected 36 h after transfection and used for ChIP assay with the indicated antibodies. The amounts of p53RE bound to the different proteins were calculated as described for Fig. 2A. The data shown are the mean values of two independent experiments. * and **, P < 0.05 and P < 0.01, respectively, versus the corresponding IgG control. The protein levels of ΔNp73a and β-actin were detected by IB (top right panel). Result is representative of two independent experiments. (D) Protein extracts (40 μg) from HFK and 38E6E7 HFK were analyzed by IB for protein levels of DNMT1, EZH2, and β-actin (left panel). Band intensities were quantified and normalized to β-actin levels (right panel). The data shown are the mean values of three independent experiments. *, P < 0.05 versus the corresponding HFK sample. (E) HFK and 38E6E7 HFK were lysed with RIPA buffer and sonicated as explained in Materials and Methods. The protein extracts (40 μg) were analyzed by IB for protein levels of histone H3Lys27m3, total histone H3, and β-actin (left panel). Band intensities were quantified as explained for Fig. 1C and normalized to β-actin levels (right panel). The data shown are the mean values of two independent experiments. The difference in the levels of histone H3Lys27m3 between 38E6E7 HFK and HFK is not statistically significant.
Colony formation assay.
For colony formation assays, the different types of cells were seeded at different dilutions (104, 103, and 102) and cultured for 7 days. Cells were then stained with crystal violet, and the average numbers of cells per colony were determined.
Gene expression silencing.
Downregulation of ΔNp73α was achieved using antisense oligonucleotide (AntiSense [AS]) as previously described (3, 14). EZH2 and DNMT1 silencing was performed using specific stealth small interfering RNA (siRNA) and scrambled siRNA as a control designed using BLOCK-iT RNAi designer (Invitrogen). The target sequence for EZH2 is 5′-AGUGGUGCUGAAGCCUCAAUGUUUA-3′, and that for DNMT1 is 5′-UUGGAGAACGGUGCUCAUGCUUACA-3′. The cells were transfected with 250 nM siRNA using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's protocol. The cells were collected 72 h after transfection or stained with crystal violet after 7 days.
Silencing of HPV38 E6E7 gene expression was obtained using either (i) the pRetroSuper construct (pRS), expressing small hairpin RNAs (shRNAs) for the polycistronic HPV38 E6 and E7 mRNA (pRS 38E6/E7) (3) or (ii) the Cre/LoxP system. For the latter approach, HFK transduced with pBabe-puro-HPV38 E6E7LoxP/LoxP retroviruses were selected for 5 days with puromycin and cultured for several weeks until immortalization. Subsequently, cells were transduced with pBabe-hygro or pBabe-hygro-CRE retrovirus and cultured in the presence of hygromycin.
RT-PCR and qPCR.
Total RNA was extracted using the Absolutely RNA miniprep kit (Stratagene, La Jolla, CA) and quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). For RT-PCR, the obtained RNA was reverse transcribed to cDNA with a RevertAid H Minus M-MuLV reverse transcriptase kit (MBI Fermentas GmbH, St. Leon-Rot, Germany) according to the manufacturer's instructions. Real-time qPCR was performed using the Mesa Green qPCR MasterMix Plus for SYBR assay (Eurogentec, San Diego, CA) with the primers listed in Table 1.
Table 1.
Primers used for RT-qPCR and ChIP-qPCR
| Method and primera | Orientation | Sequence (5′-3′) |
|---|---|---|
| RT-qPCR, cDNA | ||
| PIG3 | Forward | GCTTCAAATGGCAGAAAAGC |
| Reverse | AACCCATCGACCATCAAGAG | |
| HPV38 E6 | Forward | TCTGGACTCAAGAGGATTTTG |
| Reverse | CACTTTAAACAATACTGACACC | |
| HPV38 E7 | Forward | CAAGCTACTCTTCGTGATATAGTT |
| Reverse | CAGGTGGGACACAGAAGCCTTAC | |
| GAPDH | Forward | AAGGTGGTGAAGCAGGCGT |
| Reverse | GAGGAGTGGGTGTCGCTGTT | |
| ChIP-qPCR, gene promoters | ||
| PIG3 (−240/−399) | Forward | CCCAGGACTGCGTTTTGCCT |
| Reverse | GGTCCATTTTCCAGGCATGG | |
| PIG3 (+322/+517) | Forward | CAGGAGCTCGGGGCGGA |
| Reverse | ACAGGGCAGGGCAGGGC | |
| Survivin | Forward | TGGGTGCCCCGACGT |
| Reverse | GAAGGGCCAGTTCTTGAATGTAGA | |
| FasL | Forward | GATCCCGCTGGGCAGGC |
| Reverse | GTTCTGAAGGCTGCAGGCTC | |
| p21waf1 | Forward | TCACCATTCCCCTACCCCATGCTGCTC |
| Reverse | AAGTTTGCAACCATGCACTTGAATGTG | |
| CaN19 | Forward | GGTCCAGGATGCCCAGTC |
| Reverse | GAAGGAGAGCAAGGCAGC | |
| GAPDH | Forward | CACTGGAGCCTTCATCTCAG |
| Reverse | CTGTTGCTGGCCAGCAACTG |
The region is indicated in parentheses where applicable.
Immunofluorescence staining.
HFK were grown on cover slides and fixed with 4% paraformaldehyde, followed by permeabilization with 0.1% Triton X-100 at room temperature. Staining was performed using anti-IKKβ (Millipore; diluted 1:400) primary antibody and Alexa Fluor 488-coupled secondary antibody. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole4′) using VectaShield mounting medium (H-1200; Vector). Immunofluorescence staining was visualized with a Nikon Eclipse Ti inverted microscope (with cooled light-emitting diode [LED] excitation system).
Immunoblotting and antibodies.
For the preparation of protein extracts, cellular pellets were resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml). After quantification (Interchim BC assay), equal amounts of proteins were separated by SDS-PAGE and then transferred onto a Polyscreen polyvinylidene difluoride membrane in a Trans-Blot electrophoretic transfer cell (Bio-Rad) and subjected to immunoblotting (IB). IB for histones was performed by the lysis of cellular pellets for 10 min in ice with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with protease inhibitor tablet (Roche). The cellular extracts were sonicated twice for 10 s (30% amplitude) and centrifuged for 10 min at 4°C at 11,000 × g. Then, 40 μg of protein lysate was subjected to Western blotting as described above.
Nuclear extracts were prepared using the Panomics nuclear extraction kit (catalog no. AY2002) according to the manufacturer's instructions. The following antibodies were used for IB: anti-IKKβ (catalog no. 2684; Cell Signaling), anti-p73 (Ab-1; Calbiochem), anti-EZH2 (AC22; Cell Signaling), anti-histone H3 (catalog no. 9715; Cell Signaling), anti-H3Lys27m3 (catalog no. 4039; Epigentek), anti-DNMT1 (clone 60B1220; Abnova), anti-β-actin (C4; MP Biomedicals), anti-β-tubulin, anti-PARP (catalog no. 9542; Cell Signaling), anti-Flag (F4042; Sigma-Aldrich), and anti-HA (3F10; Roche). Images were taken using the ChemiDoc XRS+ imaging system (Bio-Rad).
