Abstract
A nationwide hepatitis B virus (HBV) vaccination program was implemented in China starting in 1992. To study the change in HBV variant prevalence with massive immunization, large HBV surface protein (LHBs) genes from HBV surface antigen (HBsAg)-positive sera were amplified and sequenced. The prevalences of LHBs mutants were compared between the 1992 and 2005 surveys in child and adult groups. The prevalence of “α” determinant mutants in the children increased from 6.5% in 1992 to 14.8% in 2005, where the G145R mutant occurred most frequently. In contrast, mutation frequencies showed little difference between 1992 (9.4%) and 2005 (9.9%) in adults. Moreover, compared to the 1992 survey, the child group surface (S) protein mutation frequency specifically increased (P = 0.005) in the 2005 survey, but the pre-S region mutation frequency did not show a significant difference (P > 0.05). However, the mutation frequency in the adult group increased in both the pre-S and S regions. Furthermore, the frequencies of the disease-related pre-S2 deletion and start codon mutations were significantly higher in the adult groups than in the child groups in both the 1992 and 2005 surveys (P < 0.01). Massive immunization enhances the HBV S protein mutation; the prevalence of LHBs mutants, particularly disease-related mutants, tends to increase with patient age.
INTRODUCTION
Hepatitis B virus (HBV) infection is a serious public health problem worldwide and a major cause of hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) (1). An estimated 350 million people are living with chronic hepatitis B worldwide (2). In China, a national serosurvey in 1992 revealed that the prevalence of hepatitis B surface antigen (HBsAg) was 9.8% in the general population (3). The most recent national serosurvey, in 2006, demonstrated that the HBsAg carrier rate is 7.2%, and the number of chronic HBV carriers is estimated to be 9.3 million (3, 4).
China began infant vaccine injections in national disease surveillance points (DSP) as early as 1986, and the four provinces in the current study were also to perform infant vaccinations at that time (5). In 1992, China formally recommended routine immunization with the hepatitis B virus vaccine for infants. Since then, the massive vaccination program has effectively decreased the risk of HBV infection, and the rate of HBsAg-positive children has significantly declined. Among children <5 years of age, the prevalence of HBsAg has dramatically decreased from 9.67% (1992) to 0.96% (2006). It is estimated that 80 million children are free from HBV infection due to the vaccination program (3).
While universal infantile HBV vaccination is very efficacious, an increase in the mutant prevalence or “vaccine escape mutation” after immunization posed a potential threat to the long-term success of massive vaccination (6–8). Hsu et al. studied the prevalence of HBV variants in children in Taiwan and demonstrated that universal vaccination had accelerated the accumulation of HBsAg “α” determinant mutants (9). The α determinant is a common immunodominant region shared by different genotypes of HBV, which spans amino acids (aa) 124 to 147 of the S protein (10). The α determinant is hydrophilic and is believed to be in the form of two major loops with cysteine-disulfide bonds (11). It is well documented that neutralizing antibodies induced by immunization against hepatitis B virus infection are targeted to the conformational epitopes of the α determinant (12). The substitution mutations within the surface protein, particularly in the α domain, allow HBV replication in vaccinated subjects, since antibodies induced by vaccines do not recognize crucial changes in the surface antigen domain (8).
Differing from the mutations in the S region that are related to vaccine escape, pre-S mutations were studied for their correlation with the progression of liver disease (13). Particularly, pre-S2 deletion and pre-S2 start codon mutations were frequently detected in cases of liver cirrhosis and HCC (14). The deletion always clustered at the site of immunologic epitopes (13). Furthermore, pre-S2 start codon mutations can impair secretion of virus. The accumulation of viral protein induces host liver cell endoplasmic reticulum stress. Such an effect is a possible mechanism for the development of HCC, as proven in studies with transgenic mice (15).
Here, large HBV surface protein (LHBs) mutations were studied to evaluate the impact of universal vaccination on HBV mutation in China. The changes in LHBs mutant prevalences between children and adults were compared as well. Some meaningful observations provide valuable information about virus mutation and broaden our understanding of HBV mutation occurrence.
MATERIALS AND METHODS
Patients and methods.
