Abstract
Genes within the E3 transcription unit of human adenoviruses modulate host immune responses to infection. A comprehensive genomics and bioinformatics analysis of the E3 transcription unit for 38 viruses within human adenovirus species D (HAdV-D) revealed distinct and surprising patterns of homologous recombination. Homologous recombination was identified in open reading frames for E3 CR1α, CR1β, and CR1γ, similar to that previously observed with genes encoding the three major structural capsid proteins, the penton base, hexon, and fiber.
TEXT
Human adenoviruses (HAdVs) infect a wide array of mucosal surfaces, including the respiratory and gastrointestinal tracts and the eye, and cause infections that vary in severity from mild to life-threatening (1–3). Sixty-nine HAdV genotypes are now formally recognized in GenBank; they have been classified into seven species (A to G), with species HAdV-D containing the most members. HAdV-D includes a substantial number of viruses identified during the first 2 decades of the AIDS epidemic (4). HAdVs are preferred candidate vectors for gene therapy trials because of their broad tissue tropism and ease of manipulation (5). These viruses are nonenveloped with an icosahedral capsid and contain a linear, double-stranded DNA genome of 34 to 36 kb. The recent completion of whole-genome sequencing and analysis for all prototype HAdV genomes (6) has contributed to renewed understanding of the role of homologous recombination in the evolution of HAdVs, particularly species D, but also other HAdV species (7–16). Genes for the three major capsid proteins, the hexon, penton base, and fiber, all readily recombine among viruses within HAdV-D (13, 17–25). However, the evolution of another region of HAdV-D genomes with relative hypervariability, the E3 transcription unit, has not been as well studied. Although E3 is labeled an early transcription region, its transcripts are expressed both early and late during viral infection (26–28). E3 gene products are not required for viral replication in cultured cells (29), but they act to inhibit cellular and cytokine-mediated host immune responses to infected cells (30–32). Specific deletions of E3 have been shown to affect the activity of bioengineered oncolytic HAdV in tumor models (33), and exogenous expression of E3 proteins can prolong xenograft transplant survival (34). These data suggest that the E3 transcription unit is important to viral pathogenesis.
The E3 transcription units in different HAdV species vary in both length and number of open reading frames (ORFs) and are areas of major sequence divergence (35, 36). The DNA sequence of the E3 transcription unit of HAdV-C2 was the first to be determined (37, 38), followed by that of HAdV-C5 (39), and consists of seven ORFs. From the 5′ end of the E3 coding region, these are 12.5K (or gp12.5 kDa), CR1α (also known as 6.7K), 19K (or gp19K), CR1β (11.6K), RIDα (10.4K), RIDβ (14.9K), and 14.7K. The RID proteins were named for their activity in receptor internalization and degradation (40–42), and CR1 proteins were named for conserved region 1 within E3 (43). The E3 transcription unit from HAdV-D contains eight ORFs, including an additional ORF not found in HAdV-C, CR1γ (30K). HAdV-D gene orthologs tend to vary in size relative to HAdV-C (43, 44).
With notable exceptions (25, 43–45), most of what is known about the functions of specific E3 proteins is derived from studies of HAdV-C. For example, HAdV-C2 E3 glycoprotein CR1α directs E3 19K to the endoplasmic reticulum (46). There, it inhibits cell lysis caused by cytotoxic T cells by binding to and retaining major histocompatibility complex (MHC) class I proteins in the endoplasmic reticulum (47), thereby blocking presentation of viral peptides within MHC class I at the cell surface (48–52). E3 19K also sequesters natural killer (NK) cell ligands (53), reducing NK cell recognition of infected cells. The HAdV-C E3 14.7K protein inhibits cell lysis by tumor necrosis factor alpha (TNF-α) and lymphotoxin (54–56). CR1α, RIDα, and RIDβ cooperate to evade TNF-α-related apoptosis through TRAIL (57–60), and the RID complex independently mediates loss of cell surface Fas (58, 61), blocks TNF-α-induced NF-κB activation (62–64), and downregulates the epidermal growth factor receptor (40, 65). HAdV-C CR1β (66), also called the adenovirus death protein, is expressed at greatest abundance late in infection (67) and is required for efficient cell lysis (68) and viral spread (69) in the final phase of viral infection. Notably, the immune evasion functions of E3 gene products are not universal to all cell types (70), and substantial differences may exist between orthologs from E3 regions of different species (45).