Chromatin immunoprecipitation.
A chromatin immunoprecipitation (ChIP) assay was performed using the Shearing ChIP and OneDay ChIP kits (Diagenode, Philadelphia, PA) according to the manufacturer's instructions. Briefly, cells were sonicated to obtain DNA fragments of 200 to 500 bp. Sheared chromatin was immunoprecipitated with isotype control IgG or the following antibodies: anti-p73 (Ab-1; Calbiochem), anti-IKKβ, anti-EZH2 (AC22; Cell Signaling), anti-DNMT1 (clone 60B1220; Abnova), and anti-H3Lys27m3 (catalog no. 4039; Epigentek). After DNA recovery, 1% of input chromatin and the precipitated DNA were analyzed by qPCR with the primers listed in Table 1. The ChIP results are presented in the histograms as the percentage of binding relative to the input (100%).
For ChIP/Re-ChIP experiments, bead-bound protein-DNA complexes obtained after the first ChIP were incubated with 50 μl of 10 mM dithiothreitol (DTT) for 30 min at 37°C with shaking at 300 rpm. Supernatant was collected after centrifugation at 12,000 × g for 1 min. Pelleted beads were incubated again with 30 μl of 10 mM DTT for 20 min and centrifuged at 12,000 × g for 1 min. Ten percent of the combined supernatants was kept as the input for the second ChIP, which was performed according to the OneDay ChIP kit (Diagenode) manufacturer's protocol.
Oligonucleotide pulldown.
Cells were lysed and sonicated in HKMG buffer (10 mM HEPES [pH 7.9], 100 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.5% Nonidet P-40) containing protease and phosphatase inhibitors. After centrifugation at 12,000 × g for 10 min, protein extracts was precleared with streptavidin-agarose beads. The PIG3 promoter was used as a template to amplify the p53RE region containing wild-type or mutated p53RE. PCR amplification was performed using a biotinylated forward primer (5′-Btn-CCCAGGACTGCGTTTTGCCT-3′) and a nonbiotinylated reverse primer (5′-GGTCCATTTTCCAGGCATGG-3′). Amplicons were extracted from agarose gel by using a MinElute gel extraction kit (Qiagen) and quantified. Then, 2 mg of prepared protein extract was incubated with 1 μg of biotin-PIG3 promoter probes and 10 μg of poly(dI-dC)·poly(dI-dC) for 16 h at 4°C. DNA-bound proteins were collected with streptavidin-agarose beads for 1 h and washed five times with HKMG buffer. DNA-bound proteins were then analyzed by IB.
Dual-luciferase reporter assay.
38E6E7 HFK were transfected with firefly luciferase pGL3-PIG3 promoter (p53REwt or p53REm) vector (0.5 μg), and pRL-TK Renilla reporter vector (15 ng) was used as an internal control. Cells were lysed after 48 h, and the luciferase activity was measured by using a dual-luciferase reporter assay system (Promega). Light emission was measured with an Optocompt luminometer (MGM Instruments). The expression of firefly luciferase relative to Renilla luciferase was compared between p53REwt and p53REm PIG3 promoters and was expressed in relative luminescence units (RLU).
Pyrosequencing.
Genomic DNA from cell lines and primary cells was prepared by proteinase K treatment, salting-out extraction, and isopropanol precipitation. Sodium bisulfite modification was performed on 500 ng of DNA using an EZ DNA Methylation-Gold kit (Zymo Research). The quality and completeness of DNA modification was assessed using control primers for modified and nonmodified DNA, respectively (Table 2). To quantify the percentage of methylated cytosine in individual CpG sites, bisulfite-converted DNA was sequenced using a pyrosequencing system (PSQ 96MA; Biotage, Sweden). This method treats each individual CpG site as a C/T polymorphism and generates quantitative data for the relative proportion of methylated versus unmethylated allele. Hot-start PCR was performed with the HotStarTaq Master Mix kit (Qiagen), and pyrosequencing was carried out in accordance with the manufacturer's protocol (Biotage). The target CpGs were evaluated by converting the resulting pyrograms into numerical values for peak heights.
Table 2.
Primers used for PCR and pyrosequencing
| Primera | Sequence (5′–3′) |
|---|---|
| PIG3 pyro (−566/−458) | |
| Forward | GGTATGGTTAGGTTTTTGGT |
| Reverse | CCAAAAATACTACTAAACATCCTAC |
| Seq1 | GGAGAAGGATGTTGG |
| Seq2 | AAGGTTTTTTAATTT |
| Seq3 | TGGATTAGTTAGTT |
| PIG3 pyro (+97/+103) | |
| Forward | GTTGGATGYGGTAGAGTAGGAT |
| Reverse | TTCTACTTAACCCCACCCC |
| Seq1 | TGGAAGTTTTTAGT |
| Seq2 | TTTTTATGTATTAGG |
| Seq3 | GGTTTTGGTTTTTG |
| PIG3 pyro (+339/+359) | |
| Forward | GAGGGAGTGTGATTTGTTTG |
| Reverse | CAACCCAACCTCAAACTAAC |
| Seq1 | TTTGATTTTTTT |
| Seq2 | TATTTTATTTTT |
| Seq3 | TAGGTAGGAGTT |
The region is indicated in parentheses where applicable. Seq, sequencing.
Statistical analysis.
Variations of sample values are represented by error bars that show the standard deviations from the mean. The statistical differences were analyzed using a two-sided Student t test. The P values were considered significant if P < 0.05 (*); P values below 0.01 were considered highly significant (**).
RESULTS
IKKβ nuclear localization and ΔNp73α accumulation are mediated mainly by HPV38 E7 oncoprotein.
Our previous work showed that IκB kinase β (IKKβ), known to play a key role in the cytoplasm as a positive regulator of the NF-κB signaling pathway, is also localized in the nuclei of human foreskin keratinocytes (HFK) immortalized by the E6 and E7 oncoproteins of HPV38 (38E6E7 HFK) (14). In these cells, the IKKβ nuclear form phosphorylated and stabilized ΔNp73α, which in turn inhibited the expression of some p53-regulated genes (14). To corroborate our findings and demonstrate that IKKβ nuclear accumulation was dependent on the expression of HPV38 E6 and E7, we generated HPV38 immortalized HFK lines in which we could silence the expression of viral genes. HFK were transduced by recombinant retroviruses in which HPV38 E6 and E7 genes are flanked by two LoxP sites. After immortalization, 38E6E7 HFK were transduced with a retrovirus expressing the recombinase Cre, which resulted in the deletion of the viral genes and consequent loss of its expression. Real-time PCR showed a decrease of ca. 50% in HPV38 E6 and E7 mRNA levels (Fig. 1A), which resulted in a significant reduction in IKKβ in the nucleus, as determined by immunofluorescence and cellular fractionation (Fig. 1B and C).