Two seroepidemiological studies were conducted with multistage cluster random sampling in the same four national DSPs during the years 1992 and 2005: Long An in Guangxi province (GX), Zheng Ding in Hebei province (HB), Xiang Tan in Hunan province (UN), and Luo He in Henan province (HN). The DSP was selected by the Chinese Center for Disease Control and Prevention to be representative of the population. Each DSP consists of one county. In the 1992 survey, persons 1 to 59 years old residing for more than 6 months in the county at the time of the survey were selected. In the 2005 survey, a house-to-house investigation was completed in the DSP by trained staff. The age of participants ranged from 3 months to 94 years. People who had resided in the DSP for 16 months at time of the survey visit were selected. The complete selection methods were described previously (16, 17). The populations recruited in the seroepidemiological surveys were divided into the following four groups: (i) children younger than 18 years of age in the 1992 survey (born during 1974 to 1992), (ii) children younger than 18 years of age in the 2005 survey (born during 1986 to 2005), (iii) adults aged between 35 and 55 years in the 1992 survey (born during 1937 to 1957), and (iv) adults aged between 35 and 55 years in the 2005 survey (born during 1950 to 1970). Sera from UN was lacking for both the 1992 and 2005 adult groups. The demographic characteristics of the subjects in the study are summarized in Table 1. The adult demographic characteristics and economic conditions at these points are not statistically different from those of the whole country. GX and UN are southern provinces, and HB and HE are northern provinces. The study population is “Han” in Hunan, Hebei, and Henan provinces, while the study population in Guangxi province is mostly “Zhuang.”
Table 1.
Demographic characteristics of the study population
| Characteristic | Value(s) for children |
Value(s) for adults |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GX | UN | HB | HN | Total | GX | HB | HN | Total | |
| 1992 survey | |||||||||
| No. (%) of male patients | 143 | 98 | 184 | 458 | 883 (48.4) | 109 | 192 | 118 | 419 (49.1) |
| No. (%) of female patients | 136 | 100 | 191 | 515 | 942 (51.6) | 108 | 200 | 128 | 433 (50.9) |
| Mean age (yr) ± SD | 9.4 | 7.6 | 9.8 | 7.2 | 8.1 ± 2.3 | 40.1 | 42.3 | 41.9 | 41.7 ± 3.7 |
| 2005 survey | |||||||||
| No. (%) of male patients | 331 | 409 | 599 | 913 | 2,252 (49.0) | 138 | 211 | 427 | 776 (48.7) |
| No. (%) of female patients | 343 | 421 | 614 | 964 | 2,344 (51.0) | 147 | 202 | 464 | 813 (51.3) |
| Mean age (yr) ± SD | 14.3 | 13.7 | 10.9 | 11.6 | 12.2 ± 2.6 | 43.1 | 41.1 | 42.3 | 41.9 ± 3.4 |
Serology analysis.
All serum specimens were tested in the National Hepatitis Laboratory, Institute for Viral Disease Control and Prevention (IVDC), Chinese Center for Disease Control and Prevention (CDC). The presence of HBsAg was determined by using a solid-phase radioimmunoassay (SPRIA). A ratio of the HBsAg level in the sample to that in the negative control (S/N ratio) of >10 was defined as positive. After assaying for serum HBV markers in each participant, the sera were stored at −30°C for later analysis.
Viral DNA extraction and sequencing.
HBsAg-positive sera were selected for extraction of viral DNA by using a viral DNA minikit (Qiagen, German). For sequence analysis, the LHBs genes were amplified by using nested PCR. First-round PCR primers were 5′-GGGTCACCATATTCTTGGG-3′ (nucleotides [nt] 2850 to 2868) and 5′-CAAAGACAAAAGAAAATTGG-3′ (nt 803 to 822). PCR was performed under the following conditions: 95°C for 5 min, 95°C for 35 s, 58°C for 35 s, and 72°C for 2 min for 30 cycles and finally 72°C for 7 min. Second-round PCR primers were 5′-GAACAAGAGCTACAGCATGGG-3′ (nt 2868 to 2888) and 5′-GGTAAAAAGGGACTCAAGATG-3′ (nt 775 to 795), and the same PCR conditions as those for the first-round reaction were used. All necessary precautions to prevent cross-contamination were taken, and negative controls were included in each assay. The nucleotide sequences of the amplified products were determined directly by using fluorescence-labeled primers (TaKaRa Bio Inc., Japan).