The recent availability of high-quality whole-genome sequences with validated annotations for all previously typed viruses within HAdV-D (6) now permits the comparative examination of the HAdV-D E3 genomic region. To form an overview of possible evolutionary relationships between E3 regions within HAdV-D, the full E3 regions of 38 viruses (Table 1) were subjected to bootstrap analysis through the neighbor-joining method, using MEGA (Molecular Evolutionary Genetics Analysis 4.0.2) (71) (Fig. 1). In numerous instances, paired viruses formed subclades on the neighbor-joining tree, for example, HAdV-D types 15 and 53, types 19a (now type 64 [23]) and 37, types 24 and 46, types 9 and 56, and types 33 and 43. These data add to information from prior analyses suggesting homologous recombination in the E3 region between types 19a (type 64) and 37 (23) and between types 9 and 56 (20). The E3 region of HAdV-D53 E3 was previously reported to be most closely related to that of type 8 (22), but at the time of that analysis, many fewer E3 regions had been sequenced, and the full E3 sequence for type 15 had not yet been released. Phylogenetic data also suggest that E3 recombination may in some cases occur for the entire E3 transcription unit, as previously suggested for HAdV-D63, which evolved as the result of a single recombination event between types 29 and 30, encompassing about 25% of their genomes and including the entire E3 and fiber ORFs (24).
Table 1.
GenBank accession numbers for viruses analyzed in this study
| Type | GenBank accession no. |
|---|---|
| HAdV-D8 | AB448767 |
| HAdV-D9 | AJ854486 |
| HAdV-D10 | JN226746 |
| HAdV-D13 | JN226747 |
| HAdV-D15 | JN226748 |
| HAdV-D19p | JQ326209 |
| HAdV-D19a | EF121005 |
| HAdV-D20 | JN226749 |
| HAdV-D22 | FJ619037 |
| HAdV-D23 | JN226750 |
| HAdV-D24 | JN226751 |
| HAdV-D25 | JN226752 |
| HAdV-D26 | EF153474 |
| HAdV-D27 | JN226753 |
| HAdV-D28 | FJ824826 |
| HAdV-D29 | JN226754 |
| HAdV-D30 | JN226755 |
| HAdV-D32 | JN226756 |
| HAdV-D33 | JN226758 |
| HAdV-D36 | GQ384080 |
| HAdV-D37 | AB448775 |
| HAdV-D38 | JN226759 |
| HAdV-D39 | JN226760 |
| HAdV-D42 | JN226761 |
| HAdV-D43 | JN226762 |
| HAdV-D44 | JN226763 |
| HAdV-D45 | JN226764 |
| HAdV-D46 | AY875648 |
| HAdV-D47 | JN226757 |
| HAdV-D48 | EF153473 |
| HAdV-D49 | DQ393829 |
| HAdV-D51 | JN226765 |
| HAdV-D53 | FJ169625 |
| HAdV-D54 | AB333801 |
| HAdV-D56 | HM770721 |
| HAdV-D58 | HQ883276 |
| HAdV-D59 | JF799911 |
| HAdV-D60 | HQ007053 |
Fig 1.
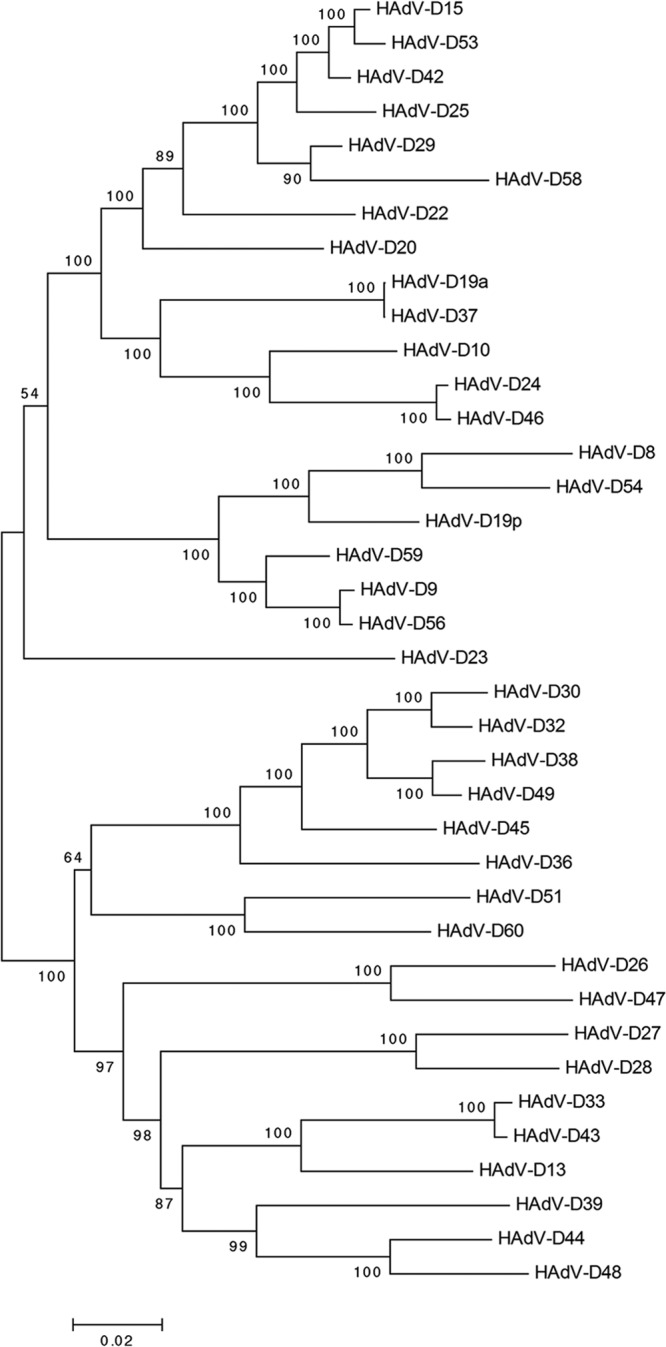
Bootstrap-confirmed (500 replicates) neighbor-joining phylogenetic tree of whole E3 transcription units from 38 viruses within HAdV-D. Bootstrap values below 80 are indicative of low confidence.
