Abstract
Cucumber Necrosis Virus (CNV) is a member of the genus Tombusvirus and has a monopartite positive-sense RNA genome packaged in a T=3 icosahedral particle. CNV is transmitted in nature via zoospores of the fungus Olpidium bornovanus. CNV undergoes a conformational change upon binding to the zoospore that is required for transmission, and specific polysaccharides on the zoospore surface have been implicated in binding. To better understand this transmission process, we have determined the atomic structure of CNV. As expected, being a member of the Tombusvirus genus, the core structure of CNV is highly similar to that of Tomato bushy stunt virus (TBSV), with major differences lying on the exposed loops. Also, as was seen with TBSV, CNV appears to have a calcium binding site between the subunits around the quasi-3-fold axes. However, unlike TBSV, there appears to be a novel zinc binding site within the β annulus formed by the N termini of the three C subunits at the icosahedral 3-fold axes. Two of the mutations causing defective transmission map immediately around this zinc binding site. The other mutations causing defective transmission and particle formation are mapped onto the CNV structure, and it is likely that a number of the mutations affect zoospore transmission by affecting conformational transitions rather than directly affecting receptor binding.
INTRODUCTION
Cucumber Necrosis Virus (CNV) is a member of the genus Tombusvirus and has a monopartite positive-sense RNA genome (1, 2). CNV is transmitted in nature via zoospores of the fungus Olpidium bornovanus (2–4). The type species of the Tombusvirus genus is Tomato bushy stunt virus (TBSV), and the structure of TBSV was the first virus structure determined by X-ray crystallography (5).
These viruses have a T=3 icosahedral protein shell with a diameter of ∼300 Å formed from 180 identical coat protein (CP) subunits. The three conformationally distinct copies of the CPs, called A, B, and C, are shown in Fig. 1. The C subunits lie next to the icosahedral 2-fold axes, while the A and B subunits surround the 5-fold axes. Each capsid protein is comprised of three domains: the first ∼60 residues at the N-terminal region (R), the shell (S) domain, and the protruding (P) domain (Fig. 2A). The first ∼90 residues are disordered in the A and B subunits, while the first ∼60 residues are disordered in the C subunit. Residues ∼60 to 78 in the C subunit form a structure called the β annulus that is thought to stabilize the capsid. The first ∼60 residues at the N-terminal region interact with the RNA interior. Between this region (R) and the S domain are ∼30 residues that act as a flexible connecting arm. The S domain is ∼170 residues and forms a tight protein shell around the RNA genome. The final ∼115 residues form the P domains, which combine with the adjacent subunit's P domains to form dimeric protrusions from the shell. The A subunits lie immediately adjacent to the icosahedral 5-fold axes, and their P domains interact with the P domains of adjacent B subunits to form 60 of the 90 dimeric protrusions. The remaining protrusions are formed from C-subunit homodimers that sit on the icosahedral 2-fold axes.
Fig 1.
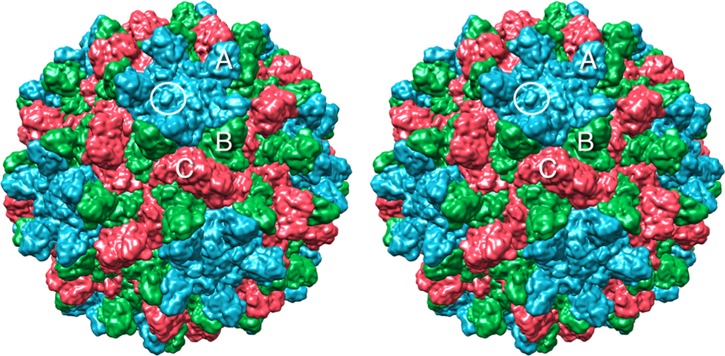
Surface rendering of CNV. The A, B, and C subunits are shown in blue, green, and red, respectively. White circle, location of the knob discussed in the text where the N109D mutation causing a transmission defect resides.
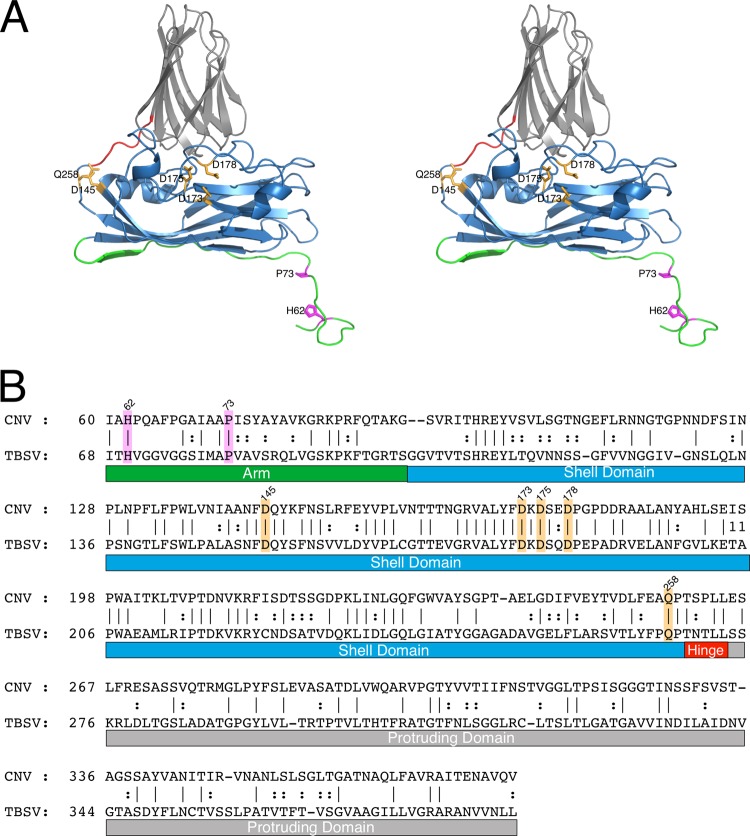
Fig 2.
Structure-based sequence alignment of CNV and TBSV using the alignment tools on the PDB server (28). (A) Stereo ribbon diagram of CNV color coded as per the various regions of the capsid protein. The arm, shell, hinge, and protruding domains are shown in green, blue, orange, and gray, respectively. (B) The primary sequences of CNV and TBSV aligned as per the atomic structure. In both panels, the putative calcium and zinc binding residues are highlighted in orange and mauve, respectively.