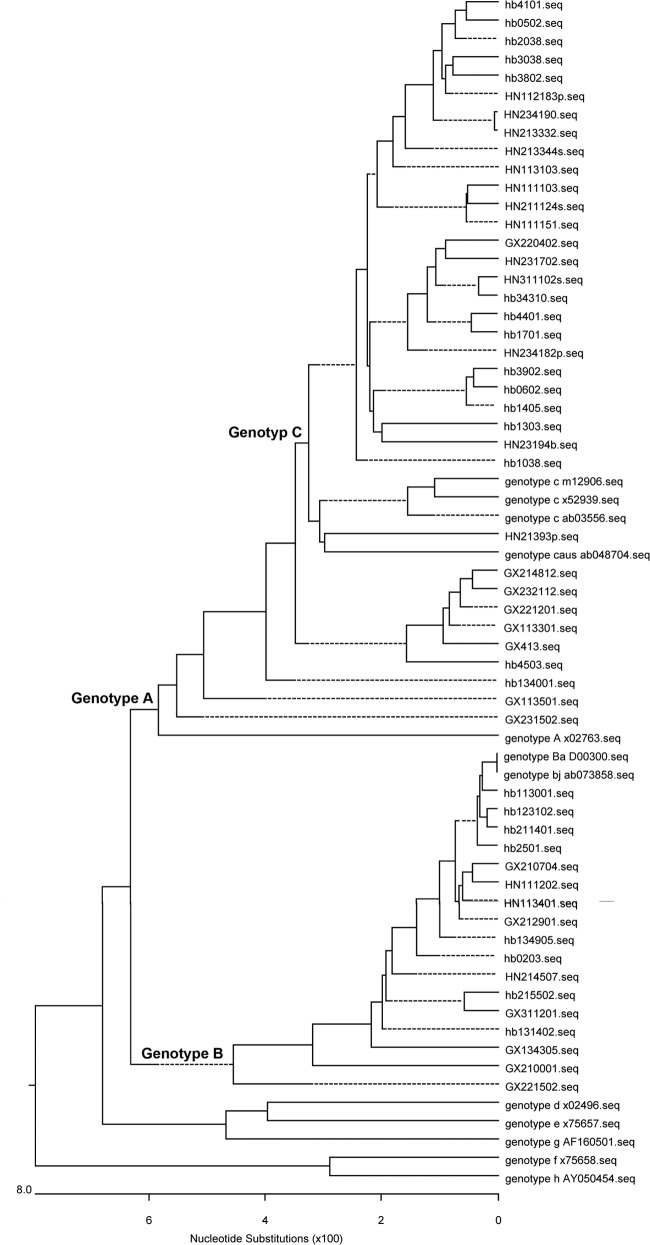
HBV genotyping and mutation identification.
HBV genotypes were determined by phylogenetic analysis using the neighbor-joining method (MEGA, v3.1), including 8 HBV strains of different genotypes obtained from GenBank with the following accession numbers: X02763 (genotype A), D00300 (genotype Ba), AB073858 (genotype Bj), AB03556, M12906, X52939 (genotype C), AB048704 (genotype Caus), X02496 (genotype D), X02496 (genotype D), X75657 (genotype E), AF160501 (genotype G), X75858 (genotype F), and AY050454 (genotype H). The reliability of the phylogenetic tree was tested by using the bootstrap test with 1,000 replicates (Fig. 1 to 4). The envelope amino acid sequence was determined by translation of the nucleotide sequence according to the LHBs open reading frame (ORF). The genotype B or C sequences were aligned with standard LHBs sequences. The mutations in target sequences were identified and recorded, which did not align with any of the standard sequences.
Fig 1.

Phylogenetic analysis of children from the 1992 survey. The first two letters represent the name of the province.
Fig 4.

Phylogenetic analysis of adults from the 2005 survey. The first two letters represent the name of the province.
Fig 2.

Phylogenetic analysis of children from the 2005 survey. The first two letters represent the name of the province.
Fig 3.
Phylogenetic analysis of adults from the 1992 survey. The first two letters represent the name of the province.
Data analysis.
