Abstract
The host defense against viral infection is acquired during the coevolution or symbiosis of the host and pathogen. Several cellular factors that restrict retroviral infection have been identified in the hosts. Feline leukemia virus (FeLV) is a gammaretrovirus that is classified into several receptor interference groups, including a novel FeLV-subgroup D (FeLV-D) that we recently identified. FeLV-D is generated by transduction of the env gene of feline endogenous gammaretrovirus of the domestic cat (ERV-DCs) into FeLV. Some ERV-DCs are replication competent viruses which are present and hereditary in cats. We report here the determination of new viral receptor interference groups and the discovery of a soluble antiretroviral factor, termed Refrex-1. Detailed analysis of FeLV-D strains and ERV-DCs showed two receptor interference groups that are distinct from other FeLV subgroups, and Refrex-1 specifically inhibited one of them. Refrex-1 is characterized as a truncated envelope protein of ERV-DC and includes the N-terminal region of surface unit, which is a putative receptor-binding domain, but lacks the transmembrane region. Refrex-1 is efficiently secreted from the cells and appears to cause receptor interference extracellularly. Two variants of Refrex-1 encoded by provirus loci, ERV-DC7 and DC16, are expressed in a broad range of feline tissues. The host retains Refrex-1 as an antiretroviral factor, which may potentially prevent reemergence of the ERVs and the emergence of novel ERV-related viruses in cats. Refrex-1 may have been acquired during endogenization of ERV-DCs and may play an important role in retroviral restriction and antiviral defense in cats.
INTRODUCTION
Feline leukemia virus (FeLV) is an exogenous retrovirus, belonging to the genus Gammaretrovirus. FeLV has been shown to induce many diseases in cats, such as lymphoma, myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), aplastic anemia, and immunodeficiency (1, 2). The primary translation product of the FeLV env gene is processed through proteolytic cleavage into two functional units: the surface unit (SU) and the transmembrane unit (TM). The entry of retroviruses into target cells is governed by the interaction of the retroviral SU with specific cell surface receptors (3). FeLV can be categorized into several FeLV subgroups based on their viral receptor interference and host range properties: FeLV subgroups and their receptors of FeLV-A, FeLV-B, FeLV-C, FeLV-AC, and FeLV-T have been identified (4–9). In addition to these FeLV subgroups, our laboratory recently identified a novel FeLV subgroup (FeLV-D), which was generated through recombination in the env region between FeLV-A and an endogenous gammaretrovirus present in the feline genome (ERV-DC) (10). FeLV-A is the common subgroup and is horizontally transmitted among cats. Other subgroups may have arisen from this variant. For instance, it has been shown that FeLV-B arose through recombination in the env region between FeLV-A and endogenous FeLV sequences (enFeLV) present in the feline genome (11, 12) and FeLV-C apparently also arose through deletion, mutation, or recombination of the FeLV-A env gene (12, 13). Therefore, endogenous retroviruses contribute to the generation of novel viruses and function as a source of emergence of novel viruses. Endogenous retroviruses (ERVs) are present in all vertebrate genomes and are thought to be the remnants of ancestral germ line infections by exogenous retroviruses. ERVs make up a significant fraction of the mammalian genome (for example, 8 to 10% of the human or mouse genomes) and are transmitted in a Mendelian fashion (14–16). Although most ERVs are not expressed in normal tissues or conditions because of deleterious mutations or a defense mounted by the host to repress proviral expression, some ERVs are transcribed and produce functional gene products (17). Several classes of endogenous retroviruses are present in cats. Of these, ERV-DCs are endogenous gammaretrovirus of domestic cats (ERV-DCs) (10). ERV-DCs are classified into genotype I, II, and III (group I, II, and III) by phylogenetic analysis. ERV-DCs infected domestic cats within the past few million years, and insertional polymorphisms of ERV-DC exist today. Most ERV-DCs are not fixed in the feline genome, whereas provirus loci ERV-DC7 and ERV-DC16 are fixed in cats (10). Some ERV-DCs have intact open reading frames (ORFs) for gag, pol, and env, and, notably, ERV-DC10 (the q12-q21 region on chromosome C1) and ERV-DC18 (q14 on chromosome D4) are replication-competent viruses which can infect a broad range of cells, including humans. ERV-DC18 may be a locus that was generated by reintegration or reinfection of ERV-DC10 (10).
The expression of ERVs can confer resistance to closely related viruses by the interference mechanism. Viral interference is a retroviral phenomenon by which cells chronically infected by one virus are resistant to superinfection with closely related viruses (18). In particular, receptor interference caused by receptor blocking or downregulation through interaction with the viral envelope protein (Env) results in superinfection resistance to viruses using the same receptor. Receptor interference caused by Env proteins of ERVs has been reported. For example, the Fv-4 gene in mice encodes a full-length Env protein which closely resembles ecotropic murine leukemia virus (MLV) and confers resistance to ecotropic MLV infection by receptor interference (19, 20). Host defense against retroviral infection is common, and feline APOBEC and BST-2 are known to function as retroviral restriction factors in cats (21–23). Interestingly, feline TRIM5 lacks antiretroviral activity (24).
In this study, we report the precise determination of a new viral receptor interference group and the discovery of a soluble antiretroviral factor, termed Refrex-1 (restriction for feline retrovirus X). Refrex-1 is a soluble truncated Env protein encoded by provirus loci ERV-DC7 and ERV-DC16, and it is expressed in a broad range of feline tissues. Refrex-1 may have been acquired during endogenization of ERV-DCs and may play an important role in retroviral restriction and antiviral defense in cats.
MATERIALS AND METHODS
Animal and samples.
A female, 2-month-old, specific-pathogen-free (SPF) cat (Kyoto-SPF1) obtained from the Nippon Institute for Biological Science was euthanized, and an autopsy performed. Tissues were stored at −80°C until DNA or RNA was extracted.
Cells.
HEK293T (25, 26), NIH 3T3 (27), P3U1 (28), KwDM (10), 3201 (29), AH927 (30), FRM (31), and CRFK (32) cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). MS4 cells (33) were cultured in DMEM with 20% FCS. HEK293T cells infected with FeLV-A/Glasgow-1 (34), FeLV-A/clone 33 (35), FeLV-B/Gardner-Arnstein (36), or FeLV-C/Sarma (37) were cultured in DMEM with 10% FCS. GPLac cells (10), an env-negative packaging cell line containing a LacZ-coding retroviral vector, were cultured in DMEM with 10% FCS.
Construction of chimeric FeLV-D.
A replication-competent chimeric FeLV-D (termed FeLV-D/c33) was constructed by ligation of the 5′ EcoRI-BamHI-digested 5-kb fragment from FeLV-A clone 33 (35) provirus DNA with the 3′ BamHI-EcoRI-digested 3.5-kb fragment from FeLV-D/ON-T provirus DNA (10).
Cloning of full-length ERV-DC.
Genomic DNA was isolated from Kyoto-SPF1 blood samples by using a QIAamp DNA blood kit (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. Flanking DNA sequences of ERV-DC16 provirus were obtained by gene walking using a Right Walk kit (Bex Co., Ltd., Tokyo, Japan). Full-length ERV-DC16 provirus was PCR amplified from the chromosomal DNA obtained from Kyoto-SPF1 blood by using the specific primers Fe-219S and Fe-44R. ERV-DC7 provirus was also molecularly isolated from chromosomal DNA obtained from Kyoto-SPF1 blood as previously described (10). KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan) was used for PCR. The PCR products were cloned into pCR-Blunt (Invitrogen, Carlsbad, CA) and sequenced.
Plasmids.
Expression plasmids were newly constructed as shown in Table 1. The genes were PCR amplified from each plasmid by using specific primers with enzyme sites, and then PCR products were digested by each restriction enzyme and cloned into the pFUΔss expression plasmid (10). The template plasmids, PCR primers, and restriction enzymes used for new construction of expression plasmids are summarized in Tables 1 and 2. The Env expression plasmids for pseudotyped virus preparations—pFUΔss A5 (FeLV-A/Glasgow-1 env), pFUΔss GB (FeLV-B/Gardner–Arnstein env), pFUΔss SC (FeLV-C/Sarma env), pFUΔss Ty2.0 (FeLV-D/Ty26 env), pFUΔss ON-T (FeLV-D/ON-T env), pFUΔss NS33-4 (FeLV-T/NS33-4 env), pFUΔss 4070A (amphotropic MLV/4070A env), and pFUΔss DC10 (ERV-DC10 env)—have been described previously (10, 12).
Table 1.
Expression plasmids constructed for this studya
| Expression plasmid | Template plasmid | Forward primer | Reverse primer |
|---|---|---|---|
| pFUΔss ON-C | p44B peL | Fe-130S | Fe-50R |
| pFUΔss 44B | pON-C | Fe-130S | Fe-50R |
| pFUΔss DC6 | pCR4 ERV-DC6 | Fe-130S | Fe-126R |
| pFUΔss DC8 | pCR4 ERV-DC8 | Fe-130S | Fe-126R |
| pFUΔss DC14 | pCR4 ERV-DC14 | Fe-130S | Fe-126R |
| pFUΔss DC19 | pCR4 ERV-DC19 | Fe-130S | Fe-126R |
| pFUΔss 4070A | pCL-Ampho | 4070A-2S | 4070A-2R |
| pFUΔss FeLIX | pCR-Blunt FeLIX | FeLIX-1F | FeLIX-1R |
| pFUΔss 3201-2D | pCR-Blunt 3201-2D | Fe-207S | Fe-190R |
| pFUΔss 3201-2A | pCR-Blunt 3201-2A | Fe-209S | Fe-187R |
Each DNA fragment was PCR amplified with the indicated primers and template plasmid and then inserted into the pFUΔss expression vector. Primer sequences are shown in Table 2.