Recent studies have started to dissect the roles of the various capsid protein domains in the assembly of CNV particles (6, 7). The internal disordered R domain appears to play a major role in particle formation that cannot be simply ascribed to neutralization of the negative charge of the RNA core by the addition of basic residues (6). If the entire R domain or C-terminal 26 amino acids of the R domain are removed, then nearly all of the particles have T=1 symmetry. The arm region that connects the disordered R domain to the S domain is apparently critical for assembly as well. If either P73 or P85 is mutated, then particles cannot assemble or assemble poorly. The exception is that a CNV mutant with the P73G mutation yields the same amount of virus as the wild type if it is propagated in Nicotiana benthamiana (6).
The cryo-transmission electron microscopy structures of both the T=1 and T=3 particles have been determined (7). As expected, the structure of the T=3 particles resembles the known structure of TBSV (5). The T=1 particles are essentially pentons formed by the A subunits of CNV. There appears to be significant internal organization in the T=3 but not the T=1 capsids, where a structured internal shell is observed. The dimensions and general features of the inner icosahedral shell agree extremely well with the findings of previous neutron diffraction experiments with TBSV (8, 9). The outermost shell (radius, 110 to 175 Å) predominantly represents the capsid protein shell and protruding domains and is essentially that portion of the capsid described by the X-ray structure of TBSV. Immediately beneath the capsid is a shell (radius, 80 to 100 Å) that is composed of RNA with little or no protein. Beneath the layer of RNA, surprisingly, is a second shell of protein (radius, 50 to 80 Å) that does not contain RNA. This neutron-scattering model agrees exceedingly well with the internal features observed in the CNV image reconstruction (7). It is tempting to speculate that the effects of some of the R- and arm-domain mutations on assembly (6, 10) are due to their participation in these internal structures.
Glycoproteins containing specific mannose or fucose residues have been implicated in zoospore-CNV binding (11). Several mannose-containing oligosaccharides were found to effectively block CNV binding to zoospores with mannotriose and mannopentose with 50% effective concentrations of ∼120 to 150 μM. The involvement of mannose- or fucose-containing oligosaccharides in zoospore binding was supported using a variety of lectin-fluorescein isothiocyanate probes, where it was found that only concanavalin A (ConA) and Tetragonolobus purpureas lectin (TPA) bound to the surface of O. bornavanus. ConA binds to mannose and/or glucose, and TPA binds to fucose. The zoospores were well labeled by these lectins, and binding was competitively inhibited by the corresponding sugars. Finally, ConA bound to O. bornovanus and O. brassicae zoospores, but TPA bound only to O. bornavanus, and this was further evidence that there was specificity in the lectin-binding reaction.
A link between vector transmission and conformational transitions in the virus has also been identified (12). Wild-type CNV becomes markedly sensitive to trypsin digestion when treated with alkaline pH buffers in the presence of EDTA or after interaction with the zoospore vector. In a manner similar to what was previously observed with human rhinovirus (13), the portions of the capsid exposed to trypsin during this swelling process are the buried N termini and arm regions. A P73G mutation in the arm region still permits the virus to swell in vitro in the presence of EDTA, but the mutant is no longer sensitive to trypsin digestion, presumably because the arm is unable to externalize. The zoospore-treated mutant was neither sensitive to trypsin nor transmitted by zoospores. Together, all of these results clearly demonstrate that the virus undergoes a conformational transition in the presence of the zoospore that both is responsible for trypsin sensitivity and is essential for vector transmission. Somehow, even though P73 is buried in the capsid, it is intimately involved in how the capsid responds during zoospore attachment.
Presented here is the atomic structure of CNV to a resolution of ∼2.9 Å. While the core structures of the various domains are highly similar to those of TBSV, the loops connecting the secondary structural elements differ in both structure and amino acid sequence. Like other members of the Tombusviridae family, there are conserved calcium binding sites near the quasi-3-fold axes that stabilize interactions between the A, B, and C subunits. However, unlike other structures of this virus family, there appears to be a Zn2+ binding site formed by three H62 residues of the C subunits at the icosahedral 3-fold axes. A number of mutations causing defective transmission were mapped onto the atomic structure, and it is likely that many are involved with the conformational transition associated with zoospore binding rather than being involved directly with receptor binding.
MATERIALS AND METHODS
Virus production and purification.
CNV was produced and purified essentially as previously described, with some modifications (7). CNV was propagated in Nicotiana benthamiana plants that were at the 5- to 7-true-leaf stage. The inoculum contained ∼50 ng/μl virus in 50 mM KPO4, pH 6.8, and 20 μl of this solution was rubbed onto the leaves that had been dusted with carborundum. The infection was allowed to proceed for 5 to 7 days, and leaves were harvested when the plants had completely wilted. The infected leaf material was stored at −80°C until processing.
The first step of virus purification was to grind ∼200 g of frozen leaf material in 1 to 2 volumes of ice-cold 0.1 M sodium acetate, pH 5.0, that contained 10 mM β-mercaptoethanol (0.7 μl/ml). Using a chilled blender, the leaves were ground until fully homogenized (1 to 2 min) and then liquefied at high speed for about 1 to 3 min. The material was then filtered through one layer of Miracloth, and the filtrate was allowed to stand on ice for 1 h. The cellular debris was removed by centrifugation at 8,000 × g for 15 min at 4°C. To the supernatant, polyethylene glycol (PEG; molecular weight, 8,000) was added to yield a final concentration of 8% (wt/vol), and the mixture was stirred for 2 h at 4°C. The precipitate was collected by centrifugation at 8,000 × g for 20 min at 4°C and resuspended in less than 60 ml of 10 mM sodium acetate, pH 5.0. This suspension was kept at 4°C overnight to ensure that the precipitate was fully resuspended.
On the next day, the debris was removed from the suspension by centrifugation at 8,000 × g for 20 min at 4°C. CsCl was added to the supernatant to yield a 2.4 M solution. The virus was then purified by ultracentrifugation in an SW41 rotor at 36,000 rpm for 36 to 48 h. The virus band appeared in the lower third of the SW41 tube and was collected via a side puncture with a syringe and hypodermic needle. The pooled bands were then dialyzed extensively for several days against 20 mM sodium acetate, pH 5.0. Allowing at least 8 h per dialysis, the virus was first dialyzed three times in the acetate buffer in the presence of 20 mM calcium chloride and then an additional three times without the calcium added. The concentration of CNV was estimated by use of the assumption that a 1-mg/ml solution has an optical density of 4.5 at 260 nm. The typical yield was ∼30 mg of pure virus from 12 plants. The purity of the virus was determined via SDS-PAGE, but the sample was first brought to a pH of between 7 and 8 prior to boiling in SDS running buffer, since the capsid protein undergoes autolysis at acidic pHs and high temperature in the presence of SDS.