Data are presented as means ± standard deviations (SD), proportions, or rates (95% confidence intervals [CIs]). To compare the values between the groups, the χ2 or Fisher exact test was applied to analyze categorical variables, and Student t tests and Mann-Whitney U tests were used for continuous variables with normal and skewed distributions, respectively. All statistical tests were 2 sided. A P value of <0.05 was considered statistically significant. All statistical analyses were performed by using SPSS 13.0 for Windows (SPSS Inc., IL).
RESULTS
Characteristics of the study population.
During the 1992 and 2005 investigations, a total of 8,862 people were enumerated, who came from the same national DSP. There were 6,421 children younger than 18 years old and 2,421 adults between the ages of 35 and 55 years (Table 1). After the SPRIA assay, HBsAg-positive sera were collected to extract HBV DNA and perform PCR. More than 1,100 base pair LHBs gene sequences were amplified. After the PCR production sequencing, the final numbers and percentages of acquired sequences were determined and are listed in Table 2.
Table 2.
Prevalence of HBV α determinant variants from HBsAg-postive personsa
| Characteristic | No. of children from indicated province |
Total no. (%) of children | No. of adults from indicated province |
Total no. (%) of adults |
|||||
|---|---|---|---|---|---|---|---|---|---|
| GX | UN | HB | HN | GX | HB | HN | |||
| 1992 survey | |||||||||
| Collected sera | 279 | 198 | 375 | 973 | 1,825 | 214 | 392 | 246 | 852 |
| HBsAg+ sera | 29 | 17 | 31 | 80 | 157 (8.6) | 22 | 33 | 23 | 78 (9.2) |
| Acquired sequences | 24 | 16 | 26 | 72 | 138 (87.9) | 14 | 23 | 16 | 53 (69.2) |
| Variants | 2 | 3 | 2 | 2 | 9 (6.5b) | 1 | 2 | 2 | 5 (9.4) |
| 2005 survey | |||||||||
| Collected sera | 674 | 832 | 1,213 | 1,877 | 4,596 | 285 | 413 | 891 | 1,589 |
| HBsAg+ sera | 22 | 19 | 28 | 47 | 116 (2.5) | 25 | 26 | 61 | 112 (7.1) |
| Acquired sequences | 19 | 17 | 24 | 41 | 101 (88.8) | 19 | 20 | 52 | 91 (81.2) |
| Acquired sequences | 7 | 3 | 2 | 3 | 15 (14.8b) | 4 | 3 | 2 | 9 (9.9) |
Percentage calculated as the lower row number/the upper row number.
χ2 = 4.48; P = 0.03.
The “α” determinant mutants in HBsAg-positive children.
To investigate the α determinant mutant virus prevalence in 1992, 157 HBsAg-positive children from four different DSPs were studied for the presence of HBV DNA by PCR (Table 2). Eventually, 138 sera were seropositive for HBV DNA. Among the 138 acquired LHBs sequences, 9 HBV DNA-positive children in 1992 harbored the mutations. The percentage of occurrence of α determinant mutations was 6.5% (95% CI, 2.4 to 10.6%). In the 2005 survey, 116 HBsAg-positive children were studied for the presence of HBV DNA by PCR. Fifteen children harbored α determinant mutant virus among the 101 children who harbored a sequenced virus. One child harbored a variant HBV strain that had double mutations. Therefore, the α determinant mutant virus percentage for the 2005 survey was 14.8% (95% CI, 7.9 to 21.8%) (15 out of 101), which was significantly higher than that for the 1992 survey (χ2 = 4.48; P = 0.03). Detailed backgrounds of the children who harbored mutant virus are summarized in Table 3.
Table 3.