Table 2.
Sequences of primers used in this study
| Primer | Sequence (5′–3′)a | Cloning site |
|---|---|---|
| Fe-44S | CGGAATTCATCGAGATGGAAGGTCC | EcoRI |
| Fe-130S | CCGAATTCTRCACAAACCCAAAATGA | EcoRI |
| Fe-184S | CCTAGGRGCTTGGBTCCYAACATTTGGTG | |
| Fe-205S | GGATCCGGATCCATGAAACCCCCAGCGGGAAT | BamHI |
| Fe-206S | GGATCCGGATCCATGAAACCCCCAACAGGAAT | BamHI |
| Fe-207S | GGATCCGGATCCATGAAACCCCCAACAGGAAT | BamHI |
| Fe-208S | GGATCCGGATCCATGAGACCCTCAGCAAGAAT | BamHI |
| Fe-209S | GGATCCGGATCCATGAAACTCCCAACAGGAAT | BamHI |
| Fe-219S | GCCACGGTCATGAAAATAAAAA | |
| FeLIX-1F | CTGGATCCATGGAAGGTCCAACGCACCCAA | BamHI |
| 4070A-2S | TCTGGCTAGCCATGGCGCGTTCAACGCTCTCAAA | NheI |
| Fe-44R | TGCAGACAGAACATACTGTGACAAA | |
| Fe-50R | TTGAATTCTCATGGTTGGTCTGGATCGTATTG | EcoRI |
| Fe-126R | CAGAATTCTCTCATTCCCCCATTTTCTTT | EcoRI |
| Fe-168R | GAAGRTAGGGTGGGGGTGTKTTAGTAAGCTA | |
| Fe-184R | GAATTCGAATTCTATTCGATTGTATCTGGCCTTT | EcoRI |
| Fe-185R | GAATTCGAATTCTTTAGGAGCCTATCTCCTGT | EcoRI |
| Fe-186R | GAATTCGAATTCTCATTCAATTGTATCTGGCCTTTCTG | EcoRI |
| Fe-187R | GGATCCGGATCCTCAAGAGGGGGAAGTTGAGTATC | BamHI |
| Fe-188R | GGATCCGGATCCTTAGGAGTCTGTCTCCCGTG | BamHI |
| Fe-190R | GGATCCGGATCCTCAGGTTGGGCCTACGGTTTGGG | BamHI |
| Fe-196R | GGATCCGGATCCTTAGGAGTCTGTCTCCCGTG | BamHI |
| FeLIX-1R | TAGAATTCTTAGCTGGGGTGAGGTATTACT | EcoRI |
| 4070A-2R | TTATGCTAGCTATGGCTCGTACTCTATAGGCT | NheI |
Restriction enzyme sites are underlined. The indicated enzyme site was used for the construction of the expression vector shown in Table 1.
Transfection.
HEK 293T cells were plated in 60-mm dishes and transfected with 8 μg of each plasmid using 20 μl of Lipofectamine 2000 reagent (Invitrogen).
Immunoprecipitation and immunoblotting.
Cell lysates were prepared by resuspending the cells in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 mM Na3VO4, and 1 μg/ml each of aprotinin and leupeptin), followed by incubation on ice for 20 min. Insoluble components were removed by top-speed centrifugation, and the protein concentrations were determined using a protein assay kit (Bio-Rad Laboratories, Carlsbad, CA). The cell supernatants were filtered through a 0.45-μm-pore-size filter and immunoprecipitated with goat anti-FeLV gp70 (National Cancer Institute [NCI], Frederick, MD) covalently conjugated with Dynabeads-protein G according to the manufacturer's instructions (Life Technologies, Carlsbad, CA). Proteins were separated by electrophoresis on 7.5% or 10 to 20% gradient Tris-glycine minigels (Oriental Instruments) under reducing conditions (3.5 × 10−2 M 2-mercaptoethanol) and then transferred electrophoretically to nitrocellulose filters for Western blotting with goat anti-FeLV gp70 antibody (NCI) or mouse anti-β actin (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were horseradish peroxidase-conjugated anti-goat IgG antibody (Santa Cruz Biotechnology) or anti-mouse IgG antibody (GE Healthcare Japan, Tokyo, Japan), followed by visualization using 20× LumiGLO (Cell Signaling Technology).
Pseudotyped virus preparation.
Preparation of pseudotyped viruses has been described previously (10). GPLac cells were transfected with each env expression plasmid in order to produce LacZ carrying pseudotype viruses. After culture with 200 μg of zeocin (InvivoGen, California)/ml and 1 μg of puromycin/ml for >2 weeks, the supernatants were collected, filtered through a 0.45-μm-pore-size filter, and stored at −80°C.
Viral interference assay.
The viral interference assay has been described previously (10, 12). Target cells were infected with each pseudotyped virus in the presence of 8 μg of Polybrene/ml for ∼48 h and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Single-cycle infectivity was titrated by counting blue-stained nuclei under the microscope.
Viral infection assay in the presence of supernatants of cell cultures.
In order to determine inhibitory effects against viral infection by cell supernatants, an infection assay was conducted in the presence of cell supernatants. The cell supernatants were collected, filtered through a 0.45-μm-pore-size filter, and stored at −20°C. An infection assay basically followed that of the viral interference assay. Essentially, 250 μl of cell supernatants and 250 μl of pseudotyped viruses were incubated with target cells (HEK293T cells in 24-well plates). Alternatively, the cell supernatants were incubated with goat anti-FeLV gp70 antibody or normal goat serum (Wako Pure Chemical Industries, Ltd., Osaka, Japan), conjugated with protein G-agarose (Santa Cruz Biotechnology) for 1 h at room temperature. The supernatants without immune complexes removed by centrifugation were also used for the viral infection assay in the same way. After 48 h of incubation in the presence of 8 μg of Polybrene/ml, the cells were stained with X-Gal, and the single-cycle infectivity was titrated by counting blue-stained nuclei under a microscope.
RT-PCR.
The total RNA was isolated from cell lines and from each Kyoto-SPF1 tissue sample using RNAiso Plus (TaKaRa, Tokyo, Japan), and the extracted RNA was treated with recombinant DNase I (RNase-free; TaKaRa). cDNA was synthesized with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa) using oligo(dT) primer. The expression of ERV-DC was detected by a PCR using the primers Fe-184S and Fe-168R, which were designed for the end of the 5′ long terminal repeat (LTR) and the start of the 3′ LTR, respectively. PCR products were cloned into pCR-Blunt (Invitrogen) and sequenced. The expression of β-actin was also detected by reverse transcription-PCR (RT-PCR) as an internal control using Hub-b-actin(DNA)s and Hub-b-actin(DNA)r (10).
Phylogenetic analysis.
Nucleotide sequences of the ERV-DC and FeLV-D env genes corresponding to the signal peptide and the surface unit (SU) region (nucleotide positions 1 to 1444 of the ERV-DC8 env gene) were used in this phylogenetic analysis. Multiple alignments of env genes were generated by using CLUSTAL W (38, 39). Nucleotide substitution models of Kimura two-parameter model (40) with discrete gamma-distributed rate variation (five categories [+G, parameter = 0.6240]) were selected using the Bayesian information criterion (41). A phylogenetic tree was constructed by using the maximum-likelihood method with robustness evaluated by bootstrapping (1,000 times). All programs used in the present study were packaged in MEGA5 (42, 43). ERV-DC6 (AB674450), ERV-DC7 from SPF cat-1 (AB807599), ERV-DC8 (AB674443), ERV-DC10 (AB674444), ERV-DC14 (AB674445), ERV-DC16 from Kyoto-SPF1 (AB807600), ERV-DC19 (AB674448), FeLV-D/ON-T (AB673426), FeLV-D/ON-C (AB673429), FeLV-D/Ty26 (AB673428), and FeLV-D/44B (AB673430) were used for phylogenetic reconstruction.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in DDBJ/EMBL/GenBank under accession numbers AB807599, AB807600, AB819753, and AB819754.
RESULTS
Interference groups of FeLV-D and ERV-DC.
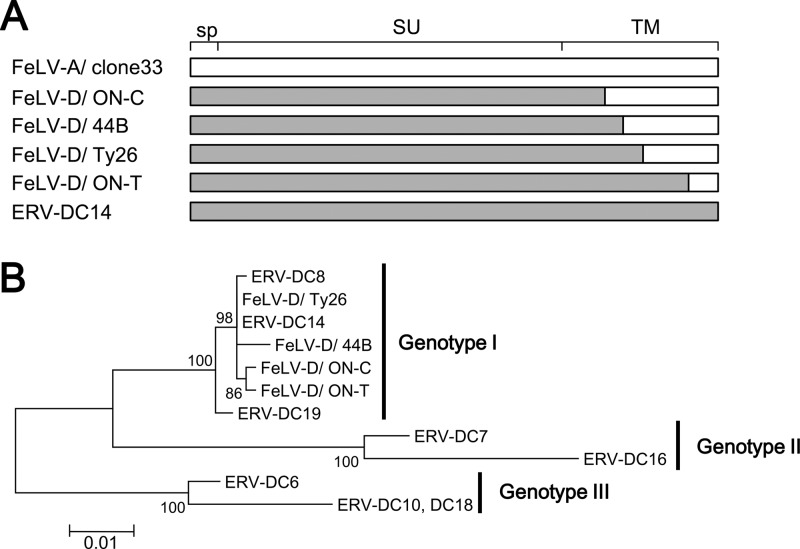
Our previous results have shown that 17 env sequences from ERV-DCs can be separated into genotypes I, II, and III (groups I, II, and III) by phylogenetic analysis (10). When env sequences from recombinant FeLV-D strains such as 44B, ON-C, ON-T, and Ty26 (Fig. 1A) were genotyped in the same way, all four FeLV-D strains were classified into ERV-DC genotype I (Fig. 1B). Thus, FeLV-D was generated by transduction of the env gene from the ERV-DC genotype I group. ERV-DC7 and ERV-DC16 belong to genotype II and do not encode complete Env proteins because of the presence of a stop codon at amino acids 252 and 298, respectively.