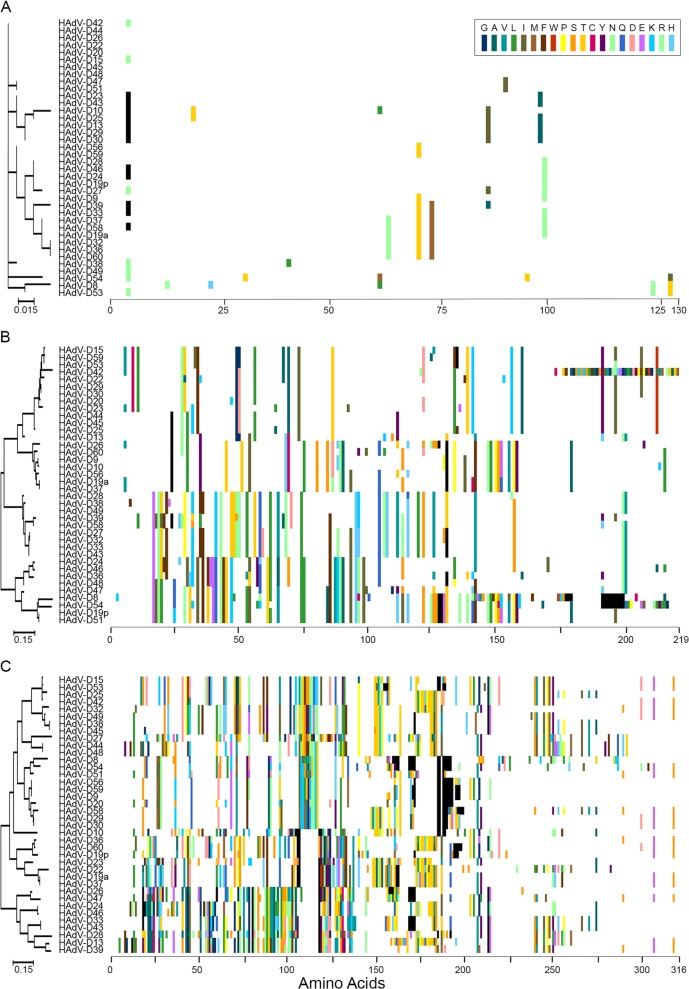
To further parse possible recombination between individual HAdV-D E3 ORFs, proteotype analysis was applied (72). In this method, amino acid alignments for all viruses under consideration are performed using MEGA with the ClustalW option to generate maximum likelihood trees for each putative protein. For visualization in the alignment, the amino acid residues at each position in the proteins of all 38 viruses are color-coded: each amino acid is arbitrarily assigned the same unique color for all occurrences across all the viruses, and then the consensus amino acid at each position in the alignment is changed to white. Gaps in the alignment are colored black. A 10% sequence divergence threshold from consensus is applied to confirm unique proteotypes (6). Those genes/proteins within a specific proteotype are assumed to have recombined with one another for that gene or region, or are all offspring of recombination with another as-yet-uncharacterized (parent) virus. Proteotype analysis was performed for all eight ORFs in the E3 region within HAdV-D. Only one proteotype was identified for each of five proteins: 12.5K, 19K, RIDα, RIDβ (data not shown), and 14.7K (Fig. 2A), consistent with very high sequence conservation for their respective genes. In contrast, CR1α, CR1β (6), and CR1γ each express multiple unique proteotypes (6, 14, and 15 proteotypes, respectively) (Fig. 2B and C), suggesting high levels of recombination in those genes. To better visualize the results of proteotyping, viruses were sorted using the CR1α proteotype as the reference and then numbered (and colored) according to proteotype (Fig. 3). Remarkably, 37 of 38 CR1α protein sequences were shared between at least two viruses, with only one unshared proteotype, that from HAdV-D13. Therefore, all but 1 of the 38 HAdV-D viruses analyzed appeared to be recombinant for the CR1α gene. Seventeen of 38 viruses appeared to be the product (or source) of recombination for the E3 region as a single unit (∼5,000 bp), and 10 viruses (5 pairs) showed evidence for a single recombination spanning the entire E3 region and adjacent fiber gene (∼6,200 bp) (Fig. 3). Based on prior analysis (6), the genomic region with the next highest degree of homologous recombination after CR1α is the penton base RGD loop, with 10 proteotypes among 38 viruses and only 3 unshared. In contrast, the hexon protein, which contains the serum neutralization determinant, was shown to have 28 proteotypes among 38 viruses.