Characteristics of HBsAg α determinant variants among children from seroepidemiological surveys in 1992 and 2005a
| Patient | Variant | Age (yr) | Sex | Genotype | Serotype | HBeAg result |
|---|---|---|---|---|---|---|
| 1992 survey | ||||||
| GX-1 | T125M | 11 | M | C | adw | # |
| GX-2 | T126S | 10 | M | B | adw | # |
| UN-1 | T126A | 5 | M | B | adw | # |
| UN-2 | Q129H | 13 | M | B | adw | # |
| UN-3 | T143M | 2 | M | B | adw | # |
| HN-1 | G130R | 7 | F | D | ayw | # |
| HN-2 | T131N | 3 | M | B | adw | # |
| HB-1 | I126S | 10 | F | C | adr | # |
| HB-2 | G145A | 14 | F | C | adr | # |
| Total | 8.33 ± 4.3 | 6 M/3 F | 3 C/5 B/1 D | 6 adw/2 adr/1 ayw | ||
| 2005 survey | ||||||
| GX-1 | T125I | 1 | F | B | adw | + |
| GX-2 | T126A | 7 | M | C | adw | − |
| GX-3 | T126A | 16 | M | C | adw | − |
| GX-4 | T126A | 16 | M | C | adw | + |
| GX-5 | T126N | 7 | F | C | adr | − |
| GX-6 | G145A | 14 | M | C | adr | − |
| GX-7 | G145A | 13 | F | C | adr | + |
| UN-1 | T131I | 17 | M | B | adw | + |
| UN-2 | G145R | 11 | M | B | adw | + |
| UN-3 | G145R | 14 | F | B | adw | + |
| HN-1 | G145A | 8 | M | C | adr | + |
| HN-2 | I126S | 10 | F | C | adr | + |
| HN-3 | Q129H | 12 | M | B | adw | + |
| HB-1 | T126S/G130N | 16 | F | C | adr | + |
| HB-2 | G145R | 6 | F | B | adw | + |
| Total | 11.2 ± 4.6 | 8 M/7 F | 9 C/6 B | 9 adw/6 adr | 11 +/4 − |
Abbreviations: M, male; F, female; HBeAg, HBV e antigen; #, not performed; −, negative; +, positive.
The α determinant mutants in HBsAg-positive adults.
As a control, the α determinant mutants virus statuses in adult groups were investigated, because massive vaccination has never been performed for adults. HBsAg-positive adults from the 1992 and 2005 surveys were studied, and the harbored viruses were amplified and sequenced. In the 1992 survey, 53 LHBs genes were successfully sequenced from 78 HBV-positive sera. A total of 5 sequences with α determinant mutations were detected. Thus, the α determinant mutation occurrence rate was 9.4% (95% CI, 1.6 to 17.3%) in the 1992 survey. In the 2005 survey, 91 LHBs genes successfully sequenced from 112 HBV-positive sera. Subsequently, 9 sequences tested positive for a mutation in the α determinant, and the mutation occurrence rate was 9.9% (95% CI, 3.8 to 16.0%). Two variant HBV sequences had double and triple mutations. Detailed backgrounds of adults harboring mutant virus are summarized in Table 4. The percentages of α determinant mutant virus in the 1992 and 2005 surveys were not significantly different (χ2 = 0.02; P = 0.9).
Table 4.
Characteristics of HBsAg α determinant variants from adults in seroepidemiologic surveys in 1992 and 2005a
| Patient | Variant | Age (yr) | Sex | Genotype | Serotype | HBeAg result |
|---|---|---|---|---|---|---|
| 1992 survey | ||||||
| GX-1 | T126A | 40 | M | B | adw | # |
| HN-1 | T126A | 38 | M | B | adw | # |
| HN-2 | T126C | 38 | F | C | adr | # |
| HB-1 | T131A | 52 | M | C | adr | # |
| HB-2 | M133L | 39 | M | B | adw | # |
| Total | 41.4 | 4 M/1 F | 2 C/3 B | 3 adw/2 adr | ||
| 2005 survey | ||||||
| GX-1 | T125I | 34 | M | B | adw | − |
| GX-2 | M133I | 24 | F | C | adr | + |
| GX-3 | M133T/T140I | 35 | F | C | adw | − |
| GX-4 | G145K | 36 | M | C | adw | + |
| HN-1 | M133T | 54 | F | C | adr | + |
| HN-2 | I126S/S132F/M134I | 21 | M | C | adr | + |
| HB-1 | F134Y | 57 | M | C | ayw | − |
| HB-2 | M133T | 57 | M | C | adr | + |
| HB-3 | F134Y | 58 | F | C | adr | + |
| Total | 41.8 | 5 M/4 F | 8 C/1 B | 3 adw/5 adr/1 ayw | 6 +/3 − |
Abbreviations: M, male; F, female; HBeAg, HBV e antigen; #, not performed; −, negative; +, positive.
Frequencies and sites of α determinant mutations are different between children and adults.