Fig 1.
Structure and phylogenetic analysis of the env gene from FeLV-D and ERV-DC. (A) Schematic representation of the env structure of FeLV-D and ERV-DC. The recombination pattern of each env is indicated. White indicates FeLV-A sequences, and gray indicates ERV-DC sequences. sp, signal peptide; SU, surface unit; TM, transmembrane. (B) Best-maximum-likelihood tree from phylogenetic analysis of env nucleotide sequences corresponding to the signal peptides and surface units from FeLV-D and ERV-DC strains.
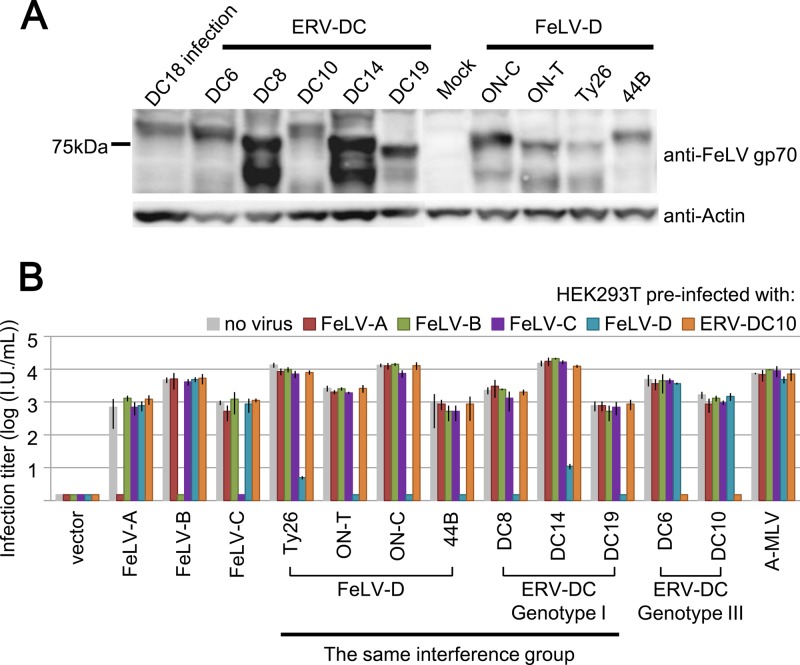
Our previous study showed that FeLV-D and ERV-DC10 belong to different receptor groups. In order to determine more detailed interference groups among FeLV-D and ERV-DCs, viral interference assays were conducted by using pseudotyped viruses. In the present study, two env expression plasmids from FeLV-D/44B and FeLV-D/ON-C and four env expression plasmids from ERV-DC6, DC8, DC14, and DC19 were newly constructed. env expression plasmids of ERV-DC10, FeLV-D/ON-T, and FeLV-D/Ty26 have been reported previously (10). HEK293T cells were transfected with each env expression plasmid. Western blot analysis showed that each Env protein from FeLV-D and ERV-DC was detected with anti-FeLV gp70 antibody, which reacts with both ERV-DC and FeLV Env proteins (10). The expression pattern seemed to differ among Env proteins and two bands at ∼75 kDa in cells expressing Env were detected in some lysates (Fig. 2A). To determine whether each Env protein functions in cell entry, an infection assay was carried out by producing pseudotyped viruses. To prepare each pseudotype, GPLac cells were transfected with env expression plasmids and supernatants of cells were used as a virus source. As shown in Fig. 2B, two pseudotyped viruses of FeLV-D/ON-C and FeLV-D/44B, as well as FeLV-D/Ty26 and FeLV-D/ON-T, could infect uninfected HEK293T cells. ERV-DC10 and DC18 are replication competent, and the pseudotyped viruses could also infect HEK293T cells (10). Four newly pseudotyped viruses of ERV-DC6, DC8, DC14, and DC19 could infect the HEK293T cells (Fig. 2B). These results indicated that these env genes were all functional for viral cell entry.
Fig 2.
Viral interference assay of LacZ pseudotyped viruses. (A) Western blot analysis with anti-FeLV Env (gp70) antibody in HEK293T cells transfected with env expression plasmids. Anti-actin antibody was also used as a control. DC18 infection; HEK293T cells were infected with a replication competent infectious virus of ERV-DC18. (B) env genes of FeLV-A/Glasgow-1, FeLV-B/Gardner–Arnstein, FeLV-C/Sarma, FeLV-D/Ty26, FeLV-D/ON-T, FeLV-D/ON-C, FeLV-D/44B, ERV-DC6, ERV-DC8, ERV-DC10, ERV-DC14, ERV-DC19, and A-MLV (Ampho-MLV/4070A) were used for preparation of LacZ pseudotyped viruses. HEK293T cells preinfected with no virus (gray), FeLV-A/clone33 (red), FeLV-B/Gardner–Arnstein (GA; green), FeLV-C/Sarma (purple), FeLV-D (FeLV-D/c33 chimeric virus; aqua blue), or ERV-DC10 (orange) were used as target cells for the interference assay. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Next, we determined more detailed viral interference groups of FeLV-D and ERV-DCs strains. HEK293T cells expressing only Env protein do not work in viral interference assays because the expression may not be sufficient. Therefore, we constructed a chimeric replication-competent infectious virus, termed FeLV-D/c33, by which the 5′LTR-gag-pol fragment from FeLV-D/ON-T provirus was replaced with that of FeLV-A/clone33 provirus because FeLV-D/ON-T was not replication competent virus due to a large deletion in the pol gene. HEK293T cells persistently infected with FeLV-D/c33 (termed 293T/FeLV-D cells) were established and FeLV-D/ON-T pseudotyped virus could not infect 293T/FeLV-D cells. However, FeLV-A, -B, and -C, and ERV-DC10 could infect 293T/FeLV-D cells. Therefore, 293T/FeLV-D cells were useful for evaluation of viral subgroups and were used for the interference assay. 4070A pseudotyped virus derived from amphotropic murine leukemia virus (MLV) 4070A was used as a control. As shown in Fig. 2B, the 4070A pseudotyped virus showed similar LacZ virus titer (104 IU/ml) in cells infected with FeLV-A/clone 33, FeLV-B/GA, FeLV-C/Sarma, and FeLV-D (FeLV-D/c33) and ERV-DC10. All four FeLV-D pseudotyped viruses (Ty26, ON-T, ON-C, and 44B) could infect HEK293T cells preinfected with FeLV-A, FeLV-B, FeLV-C, and ERV-DC10, but not cells infected with FeLV-D. FeLV-A, FeLV-B, and FeLV-C pseudotyped viruses could infect HEK293T/FeLV-D cells, but not HEK293T cells infected with each virus of the same group. ERV-DC8, -DC14, and -DC19 could infect HEK293T cells preinfected with FeLV-A, FeLV-B, FeLV-C, and ERV-DC10 but not FeLV-D. In contrast, ERV-DC6 and ERV-DC10 could infect HEK293T cells preinfected with FeLV-A, FeLV-B, FeLV-C, and FeLV-D, but not ERV-DC10. These results indicated that ERV-DC8, -DC14, and -DC19 could be classified as the same interference group and were distinct from the group of ERV-DC6 and DC10/18. Furthermore, ERV-DC8, -DC14, and -DC19 could be classified as the same interference group with FeLV-D strain, such as Ty26, ON-T, ON-C, and 44B. Therefore, our data established that there were two interference groups for FeLV-D and ERV-DC that correlate with their genotypes. The interference group of ERV-DC genotype II, such as ERV-DC7 and ERV-DC16, could not be determined because they encode defective and truncated Env proteins.
Presence of inhibitory factors in culture supernatant of feline cells.