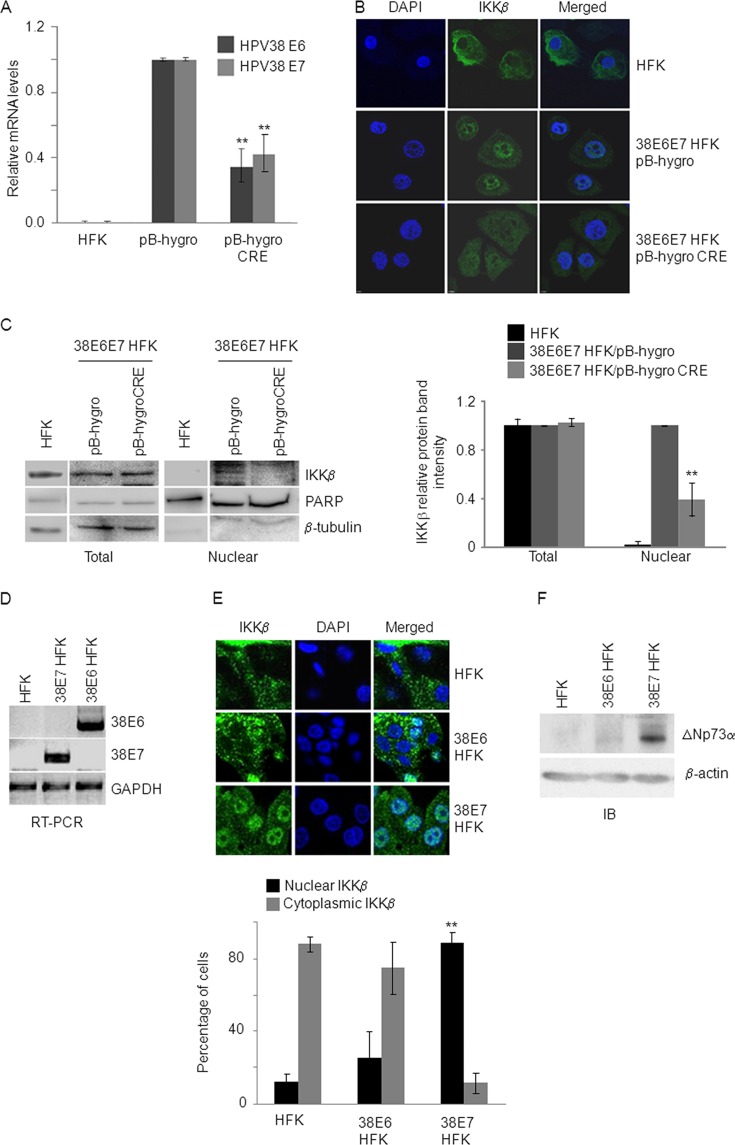
Fig 1.
HPV38 E7 expression promotes IKKβ nuclear accumulation. (A, B, and C) HFK were first transduced with the retrovirus pBabe-puro-38E6E7LoxP/LoxP. After selection in puromycin, cells were transduced with retrovirus carrying either pBabe-hygro-CRE recombinase or pBabe-hygro as a negative control and selected for hygromycin resistance. The results were compared to the parental untransduced HFK. (A) Total RNA from indicated control HFK and transduced HFK was extracted, and HPV38 E6 and E7 expression levels were measured by RT-qPCR. The data shown are the mean values of three independent experiments. **, P < 0.01 pB-hygro-CRE versus the respective pBabe-hygro control sample. (B) HFK, pBabe-hygro-CRE and pBabe-hygro 38E6E7 HFK were seeded on cover slides and stained with anti-IKKβ antibody. Cellular nuclei were counterstained with DAPI. The results are representative of two independent experiments. (C) Twenty micrograms of nuclei extracts and 20 μg of total lysate from HFK, pBabe-hygro-CRE, and pBabe-hygro HFK were analyzed by IB with the indicated antibodies (left panel). Protein bands were quantified using Bio-Rad Image Lab software (right panel). The histogram represents the levels of IKKβ in the total and nuclear extract after normalization to the levels of β-tubulin and PARP, respectively. The data shown are the mean values of three independent experiments. **, P < 0.01 pB-hygro-CRE versus the pB-hygro control sample. (D, E, and F) HFK expressing HPV38 E6 (38E6 HFK) or HPV38 E7 (38E7 HFK) were generated by retroviraltransduction. HFK transduced with the pLXSN empty vector (HFK) were used as a negative control. After transduction, HFK were selected in neomycin, harvested and processed for the preparation of total RNA and protein extracts. (D) The expression of HPV38 E6, E7, and GAPDH was determined by RT-PCR. The results are representative of two independent experiments. (E) HFK, 38E6 HFK, and 38E7 HFK were seeded on cover slides and stained with anti-IKKβ antibody and DAPI (upper panel). In each staining, the percentage of cells with nuclear or cytoplasmic staining was determined by counting 100 cells/field in three different fields. Cells with nuclear or cytoplasmic staining for IKKβ were represented as the ratio to total cell number (100%). The results shown in the histogram (lower panel) are the average of two independent experiments. The difference in the percentage of cells with nuclear IKKβ in 38E7 HFK versus HFK is significant (**, P < 0.01). (F) Portions (40 μg) of total protein extracts from HFK, 38E6 HFK, and 38E7 HFK were analyzed by IB with the indicated antibodies. The results are representative of two independent experiments.
Next, we determined whether IKKβ nuclear translocation and ΔNp73α accumulation were induced by one or both viral oncoproteins and generated HFK expressing HPV38 E6 (38E6 HFK) or HPV38 E7 (38E7 HFK) (Fig. 1D). Immunofluorescence staining showed that IKKβ is localized mainly in the cytoplasm in HFK and 38E6 HFK, whereas it is accumulated in the nucleus of 38E7 HFK (Fig. 1E). Accordingly, ΔNp73α protein levels were elevated in 38E7 HFK compared to 38E6 HFK and mock cells (Fig. 1F). In addition, as expected, inhibition of IKKβ by the chemical compound Bay-11-7082 led to a significant decrease in ΔNp73α protein levels (data not shown). Thus, HPV38 E7 is directly involved in IKKβ nuclear translocation and the resulting ΔNp73α accumulation.
IKKβ and ΔNp73α are part of the same transcriptional regulatory complex.
Previous findings highlighted a nuclear role for IKKβ being directly involved in transcriptional regulation (28). Indeed, in the nuclei of B cells, IKKβ is recruited together with the receptor of B-cell activating factor (BAFF) to specific promoters, where it phosphorylates histone H3, inducing chromatin remodeling and transcription initiation (28). Our previous study showed that IKKβ induces the recruitment of ΔNp73α to p53-regulated promoters but did not rule out the possibility that IKKβ could be part of a transcriptional regulatory complex and bind the DNA. Thus, we first determined whether, similarly to ΔNp73α, IKKβ is recruited to p53-regulated promoters. ChIP experiments showed that in 38E6E7 HFK, IKKβ is able to bind the −240/−400 region of the PIG3 promoter, which comprises a p53RE (Fig. 2A), whereas no binding was observed on the same promoter in primary keratinocytes. In addition, silencing of viral gene expression by shRNA (Fig. 2B) significantly affected the recruitment of IKKβ and ΔNp73α to the PIG3 promoter (Fig. 2A) and restored PIG3 expression (Fig. 2C) as a consequence of the loss of the negative regulatory complex on the promoter. ChIP/Re-ChIP experiments confirmed that IKKβ and ΔNp73α are part of the same transcriptional regulatory complex (Fig. 2D). To corroborate these findings, we performed oligonucleotide pulldown experiments using biotinylated DNA probes that contain a region of the PIG3 promoter encompassing the wild-type or mutated p53RE (the −240/−400 region). 38E6E7 HFK were transiently transfected with vectors expressing Flag-IKKβ or hemagglutinin (HA)-tagged ΔNp73α, and cellular extracts were prepared and used for the oligonucleotide pulldown experiments. We observed that the PIG3 promoter fragment containing the wild-type p53RE coprecipitated with Flag-IKKβ and HA-ΔNp73α, whereas mutation of the p53RE significantly decreased precipitation of the two cellular proteins (Fig. 3A). Similar results were obtained using cellular extracts of 38E6E7 HFK expressing endogenous levels of IKKβ and ΔNp73α (Fig. 3C).