Crystallization.
CNV was crystallized using the vapor diffusion, hanging-drop method (14). The virus concentration was adjusted to 10 mg/ml in 20 mM sodium acetate buffer at pH 5.0. If the virus needed to be concentrated, low-speed centrifuge concentrators (Centricons) were used, rather than pelleting of the virus via ultracentrifugation. The largest crystals were obtained when the reservoir contained 0.16 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer and 3.2 M sodium formate, pH 6.0. The volume of the reservoir was 1 ml, and the drops were composed of 10 μl of the virus solution and 10 μl of the reservoir. Crystals started to appear within 5 days and grew to maximum size in 2 to 3 weeks.
Diffraction data collection.
Diffraction data were collected at room temperature using a Proteum R Smart 6000 charge-coupled-device detector connected to a Bruker-Nonius FR591 rotating anode generator. While numerous attempts were made to freeze the crystals for diffraction, all of the cryoprotectants and methods employed destroyed the crystals or limited the diffraction resolution to less than 4 Å. Therefore, the rather fragile crystals greatly benefited from the use of MicroRT X-ray capillaries (MiTeGen Corporation) for room temperature data collection. An oscillation angle of 0.3° with an exposure time of 10 min per frame was used. Approximately 30 to 40° of data could be collected before there was a noticeable decay in the diffraction resolution. While reflections were observed at higher resolution, data were integrated and scaled to 2.9 Å. Diffraction intensities from four crystals were integrated, merged, and scaled using the Proteum software suite. The program SHEL-X was used to scale and merge the various data sets. The crystals belong to the space group I23 with unit cell dimensions of a, b, and c of 384 Å and α, β, and γ of 90°. As data from the crystals were combined, care was taken to test the two permutations for axes (h, k, and l or k, h, and −l). This was most readily done by calculating the self-rotation functions of the data sets to determine which of the two conventions matched the base data set.
Structure determination.
From the cell volume, the CNV particle had to be sitting on a special position in the I23 lattice with two possible orientations that differed by a 90° rotation. In this unit cell, there was a single icosahedral penton in the crystallographic asymmetric unit. Using the self-rotation function in the CCP4 suite as a guide (15), the h, k, and l indexing was consistent with the icosahedral setting used by the Viper database (http://viperdb.scripps.edu): z(2)-3-5-x(2). In this orientation, the two icosahedral 2-fold axes coincide with the z- and x-coordinate axes, while a set of icosahedral 3-fold and 5-fold axes lie, in that order, between the z and x axes in the x-z plane.
The atomic model for TBSV (5) was used for molecular replacement. When a TBSV penton was placed into the I23 asymmetric unit and briefly refined using Phenix (16), the Rmodel quickly dropped to the low 30% range. The model was refined with multiple cycles of refinement and rebuilding using omit (Fo-Fc) and 5-fold-averaged electron density maps. The current model has an Rmodel of 21.3% and an Rfree of 24.1% using all data to 2.9 Å (Fig. 2 and Table 1).
Table 1.
Data collection and refinement statistics
| Parametera | Value for CNVb |
|---|---|
| PDB accession no. | 4LLF |
| Data collection statistics | |
| Wavelength (Å) | 1.5418 |
| Data resolution (Å) | 75.3–2.9 (3.0–2.9) |
| Total no. of reflections | 208,393 |
| No. of independent reflections | 134,168 |
| Completeness (%) | 64 (33) |
| Redundancy | 8.2 |
| Avg I/σ〈I〉 | 6.3 (0.5) |
| Rsym (%) | 9.3 (49.4) |
| Refinement statistics | |
| Resolution (Å) | 75.3–2.9 (1.89–1.80) |
| No. of: | |
| Nonsolvent atoms | 34,500 |
| Zn2+ atoms | 10 |
| Ca2+ atoms | 15 |
| Water molecules | 72 |
| Rwork | 21.3 (45.6) |
| No. of reflections | 15,078 (2,487) |
| Rfree (%) | 24.1 (47.3) |
| Avg B factor (Å2) | |
| Nonsolvent atoms | 52.6 |
| Zn2+ | 34.1 |
| Ca2+ | 86.0 |
| Water | 37.3 |
| RMSD | |
| Bond length (Å) | 0.011 |
| Bond angles (°) | 1.5 |
| Ramachandran analysis (%) | |
| Most favored region | 87.6 |
| Additionally allowed | 11.6 |
| Generously allowed | 0.8 |
| Disallowed | 0 |
Rsym, Σj|I(h)j − <I(h)>|/Σj I(h)j; I, intensity of a reflection; RMSD, root mean square deviation.
The values in parentheses represent the highest-resolution shell for CNV.
CNV mutagenesis.
Oligonucleotide-directed in vitro mutagenesis was used to produce CNV mutants with CP mutations. To produce mutants with T292A, D293A, W296A, Q297A, S338A, S338F, S338T, S339G, L353A, P63G, P63A, and P85A mutations, an EcoRI/NcoI fragment encompassing the CNV CP and flanking regions in a full-length infectious cDNA clone of CNV (pK2/M5) (12) was subcloned into EcoRI/NcoI-digested pT7 Blue (Novagen) and used as a template for in vitro mutagenesis. The oligonucleotide primers used for mutagenesis are available upon request. The mutations were confirmed by sequencing. Selected plasmid DNA was then digested with EcoRI/NcoI, and the fragment containing the mutation was cloned into similarly digested pK2/M5. pK2/M5 was used as the template for the production of the P67G, P67A, P259G, P259A, P262G, and P262A mutations. Following mutagenesis, plasmid DNA was digested with either EcoRI/BglII (in the case of P67G and P67A) or EcoRI/NcoI (in the case of P259G, P259A, P262G, and P262A), and the mutated fragment was cloned back into pK2/M5 to obtain a full-length clone containing the mutation. Preparation of T7 polymerase runoff transcripts and inoculation of plants were done as previously described (12). Partially purified virions were tested for their ability to be transmitted by O. bornovanus zoospores as described previously (12).
Protein structure accession number.
The coordinates for CNV have been deposited in the Protein Data Bank (PDB) database (PDB accession number 4LLF).