Fig 2.
Proteotyping analysis for select open reading frames of the E3 transcription unit of HAdV-D: (A) E3 14.7K; (B) CR1α; (C) CR1γ. Proteotyping analysis of CR1β was previously published (6). Maximum likelihood phylogenetic trees are shown to the left for each putative protein, and amino acid signatures are on the right. The scale bar at the bottom left of each panel denotes the phylogenetic distance reflected in the horizontal dimension of the corresponding tree. To construct the amino acid signatures shown, each amino acid was assigned a unique color (see the legend in the upper right corner), consensus amino acids at each position across all 38 viruses were assigned white, and gaps in the alignment are shown in black.
Fig 3.

Assortment analysis of putative proteotypes for hexon, E3 CR1, and fiber open reading frames, organized according to the putative CR1α protein. Each unique proteotype was assigned a number and color for accounting and visual representation, respectively. The seven vertical red bars on the left side of the figure denote those virus groups that demonstrate homologous recombination across the entire E3 transcription unit. The five blue bars on the right side of the figure identify those virus groups with evidence for recombination inclusive of all E3 and adjacent fiber genes. Inclusion of a protein in a given proteotype required an amino acid content that was <10% different from other members of the proteotype. The number recombined reflects the total number of viruses within any proteotype with at least two members.
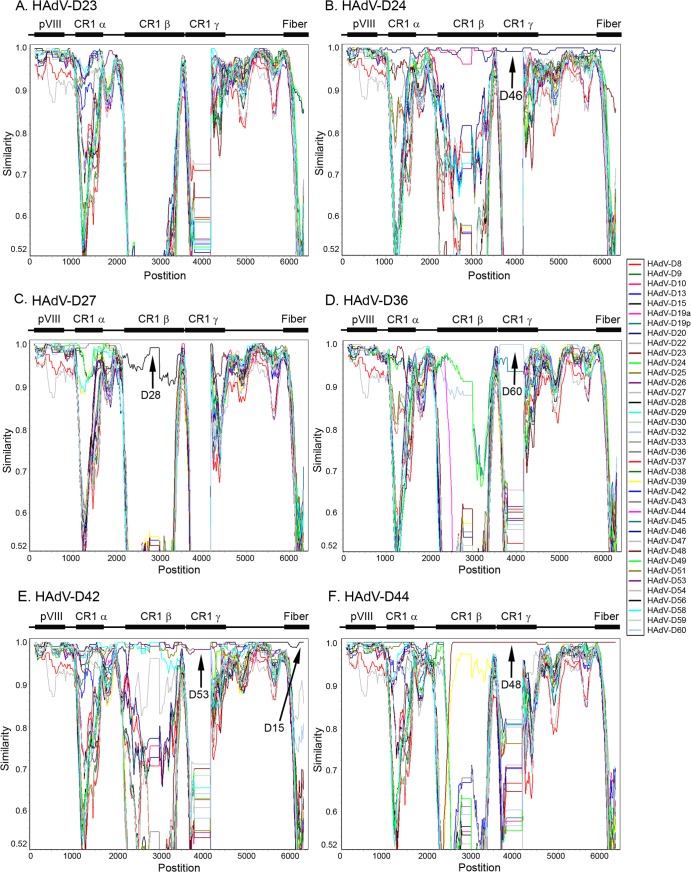
When considered together, phylogenetic and proteotype analyses provide strong evidence for E3 recombination among viruses within HAdV-D. To further explore this, genome sequence recombination analysis using SimPlot (http://sray.med.som.jhmi.edu/SCRoftware/simplot/) (73) was performed for the entire E3 transcription unit along with 800 additional nucleotides on either end of E3, which included the pVIII and (partial) fiber genes on the 5′ and 3′ ends of the E3 region, respectively (representative examples are shown in Fig. 4). This software compares multiple reference genomic sequences to a query sequence to allow visualization of possible recombined regions on the basis of high nucleotide similarity between two or more sequences. The SimPlot for HAdV-D23 (Fig. 4A) demonstrated relatively high similarity with multiple other viruses for CR1α but a unique sequence for CR1β and -γ. These data are consistent with proteotype analysis that showed the CR1α amino acid sequence of HAdV-D23 was shared among 12 viruses (Fig. 3). SimPlot analysis for HAdV-D24 demonstrated high similarity with type 46 across all three CR1 ORFs and the adjacent fiber gene (Fig. 4B), consistent with phylogenetic and proteotype analyses (Fig. 1 and 3). HAdV-D27 (Fig. 4C) appeared to be recombinant in CR1α (multiple) and possibly CR1β (with type 28), but not in CR1γ. HAdV-D36 (Fig. 4D), which has been associated with obesity (74), showed evidence for recombination in CR1α (with multiple viruses) and CR1γ (with type 60 and possibly type 19p), but not CR1β. HAdV-D42 (Fig. 4E) demonstrated recombination with type 15 across the entire E3 transcription unit and the adjacent fiber gene and also recombination with type 53 for the entire E3 region but not the fiber gene. Notably, types 42, 15, and 53 all appeared closely related by phylogenetic analysis (Fig. 1). SimPlot analysis results for HAdV-D44 (Fig. 4F) also were consistent with the phylogenetic and proteotype analyses, demonstrating recombination with type 48 for CR1β, CR1γ, and the fiber gene.