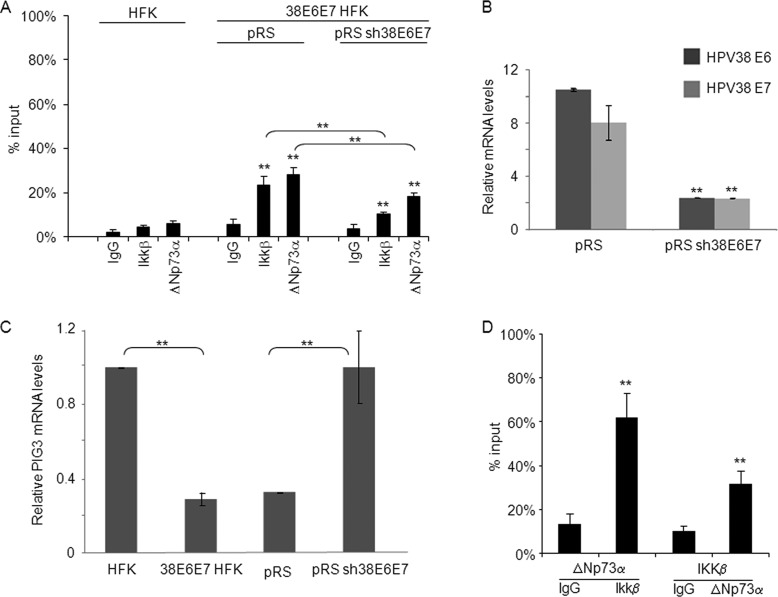
Fig 2.
ΔNp73α and IKKβ co-bind on the PIG3 promoter. (A) HFK and 38E6E7 HFK transduced with pRetroSuper (pRS) or shRNA-HPV38E6E7 (pRS sh38E6E7) were fixed with formaldehyde and processed for ChIP using anti-IKKβ, anti-p73α and nonspecific IgG. The eluted DNA was analyzed by qPCR with primers flanking the p53RE within the PIG3 promoter. Part of the total chromatin fraction (1/10th) was used as input. The histogram shows the amount of promoter specifically bound by each protein and expressed as a percentage of the amount of the PIG3 promoter in the input (100%). The results shown in the histogram are the average of three independent experiments performed in duplicate. The differences in the percentage of binding between IKKβ or ΔNp73α ChIP and the corresponding IgG control are significant (**, P < 0.01) in pRS and pRS sh38E6E7 samples. In addition, the decrease in IKKβ or ΔNp73α binding affinity to the PIG3 promoter in pRS sh38E6E7 versus pRS is also significant (**, P < 0.01). (B) 38E6E7 HFK were retrovirally transduced with pRS empty vector or pRS sh38E6E7. After puromycin selection, cells were collected for RNA extraction. The mRNA levels of HPV38 E6 and E7 were determined by RT-qPCR. The results are the averages of three independent experiments performed in duplicate. **, P < 0.01 for pRS sh38E6E7 versus the corresponding pRS sample. (C) Total RNA was extracted from HFK, 38E6E7 HFK, and 38E6E7 HFK transduced with pRS and pRS sh38E6E7 and analyzed by RT-qPCR for PIG3 expression levels and normalized to GAPDH levels. The results are the average of three independent experiments performed in duplicate. **, P < 0.01 between the indicated samples. (D) 38E6E7 HFK were processed for ChIP with ΔNp73α or IKKβ antibodies. The eluted DNA was subjected to Re-ChIP with IKKβ or ΔNp73α antibodies, respectively. IgG antibody was included as negative control. Input and coimmunoprecipitation DNA were analyzed by qPCR for the p53RE region of the PIG3 promoter. The results are the average of three independent experiments performed in duplicate. **, P < 0.01 versus the corresponding IgG control.
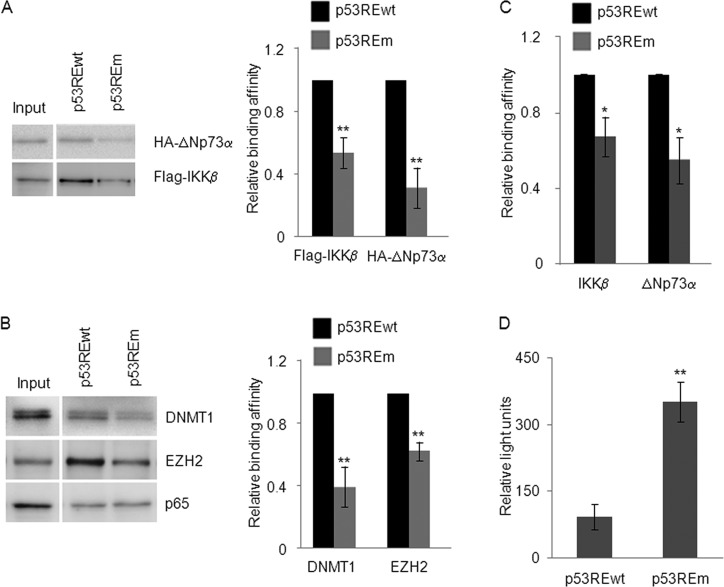
Fig 3.
EZH2 and DNMT1, like ΔNp73α and IKKβ, bind to p53RE on the PIG3 promoter. (A and B) 38E6E7 HFK transiently expressing HA-ΔNp73α and Flag-IKKβ were processed for the oligonucleotide pulldown assay. Two milligrams of cell lysate was incubated with biotinylated probes containing the p53RE of the PIG3 promoter, either wild-type (p53REwt) or mutated (p53REm). DNA-associated proteins were recovered by precipitation with streptavidin beads and analyzed by IB with the indicated antibodies (left panel). Protein band intensities of three independent IB for each experiment were quantified by Image Lab (Bio-Rad) and shown in the right panels. The results are the average of three independent experiments. The differences in the binding affinity of IKKβ and ΔNp73α to the p53REwt versus p53REm probes are significant (**, P < 0.01). (C) The oligonucleotide pulldown assay was performed using 2 mg of cell lysate from untransfected 38E6E7 HFK, as explained for panels A and B. The results are the average of two independent experiments. *, P < 0.05 for p53REm versus the corresponding p53REwt sample. (D) 38E6E7 HFK were transfected with PIG3 promoter-firefly luciferase reporter construct harboring either a wild-type or mutated p53RE and with a Renilla construct. At 24 h after transfection, cells were processed for the luciferase assay. Relative light units (RLU) values are represented in the histogram. The data shown are the mean values of three independent experiments performed in triplicate. The differences in the luciferase activity between mutated and wild-type promoter are significant (**, P < 0.01).