RESULTS
Not surprisingly, the structure of CNV is highly similar to that of TBSV (Fig. 2). The core structures of the shell and protruding domains are nearly identical, with the major differences lying in the loops that connect the secondary structural elements.
Since the CNV and TBSV structures appear to be more similar than the level of similarity inferred by their sequence similarity, the structures were used to align the amino acid sequences (Fig. 2). Figure 2A shows the C subunit of CNV colored according to the key domains of the tombusviruses. The results of the structure-driven sequence alignment are shown in Fig. 2B. What is immediately apparent is that the residues in contact with the putative zinc and calcium ions, as described below, are perfectly conserved. In addition, it is interesting to note that P73, which forms the dome over the bound zinc, is one of the few conserved residues in this N-terminal region. It may be that proline at this position helps to twist the termini around to form the β annulus.
Potential calcium binding sites.
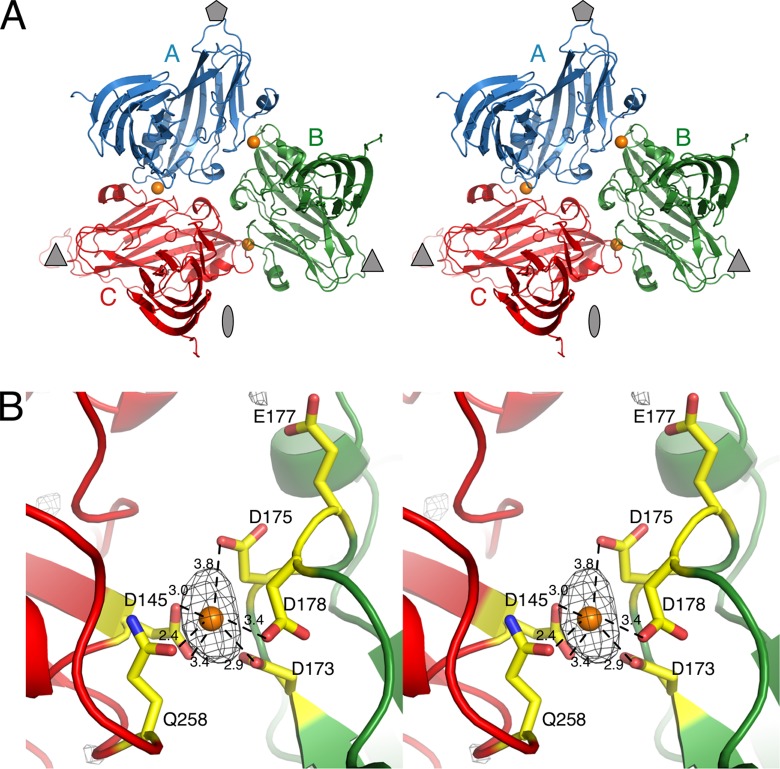
During the refinement process, several strong peaks appeared in the Fo-Fc electron density maps, some of which were assumed to be calcium ions because of the binding environment (Fig. 3). There was one strong peak between each of the subunit interfaces: A-B, B-C, and A-C. From one subunit, residues Q258 and D145 were in close contact with the modeled calcium ion, while residues D175, D178, and D173 contributed interactions from the adjacent subunit. The location for the bound calcium ions is essentially the same as what was observed for TBSV (17) and melon necrotic spot virus (18). E177 is also shown, since it was found to be important for zoospore transmission (Table 2). The refined B values were higher for the putative calcium ions than the average for the protein atoms, suggesting that all of the sites might not be occupied in the structure.
Fig 3.
Location of putative calcium ions in CNV. (A) Stereo ribbon diagram of the structure of a CNV icosahedral unit. The A, B, and C subunits are displayed in blue, green, and red, respectively. The approximate location of the 5-, 3-, and 2-fold axes are denoted as a gray pentagon, triangle, and ellipse, respectively. The locations of calcium ions are denoted by orange spheres. (B) Details of the interactions for the bound ion. The black mesh represents the peak from the Fo-Fc electron density contoured at 4σ. This particular ion is at the B (green) and C (red) subunit interface.
Table 2.
Location of mutations due to passaging in plants and effects on zoospore transmission and bindinga
| Isolate | Mutation | Location | % transmissionb | % binding |
|---|---|---|---|---|
| Wild type | 96 (49/51) | 100 | ||
| LLK00 | F66C | Arm | 100 (10/10) | 130 ± 8.5 |
| LLK85 | N109D | Shell | 76 (19/25) | 53 ± 12 |
| LLK84 | G221V | Shell | 50 (10/20) | 53 ± 7.5 |
| LLK8 | L294F | Protrusion | 21 (3/14) | 68 ± 18 |
| LLK26 | L294F | Protrusion | 10(1/10) | NDc |
| LLK10 | V295A | Protrusion | 27 (4/15) | 39 ± 22 |
| LLK63 | S338G | Protrusion | 14 (3/21) | 21 ± 17 |
| LLK82 | G357E | Protrusion | 75 (15/20) | 64 ± 24 |
| LL5K8 | E177K + L294F | Shell + protrusion | 0 (0/8) | 22 ± 7 |
Data are summarized from Kakani et al. (21).
Data in parentheses represent the number of assays where CNV was transmitted by the zoospores/the total number of times the assay was performed for that mutant.
ND, not determined.
It is important to note that early in the crystallization trials, CNV was purified via CsCl isopycnic density gradient ultracentrifugation and dialyzed directly against the 20 mM sodium acetate buffer, pH 5.0. While the material did form crystals, their dimensions were at most ∼0.2 mm in length. Because it was possible that the very high concentrations of CsCl used for isopycnic ultracentrifugation might displace calcium and affect particle stability, the protocol was changed to include calcium chloride during the initial dialysis (see Materials and Methods). With that one change, large crystals (∼1 mm) were reproducibly obtained. The addition of calcium may have either made for more homogeneous particles or stabilized the virions during storage and crystallization.
Potential zinc binding site.