In the 1992 child group, the most frequent sequence mutation site was T126 (3 out of 138), and only one strain had a G145A mutation. However, in the 2005 child group, the most frequent sequence mutation site was G145 (6 out of 101), and 5 mutations occurred at T126 (Fig. 5B). Even though there was no significant difference, the T126 mutation rates were higher in 2005 than in the 1992 survey (Fig. 5B). It was reported that the T126A mutant was found in an infant with chronic infection despite immunoprophylaxis, meaning that T126 seems to affect the recognition properties of the surface antigen (18). G145 mutations were of relatively low frequency in the 1992 child group and the two adult groups (Fig. 5). However, its frequency significantly increased in the 2005 child group (P = 0.04). As the most commonly found mutant, G145R has been reported to alter the projecting loop (residues 139 to 147) of the α determinant. Consequently, neutralizing antibody induced by vaccination no longer recognizes the mutated epitope, which is hence termed a vaccine escape mutant (11, 19).
Fig 5.
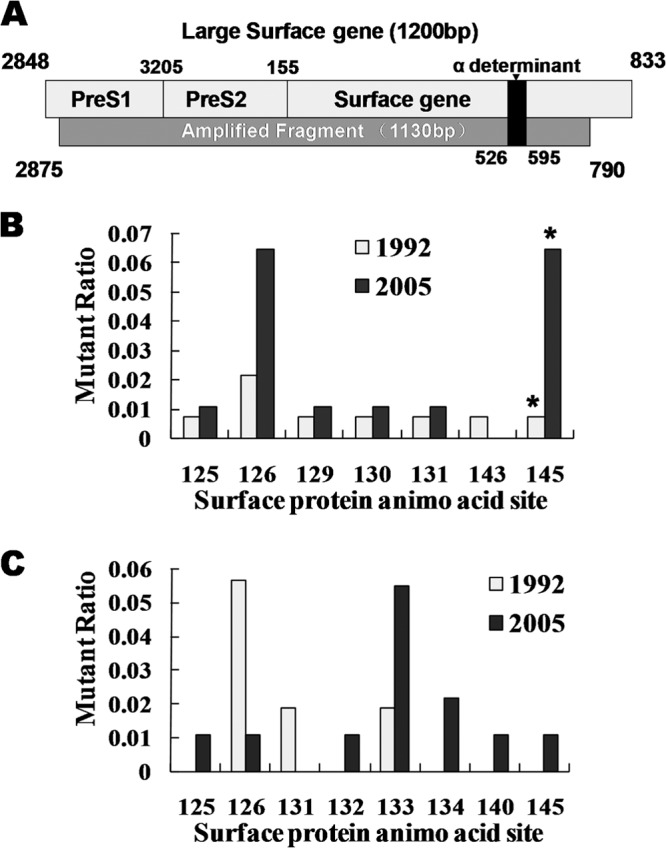
Mutations in the α determinant. (A) Schematic representation showing LHBs and amplified sequences. The start and end residues of the pre-S1, pre-S2, and S proteins and the α determinant of HBsAg are indicated. The bottom rectangle represents the amplified sequence, with the start and end positions labeled. (B) Mutations in the α determinant of child groups. (C) Mutations in the α determinant of adult groups. The y axis represents the mutation ratio (number of mutations/number of all indicated sequences). The x axis represents the amino acid site of the S protein where the indicated mutations occurred.
Importantly, the frequently mutated sites in the adult groups were different from those in the child groups. Residues M133 and F134 have high mutation frequencies in adults (Fig. 5C), and such mutations are implicated in the development of hepatocellular carcinoma (20, 21). Both of the mutations are located in the first loop of the α determinant (residues 124 to 138). Furthermore, the M133T mutation can change HBV secretion (22). Lastly, the adult group's mutation panel is similar to the one previously reported for nonimmunized Chinese carriers (23).
Change in the LHBs amino acid substitution frequency in children.