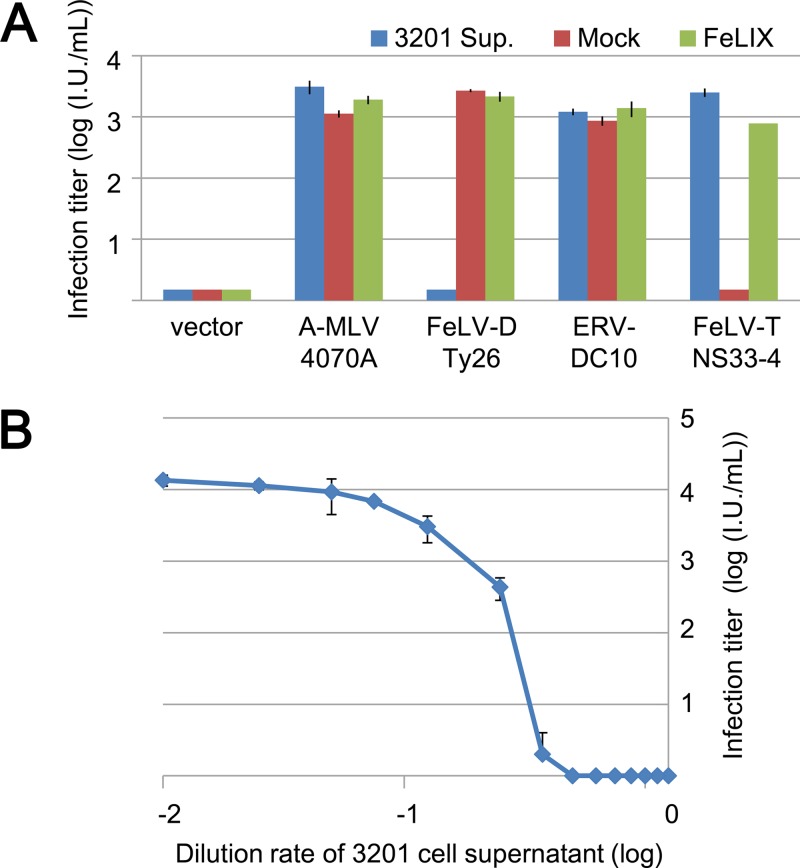
T-lymphotropic of FeLV (FeLV-T) is a subgroup of FeLV which requires a soluble cofactor, called FeLIX, for infection. FeLIX is a truncated Env protein of enFeLV corresponding to a receptor-binding domain (RBD) (9). To determine whether FeLV-D and ERV-DC belong to the FeLV-T subgroup, the supernatant of 3201 cells as a source of a cofactor, FeLIX, for FeLV-T was used in an additional viral infection assay. As shown in Fig. 3A, FeLV-T/NS33-4 (12) infects HEK293T cells only in the presence of supernatant of 3201 cells or FeLIX, which is derived from the supernatant of HEK293T cells transfected with a FeLIX expression plasmid. Unexpectedly, FeLV-D/Ty26 infection of HEK293T cells was inhibited by the supernatant of 3201 cells. However, FeLIX did not inhibit FeLV-D/Ty26 infection. ERV-DC10 and 4070A MLV infection was not inhibited by the supernatant of 3201 cells or FeLIX (Fig. 3A). Thus, FeLV-D/Ty26 and ERV-DC10 do not belong to the FeLV-T subgroup.
Fig 3.
Viral infection assay of lacZ pseudotyped virus in the presence of supernatant of 3201 cells or FeLIX. (A) env genes from Ampho-MLV/4070A (A-MLV/4070A), FeLV-D/Ty26, ERV-DC10, and FeLV-T/NS33-4 were used for pseudotyped virus preparation. HEK293T cells were used as target cells for the viral infection assay. The supernatant of 3201 cells (3201 Sup.; blue) or HEK293T cells transfected with FeLIX expression vector (green) or empty vector (Mock; red) was added in the culture when HEK293T cells were infected with each pseudotyped virus. (B) Dose-dependent inhibition by the supernatant of 3201 cells in the viral infection assay. Supernatant of 3201 cells was added to the culture and tested for infection using FeLV-D/Ty26 pseudotyped virus. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
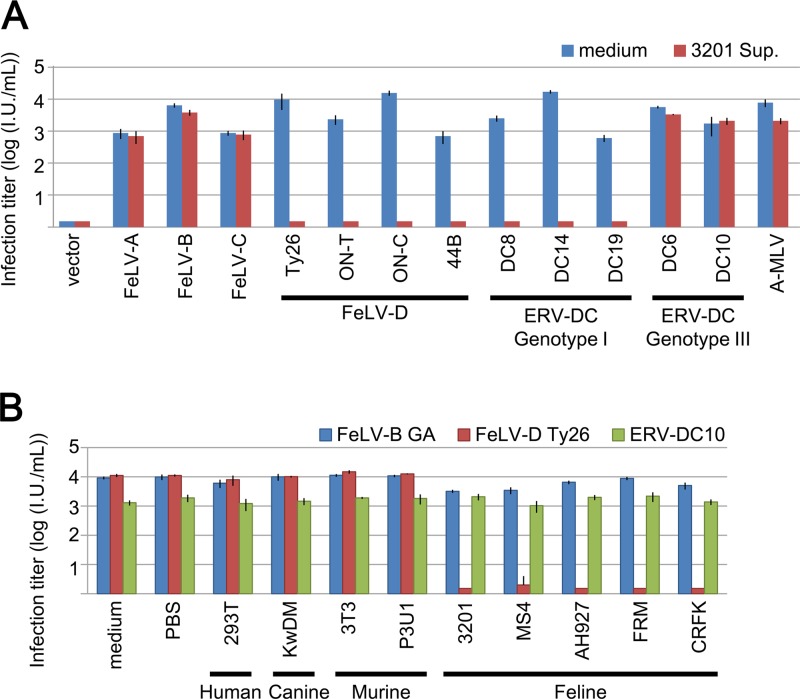
We determined whether supernatant from 3201 cells had a dose-dependent inhibitory effect. As shown in Fig. 3B, infection of HEK293T cells by FeLV-D/Ty26 pseudotyped virus was inhibited in a dose-dependent manner. Therefore, the supernatant of 3201 cells contains inhibitory factors for FeLV-D/Ty26 pseudotyped virus, and these factors are not FeLIX. We next determined the effect of the supernatant of 3201 cells on viral strains. As shown in Fig. 4A, infection of HEK293T cells by all four FeLV-D strains (Ty26, ON-T, ON-C, and 44B) was completely inhibited by the supernatant from 3201 cells. Furthermore, infection of HEK293T cells by ERV-DC8, DC14, and DC19 was also inhibited by the supernatant from 3201 cells. However, these inhibitory effects were not seen for infection with ERV-DC6 or DC10, FeLV-A, -B, or -C, or ampho-MLV/4070A (Fig. 4A). These results indicated that Env-pseudotyped viruses derived from FeLV-D and ERV-DC genotype I were affected by inhibitory factors but that other Env-pseudotyped viruses were not.
Fig 4.
Viral infection assay of LacZ pseudotyped viruses in the presence of supernatant from several cell lines. (A) Env-pseudotyped viruses FeLV-A/Glasgow-1, FeLV-B/Gardner–Arnstein(GA), FeLV-C/Sarma, FeLV-D/Ty26, FeLV-D/ON-T, FeLV-D/ON-C, FeLV-D/44B, ERV-DC6, ERV-DC8, ERV-DC10, ERV-DC14, ERV-DC18, ERV-DC19, and A-MLV (Ampho-MLV/4070A) were tested for infection in the presence of the supernatant of 3201 cells (3201 Sup.; red) or medium (control; blue). HEK293T cells were used as target cells in this assay. (B) Supernatant of 293T (HEK293T), KwDM, 3T3 (NIH 3T3), P3U1, MS4, AH927, FRM, and CRFK cells was added to the culture when 293T cells were infected with each pseudotyped virus, FeLV-B/Gardner–Arnstein (GA) (blue), FeLV-D/Ty26 (red), or ERV-DC10 (green). X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Next, we tested the inhibitory effect by using the supernatant of several cell lines such as human (HEK293T), canine (KwDM), murine (NIH 3T3 and P3U1), and feline (MS4, AH927, FRM, and CRFK) cells. FeLV-B, FeLV-D/Ty26, and ERV-DC10 Env-pseudotyped viruses were used for the inhibition assay. As shown in Fig. 4B, supernatants from feline cell lines showed complete inhibition of infection with FeLV-D, but not FeLV-B or ERV-DC10. Interestingly, supernatants from other cell lines, including human, canine, and murine cells did not inhibit FeLV-D, FeLV-B, and ERV-DC10 infection. These results indicated that feline cells produced inhibitory factors for which infection with pseudotyped viruses from FeLV-D and ERV-DC genotype I were inhibited. We termed this factor as Refrex-1 (restriction for feline retrovirus X).
Possible mechanism for the effect of Refrex-1.
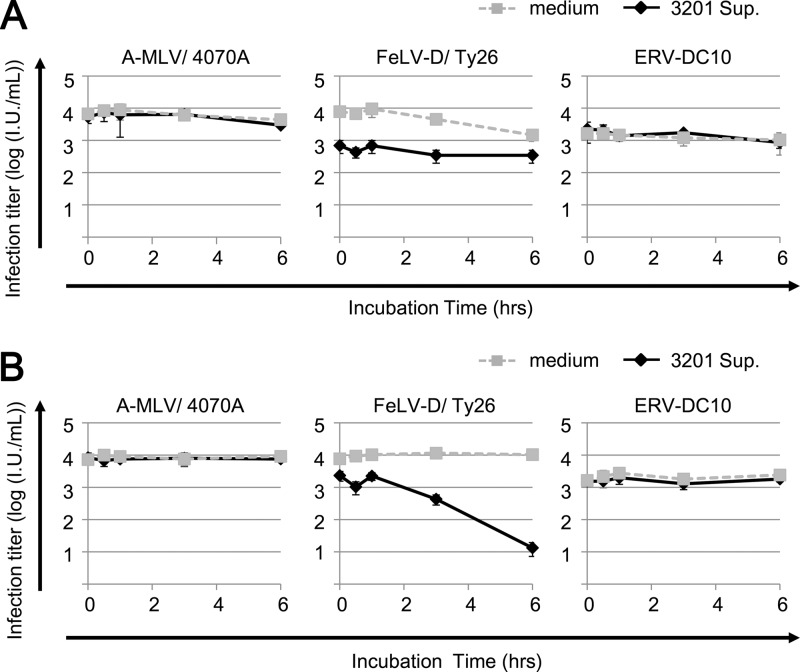
In order to determine whether Refrex-1 has an effect on the target cells or on the viruses, we carried out an infection assay by preincubation as follows. The target cells or the viruses were pretreated with 10-fold-diluted supernatant (50 μl) of 3201 cells for 30 s, 30 min, 1 h, 3 h, or 6 h at 37°C before viral inoculation. As shown in Fig. 5A, FeLV-D/Ty26 pseudotyped virus was incubated with Refrex-1 (the supernatant of 3201 cells) at 37°C for 6 h, and each reaction was inoculated into HEK293T cells. The infection titer of FeLV-D/Ty26 showed an ∼10-fold reduction compared to the controls (without supernatant). Preincubation of ERV-DC10 or Ampho-MLV/4070A with Refrex-1 did not decrease the viral infection titer (Fig. 5A). In contrast to preincubating Refrex-1 with viruses, HEK293T cells were preincubated with Refrex-1 at 37°C for the exposure time described above, washed with phosphate-buffered saline (PBS), and then infected with FeLV-D/Ty26, ERV-DC10, or Ampho-MLV/4070A. After 48 h, each titer of viral infection was determined. As shown in Fig. 5B, infection titer of FeLV-D/Ty26 showed a dramatic reduction in an incubation-time-dependent manner by 6 h. However, the inhibitory effect of this strategy was not observed for ERV-DC10 or Ampho-MLV/4070A. These results suggest that Refrex-1 might interfere with viral entry mediated by a receptor for FeLV-D extracellularly rather than viral inactivation by Refrex-1.