Due to the inhibitory function of IKKβ and ΔNp73α on the PIG3 promoter, we next determined whether enzymes inducing epigenetic changes were also present in the complex. Therefore, we analyzed the precipitated complexes in oligonucleotide pulldown assays by IB using several antibodies against epigenetic enzymes (data not shown). The IKKβ/ΔNp73α complex coprecipitated with the polycomb group (PcG) 2 member EZH2 and DNA methyltransferase DNMT1 (Fig. 3B). As observed for IKKβ and ΔNp73α, mutation of the PIG3 p53RE affected precipitation of EZH2 and DNMT1 (Fig. 3B). As a control, we also determined the presence of p65 since the promoter region used in the assay also includes a putative NF-κB RE. As expected, p65 binding to the DNA probe was not affected by mutation of the p53RE (Fig. 3B). Finally, transient transfection experiments in 38E6E7 HFK using constructs containing the PIG3 promoter cloned in front of the luciferase reporter gene showed that mutation of the p53RE strongly increased luciferase activity (Fig. 3D), providing additional evidence for the transcriptional inhibitory role of the p53RE in these cells.
To further evaluate the possibility that the four cellular proteins are part of the same complex, we performed ChIP/Re-ChIP experiments. We first carried out ChIP with DNMT1, EZH2, or ΔNp73α antibodies, followed by Re-ChIP using an IKKβ antibody. The data showed that DNMT1, EZH2, and ΔNp73α can associate with IKKβ (Fig. 4A), further confirming the data obtained with the oligonucleotide pulldown assay shown in Fig. 3A and B. ChIP experiments carried out in cells expressing either HPV38 E6 or E7 showed that DNMT1, EZH2, ΔNp73α, and IKKβ bind to the PIG3 promoter in the presence of E7, but not in the presence of E6 (Fig. 4B), a finding in agreement with previous results showing that HPV38 E7 induces high levels of ΔNp73α and IKKβ nuclear accumulation (Fig. 1E and F). In addition, downregulation of ΔNp73α expression in 38E6E7 HFK by transfection of ΔNp73α AntiSense oligonucleotide (AS) led to loss of the tetrameric complex on the PIG3 promoter (Fig. 4C). These results were consistent with previous findings from our group showing that ΔNp73α AS treatment of 38E6E7 HFK led to rescue of the mRNA levels of different p53-regulated genes, including PIG3 (3). We also observed that 38E6E7 HFK displayed increased levels of EZH2 compared to primary keratinocytes (Fig. 4D). However, total trimethylation at lysine 27 of histone H3 (H3Lys27m3), an event induced by EZH2, did not significantly change in the presence of HPV38 E6 and E7 (Fig. 4E), showing that the events described here are not the results of a general effect of HPV38 E6 and E7 on the enzymatic activity of EZH2, but are specific for a subset of gene promoters. Together, these findings demonstrate the existence in 38E6E7 HFK of a tetrameric complex able to inhibit the expression of p53-regulated genes, such as PIG3.
The IKKβ/ΔNp73α/DNMT1/EZH2 tetrameric complex induces epigenetic changes on the PIG3 promoter, inhibiting its activity.
We further characterized the role of the novel IKKβ/ΔNp73α/DNMT1/EZH2 tetrameric complex in transcriptional regulation. ChIP experiments revealed H3Lys27 hypermethylation in the PIG3 promoter region surrounding the p53RE, as well as in the PIG3 coding sequence region (+322/+492), which contains a microsatellite region that has been proposed as a p53 binding site (29) (Fig. 5A). Interestingly, H3Lys27m3 was not observed in the promoter of another p53-regulated gene, Survivin (Fig. 5A), indicating that the IKKβ/ΔNp73α complex does not bind to the p53RE of all p53-regulated promoters. To further explore this issue, we performed ChIP for IKKβ, ΔNp73α, DNMT1, and EZH2 and investigated their ability to bind to different p53-regulated promoters. The complex was found to be associated with most of the examined p53-regulated promoters, except the Survivin promoter (Fig. 5B).
Fig 5.
The PIG3 promoter is silenced by DNA methylation and histone H3 Lys27 trimethylation. (A) 38E6E7 HFK were harvested and processed for ChIP using antibodies for the indicated proteins and nonspecific IgG control. The bound DNA was analyzed by qPCR with primer sets specific for two regions of the PIG3 gene (the p53RE region of the PIG3 promoter and the PIG3 ORF +322/+492 region), as well as primer for Survivin or the GAPDH promoter. The amount of promoter specifically bound by each protein was expressed as a percentage of the input sample. The data shown are the mean values of three independent experiments performed in duplicate. **, P < 0.01 versus the corresponding IgG control. (B) 38E6E7 HFK were subjected to ChIP using antibodies for the indicated proteins and nonspecific IgG control. PCR was performed with primers flanking the p53REs of the indicated gene promoters. Input indicates 10% of total DNA per IP. PCR products were loaded on 2% agarose gel. The data shown are representative of two independent experiments. (C) DNA of HFK, 38E6E7 HFK, and 38E6E7 HFK treated with 5-Aza was extracted, and the methylation status of indicated PIG3 promoter regions was analyzed by pyrosequencing. Assays were designed next to the p53RE, TSS and microsatellite regions. Forward (F) and reverse (R) primers used for pyrosequencing are indicated. Average methylation is shown for the different CpG sites analyzed. CpG sites are named relative to the distance to TSS. The data shown are representative of two independent experiments. The differences in the methylation levels in the −566/−458 region of the PIG3 promoter in 38E6E7 HFK versus 38E6E7 HFK + 5-Aza, and in 38E6E7 HFK versus HFK are statistically significant (P values of 0.02 and 0.003, respectively). No statistically significant differences were observed in other regions (+97/+103 and + 339/+359). (D) 38E6E7 HFK were treated for 72 h with DNA methylation inhibitor 5-Aza or DMSO alone (Control). PIG3 mRNA levels were analyzed by RT-qPCR. The data shown are the mean values of three independent experiments. *, P < 0.05 versus the untreated control sample. (E) 38E6E7 HFK were transfected with a control siRNA (Scramble) or stealth siRNAs specific for EZH2 and DNMT1. At 72 h after transfection, the protein levels of DNMT1, EZH2, and β-actin were analyzed by IB (left panel), and PIG3 mRNA levels were determined by RT-qPCR (right panel). The results are representative of two independent experiments. * and **, P < 0.05 and P < 0.01, respectively, for siRNAs versus the Scramble sample. (F) 38E6E7 HFK were transfected with a control siRNA (Scramble) or stealth siRNAs specific for EZH2 and DNMT1. The day after transfection, 104 cells were seeded to perform colony formation assays. Cells were cultured for 7 days and then stained with crystal violet. The results are representative of two independent experiments.