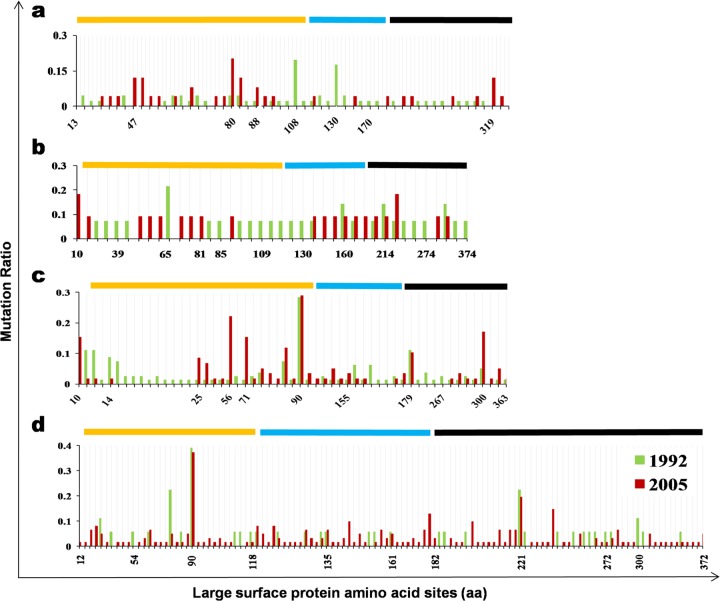
The substitution mutations in LHBs were identified according to standard strains of genotype B or C. Child group mutations were compared and analyzed between the 1992 and 2005 surveys. In the 1992 survey, there were 46 genotype B sequences, with a total number of 7,314 residues in the pre-S region. Among the 7,314 residues, 42 substitution mutations (0.58%) occurred. In 2005, there were 44 mutations out of 3,975 total residues (0.73%) in the same region. Likewise, in the 1992 survey, 46 genotype B sequences had 9,660 total residues in the S region, and 8 mutations (0.08%) were identified. In the 2005 survey, 8 mutations were found out of 5,250 residues (0.15%). For genotype C, 121 mutations were identified out of 12,879 residues (0.94%) and 100 mutations were identified out of 9,381 (1.07%) residues in the pre-S region in the 1992 and 2005 surveys, respectively. In the S region, the mutation frequencies were 0.16% (27/17,010) in the 1992 survey and 0.28% (35/12,390) in the 2005 survey (Table 5). The mutation sites and frequencies are summarized in Fig. 6.
Table 5.
Amino acid substitution frequencies in S and pre-S regionsa
| Parameter | Value(s) for genotype B |
Value(s) for genotype C |
Total value(s) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1992 | 2005 | χ2 | P | 1992 | 2005 | χ2 | P | 1992 | 2005 | χ2 | P | |
| Children | ||||||||||||
| Total no. of sequences | 46 | 25 | 81 | 59 | 127 | 84 | ||||||
| No. of pre-S residues | 7,314 | 3,975 | 12,879 | 9,381 | 20,193 | 13,356 | ||||||
| No. (%) of pre-S mutations | 42 (0.58) | 29 (0.73) | 0.99 | 0.32 | 121 (0.94) | 100 (1.07) | 0.88 | 0.35 | 163 (0.81) | 129 (0.97) | 2.35 | 0.13 |
| No. of S residues | 9,660 | 5,250 | 17,010 | 12,390 | 26,670 | 17,640 | ||||||
| No. (%) of S mutations | 8 (0.08) | 8 (0.15) | 1.54 | 0.22 | 27 (0.16) | 35 (0.28) | 5.22 | 0.02* | 35 (0.13) | 43 (0.24) | 7.65 | 0.005** |
| Adults | ||||||||||||
| Total no. of sequences | 14 | 11 | 18 | 62 | 32 | 73 | ||||||
| No. of pre-S residues | 2,226 | 1,749 | 2,862 | 9,858 | 5,088 | 11,607 | ||||||
| No. (%) of pre-S mutations | 20 (0.90) | 17 (0.97) | 0.06 | 0.81 | 30 (1.05) | 151 (1.53) | 3.70 | 0.05 | 50 (0.98) | 168 (1.45) | 5.92 | 0.015* |
| No. of S residue | 2,940 | 2,310 | 3,780 | 13,020 | 6,720 | 15,330 | ||||||
| No. (%) of S mutations | 9 (0.31) | 6 (0.26) | 0.10 | 0.75 | 13 (0.34) | 77 (0.59) | 3.37 | 0.07 | 22 (0.33) | 83 (0.54) | 4.52 | 0.034* |
Percentage calculated as the lower row number/the upper row number. *, P < 0.05; **, P < 0.01.