Fig 5.
Incubation of Refrex-1 with target cells before viral inoculation blocks viral infection. (A) Each pseudotyped virus—A-MLV/4070A (Ampho-MLV/4070A), FeLV-D/Ty26, or ERV-DC10—was incubated with the supernatant of 3201 cells or medium for 30 s, 30 min, 1 h, 3 h, or 6 h at 37°C before viral inoculation. Each mixture was inoculated into HEK293T cells. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection. (B) HEK293T cells were incubated with supernatant of 3201 cells or medium for 30 s, 30 min, 1 h, 3 h, or 6 h at 37°C. After incubation, the cells were washed with PBS and infected with each indicated pseudotyped virus. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Identification of Refrex-1 as a restriction factor.
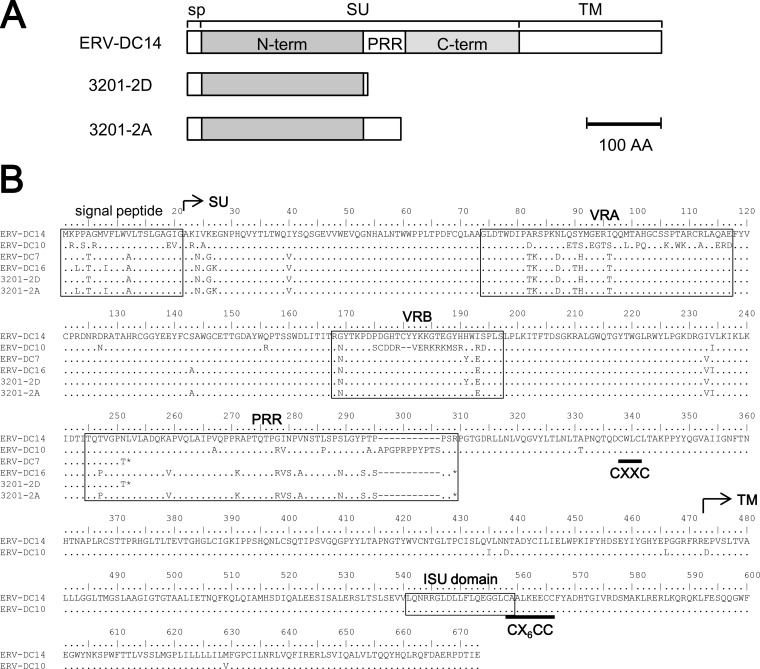
We attempted to identify Refrex-1 based on a possible mechanism of receptor-mediated interference as shown in Fig. 5. First, cDNAs synthesized from RNAs of 3201 and CRFK cells were PCR amplified with specific primers, Fe-184S and Fe-168R, which were designed for the end of the 5′ LTR and the start of the 3′ LTR. PCR products were cloned and sequenced. Eight different clones of 1.0 to 6.5 kb in size were obtained. These were spliced forms of ERV-DCs, which were classified into genotype II or III. Next, we constructed the expression plasmids of nine ORFs from eight cDNA clones. The supernatants from HEK293T cells transfected with each expression plasmid were tested for Refrex-1 activity by viral infection assay. Two of these clones, 3201-2A and 3201-2D, were positive for a screening experiment (data not shown). Therefore, we further characterized these clones as candidates for Refrex-1. As shown in Fig. 6A, 3201-2A and 3201-2D, which corresponded to the env subgenomic gene of ERV-DCs, were isolated with an approximate size of 2.5 kb. 3201-2A and 3201-2D encoded truncated Env proteins of ERV-DCs that were 297 and 251 amino acids long, respectively. 3201-2A and 3201-2D contained a stop codon in the middle of the env ORF, which resulted in a point mutation (CGA to TGA) and a frameshift due to a 1-bp deletion, respectively (Fig. 6B). As shown in Fig. 6, 3201-2A and 3201-2D retained the signal peptide and N-terminal region of SU, which is a putative RBD, but it lacked the C-terminal region and TM. 3201-2A and 3201-2D had two or one putative N-linked glycosylation sites (amino acid positions 169 and 283 in 3201-2A and 169 in 3201-2D), respectively. Judging from their structures, 3201-2A and 3201-2D were secretory proteins.
Fig 6.
Identification of Refrex-1 from 3201 cells. (A) Schematic representation of Refrex-1 structure from two clones, 3201-2D (ERV-DC7) and 3201-2A (ERV-DC16), compared to ERV-DC14 Env. (B) Alignment of amino acid sequences of Env from ERV-DC and 3201-2D and 3201-2A. SU, surface unit; N-term, N-terminal region of SU; C-term, C-terminal region of SU; TM, transmembrane unit; VRA, variable region A; VRB, variable region B; PRR, proline-rich region; ISU domain, immunosuppressive domain; CXXC and CX6CC indicate sites of covalent interaction.
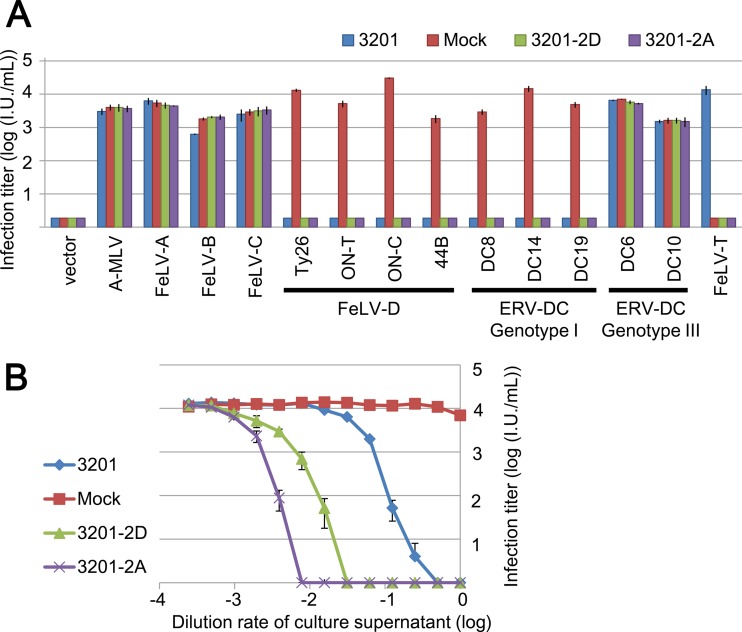
Next, we determined in detail whether these proteins act as Refrex-1 or not. As the sources of these proteins, the supernatants of HEK293T transfected with the 3201-2A or 3201-2D expression plasmids were used. As shown in Fig. 7A, 3201-2A and 3201-2D specifically inhibited FeLV-D and ERV-DC genotype I, as did the supernatant of 3201 cells. Furthermore, serially diluted supernatants of 3201-2A and 3201-2D inhibited FeLV-D/Ty26 in a dose-dependent manner (Fig. 7B). Thus, we identified that 3201-2A and 3201-2D functioned as bona fide restriction factors. On the other hand, 3201-2A and 3201-2D did not enhance FeLV-T infection (Fig. 7A). This result indicates that 3201-2A and 3201-2D do not work as cofactors for FeLV-T infection like FeLIX.
Fig 7.
Effect of Refrex-1 in the infection assay. (A) Supernatant of 3201 (blue) or 293T cells transfected with 3201-2D (green), 3201-2A (purple) or empty vector (Mock; red) was added to the culture when HEK293T cells were infected with each indicated pseudotype virus. Empty vector is used as a control. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection. (B) Dose-dependent inhibitory effect of Refrex-1on viral infection. Supernatant of 3201 (blue) or HEK293T cells transfected with 3201-2D (green), 3201-2A (purple) or empty vector (Mock; red) was diluted with medium, and each 250 μl of the supernatant was added to the culture when HEK293T cells were infected with FeLV-D/Ty26 pseudotyped virus. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Refrex-1 is encoded by ERV-DC7 and ERV-DC16 loci.