Next, we evaluated the role of the DNA methyltransferase DNMT1 in silencing PIG3 expression. The PIG3 promoter is located in a CpG-rich region containing the p53RE 320 bp upstream of the transcription start site (TSS). This CpG-rich region also includes a bona fide CpG island overlapping the first exon of the gene containing the microsatellite region (Fig. 5C). Pyrosequencing assays were performed in bisulfite-modified DNA obtained from HFK, as well as 38E6E7 HFK cultured in the presence or absence of 5-Aza. Our analysis revealed that the CpGs on the PIG3 promoter region around the p53RE are highly methylated in 38E6E7 HFK (close to 100%) and less methylated in mock cells (HFK) (average, 46%). No methylation was observed at the level of the microsatellite region and the TSS (Fig. 5C). Intermediate methylation levels were obtained after 5-Aza treatment of 38E6E7 HFK. In accordance with this observation, exposure of 38E6E7 HFK to DNA 5-Aza led to the rescue of PIG3 expression (Fig. 5D). The inhibition of DNMT1 intracellular levels in 38E6E7 HFK by specific siRNA also resulted in increased PIG3 expression (Fig. 5E), confirming the specificity of the results obtained with 5-Aza treatment. Gene silencing of EZH2 by siRNA also led to a rescue of PIG3 expression levels (Fig. 5E). In addition, knockdown of either DNMT1 or EZH2 resulted in the inhibition of 38E6E7 HFK growth ability (Fig. 5F). In summary, the epigenetic enzymes EZH2 and DNMT1, via their interaction with the ΔNp73α/IKKβ complex, appear to be directly involved in the repression of p53-regulated promoters inducing H3Lys27m3 and DNA methylation, respectively, and play a positive role in cellular proliferation.
IKKβ plays a key role in the proliferation of 38E6E7 HFK independent of NF-κB signaling.
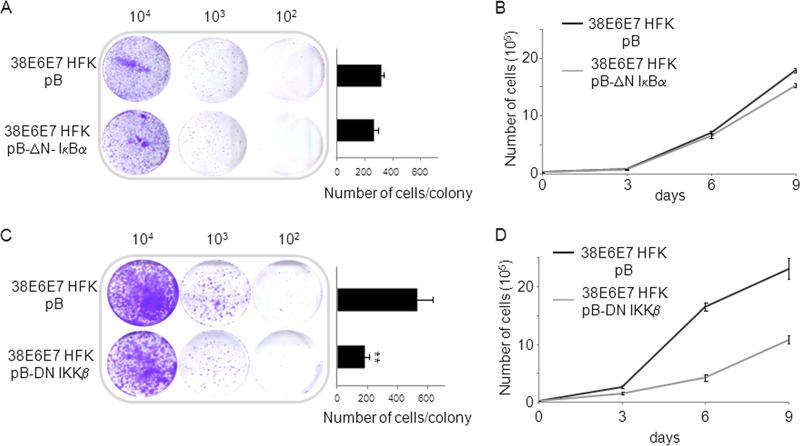
Our data highlighted a novel role of IKKβ in the nucleus of 38E6E7 HFK. In addition, we previously showed that HPV38 E6 and E7 stimulate IKKβ activity in the cytoplasm, promoting IκBα phosphorylation and degradation, leading to activation of NF-κB signaling (14, 30). We therefore determined whether IKKβ could contribute to proliferation of 38E6E7 HFK, independent of the activation of NF-κB signaling. We first stably expressed in 38E6E7 HFK a deletion mutant of IκBα (ΔN-IκBα) that lacks the first 36 amino acids at the N terminus encompassing the IKKβ phosphorylation sites. ΔN-IκBα is no longer regulated by IKKβ and is able to constitutively inhibit the NF-κB canonical pathway by sequestering p65 in the cytoplasm (14, 30). As shown by colony formation assays and short-term growth curves, inhibition of the NF-κB canonical pathway only marginally affected the proliferation of 38E6E7 HFK (Fig. 6A and B). In contrast, constitutive expression of a dominant-negative mutant of IKKβ (Flag-DN-IKKβ) in 38E6E7 HFK significantly decreased their proliferation (Fig. 6C and D). Similar experiments performed in HFK showed that blocking IKKβ, but also NF-κB, negatively affected their growth ability (data not shown). Taken together, these data show that IKKβ, independently of the NF-κB signaling, plays an additional role in promoting proliferation. However, in 38E6E7 HFK the stronger effect of DN-IKKβ on proliferation in comparison to ΔN-IκBα is partly explained by the ability of IKKβ to promote ΔNp73α accumulation in the nucleus (14), an event that does not occur in primary HFK.
Fig 6.
IKKβ plays an NF-κB-independent role in 38E6E7 HFK proliferation. (A and C) 38E6E7 HFK were retrovirally transduced with pBabe-puro, pBabe-puro-dominant-negative IKKβ (pB-DN-IKKβ), or truncated form of IκBα (pB-ΔN-IκBα) and seeded at different dilution (104, 103, and 102) to perform colony formation assays. Cells were cultured for 7 days and then stained with crystal violet (left panel). The average numbers of cells forming colonies were counted and reported in the histograms (right panel). The data shown are the mean values of three independent experiments. **, P < 0.01 38E6E7 HFK pB-DN-IKKβ versus 38E6E7 HFK pB sample. (B and D) After retroviral transduction, 104 38E6E7 HFK expressing DN-IKKβ, ΔN-IκBα, or with pBabe control vector were seeded (day 0) and cultured for 9 days. The total numbers of cells were counted at the indicated time points and reported in the growth curve. The data shown are the mean values of three independent experiments.
DISCUSSION
Several viruses are able to inactivate p53 functions, probably as part of their strategies to guarantee the completion of the viral life cycle. Mucosal high-risk HPV types induce p53 degradation via the E6 oncoprotein. Although HPV49 has the same mechanism as the mucosal high-risk human papillomavirus types in inactivating p53 (31), other beta HPV types target this cellular tumor suppressor by alternative strategies (3, 7, 9, 14). We previously showed that the expression of HPV38 E6 and E7 oncoproteins in primary keratinocytes promoted accumulation of the p53 antagonist ΔNp73α and cellular immortalization (3, 4, 6, 14). Downregulation of ΔNp73α expression in 38E6E7 HFK significantly reduced cellular viability and resulted in activation of p53-dependent apoptosis (3). Similarly, knocking down ΔNp73α expression in HPV38 E6 and E7 transgenic mice partially restored p53 functions (15). Thus, data obtained in in vitro and in vivo experimental models highlighted the key role of ΔNp73α accumulation in cellular proliferation and transformation.