Even at the early stages of building and refinement, large, oblong peaks appeared in the difference (Fo-Fc) electron density maps (Fig. 4) at the icosahedral 3-fold axes. It was immediately apparent that the icosahedrally related 3-fold H62 residues clustered around this density in a manner reminiscent of a zinc binding protein (e.g., ZnuA from Synechocystis sp. strain 6803 [19]). However, the density in the difference map was not spherical for the putative zinc atom, and there is usually a fourth ligating residue (or water molecule) above the plane formed by the three chelating histidine residues. While it was considered that a water molecule might be associated with the zinc but was just not visible at this resolution, it was also possible that there might be two conformations for the zinc and chelating histidine residues. However, since there was insufficient space to accommodate two zinc atoms, the elongated shape was likely due to two different conformations for the bound zinc. To that end, two zinc atoms associated with the C domain were given an occupancy of 1/6, each, to compensate for there being two conformations for a zinc atom at the icosahedral (and I23) 3-fold axis. In addition, the side chain for H62 was also assigned two conformations so that both zinc conformations could interact with Nε2 atoms in the side chain. For both conformations, the Zn2+-Nε2 distance was between 2.1 and 2.2 Å, typical for Zn2+ interactions with histidine (19). Not only does this dual position for the bound zinc fit well into the electron density, but it also agrees well with the scattering in that region. Even with an occupancy of 1/6 for each Zn2+ atom, the refined, average B factor was ∼34 Å2. This value is actually lower than that for the protein atoms in the immediate vicinity and significantly lower than the average B value for all nonsolvent atoms (Table 1).
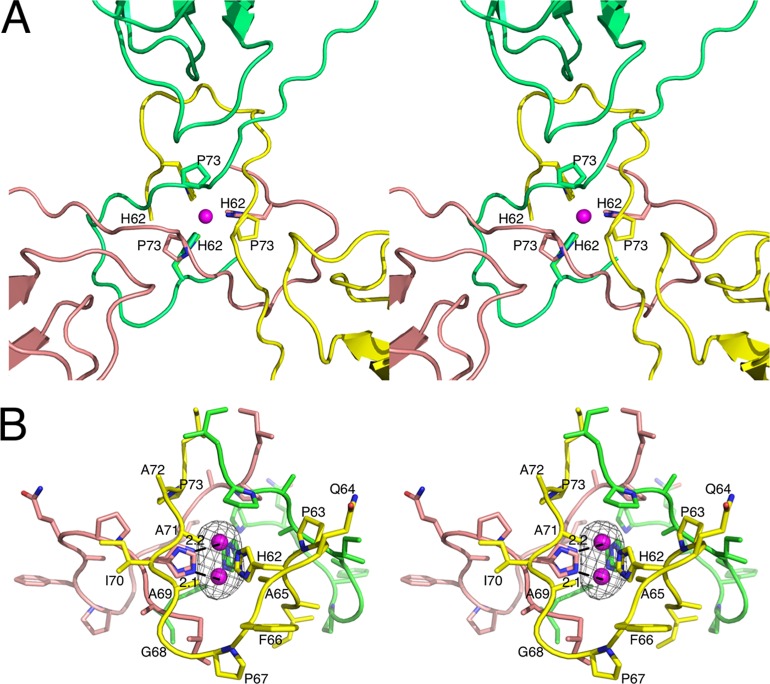
Fig 4.
Location and binding environment of putative zinc ion. (A) The putative zinc ion (mauve sphere) binding site is located at the icosahedral 3-fold axes. The three icosahedrally related subunits are colored yellow, pink, and green. The view is from the exterior of the capsid looking into the 3-fold axis. The ligating residue H62 and the location of the P73 mutation for defective transmission are also noted. (B) Side-on view of the putative zinc binding site. The black mesh represents the Fo-Fc map contoured at 4σ. The two partially occupied zinc atoms are shown along with distance between one of the two bound zinc atoms and the closest ε2 nitrogen atom of the two alternative histidine side chain conformations.
The location of the putative Zn2+ atom is within the β annulus (20) formed by the N termini of three C subunits at the icosahedral 3-fold axes. As mentioned above, it is common to see at least one additional zinc binding atom (e.g., water) above and/or below the plane formed by the chelating histidine imidazole rings. However, additional Zn2+ binding interactions are not obvious in this case. Immediately above the zinc atom is P73, which forms a dome. This is possibly important with regard to zoospore transmission, as discussed below. Directly beneath the bound zinc is some density that is likely due to residue 59 that was too disordered to be built into this structure. However, this density is too far away to interact with the zinc atom. At this resolution, there may be a water molecule in this zinc binding cavity that is too weak to observe. It is also possible that the zinc and chelating histidine residues have two conformations because there is not a fourth binding interaction to fix the positions of the zinc and imidazole rings.
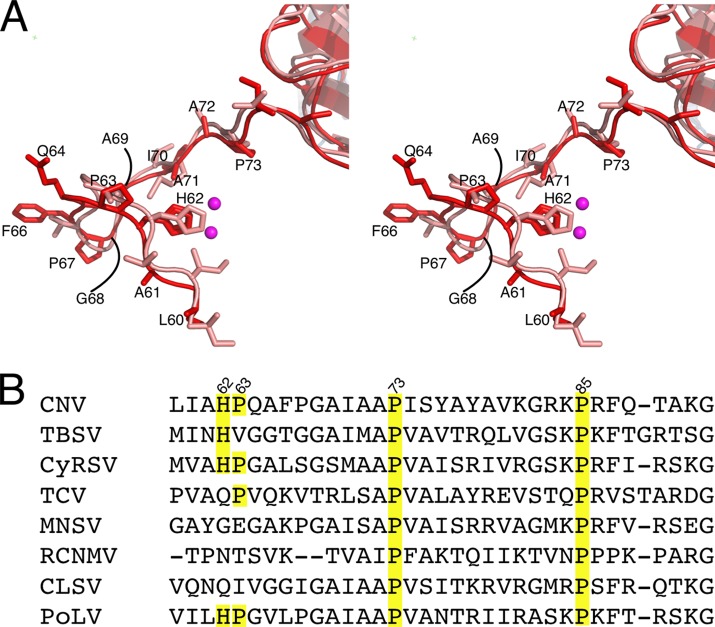
In light of this possible zinc binding site in CNV, the structures of the N termini of the C subunits of CNV and TBSV were compared. As shown in Fig. 5, the N termini of the CNV and TBSV C subunits have similar backbone structures, in spite of having very different amino acid sequences. This disparity in sequence makes it more notable that H62 and P73 are conserved in both viruses and have the same position with respect to the zinc seen in CNV. It is not clear why zinc was not observed in the structure of TBSV since the zinc binding site is essentially identical to that of CNV. Zinc was not added to CNV and therefore was likely picked up during particle formation in the plant tissue. This is similar to what occurred with the bacterial zinc binding protein ZnuA (19). Since both TBSV and CNV were purified in similar ways, it seems likely that zinc was not observed in TBSV due to limitations in resolution. As also shown in Fig. 5, the His residue at position 62 in CNV appears to be conserved in the tombusviruses TBSV and Cymbidium ring spot virus (CyRSV) and in the aureusvirus Pathos latent virus (PoLV). Indeed, H62 is conserved in 9 of the 12 definitive tombusviruses, in the aureusvirus PoLV, as well as the tentative aureusvirus sesame necrotic mosaic virus. In addition, P73 and P85 are found in all of the members, representing 4 genera, of the Tombusviridae listed.