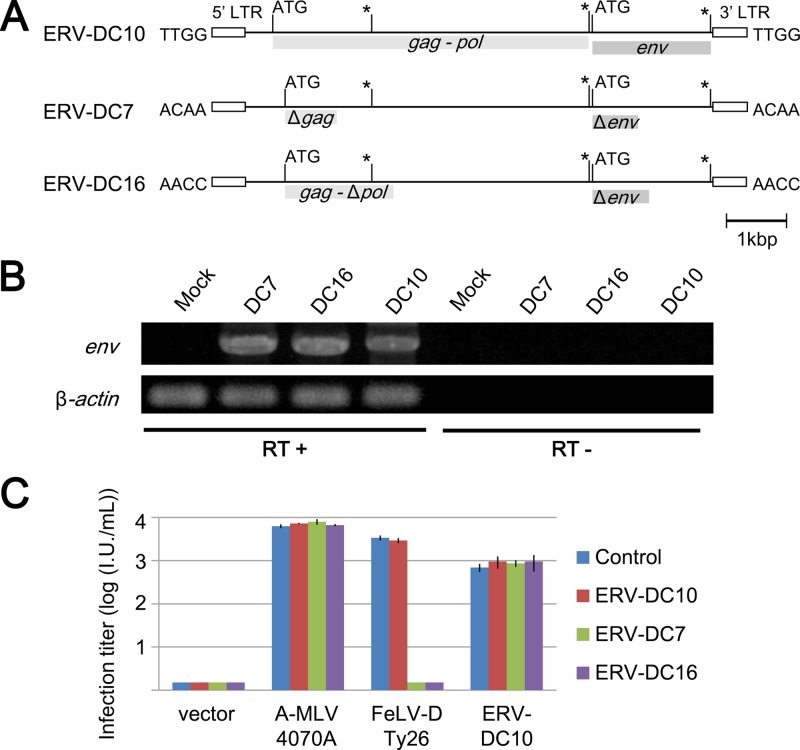
As shown in Fig. 6B, amino acid sequences of 3201-2D and 3201-2A completely corresponded to that of ERV-DC7 and ERV-DC16 loci, respectively. Each stop codon of 3201-2D and 3201-2A was found at the same position in ERV-DC7 and ERV-DC16. Therefore, we determined whether ERV-DC7 and ERV-DC16 worked as Refrex-1 by an infection assay. Because ERV-DC16 provirus has partially been isolated in our previous report, full-length ERV-DC16 provirus was newly isolated in this experiment as described in Materials and Methods. Schematic representations of structures of these proviruses are shown in Fig. 8A. ERV-DC7 and ERV-DC16 encoded defective ORFs of gag, pol, and env. We do not know whether ERV-DC7 and ERV-DC16 proviruses are inactivated or not. To address this question, HEK293T cells were transfected with provirus clones ERV-DC7, ERV-DC16, and ERV-DC10 (10), and then total RNA was isolated and RT-PCR was performed to detect subgenomic env genes by using primers Fe-184S and Fe-168R. Approximately 2.5 kb of subgenomic env gene was detected in cells transfected with each clone, but not in HEK293T cells or reverse transcriptase negative samples (Fig. 8B). Sequence analysis confirmed that a 2.5-kb fragment corresponded to the env genes, indicating that ERV-DC7 and ERV-DC16, as well as ERV-DC10, were not inactivated in transfected cells. Next, the supernatants of the transfected cells were used in the infection assay to assess whether or not they contained the biological activity of Refrex-1. As a control, we used ERV-DC10, which is replication competent and was confirmed to be expressed. As shown in Fig. 8C, the supernatants of the cells transfected with ERV-DC7 or ERV-DC16 proviruses specifically inhibited infection of FeLV-D/Ty26; however, ERV-DC10 did not. These results indicated that Refrex-1 is a soluble truncated Env protein encoded by provirus loci, ERV-DC7, and ERV-DC16.
Fig 8.
ERV-DC7 and ERV-DC16 loci encode Refrex-1. (A) Structures of the genomes of full-length ERV-DC7 and ERV-DC16 in comparison to ERV-DC10, which is a replication-competent virus. gag, pol, and env genes are illustrated, together with the 5′ and 3′ LTRs and the positions of the gag and env translational initiation codons (ATG). Asterisks indicate conserved stop codons. Gag and Pol proteins may be synthesized as a large single polypeptide precursor via termination suppression. Flanking 4-bp target site duplication (TSD) sequences are shown for each provirus. (B) Detection of env subgenomic DNA by RT-PCR from total RNA from HEK293T cells transfected with ERV-DC7, ERV-DC16, or ERV-DC10 provirus clones. RT+ or RT−, presence or absence of reverse transcriptase during cDNA synthesis. PCR products were run on 1% agarose gel and stained with ethidium bromide. (C) Supernatant from HEK293T cells transfected with ERV-DC7 (green), ERV-DC16 (purple), ERV-DC10 (red), or untransfected HEK293T cells (control; blue) was tested for viral infection using pseudotyped viruses Ampho-MLV/4070A, FeLV-D/Ty26, or ERV-DC10. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Detection of Refrex-1 in cell culture supernatant.
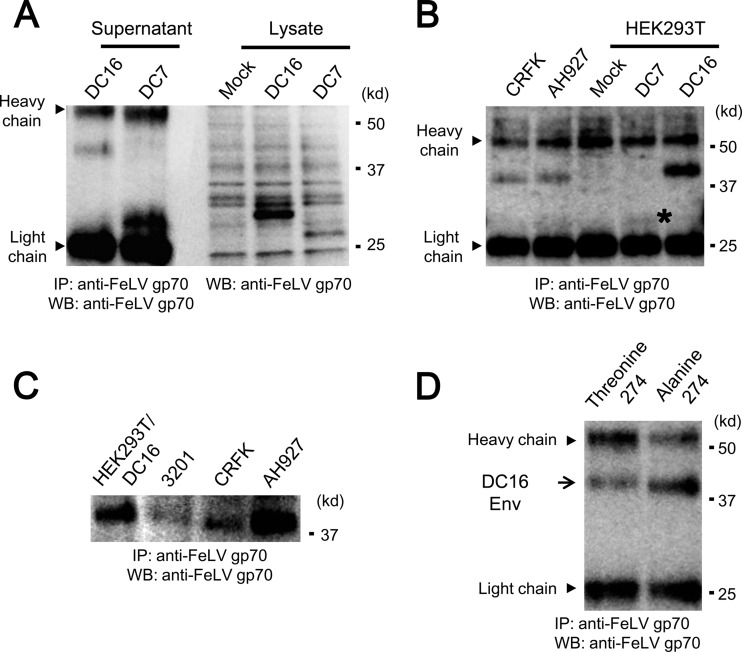
To characterize Refrex-1, ERV-DC7 and ERV-DC16 env expression plasmids were transfected into HEK293T cells, and cell lysates were immunoblotted with goat anti-FeLV gp70. ERV-DC7 and ERV-DC16 encoded ∼28- and ∼32-kDa proteins, respectively (Fig. 9A). When supernatants from transfected cells were immunoprecipitated with goat anti-FeLV gp70 antibody, extracellular forms of ERV-DC7 and ERV-DC16 were detected as 30- and 40-kDa proteins, respectively (Fig. 9A and B). This antibody weakly bound to ERV-DC7 Env protein and seemed to have a predilection for ERV-DC16 Env, which may have been due to the specificity of the antibody or the protein stability. The difference in size between the intracellular and extracellular forms of these proteins, especially ERV-DC16 Env, may be due to modifications such as glycosylation. We next carried out experiments to detect Refrex-1 in culture supernatants of feline cells. As shown in Fig. 9B and C, a 40-kDa protein corresponding to ERV-DC16 Env was detected in culture supernatants of 3201, CRFK, and AH927 cells. However, there was a small difference in the size of the Env protein expressed in some feline cell lines compared to cells transfected with ERV-DC16. This could be due to a single-nucleotide polymorphism [GCT (alanine) or ACT (threonine)] in ERV-DC16 Env at amino acid position 274, which slightly altered the molecular mass of the protein when two ERV-DC16 Env proteins were analyzed in supernatants of HEK293T cells expressing ERV-DC16 (alanine at 274) or ERV-DC16 (threonine at 274) (Fig. 9C and D). We could not detect ERV-DC7 Env in the culture supernatants of feline cells. This might have been due to the antibody specificity, the expression (low or none), or the stability of ERV-DC7 Env. The transcript of subgenomic env gene from ERV-DC7 was detected in 3201 cells, but was not detected in AH927 and CRFK cells (data not shown).
Fig 9.
Western blot analysis of Refrex-1. (A) Detection of Refrex-1 in cell culture supernatants (left) and in cell lysates (right) from HEK293T expressing Refrex-1. Cell culture supernatants were immunoprecipitated with goat anti-FeLV gp70 antibody. Immunoprecipitates of 10- and 4-ml supernatants, respectively, from HEK293T cells transfected with ERV-DC7 or ERV-DC16 env expression plasmid, and cell lysates were immunoblotted with the same antibody. Mock, HEK293T cells with no transfection. (B) Detection of Refrex-1 in the supernatants of feline cell lines, AH927 and CRFK. Immunoprecipitates of supernatants (2 ml) from AH927 and CRFK cells were immunoblotted with goat anti-FeLV gp70 antibody. Portions (2 ml) of supernatant from HEK293T cells transfected with ERV-DC7 or ERV-DC16 env expression plasmid were used for immunoprecipitation. Mock, HEK293T cells with no transfection. An asterisk indicates the position of ERV-DC7 Env (faint band). (C) Comparison of ERV-DC16 Env from cell culture supernatants. Immunoprecipitates of supernatants from indicated cells were immunoblotted with goat anti-FeLV gp70 antibody. ERV-DC16 env expression plasmid derived from 3201-2A was transfected into HEK293T cells (HEK293T/DC16). (D) Comparison of two ERV-DC16 Env proteins in supernatants of HEK293T cells expressing ERV-DC16 (alanine at 274) or ERV-DC16 (threonine at 274) derived from CRFK or 3201 cells, respectively. Immunoprecipitates of supernatants from indicated cells were immunoblotted with goat anti-FeLV gp70 antibody. Immunoprecipitation was performed with goat anti-FeLV gp70 antibody.