One of the key events in ΔNp73α accumulation in 38E6E7 HFK is the translocation of IKKβ into the nucleus, where it phosphorylates and stabilizes ΔNp73α (14). Here, we show that these events are mediated by HPV38 E7. The underlying mechanism is still unknown. Several viruses have been demonstrated to interfere with the NF-κB pathway (32, 33); for instance, HTLV1 is known to activate the IKK complex by direct binding of its transforming protein, Tax, to IKKγ (34). Preliminary studies from our group have shown that, similarly to Tax, HPV38 E7 can bind to IKKβ (D. Saidj, unpublished data). Whether this event triggers nuclear accumulation of IKKβ is still under investigation. Interestingly, independent studies have described novel IKKβ functions in the nucleus, although none of them were linked to ΔNp73α (35–37). For instance, Fu et al. reported that IKKβ forms a positive transcriptional regulatory complex with the receptor of B-cell activating factor (BAFF) (28). Findings of the current study show that, similarly to the BAFF complex, IKKβ is intrinsically part of the transcriptional regulatory complex that inhibits the expression of some p53-regulated genes. Most importantly, our data showed for the first time that two epigenetic enzymes, EZH2 and DNMT1, are part of the IKKβ/ΔNp73α complex. EZH2 is a component of the polycomb complex 2 and induces H3Lys27m3 and consequent closure of chromatin (38). Accordingly, ChIP experiments performed in 38E6E7 HFK demonstrated the presence of H3Lys27m3 on p53-regulated promoters.
The presence of DNMT1 in the IKKβ/ΔNp73α complex is consistent with previous findings that documented its ability to bind to EZH2 (38) and can account for the high levels of methylation detected in the region surrounding the p53RE in 38E6E7 HFK. EZH2 and DNMT1 binding to the IKKβ/ΔNp73α complex could contribute to its ability to block p53-mediated transcription. According to the current model, ΔNp73α exerts its dominative-negative function toward p53 via competition for the same RE, since the two proteins share a homologous DNA-binding domain (39). Due to its lack of a transactivation TA domain, binding of ΔNp73α to the promoter of certain genes results in their transcriptional silencing (16, 17). Our previous data confirm this model; in fact, we showed that in 38E6E7 HFK, ΔNp73α bound to the p53RE in the PIG3 promoter and that silencing of ΔNp73α expression restored p53 binding to the promoter and reactivated PIG3 expression (3, 14).
Consistent with the previous findings, in the present study we observed that the “naked” DNA construct, containing the luciferase gene driven by the wild-type PIG3 promoter, has low activity if transfected into 38E6E7 HFK, whereas it can be strongly activated if ΔNp73α binding to p53RE is hampered. However, we described here that ΔNp73α is able to repress transcription also by facilitating the recruitment of DNMT1 and EZH2 to specific p53-regulated promoters, which in turn induce epigenetic changes. It is likely that these two enzymes, together with ΔNp73α and IKKβ, contribute to prolonged silencing of the expression of some p53-regulated genes under certain conditions, for instance during cellular transformation. Indeed, EZH2 and DNMT1, like ΔNp73α, are often upregulated in cancer (40–44), where they appear to target specific genes for methylation (45). In addition, our data show that silencing the expression of EZH2 and DNMT1 in 38E6E7 HFK leads to a rescue of PIG3 levels and to a reduction in cell growth ability, further confirming the key role of EZH2 and DNMT1 in HPV38-mediated inhibition of p53-regulated expression.
The mechanisms by which EZH2 and DNMT1 can be recruited to specific promoters are still poorly characterized (46). Our findings show that ΔNp73α is one of these mechanisms. Indeed, downregulation of ΔNp73α in 38E6E7 HFK led to disruption of the complex and release of the two epigenetic enzymes, which are no longer recruited to the PIG3 promoter.
We previously showed that ΔNp73α and IKKβ colocalize in the nuclei of cancer-derived cells, where they are responsible for inhibition of p53 proapoptotic pathways (14). Downregulation of ΔNp73α or IKKβ levels in these cancer cells led to an increase in their sensitivity to chemotherapeutic agents by restoring p53-mediated apoptosis (our unpublished data). Since EZH2 and DNMT1 both accumulate in cancer cells, it will be of interest to explore whether, as observed in 38E6E7 HFK, they can form a multimeric complex with ΔNp73α and IKKβ in cancer-derived cell lines in order to constitutively silence p53-regulated transcription.
In summary, here we show for the first time that the E7 oncoprotein of HPV38 promotes the formation of a multimeric complex that negatively regulates the expression of some p53 target gene promoters. Whether this is a specific event exclusively occurring in HPV38-infected cells or a more common mechanism taking place in human carcinogenesis remains to be further investigated.
ACKNOWLEDGMENTS
We are grateful to all of the members of our laboratories for their cooperation.
The study was partly supported by the European Commission, grant HPV-AHEAD (FP7-HEALTH-2011-282562).
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Bouvard V, Gabet AS, Accardi R, Sylla SB, Tommasino M. 2006. The cutaneous human papillomavirus types and non-melanoma-skin cancer, p 269–277 In Campo MS. (ed), Papillomavirus research: from natural history to vaccine and beyond. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 2.Pfister H, Fuchs PG, Majewski S, Jablonska S, Pniewska I, Malejczyk M. 2003. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch. Dermatol. Res. 295:273–279 [DOI] [PubMed] [Google Scholar]
- 3.Accardi R, Dong W, Smet A, Cui R, Hautefeuille A, Gabet AS, Sylla BS, Gissmann L, Hainaut P, Tommasino M. 2006. Skin human papillomavirus type 38 alters p53 functions by accumulation of ΔNp73. EMBO Rep. 7:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldeira S, Zehbe I, Accardi R, Malanchi I, Dong W, Giarre M, de Villiers EM, Filotico R, Boukamp P, Tommasino M. 2003. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 77:2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong W, Kloz U, Accardi R, Caldeira S, Tong WM, Wang ZQ, Jansen L, Durst M, Sylla BS, Gissmann L, Tommasino M. 2005. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 79:14899–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabet AS, Accardi R, Bellopede A, Popp S, Boukamp P, Sylla BS, Londono-Vallejo JA, Tommasino M. 2008. Impairment of the telomere/telomerase system and genomic instability are associated with keratinocyte immortalization induced by the skin human papillomavirus type 38. FASEB J. 22:622–632 [DOI] [PubMed] [Google Scholar]
- 7.Howie HL, Koop JI, Weese J, Robinson K, Wipf G, Kim L, Galloway DA. 2011. Beta-HPV 5 and 8 E6 promote p300 degradation by blocking AKT/p300 association. PLoS Pathog. 7:e1002211. 10.1371/journal.ppat.1002211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel A, Kopp-Schneider A, Zentgraf H, Gruber AD, de Villiers EM. 2006. E6/E7 expression of human papillomavirus type 20 (HPV-20) and HPV-27 influences proliferation and differentiation of the skin in UV-irradiated SKH-hr1 transgenic mice. J. Virol. 80:11153–11164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muench P, Probst S, Schuetz J, Leiprecht N, Busch M, Wesselborg S, Stubenrauch F, Iftner T. 2010. Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res. 70:6913–6924 [DOI] [PubMed] [Google Scholar]