Fig 4.
SimPlot analysis of the E3 transcription unit, including 800 nucleotides on either side, for 38 viruses within HAdV-D. (A) HAdV-D23; (B) HAdV-D24; (C) HAdV-D27; (D) HAdV-D36; (E) HAdV-D42; (F) HAdV-D44. A window size of 200 bp and step size of 20 bp were used for the analysis. The color code for each virus in the analysis is shown on the right margin. Arrows point to examples of homologous recombination.
These analyses of the E3 transcription unit from 38 fully sequenced and annotated viruses within HAdV-D represent powerful evidence that evolution of the CR1 ORFs is driven by homologous recombination, and the results provide new insights into HAdV-D evolution. The HAdV-D E3 CR1 genes are among the most highly recombinant in the entire HAdV-D genome. Given that the E3 transcription unit of HAdV-C is not essential to viral replication in vitro (29), yet individual components can facilitate immune evasion by infected cells, these data suggest that evolution of E3 CR1 genes in HAdV-D may be driven by host factors that encourage the evolution of unique proteotypes for these genes. Considering that E3 genes do not encode proteins known to elicit neutralizing antibodies, the selection pressure imposed on this region likely occurs at the cellular level. Studies of E3 CR1 functions have focused almost entirely on HAdV-C, in which CR1α contributes to inhibition of TNF-α-induced apoptosis (57) and CR1β appears to assist in cell lysis at the final stages of infection (68). Not as much is known about the function of CR1 gene products from HAdV-D. It is noteworthy that the CR1β gene of HAdV-D codes for a much larger protein (49K) than in HAdV-C (11.6K) (25, 44). Also, CR1γ, which is not present in HAdV-C, has no known function (75). A stereotypical flux in GC nucleotide content was recently proposed as a potential marker for homologous recombination sites in HAdV-D (6), including the E3 region, but the exact mechanism for homologous recombination in adenoviruses has yet to be determined.
Finally, recognition of promiscuous recombination among CR1 genes in HAdV-D E3 should be of interest to those investigators using HAdV-D for development of viral gene vectors (76–79), particularly when the transgene is placed within the E3 cassette (77, 80–84). HAdV-D-based gene vectors with a transgene within the E3 region might be susceptible to recombination into wild-type HAdV-D during a coincidental coinfection, resulting in an infectious, replication-competent virus with potential expression of the transgene.
ACKNOWLEDGMENTS
This work was supported by NIH grants EY013124 and P30EY014104, a Senior Scientific Investigator Award (to J.C.) from Research to Prevent Blindness, Inc., New York, NY, and the Massachusetts Lions Eye Research Fund.
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Harding SP, Mutton KJ, van der Avoort H, Wermenbol AG. 1988. An epidemic of keratoconjunctivitis due to adenovirus type 37. Eye 2:314–317 [DOI] [PubMed] [Google Scholar]
- 2.Wood DJ. 1988. Adenovirus gastroenteritis. Br. Med. J. Clin. Res. 296:229–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingle JH, Langmuir AD. 1968. Epidemiology of acute, respiratory disease in military recruits. Am. Rev. Respir. Dis. 97(Suppl):1–65 [DOI] [PubMed] [Google Scholar]
- 4.De Jong JC, Wermenbol AG, Verweij-Uijterwaal MW, Slaterus KW, Wertheim-Van Dillen P, Van Doornum GJ, Khoo SH, Hierholzer JC. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jager L, Ehrhardt A. 2007. Emerging adenoviral vectors for stable correction of genetic disorders. Curr. Gene Ther. 7:272–283 [DOI] [PubMed] [Google Scholar]
- 6.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci. Rep. 3:1812. 10.1038/srep01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a reemergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J, Seto D. 2013. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh MP, Seto J, Liu EB, Dehghan S, Hudson NR, Lukashev AN, Ivanova O, Chodosh J, Dyer DW, Jones MS, Seto D. 2011. Computational analysis of two species C human adenoviruses provides evidence of a novel virus. J. Clin. Microbiol. 49:3482–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajon AE, Dickson LM, Murtagh P, Viale D, Carballal G, Echavarria M. 2010. Molecular characterization of an adenovirus 3-16 intertypic recombinant isolated in Argentina from an infant hospitalized with acute respiratory infection. J. Clin. Microbiol. 48:1494–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajon AE, Wadell G. 1996. Sequence analysis of the E3 region and fiber gene of human adenovirus genome type 7h. Virology 215:190–196 [DOI] [PubMed] [Google Scholar]
- 12.Bruzzone M, Barro M, Spencer E. 2001. Identification of adenovirus 7h heterogeneity in the E3 region. Biol. Res. 34:75–82 [DOI] [PubMed] [Google Scholar]