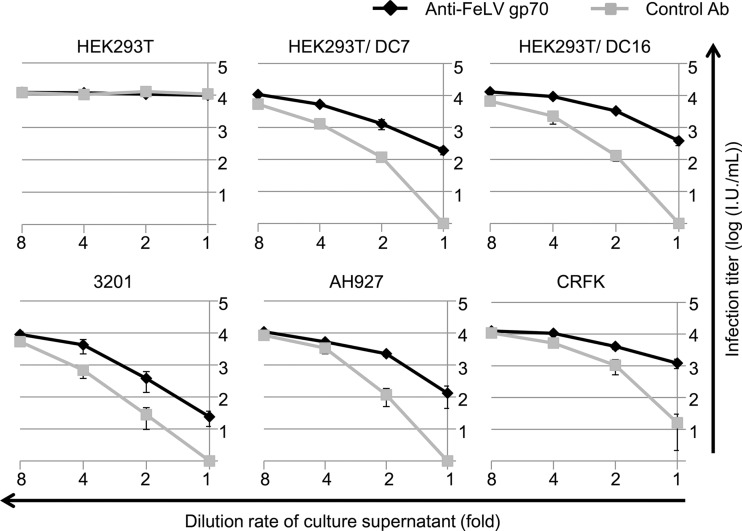
To confirm the presence of Refrex-1 activity in culture supernatants of feline cells, Refrex-1 was depleted from the supernatants using goat anti-FeLVgp70 antibody, and Refrex-1 activity was tested. To test whether the depletion analysis worked, the culture supernatants from HEK293T cells stably expressing ERV-DC7 or ERV-DC16 Env were immunoprecipitated with goat anti-FeLV gp70 antibody or normal goat serum. After removing the immune complexes by protein G-agarose, the supernatants were tested for Refrex-1 activity by viral infection assay with FeLV-D. As shown in Fig. 10, depletion of ERV-DC7 and ERV-DC16 by anti-FeLV antibody reduced Refrex-1 activity in a dose-dependent manner compared to the effect of normal goat serum, indicating that the depletion analysis successfully worked in transfectants. To determine the effect of Refrex-1 in feline cells, Refrex-1 was depleted from the culture supernatants of 3201, AH927, and CRFK cells. Depletion of Refrex-1 from the culture supernatants of feline cells reduced the effect of Refrex-1 in a dose-dependent manner, similar to the transfectants. These results confirmed that Refrex-1 was present in the culture supernatant of feline cells and that it was, at least, a truncated soluble Env protein from ERV-DC16.
Fig 10.
Depletion of Refrex-1 from cell culture supernatants. Culture supernatants from HEK293T, HEK293T expressing ERV-DC7 Env (HEK293T/DC7), HEK293T expressing ERV-DC16 Env (HEK293T/DC16), and the feline cell lines 3201, AH927, and CRFK were treated with goat anti-FeLV gp70 antibody or normal goat serum (control), and Refrex-1 was depleted from the supernatants. The remaining supernatants were diluted with medium, and 250 μl of each supernatant was added to the culture when HEK293T cells were infected with FeLV-D/Ty26 pseudotyped virus. X-Gal-positive cells were counted as infectious units (I.U.) at 48 h postinfection.
Expression of ERV-DC7 and ERV-DC16 env genes in feline tissues.
We determined the expression of ERV-DC7 and ERV-DC16 env genes in feline tissues. As mentioned above, a 2.5-kb subgenomic RNA corresponding to spliced env genes was amplified by RT-PCR with primers Fe-184S and Fe-168R. The 2.5-kb fragments were cloned and sequenced. Table 3 summarizes the env expression pattern of ERV-DCs. Many tissues express env genes of ERV-DC7 and ERV-DC16, but no detection was observed in the liver, bone marrow, small intestine, skeletal muscle, and ocular structures. The expression pattern of ERV-DCs differs among tissues, suggesting that the LTR from ERV-DC may show tissue-specific promoter activity. ERV-DC7 and ERV-DC16 are widely expressed in tissues.
Table 3.
Expression of Refrex-1 in various feline tissues from an SPF cat
| Tissue type |
env expressiona |
|
|---|---|---|
| ERV-DC7 | ERV-DC16 | |
| PBMC | + | + |
| Spleen | + | + |
| Thymus | + | ND |
| Tonsil | + | ND |
| Mandibular lymph node | + | + |
| Superficial cervical lymph node | + | + |
| Tongue | ND | + |
| Esophagus | + | + |
| Stomach | ND | + |
| Colon | + | ND |
| Uterus | + | + |
| Ovary | + | + |
| Cerebrum | + | ND |
| Pituitary gland | + | + |
| Cerebellum | + | ND |
| Medulla oblongata | + | ND |
| Diaphragm | + | + |
| Myocardium | ND | + |
| Thyroid gland | + | ND |
| Adrenal gland | + | ND |
| Pancreas | + | + |
| Mandibular gland | ND | + |
| Urinary bladder | + | ND |
| Subcutaneous tissue | – | – |
| Bone marrow | – | – |
| Liver | – | – |
| Small intestine | – | – |
| Ocular structures | – | – |
| Lung | – | – |
| Skeletal muscle | – | – |
+, Positive Refrex-1 expression; –, negative Refrex-1 expression. ND, Refrex-1 expression was not determined.
DISCUSSION
Our previous study showed that FeLV-D could not interfere with replication-competent viruses ERV-DC10/DC18 on receptor usage (10). In the present study, we clearly identified two receptor interference groups among ERV-DC and FeLV-D, which were distinct from FeLV-A, FeLV-B, FeLV-C, FeLV-T, and amphotropic MLV/4070A subgroups. Receptor interference groups among ERV-DC and FeLV-D seemed to correlate with their genotypes of env genes; one included ERV-DC genotype I, and the other included ERV-DC genotype III. All FeLV-D strains were shown to be generated by the transduction of ERV-DC env genes classified into genotype I, and thus the receptor interference group of all FeLV-D strains was similar to that of ERV-DC genotype I, such as ERV-DC8 and ERV-DC14, but not genotype III, such as ERV-DC10 and ERV-DC18. As shown in Fig. 6, amino acid differences of Env proteins between ERV-DC genotype I (ERV-DC14) and genotype III (ERV-DC10) were clustered in several regions that correspond to the putative variable region A (VRA), variable region B (VRB), and proline-rich region (PRR) of FeLV. These regions of ERV-DC may determine receptor specificity like those of FeLV and MLV, and amino acid differences in these regions among ERV-DC genotype I and genotype III may be reflected in the differences of their receptor interference groups.
During the course of the receptor interference studies, we discovered a soluble restriction factor, termed Refrex-1. Refrex-1 was present in the supernatants from feline cell lines and specifically inhibited infection of ERV-DC genotype I and FeLV-D. Refrex-1 is a truncated Env protein of ERV-DC and has the signal peptide (SP) and N-terminal region of SU, which is a putative receptor-binding domain (RBD), but it lacks the C-terminal region of SU and TM because of a stop codon in the middle of the env gene. Refrex-1 is efficiently secreted from cells and appears to cause receptor interference extracellularly because of its structure. Such a truncated Env protein lacking TM is not tethered to the cell membrane and is therefore effectively secreted from cells (44, 45). It is not entirely clear how Refrex-1 inhibits viral infection. One possible mechanism is that Refrex-1 competes with virus by binding to the viral receptor. It has been shown that FeLIX and a soluble RBD artificially constructed from FeLV-B or MLV bind to their receptor and inhibit the infection of cells (46, 47). Expression of ERVs env genes can confer resistance to viral infection by a receptor interference mechanism. Fv-4, Rmcf, and Rmcf2 in the mouse inhibit the infection of ecotropic, polytropic, and xenotropic MLV, respectively (20, 48, 49). Although these genes are similar to Refrex-1, they encode full-length Env proteins, including TM, and so they are mainly present on the cellular membrane and protect the cells expressing them. On the other hand, Refrex-1 can protect not only the cell expressing it but also cells without expressing it because Refrex-1 is a secreted protein and exerts its effect on cells from the outside.
FeLIX is a truncated Env protein of enFeLV corresponding to an RBD, and a large amount is secreted from 3201 cells (data not shown). FeLIX is thought to play a role in natural resistance of FeLV-B (46). Although our study showed that the culture supernatant of 3201 cells slightly inhibited FeLV-B infection (2- to 5-fold inhibition [P < 0.03]) (see, for example, Fig. 7A), it did not result in a substantial reduction, as shown by others (9). Unlike the effect of Refrex-1, FeLIX has a potential inhibitory effect against FeLV-B, as previously described (47). It will be important to elucidate the different effects between Refrex-1 and FeLIX if Refrex-1 causes receptor-mediated interference. However, the similarity of the structure and function of FeLIX and Refrex-1 and their inhibition of retroviral infection indicate an example of parallel evolution of these molecules in domestic cats. We speculate that it may be a consequence of retroviral endogenization or retroviral infectious disease.
Full-length env that was similar to that of ERV-DC14 and that belonged to genotype I slightly inhibited infection with FeLV-D (data not shown). However, supernatant from HEK293T cells transfected with the infectious molecular clone, ERV-DC10, did not inhibit viral infection of ERV-DC10. This may be a reason why full-length Env protein is not produced in great amounts in the supernatant, although the full-length env gene is well expressed in the cells.