Fig 5.
Conservation of the zinc binding structure at the β annulus. (A) The N termini of the C subunit around the putative zinc binding site for CNV (red) and TBSV (pink). The mauve spheres represent the zinc ions. Not only is the position of H62 highly conserved, but the site that affects transmission, P73, is also conserved. (B) Sequence alignment of this region in Cymbidium ring spot virus (CyRSV; a tombusvirus), Turnip crinkle virus (TCV; a carmovirus), Melon necrotic spot virus (MNSV; a carmovirus), Red clover necrotic mosaic virus (RCNMV; a dianthovirus), Cucumber leaf spot virus (CLSV; an aureusvirus), and Pathos latent virus (PoLV; an aureusvirus).
Location of mutations causing defective transmission.
Mechanically passaged CNV isolates were screened for mutants that were defective in fungal transmission and then the mutations were confirmed by site-directed mutagenesis (21). With the atomic structure of CNV, the location of these mutations can be mapped to try to understand how they might affect zoospore binding and transmission (Fig. 6A and Table 2). The only mutation in this list that did not affect transmission was F66C, which is near the H62 that binds to Zn2+.
Fig 6.
(A) Locations of mutations causing defective transmission in CNV isolated via mechanical passaging. Shown here is the C subunit of CNV, with the mutation sites denoted by spheres. The orientation here is with the RNA at the bottom and the icosahedral 3-fold axis running vertically on the right side. The various mutations and their effects are shown in Table 2. (B) Site-directed mutagenesis of CNV around the locations of the naturally occurring mutants. The various mutations and their effects are summarized in Table 3. Red asterisks, the three sites where mutations affected transmission, but not particle formation.
G221 is right at the quasi-3-fold axis (Fig. 3 and 6). If the glycine is simply mutated to a valine, then the side chain clashes with the side chain of D222 from the immediately adjacent subunit. D222, E177, and D175 are all clustered together and immediately adjacent to the calcium binding site and K224 (Fig. 3). The G221V mutation could greatly destabilize the A-B-C interactions at the quasi-3-fold axis or could affect how the quasi-3-fold axes expand in response to pH changes or zoospore binding.
N109 lies on a small knob immediately adjacent to the icosahedral 5-fold and 3-fold axes (Fig. 1 and 6). The side chain is fully exposed to the solvent and does not interact with any adjacent subunits. Therefore, the fact that the fairly conserved mutation of N109D blocks ∼25% transmission and 50% binding to zoospores suggests that it could be directly involved in receptor binding.
L294 and V295 lie on the portion of the P-domain β sheet that is on the side opposite the P domain-dimer interface. Both the L294F and V295A mutations decrease transmission to a level of ∼20% of that of the wild type. However, the L294F mutation bound to the zoospores at a level of ∼70% of that of the wild type, whereas V295A bound ∼40% as well as the wild type. This suggests that there can be a disconnect between binding and transmission whereby the effects of a particular mutation can be greater on transmission than on binding. The side chain for L294 faces toward the inner core of the P domain and, with residues like W296, I308, and F310, contributes to the hydrophobic interface between the β sheets. The limited space around the L294 side chain makes it unlikely that the L294F mutation would not cause some distortion in the β barrel. V295 and V288 form a small hydrophobic patch on the outer surface of the P domain. It seems unlikely that the V295A mutation would cause major conformational changes in the P domain. It is important to note that neither mutation drastically affects capsid stability or assembly (21). Interestingly, these residues are on the P domain surface that rises up immediately adjacent to the knob on the shell domain that contains the N109D mutation.
The S338G mutation lies on a loop at the tip of the P domain, is essentially exposed to solvent, and leads into the P domain-dimer interface. This mutation has a profound effect on zoospore binding and transmission. Since this particular loop is already fairly flexible, as per its relatively high B value, it seems unlikely that the S338G mutation would have a profound effect on the P-domain structure. Similarly, G357 is fully exposed on a relatively disordered loop, and a simple mutation to a Glu would place the side chain into the solvent. It may be that these residues directly interact with the zoospore surface.
Finally, the E177K/L294F double mutant is not transmitted by zoospores and binds to zoospores only 22% as well as the wild type. E177, as noted above and shown in Fig. 6, is immediately adjacent to the calcium binding site and lies in a cluster of acidic amino acids. At a minimum, the E177K mutation would disrupt the negatively charged field in that area and could disrupt calcium binding and thus subunit interactions.
Site-directed mutagenesis of CNV.
As reviewed above, the naturally occurring mutations causing defective transmission were located all over the capsid protein. To try to narrow down regions that are essential for zoospore interactions versus associated conformational changes, a number of residues were targeted via site-directed mutagenesis (Table 3 and Fig. 6). Of all of the mutations tested, only P63G, P73G, and P259G affected transmission without being deleterious to particle formation. It should be noted that the selection of mutants was done in parallel with the structure determination and therefore was not benefited by the structural information described above.
Table 3.
Site-directed mutagenesis of the capsid protein and effects on particle assembly and transmission
| Mutation | Location | Particle assembly | % transmissiona |
|---|---|---|---|
| Wild type | + | 87 (34/39) | |
| P63G | Arm | + | 10 (2/20) |
| P63A | Arm | + | 100 (5/5) |
| P67G | Arm | + | 100 (5/5) |
| P67A | Arm | + | 100 (4/4) |
| P73G | Arm | + | 4 (1/25) |
| P85A | Arm | + | 100 (4/4) |
| P259G | Shell | + | 0 (0/25) |
| P259A | Shell | + | 93 (14/15) |
| P262G | Hinge | + | 100 (5/5) |
| P262A | Hinge | + | 100 (5/5) |
| T292A | Protrusion | + | 100 (5/5) |
| D293A | Protrusion | − | Not tested |
| W296A | Protrusion | − | Not tested |
| Q297A | Protrusion | + | 100 (5/5) |
| S338A | Protrusion | + | 100 (5/5) |
| S338F | Protrusion | − | Not tested |
| S338T | Protrusion | − | Not tested |
| S339G | Protrusion | + | 100 (5/5) |
| L353A | Protrusion | − | Not tested |
Data in parentheses represent the number of assays where CNV was transmitted by the zoospores/the total number of times the assay was performed for that mutant.