- 13.Lukashev AN, Ivanova OE, Eremeeva TP, Iggo RD. 2008. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 89:380–388 [DOI] [PubMed] [Google Scholar]
- 14.Matsushima Y, Shimizu H, Phan TG, Ushijima H. 2011. Genomic characterization of a novel human adenovirus type 31 recombinant in the hexon gene. J. Gen. Virol. 92:2770–2775 [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Sleigh M, Engler JA, Broker TR. 1980. The evolution of the adenoviral genome. Ann. N. Y. Acad. Sci. 354:426–452 [DOI] [PubMed] [Google Scholar]
- 16.Madisch I, Harste G, Pommer H, Heim A. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 79:15265–15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu EB, Ferreyra L, Fischer SL, Pavan JV, Nates SV, Hudson NR, Tirado D, Dyer DW, Chodosh J, Seto D, Jones MS. 2011. Genetic analysis of a novel human adenovirus with a serologically unique hexon and a recombinant fiber gene. PLoS One 6(9):e24491. 10.1371/journal.pone.0024491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu EB, Wadford DA, Seto J, Vu M, Hudson NR, Thrasher L, Torres S, Dyer DW, Chodosh J, Seto D, Jones MS. 2012. Computational and serologic analysis of novel and known viruses in species human adenovirus D in which serology and genomics do not correlate. PLoS One 7(3):e33212. 10.1371/journal.pone.0033212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J. Virol. 83:8980–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson CM, Zhou X, Rajaiya J, Yousuf MA, Singh G, Deserres JJ, Walsh MP, Wong S, Seto D, Dyer DW, Chodosh J, Jones MS. 2013. Predicting the next eye pathogen: analysis of a novel adenovirus. mBio 4(2):e00595–12. 10.1128/mBio.00595-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4(6):e5635. 10.1371/journal.pone.0005635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Robinson CM, Rajaiya J, Dehghan S, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest. Ophthalmol. Vis. Sci. 53:2804–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh G, Robinson CM, Dehghan S, Schmidt T, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Overreliance on the hexon gene leading to misclassification of human adenoviruses. J. Virol. 86:4693–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blusch JH, Deryckere F, Windheim M, Ruzsics Z, Arnberg N, Adrian T, Burgert HG. 2002. The novel early region 3 protein E3/49K is specifically expressed by adenoviruses of subgenus D: implications for epidemic keratoconjunctivitis and adenovirus evolution. Virology 296:94–106 [DOI] [PubMed] [Google Scholar]
- 26.Bhat BM, Wold WS. 1986. Genetic analysis of mRNA synthesis in adenovirus region E3 at different stages of productive infection by RNA-processing mutants. J. Virol. 60:54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow LT, Broker TR. 1978. The spliced structures of adenovirus 2 fiber message and the other late mRNAs. Cell 15:497–510 [DOI] [PubMed] [Google Scholar]
- 28.Chow LT, Gelinas RE, Broker TR, Roberts RJ. 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12:1–8 [DOI] [PubMed] [Google Scholar]
- 29.Morin JE, Lubeck MD, Barton JE, Conley AJ, Davis AR, Hung PP. 1987. Recombinant adenovirus induces antibody response to hepatitis B virus surface antigen in hamsters. Proc. Natl. Acad. Sci. U. S. A. 84:4626–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horwitz MS. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 6(Suppl 1):S172–S183 [DOI] [PubMed] [Google Scholar]
- 31.Windheim M, Hilgendorf A, Burgert HG. 2004. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 273:29–85 [DOI] [PubMed] [Google Scholar]
- 32.Lichtenstein DL, Toth K, Doronin K, Tollefson AE, Wold WS. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23:75–111 [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Hallden G, Hill R, Anand A, Liu TC, Francis J, Brooks G, Lemoine N, Kirn D. 2003. E3 gene manipulations affect oncolytic adenovirus activity in immunocompetent tumor models. Nat. Biotechnol. 21:1328–1335 [DOI] [PubMed] [Google Scholar]
- 34.Toth K, Doronin K, Kuppuswamy M, Ward P, Tollefson AE, Wold WS. 2005. Adenovirus immunoregulatory E3 proteins prolong transplants of human cells in immunocompetent mice. Virus Res. 108:149–159 [DOI] [PubMed] [Google Scholar]
- 35.Robinson CM, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Molecular evolution of human species D adenoviruses. Infect. Genet. Evol. 11:1208–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson CM, Shariati F, Gillaspy AF, Dyer DW, Chodosh J. 2008. Genomic and bioinformatics analysis of human adenovirus type 37: new insights into corneal tropism. BMC Genomics 9:213. 10.1186/1471-2164-9-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herisse J, Courtois G, Galibert F. 1980. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 8:2173–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herisse J, Galibert F. 1981. Nucleotide sequence of the EcoRI E fragment of adenovirus 2 genome. Nucleic Acids Res. 9:1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cladaras C, Wold WS. 1985. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology 140:28–43 [DOI] [PubMed] [Google Scholar]