Although there are two variants of Refrex-1, they seem to have independently arisen from env genes which encode full-length Env proteins, judging from the positions of their stop codons (Fig. 6A). These two variants of Refrex-1 are encoded by ERV-DC7 and ERV-DC16 loci mapped on chromosomes X and A2, respectively, according to the UCSC genome browser (http://genome.ucsc.edu/). Interestingly, ERV-DC7 and ERV-DC16 are fixed loci in the genomes of domestic cats (10), although most ERV-DCs exhibit insertionally polymorphic integrations because they have been recently integrated into cat genomes (10). We hypothesize that ERV-DC7 and ERV-DC16 have acquired a function as Refrex-1 due to their mutations, and they have been distributed among cats and became fixed through virus-host coevolution. Fixation of these proviruses in cat genomes may have contributed to the endogenization of ERV-DC genotype I. Interestingly, they seem to not be replication-competent viruses, even though ERV-DC8 and ERV-DC14, classified into ERV-DC genotype I, contain intact ORFs of gag, pol, and env genes. It is possible that Refrex-1 may prevent reemergence of ERV infectious diseases and may also contribute to inactivation of ERV-DCs. We currently plan to determine the evolutionary diversity of ERV-DCs and Refrex-1 among Felis spp. to understand their role in retroviral infectious diseases.
Refrex-1 inhibits ERV-DC genotype I infection but does not inhibit that of ERV-DC genotype III because ERV-DC genotype I (or FeLV-D) and genotype III use different receptors from each other and Refrex-1 may interfere only with the receptor for genotype I. Although it is unknown whether or not Refrex-1 was already present in domestic cats when ERV-DC genotype III was generated, ERV-DC genotype III seems to be an escape mutant from Refrex-1, which was generated by acquisition of a novel receptor tropism. Moreover, replication competent viruses ERV-DC10 and ERV-DC18 are not inhibited by Refrex-1 because they belong to ERV-DC genotype III. Although it is unknown whether these viruses horizontally transmit among cats, if they do not, there may be other mechanisms of restriction against ERV-DC10 and ERV-DC18 in cats.
Interaction between virus and receptor can affect the physiological function of the receptor. For example, interaction between FeLV-A Env protein and its receptor, thiamine transporter (THTR1), blocks thiamine uptake mediated by THTR1 and induces growth arrest of cells (50). It has also been shown that interaction between FeLV-C Env protein and its receptor, FLVCR-1, causes abnormal erythroid differentiation (51). Similarly, expression of truncated Env proteins encoded by ERV-DC7 and ERV-DC16 may also affect the physiological function of their receptor, and so they can possibly function like cytokines in addition to causing viral interference as Refrex-1. In the present study, we confirmed that Refrex-1 is expressed in a broad range of tissues in cats. Interestingly, the LTRs of ERV-DC7 and ERV-DC16 might exhibit tissue specific promoter activity. The physiological functions of ERV-DC and Refrex-1will be elucidated with further studies.
Finally, hosts have been invaded by retroviruses not only in modern times but also in ancient times. These host-virus conflictions have triggered the emergence and adaptation of restriction factors. The newly discovered restriction factor, Refrex-1, reported here may be an example of adaptive evolution, reflecting the arms race that has taken place at the host-virus interface during the evolution of species.
In conclusion, our findings provide evidence for the existence of a novel retroviral restriction factor in the domestic cat and may be a model case of the coevolution of the host and pathogen. Our findings may provide a novel insight to develop an effective antiretroviral therapy.
ACKNOWLEDGMENTS
We thank Sandra Ruscetti (NCI, Frederick, MD) and Takako Suzuki (National Institute of Animal Health, Japan) for helpful discussions.
This study was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant 22380168 to K.N.).
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Hisasue M, Nagashima N, Nishigaki K, Fukuzawa I, Ura S, Katae H, Tsuchiya R, Yamada T, Hasegawa A, Tsujimoto H. 2009. Myelodysplastic syndromes and acute myeloid leukemia in cats infected with feline leukemia virus clone33 containing a unique long terminal repeat. Int. J. Cancer 124:1133–1141 [DOI] [PubMed] [Google Scholar]
- 2.Hartmann K. 2011. Clinical aspects of feline immunodeficiency and feline leukemia virus infection. Vet. Immunol. Immunopathol. 143:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overbaugh J, Miller AD, Eiden MV. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099 [PubMed] [Google Scholar]
- 5.Tailor CS, Willett BJ, Kabat D. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 73:6500–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J. Virol. 80:3378–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalev Z, Duffy SP, Adema KW, Prasad R, Hussain N, Willett BJ, Tailor CS. 2009. Identification of a feline leukemia virus variant that can use THTR1, FLVCR1, and FLVCR2 for infection. J. Virol. 83:6706–6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830 [DOI] [PubMed] [Google Scholar]
- 10.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J. Virol. 86:8634–8644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overbaugh J, Riedel N, Hoover EA, Mullins JI. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731–734 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. 10.1371/journal.pone.0061009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigby MA, Rojko JL, Stewart MA, Kociba GJ, Cheney CM, Rezanka LJ, Mathes LE, Hartke JR, Jarrett O, Neil JC. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J. Gen. Virol. 73:2839–2847 [DOI] [PubMed] [Google Scholar]
- 14.Boeke JD, Stoye JP. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p 343–436 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 15.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 16.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 [DOI] [PubMed] [Google Scholar]
- 17.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13:283–296 [DOI] [PubMed] [Google Scholar]
- 18.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda H, Odaka T. 1983. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology 128:127–139 [DOI] [PubMed] [Google Scholar]
- 20.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 55:768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O'Brien SJ, Lochelt M, Yuhki N. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietrich I, McMonagle EL, Petit SJ, Vijayakrishnan S, Logan N, Chan CN, Towers GJ, Hosie MJ, Willett BJ. 2011. Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J. Virol. 85:5840–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuma A, Abe M, Morikawa Y, Miyazawa T, Yasuda J. 2011. Cloning and characterization of the antiviral activity of feline Tetherin/BST-2. PLoS One 6:e18247. 10.1371/journal.pone.0018247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEwan WA, Schaller T, Ylinen LM, Hosie MJ, Towers GJ, Willett BJ. 2009. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 83:8270–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 26.Kim CH, Oh Y, Lee TH. 1997. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene 199:293–301 [DOI] [PubMed] [Google Scholar]
- 27.Jainchill JL, Aaronson SA, Todaro GJ. 1969. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J. Virol. 4:549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unkeless JC. 1979. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J. Exp. Med. 150:580–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder HW, Jr, Hardy WD, Jr, Zuckerman EE, Fleissner E. 1978. Characterisation of a tumour-specific antigen on the surface of feline lymphosarcoma cells. Nature 275:656–658 [DOI] [PubMed] [Google Scholar]
- 30.Rasheed S, Gardner MB. 1980. Characterization of cat cell cultures for expression of retrovirus, FOCMA and endogenous sarc genes, p 393–400 In Hardy WD, Jr, Essex M, McClelland AJ. (ed), Proceedings of the third international feline leukemia virus meeting. Elsevier/North-Holland Publishing Co, New York, NY [Google Scholar]
- 31.Muleya JS, Nakaichi M, Sugahara J, Taura Y, Murata T, Nakama S. 1998. Establishment and characterization of a new cell line derived from feline mammary tumor. J. Vet. Med. Sci. 60:931–935 [DOI] [PubMed] [Google Scholar]
- 32.Crandell RA, Fabricant CG, Nelson-Rees WA. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185 [DOI] [PubMed] [Google Scholar]
- 33.Mochizuki H, Takahashi M, Nishigaki K, Ide T, Goto-Koshino Y, Watanabe S, Sato H, Sato M, Kotera Y, Fujino Y, Ohno K, Uchida K, Tsujimoto H. 2011. Establishment of a novel feline leukemia virus (FeLV)-negative B-cell cell line from a cat with B-cell lymphoma. Vet. Immunol. Immunopathol. 140:307–311 [DOI] [PubMed] [Google Scholar]
- 34.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J. Virol. 58:825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishigaki K, Hanson C, Thompson D, Yugawa T, Hisasue M, Tsujimoto H, Ruscetti S. 2002. Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus. J. Virol. 76:1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunberg JH, Williams ME, Innis MA. 1984. Nucleotide sequences of the envelope genes of two isolates of feline leukemia virus subgroup B. J. Virol. 49:629–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riedel N, Hoover EA, Gasper PW, Nicolson MO, Mullins JI. 1986. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J. Virol. 60:242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 41.Schwarz G. 1978. Estimating the dimension of a model. Ann. Stat. 6:461–464 [Google Scholar]
- 42.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum-parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heard JM, Danos O. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 65:4026–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battini JL, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDougall AS, Terry A, Tzavaras T, Cheney C, Rojko J, Neil JC. 1994. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J. Virol. 68:2151–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnett AL, Wensel DL, Li W, Fass D, Cunningham JM. 2003. Structure and mechanism of a coreceptor for infection by a pathogenic feline retrovirus. J. Virol. 77:2717–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung YT, Lyu MS, Buckler-White A, Kozak CA. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 76:8218–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu T, Yan Y, Kozak CA. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J. Virol. 79:9677–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendoza R, Miller AD, Overbaugh J. 2013. Disruption of thiamine uptake and growth of cells by feline leukemia virus subgroup A. J. Virol. 87:2412–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. 2004. Identification of a human heme exporter that is essential for erythropoiesis. Cell 118:757–766 [DOI] [PubMed] [Google Scholar]