While the naturally occurring mutation F66C did not appear to affect CNV binding or transmission, P63 and P67 were targeted to see whether these internal portions of the capsid that become exposed upon contact with the zoospore (12) can affect transmission. P73 and P85 were modified to see if they might control particle assembly. Of all of the site-directed mutations in the arm domain, only the mutants with the P63G and P73G mutations were transmission defective. Since the P63A mutation had no effect on transmission, the P63G mutation appears to be exerting its effects by increasing the flexibility at that position. This portion of the capsid is ordered only in the C subunit, and therefore, these mutations in the A and B subunits could affect the formation of the inner core (7), how the N termini are exposed upon zoospore binding, or possibly some structure that the termini might form upon extrusion from the capsid. Alternatively, these mutations could be affecting the structures observed in the N termini of the C subunits at the icosahedral 3-fold axes. P63 is immediately adjacent to the H62 that is bound to a zinc ion, and P73 forms the dome-like structure over the bound zinc. Therefore, it may be that these mutations are disrupting the Zn2+ binding site. It should be noted that while the mutant with the P73G mutation yielded as much virus as the wild type, the P73A, P73C, and P73L mutations all had profound effects on virus yield. Therefore, while a more flexible Gly residue allows particle formation, unlike all of the other residues used for mutagenesis at this position, it disrupts something essential for vector transmission, such as proper extrusion of the arm upon particle swelling (12).
Since there seemed to be the largest effects when proline residues were changed, P259 and P262, which lie in the shell domain, were mutated. Mutations in P262 did not affect transmission, but the P259G mutation essentially eliminated zoospore transmission. P259 is in the shell domain and not far from the hinge region connecting the P and S domains. P259 is buried under the P domain and contacts an adjacent subunit. As with P63, only mutating P259 to the more flexible glycine residue affected transmission.
The P-domain mutations were made because they were in or near regions in the P domain that appeared to be important for fungus transmission, on the basis of previous studies by Kakani et al. (21) (Fig. 6B). However, unlike the mutants with the naturally occurring P-domain mutations, half of the mutants did not form particles and the other half were still transmitted by the zoospores. While the naturally occurring L294F and V295A mutants were poorly transmitted (∼20% compared to wild type), the W296A mutant did not form particles and the Q297A mutation did not affect transmission. The W296A mutation likely destabilizes the P domain, in general, by eliminating a large hydrophobic side chain in the β-barrel core. This is likely similar to the effects of the L353A mutation, which also does not allow particle formation.
The naturally occurring S338G mutation allowed particle formation but interfered with transmission. S338, being at the tip of the P domain-dimer interface, is apparently at a very sensitive location since even a conserved mutation to Thr blocked particle formation. Similarly, mutating S338 to the large Phe residue blocked particle formation. The effects of the S338G mutation on transmission is again likely due to some effect of making that site more flexible, since the S338A mutation had no effect on either particle formation or zoospore binding. However, mutating S339, which points away from the P domain-dimer interface, to a glycine had no effect on either particle formation or transmission.
DISCUSSION
The atomic structure of CNV is, as expected, highly similar to that of TBSV. The majority of the 37% amino acid identity that they share lies in the shell domain, with the loops in the P domains showing the greatest divergence in structure and sequence. One of the most notable findings was that CNV appears to have a Zn2+ ion bound to the arm regions of the C subunits. What is unusual is that the metal sits on the icosahedral 3-fold axes and, from the shape of the density, appears to bind in above and below the plane made by the three H62 residues, with partial occupancies. The binding environment observed here is almost identical to that for other zinc binding proteins, such as the zinc transporter protein ZnuA. Since zinc was not added during the purification process, it appears that the metal has an extremely high affinity for this binding site. Interestingly, a His residue is found at that position in the sequences of TBSV, CyRSV, and PoLV and has the same conformation in the TBSV crystal structure. The function of the bound zinc is not at all clear, but it could play a role in stabilizing the capsid at the β annulus or could help drive assembly in the cell. Interestingly, a nearly identical zinc binding site was found in the major capsid protein VP6 of rotavirus and has been shown to be important for stability and assembly (22, 23). Zinc was also found in the α-helical triple-coiled-coil domain of the avian reovirus S1133 fiber (24) and may be part of an important hinge region between fiber domains.
It is clear that CNV transmission is not simply a process by which the virus binds to the zoospore and is dragged into the plant host. When CNV interacts with the zoospores, the capsid expands and the buried N termini are extruded and can be cleaved by trypsin. While at first glance this would seem to be merely a function of the extensive capsid swelling, this cannot be wholly true, since the P73G mutation still allows swelling but not the associated exposure of the N termini (12). The mutations that block virus binding to and transmission by zoospores should help to narrow down the receptor binding site. However, the locations of a number of the mutations and the zoospore-associated conformational changes complicate interpretation. For example, we found that the two proline residues (P63 and P73) immediately surrounding the conserved H62 that binds to a Zn2+ ion play a significant role in particle assembly and zoospore transmission. However, this region is ordered only in the C subunit and is buried under the capsid. Is the abrogation of zoospore binding by the P63G and P73G mutations due to effects on this Zn2+ binding site via the conformational changes essential for zoospore transmission, or do these mutations affect the disordered regions in the A and B subunits that are extruded upon zoospore exposure?
The mutations that might be directly involved in zoospore binding are those that are exposed or on the portions of the P domain that do not appear to be involved in assembly or the zoospore-associated conformational changes. N109 is highly exposed and lies in the knob-like structure around the 5-fold and 3-fold axes. Therefore, the conservative N109D mutation is unlikely to have profound effects on the structure, and for this reason, it would be a good candidate for receptor binding. Similarly, albeit not as clear, L294, V295, and G357 all lie on the external portion of the P-domain β barrel, and mutations at these sites have profound effects on binding and transmission. While S338 is also on the tip of the P domain, CNV does not tolerate changes here, as per the marked loss of viral yield. Further, only the S338G mutation affected transmission without a concomitant loss in virus production. Nevertheless, the region on the P domain around V295, L294, and G357 seems to be a logical possibility for a location for saccharide binding. It not only represents a highly exposed portion of capsid but also is the largest cluster of mutations resulting in defective binding. Since saccharide binding sites on proteins are as diverse as carbohydrates themselves, it is difficult to suggest that this region is similar to other carbohydrate binding sites (e.g., see reference 25). However, it is interesting to note that a region of the cucumber mosaic virus apparently involved in aphid transmission lies on the outermost portion of the capsid (26, 27).