- 40.Tollefson AE, Stewart AR, Yei SP, Saha SK, Wold WS. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to downregulate the epidermal growth factor receptor. J. Virol. 65:3095–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tollefson AE, Krajcsi P, Yei SP, Carlin CR, Wold WS. 1990. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J. Virol. 64:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tollefson AE, Krajcsi P, Pursley MH, Gooding LR, Wold WS. 1990. A 14,500 MW protein is coded by region E3 of group C human adenoviruses. Virology 175:19–29 [DOI] [PubMed] [Google Scholar]
- 43.Deryckere F, Burgert HG. 1996. Early region 3 of adenovirus type 19 (subgroup D) encodes an HLA-binding protein distinct from that of subgroups B and C. J. Virol. 70:2832–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Windheim M, Burgert HG. 2002. Characterization of E3/49K, a novel, highly glycosylated E3 protein of the epidemic keratoconjunctivitis-causing adenovirus type 19a. J. Virol. 76:755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J, Li L, Bouvier M. 2011. Adenovirus E3-19K proteins of different serotypes and subgroups have similar, yet distinct, immunomodulatory functions toward major histocompatibility class I molecules. J. Biol. Chem. 286:17631–17639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson-Rawls J, Deutscher SL, Wold WS. 1994. The signal-anchor domain of adenovirus E3-6.7K, a type III integral membrane protein, can direct adenovirus E3-gp19K, a type I integral membrane protein, into the membrane of the endoplasmic reticulum. Virology 201:66–76 [DOI] [PubMed] [Google Scholar]
- 47.Jefferies WA, Burgert HG. 1990. E3/19K from adenovirus 2 is an immunosubversive protein that binds to a structural motif regulating the intracellular transport of major histocompatibility complex class I proteins. J. Exp. Med. 172:1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgert HG, Maryanski JL, Kvist S. 1987. “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proc. Natl. Acad. Sci. U. S. A. 84:1356–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgert HG, Kvist S. 1987. The E3/19K protein of adenovirus type 2 binds to the domains of histocompatibility antigens required for CTL recognition. EMBO J. 6:2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson M, McMichael A, Peterson PA. 1987. Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 138:3960–3966 [PubMed] [Google Scholar]
- 51.Burgert HG, Kvist S. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41:987–997 [DOI] [PubMed] [Google Scholar]
- 52.Cox JH, Bennink JR, Yewdell JW. 1991. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J. Exp. Med. 174:1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McSharry BP, Burgert HG, Owen DP, Stanton RJ, Prod'homme V, Sester M, Koebernick K, Groh V, Spies T, Cox S, Little AM, Wang EC, Tomasec P, Wilkinson GW. 2008. Adenovirus E3/19K promotes evasion of NK cell recognition by intracellular sequestration of the NKG2D ligands major histocompatibility complex class I chain-related proteins A and B. J. Virol. 82:4585–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gooding LR, Elmore LW, Tollefson AE, Brady HA, Wold WS. 1988. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell 53:341–346 [DOI] [PubMed] [Google Scholar]
- 55.Rawle FC, Tollefson AE, Wold WS, Gooding LR. 1989. Mouse anti-adenovirus cytotoxic T lymphocytes. Inhibition of lysis by E3 gp19K but not E3 14.7K. J. Immunol. 143:2031–2037 [PubMed] [Google Scholar]
- 56.Horton TM, Ranheim TS, Aquino L, Kusher DI, Saha SK, Ware CF, Wold WS, Gooding LR. 1991. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J. Virol. 65:2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benedict CA, Norris PS, Prigozy TI, Bodmer JL, Mahr JA, Garnett CT, Martinon F, Tschopp J, Gooding LR, Ware CF. 2001. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 276:3270–3278 [DOI] [PubMed] [Google Scholar]
- 58.Elsing A, Burgert HG. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. U. S. A. 95:10072–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tollefson AE, Hermiston TW, Lichtenstein DL, Colle CF, Tripp RA, Dimitrov T, Toth K, Wells CE, Doherty PC, Wold WS. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726–730 [DOI] [PubMed] [Google Scholar]