- 10.Muschik D, Braspenning-Wesch I, Stockfleth E, Rosl F, Hofmann TG, Nindl I. 2011. Cutaneous HPV23 E6 prevents p53 phosphorylation through interaction with HIPK2. PLoS One 6:e27655. 10.1371/journal.pone.0027655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaper ID, Marcuzzi GP, Weissenborn SJ, Kasper HU, Dries V, Smyth N, Fuchs P, Pfister H. 2005. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res. 65:1394–1400 [DOI] [PubMed] [Google Scholar]
- 12.Wallace NA, Robinson K, Howie HL, Galloway DA. 2012. HPV 5 and 8 E6 abrogate ATR activity resulting in increased persistence of UVB induced DNA damage. PLoS Pathog. 8:e1002807. 10.1371/journal.ppat.1002807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viarisio D, Mueller-Decker K, Kloz U, Aengeneyndt B, Kopp-Schneider A, Grone HJ, Gheit T, Flechtenmacher C, Gissmann L, Tommasino M. 2011. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog. 7:e1002125. 10.1371/journal.ppat.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Accardi R, Scalise M, Gheit T, Hussain I, Yue J, Carreira C, Collino A, Indiveri C, Gissmann L, Sylla BS, Tommasino M. 2011. IκB kinase beta promotes cell survival by antagonizing p53 functions through ΔNp73α phosphorylation and stabilization. Mol. Cell. Biol. 31:2210–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong W, Arpin C, Accardi R, Gissmann L, Sylla BS, Marvel J, Tommasino M. 2008. Loss of p53 or p73 in human papillomavirus type 38 E6 and E7 transgenic mice partially restores the UV-activated cell cycle checkpoints. Oncogene 27:2923–2928 [DOI] [PubMed] [Google Scholar]
- 16.Buhlmann S, Putzer BM. 2008. DNp73 a matter of cancer: mechanisms and clinical implications. Biochim. Biophys. Acta 1785:207–216 [DOI] [PubMed] [Google Scholar]
- 17.Melino G, De Laurenzi V, Vousden KH. 2002. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer 2:605–615 [DOI] [PubMed] [Google Scholar]
- 18.Melino G, Lu X, Gasco M, Crook T, Knight RA. 2003. Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci. 28:663–670 [DOI] [PubMed] [Google Scholar]
- 19.Petrenko O, Zaika A, Moll UM. 2003. deltaNp73 facilitates cell immortalization and cooperates with oncogenic Ras in cellular transformation in vivo. Mol. Cell. Biol. 23:5540–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM. 2002. Transactivation-deficient ΔTA-p73 acts as an oncogene. Cancer Res. 62:3598–3602 [PubMed] [Google Scholar]
- 21.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. 2010. Isoform-specific p73 knockout mice reveal a novel role for ΔNp73 in the DNA damage response pathway. Genes Dev. 24:549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casciano I, Banelli B, Croce M, Allemanni G, Ferrini S, Tonini GP, Ponzoni M, Romani M. 2002. Role of methylation in the control of ΔNp73 expression in neuroblastoma. Cell Death Differ. 9:343–345 [DOI] [PubMed] [Google Scholar]
- 23.Fillippovich I, Sorokina N, Gatei M, Haupt Y, Hobson K, Moallem E, Spring K, Mould M, McGuckin MA, Lavin MF, Khanna KK. 2001. Transactivation-deficient p73α (p73Δexon2) inhibits apoptosis and competes with p53. Oncogene 20:514–522 [DOI] [PubMed] [Google Scholar]
- 24.Stiewe T, Putzer BM. 2002. Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ. 9:237–245 [DOI] [PubMed] [Google Scholar]
- 25.Accardi R, Fathallah I, Gruffat H, Mariggio G, Le Calvez-Kelm F, Voegele C, Bartosch B, Hernandez-Vargas H, McKay J, Sylla BS, Manet E, Tommasino M. 2013. Epstein-Barr virus transforming protein LMP-1 alters B cells gene expression by promoting accumulation of the oncoprotein ΔNp73α. PLoS Pathog. 9:e1003186. 10.1371/journal.ppat.1003186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allart S, Martin H, Detraves C, Terrasson J, Caput D, Davrinche C. 2002. Human cytomegalovirus induces drug resistance and alteration of programmed cell death by accumulation of ΔN-p73α. J. Biol. Chem. 277:29063–29068 [DOI] [PubMed] [Google Scholar]
- 27.Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L, Lin-Lee YC, Pham LV, Tamayo AT, Yoshimura LC, Ford RJ. 2009. BAFF-R promotes cell proliferation and survival through interaction with IKKβ and NF-κB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood 113:4627–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30:315–320 [DOI] [PubMed] [Google Scholar]
- 30.Hussain I, Fathallah I, Accardi R, Yue J, Saidj D, Shukla R, Hasan U, Gheit T, Niu Y, Tommasino M, Sylla BS. 2011. NF-κB protects human papillomavirus type 38 E6/E7-immortalized human keratinocytes against tumor necrosis factor alpha and UV-mediated apoptosis. J. Virol. 85:9013–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornet I, Bouvard V, Campo MS, Thomas M, Banks L, Gissmann L, Lamartine J, Sylla BS, Accardi R, Tommasino M. 2012. Comparative analysis of transforming properties of E6 and E7 from different beta human papillomavirus types. J. Virol. 86:2366–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun SC, Cesarman E. 2011. NF-κB as a target for oncogenic viruses. Curr. Top. Microbiol. Immunol. 349:197–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tato CM, Hunter CA. 2002. Host-pathogen interactions: subversion and utilization of the NF-κB pathway during infection. Infect. Immun. 70:3311–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC, Harhaj EW, Xiao G, Good L. 2000. Activation of I-κB kinase by the HTLV type 1 Tax protein: mechanistic insights into the adaptor function of IKKγ. AIDS Res. Hum. Retrovir. 16:1591–1596 [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Meng Q, Kao W, Xia Y. 2011. IκB kinase beta regulates epithelium migration during corneal wound healing. PLoS One 6:e16132. 10.1371/journal.pone.0016132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irelan JT, Murphy TJ, DeJesus PD, Teo H, Xu D, Gomez-Ferreria MA, Zhou Y, Miraglia LJ, Rines DR, Verma IM, Sharp DJ, Tergaonkar V, Chanda SK. 2007. A role for IκB kinase 2 in bipolar spindle assembly. Proc. Natl. Acad. Sci. U. S. A. 104:16940–16945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchiya Y, Asano T, Nakayama K, Kato T, Jr, Karin M, Kamata H. 2010. Nuclear IKKβ is an adaptor protein for IκBα ubiquitination and degradation in UV-induced NF-κB activation. Mol. Cell 39:570–582 [DOI] [PubMed] [Google Scholar]
- 38.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871–874 [DOI] [PubMed] [Google Scholar]
- 39.Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. 2000. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113(Pt 10):1661–1670 [DOI] [PubMed] [Google Scholar]
- 40.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 100:11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, Quintanilla-Martinez L, Kovar H, Staege MS, Muller-Tidow C, Burdach S. 2009. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. U. S. A. 106:5324–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33:61–65 [DOI] [PubMed] [Google Scholar]
- 43.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624–629 [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, Wang X, Ghosh D, Shah RB, Varambally S, Pienta KJ, Chinnaiyan AM. 2007. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 67:10657–10663 [DOI] [PubMed] [Google Scholar]
- 45.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. 2007. Epigenetic stem cell signature in cancer. Nat. Genet. 39:157–158 [DOI] [PubMed] [Google Scholar]
- 46.Paschos K, Allday MJ. 2010. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]