Further studies are clearly needed to delineate the regions of the capsid directly involved in zoospore binding and separate them from those necessary for the associated conformational changes. For example, infectivity studies are required to assess the fitness of mutants with mutations at P259 and P63, and experiments need to be designed to assess potential changes in conformation induced by these substitutions. Do these mutants bind to zoospores efficiently using the in vitro binding assay, and do these mutants undergo the proper conformational changes upon zoospore binding? As another approach, work is under way to clone and express the isolated P domain. By working with the isolated P domain, we can determine whether it is sufficient for zoospore and/or polysaccharide binding. The P-domain mutants can be methodically analyzed for folding and binding properties without concern for the extrusion of the N termini or swelling of the capsid. Essentially, at this time it is unclear whether the internal portions of the capsid that are exposed upon interaction with the zoospores are directly involved with binding or whether they are just involved with the conformational changes necessary for transmission.
ACKNOWLEDGMENTS
This work was supported by start-up funds from the Donald Danforth Plant Science Center (to T.J.S.) and by NSERC research grant RG 43840 (to D.R.).
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Rochon DM, Tremaine JH. 1989. Complete nucleotide sequence of the cucumber necrosis virus genome. Virology 169:251–259 [DOI] [PubMed] [Google Scholar]
- 2.Rochon D, Kakani K, Robbins M, Reade R. 2004. Molecular aspects of plant virus transmission by Olpidium and Plasmodiophorid vectors. Annu. Rev. Phytopathol. 42:211–241 [DOI] [PubMed] [Google Scholar]
- 3.Campbell RN. 1996. Fungal transmission of plant viruses. Annu. Rev. Phytopathol. 34:87–108 [DOI] [PubMed] [Google Scholar]
- 4.Dias HF. 1970. The relationship between cucumber necrosis virus by Olpidium cucurbitacaerum. Virology 40:828–839 [DOI] [PubMed] [Google Scholar]
- 5.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. 1978. Tomato bushy stunt virus at 2.9Å resolution. Nature 276:368–373 [DOI] [PubMed] [Google Scholar]
- 6.Kakani K, Reade R, Katpally U, Smith TJ, Rochon D. 2008. Induction of particle polymorphism in cucumber necrosis virus coat protein mutants in vivo. J. Virol. 82:1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katpally U, Kakani K, Reade R, Rochon D, Smith TJ. 2007. Structures of T=1 and T=3 Cucumber necrosis virus particles: evidence of internal scaffolding. J. Mol. Biol. 365:502–512 [DOI] [PubMed] [Google Scholar]
- 8.Chauvin C, Witz J, Jacrot B. 1978. Structure of the tomato bushy stunt virus: a model for protein-RNA interactions. J. Mol. Biol. 124:641–651 [DOI] [PubMed] [Google Scholar]
- 9.Timmins PA, Wild D, Witz J. 1994. The three-dimensional distribution of RNA and protein in the interior of tomato bushy stunt virus: a neutron low-resolution single-crystal diffraction study. Structure 2:1191–1201 [DOI] [PubMed] [Google Scholar]
- 10.Reade R, Kakani K, Rochon D. 2010. A highly basic KGKKGK sequence in the RNA-binding domain of the Cucumber necrosis virus coat protein is associated with encapsidation of full-length CNV RNA during infection. Virology 403:181–188 [DOI] [PubMed] [Google Scholar]
- 11.Kakani K, Robbins M, Rochon D. 2003. Evidence that binding of cucumber necrosis virus to vector zoospores involves recognition of oligosaccharides. J. Virol. 77:3922–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakani K, Reade R, Rochon D. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J. Mol. Biol. 338:507–517 [DOI] [PubMed] [Google Scholar]
- 13.Lewis JK, Bothner B, Smith TJ, Siuzdak G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. U. S. A. 95:6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McPherson A. 1990. Current approaches to macromolecular crystallization. Eur. J. Biochem. 189:1–23 [DOI] [PubMed] [Google Scholar]
- 15.Bailey S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 16.Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. The Phenix refinement framework. CCP4 Newsl. 42:Contribution 8 [Google Scholar]
- 17.Hogle J, Kirchhausen T, Harrison SC. 1983. Divalent cation sites in tomato bushy stunt virus. Difference maps at 2.9 Å resolution. J. Mol. Biol. 171:95–101 [DOI] [PubMed] [Google Scholar]
- 18.Wada Y, Tanaka H, Yamashita E, Kubo C, Ichiki-Uehara T, Nakazono-Nagaoka E, Omura T, Tsukihara T. 2008. The structure of melon necrotic spot virus determined at 2.8Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64(Pt 1):8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S, Wei B, Bhattacharyya-Pakrasi M, Pakrasi HB, Smith TJ. 2003. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. J. Mol. Biol. 333:1061–1069 [DOI] [PubMed] [Google Scholar]
- 20.Harrison SC. 1983. Virus structure: high-resolution perspectives. Adv. Virus Res. 28:175–240 [DOI] [PubMed] [Google Scholar]
- 21.Kakani K, Sgro J-Y, Rochon D. 2001. Identification of specific cumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J. Virol. 75:5576–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BVV, Cohen J, Rey FA. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 20:1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erk I, Huet J-C, Duarte M, Duquerroy S, Rey FA, Cohen J, Lepault J. 2003. A zinc ion controls assembly and stability of the major capsid protein of rotavirus. J. Virol. 77:3595–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guardado-Calvo P, Fox GC, Llamas-Saiz AL, van Raaij MJ. 2009. Crystallographic structure of the α-helical triple coiled-coil domain of avian reovirus S1133 fibre. J. Gen. Virol. 90:672–677 [DOI] [PubMed] [Google Scholar]
- 25.Koropatkin N, Martens EC, Gordon JI, Smith TJ. 2009. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry 48:1532–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry KL, Zhang L, Shintaku MH, Palukaitis P. 1994. Mapping determinants in cucumber mosaic virus for transmission by Aphis gossypii. Virology 205:591–595 [DOI] [PubMed] [Google Scholar]
- 27.Smith TJ, Chase E, Schmidt T, Perry KL. 2000. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J. Virol. 74:7578–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prlić A, Bliven S, Rose PW, Bluhm WF, Bizon C, Godzik A, Bourne PE. 2010. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics 26:2983–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]