- 60.Lichtenstein DL, Doronin K, Toth K, Kuppuswamy M, Wold WS, Tollefson AE. 2004. Adenovirus E3-6.7K protein is required in conjunction with the E3-RID protein complex for the internalization and degradation of TRAIL receptor 2. J. Virol. 78:12297–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shisler J, Yang C, Walter B, Ware CF, Gooding LR. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J. Virol. 71:8299–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedman JM, Horwitz MS. 2002. Inhibition of tumor necrosis factor alpha-induced NF-kappa B activation by the adenovirus E3-10.4/14.5K complex. J. Virol. 76:5515–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNees AL, Garnett CT, Gooding LR. 2002. The adenovirus E3 RID complex protects some cultured human T and B lymphocytes from Fas-induced apoptosis. J. Virol. 76:9716–9723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gooding LR, Sofola IO, Tollefson AE, Duerksen-Hughes P, Wold WS. 1990. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J. Immunol. 145:3080–3086 [PubMed] [Google Scholar]
- 65.Carlin CR, Tollefson AE, Brady HA, Hoffman BL, Wold WS. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 57:135–144 [DOI] [PubMed] [Google Scholar]
- 66.Wold WS, Cladaras C, Magie SC, Yacoub N. 1984. Mapping a new gene that encodes an 11,600-molecular-weight protein in the E3 transcription unit of adenovirus 2. J. Virol. 52:307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tollefson AE, Scaria A, Saha SK, Wold WS. 1992. The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J. Virol. 66:3633–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tollefson AE, Scaria A, Hermiston TW, Ryerse JS, Wold LJ, Wold WS. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doronin K, Toth K, Kuppuswamy M, Krajcsi P, Tollefson AE, Wold WS. 2003. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology 305:378–387 [DOI] [PubMed] [Google Scholar]
- 70.Routes JM, Cook JL. 1990. Resistance of human cells to the adenovirus E3 effect on class I MHC antigen expression. Implications for antiviral immunity. J. Immunol. 144:2763–2770 [PubMed] [Google Scholar]
- 71.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 72.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580 [DOI] [PubMed] [Google Scholar]
- 73.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atkinson RL, Dhurandhar NV, Allison DB, Bowen RL, Israel BA, Albu JB, Augustus AS. 2005. Human adenovirus-36 is associated with increased body weight and paradoxical reduction of serum lipids. Int. J. Obes. (Lond.) 29:281–286 [DOI] [PubMed] [Google Scholar]
- 75.Robinson CM, Rajaiya J, Zhou X, Singh G, Dyer DW, Chodosh J. 2011. The E3 CR1-gamma gene in human adenoviruses associated with epidemic keratoconjunctivitis. Virus Res. 160:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thirion C, Lochmuller H, Ruzsics Z, Boelhauve M, Konig C, Thedieck C, Kutik S, Geiger C, Kochanek S, Volpers C, Burgert HG. 2006. Adenovirus vectors based on human adenovirus type 19a have high potential for human muscle-directed gene therapy. Hum. Gene Ther. 17:193–205 [DOI] [PubMed] [Google Scholar]
- 77.Ruzsics Z, Wagner M, Osterlehner A, Cook J, Koszinowski U, Burgert HG. 2006. Transposon-assisted cloning and traceless mutagenesis of adenoviruses: development of a novel vector based on species D. J. Virol. 80:8100–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lemckert AA, Grimbergen J, Smits S, Hartkoorn E, Holterman L, Berkhout B, Barouch DH, Vogels R, Quax P, Goudsmit J, Havenga MJ. 2006. Generation of a novel replication-incompetent adenoviral vector derived from human adenovirus type 49: manufacture on PER.C6 cells, tropism and immunogenicity. J. Gen. Virol. 87:2891–2899 [DOI] [PubMed] [Google Scholar]
- 79.Teigler JE, Iampietro MJ, Barouch DH. 2012. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J. Virol. 86:9590–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hawkins LK, Hermiston T. 2001. Gene delivery from the E3 region of replicating human adenovirus: evaluation of the E3B region. Gene Ther. 8:1142–1148 [DOI] [PubMed] [Google Scholar]
- 81.Hawkins LK, Hermiston TW. 2001. Gene delivery from the E3 region of replicating human adenovirus: evaluation of the ADP region. Gene Ther. 8:1132–1141 [DOI] [PubMed] [Google Scholar]
- 82.Hawkins LK, Johnson L, Bauzon M, Nye JA, Castro D, Kitzes GA, Young MD, Holt JK, Trown P, Hermiston TW. 2001. Gene delivery from the E3 region of replicating human adenovirus: evaluation of the 6.7K/gp19K region. Gene Ther. 8:1123–1131 [DOI] [PubMed] [Google Scholar]
- 83.Pham L, Nakamura T, Gabriela Rosales A, Carlson SK, Bailey KR, Peng KW, Russell SJ. 2009. Concordant activity of transgene expression cassettes inserted into E1, E3 and E4 cloning sites in the adenovirus genome. J. Gene Med. 11:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ilan Y, Droguett G, Chowdhury NR, Li Y, Sengupta K, Thummala NR, Davidson A, Chowdhury JR, Horwitz MS. 1997. Insertion of the adenoviral E3 region into a recombinant viral vector prevents antiviral humoral and cellular immune responses and permits long-term gene expression. Proc. Natl. Acad. Sci. U. S. A. 94:2587–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]