Fig 6.
Distribution of LHBs substitution mutations. (A) Mutation of genotype B in child groups (1992 survey, n = 46; 2005 survey, n = 25). (B) Mutation of genotype B in adult groups (1992 survey, n = 14; 2005 survey, n = 11). (C) Mutation of genotype C in child groups (1992 survey, n = 81; 2005 survey, n = 59). (D) Mutation of genotype C in adult groups (1992 survey, n = 18; 2005 survey, n = 62). The y axis represents the mutation ratio (number of mutations/number of all indicated sequences). The x axis represents the amino acid site of the LHBs protein where the indicated mutations occurred. The yellow line represents the pre-S1 region. The blue line represents the pre-S2 region. The black line represents the S region.
Combining the genotype B and the genotype C results, there were no differences in the pre-S region between the 1992 (163/20,193; 0.81%) and 2005 (129/13,356; 0.97%) surveys (χ2 = 2.35; P = 0.13). However, a significant increase in the mutation frequency was shown in the S region by comparing the 1992 (0.13%) and 2005 (0.24%) surveys (χ2 = 7.65; P = 0.005). Therefore, the child group of the 2005 survey had a specifically higher mutation frequency in the S region.
Change of the LHBs amino acid substitution frequency in adults.
The LHBs mutations in adult groups were also investigated and compared between the 1992 and 2005 surveys. For genotype B, the mutation frequency in the 1992 survey was 0.90% (20/2,226) in the pre-S region, versus 0.91% (16/1,749) in the 2005 survey. In the S region, the mutation frequencies were 0.49% and 0.34% in the 1992 and 2005 surveys, respectively. For genotype C, 30 mutations were identified in the pre-S region from 2,862 residues (1.05%) in 1992, compared to 151 mutations from 9,699 total residues (1.53%) in the 2005 survey. In the S region, the mutation frequencies were 0.48% in the 1992 survey and 0.62% in the 2005 survey.
Combining the results for genotype B and genotype C, the mutation frequencies in the 2005 survey were significantly higher than those in the 1992 survey in both the pre-S region (χ2 = 5.92; P = 0.02) and the S region (χ2 = 4.52; P = 0.03) (Table 5). Therefore, the mutation frequencies in the 2005 adult group were higher than those in the 1992 survey for both the pre-S and S regions.
Pre-S2 deletion and start codon mutation frequencies were significantly higher in the adult group than in the child group.
Previous studies have shown that pre-S2 deletion and start codon mutations are related to progressive liver disease (13, 24). Therefore, the prevalences of pre-S2 deletion and start codon mutations were compared between the child and adult groups. In both the 1992 and 2005 child groups, neither pre-S2 start codon mutations nor deletions were observed. However, in the adult group, pre-S2 start codon mutation occurrence frequencies were 6.3% (2 out of 32) in 1992 and 8.2% (6 out of 73) in the 2005 survey (Table 6). The deletion mutation occurrence frequency was identical to the start codon mutation frequency. Specific pre-S2-deleted residues (counting from the start codon of LHBs) found were residues 123 to 141 (1992), 130 to 141 (1992), 127 to 141 (2 strains in 2005), 132 to 141 (2 strains in 2005), 132 to 143 (2005), and 140 to 144 (2005). Moreover, three strains had both deletion and start codon mutations. These results demonstrate a significantly high increase in the frequency of occurrence of pre-S2 deletion and start codon mutations among the adult group (P < 0.01).
Table 6.
Prevalence of HBV with pre-S2 deletion and start codon mutationsa
| Parameter | Value for group |
P | |
|---|---|---|---|
| Children | Adults | ||
| 1992 | |||
| No. of sequences | 127 | 32 | |
| No. (%) of pre-S2 start codon mutations | 0 | 2 (6.3) | 0.04 |
| No. (%) of pre-S2 deletion mutations | 0 | 2 (6.3) | 0.04 |
| 2005 | |||
| No. of sequences | 84 | 73 | |
| No. (%) of pre-S2 start codon mutations | 0 | 6 (8.2) | 0.01 |
| No. (%) of pre-S2 deletion mutations | 0 | 6 (8.2) | 0.01 |
| Total | |||
| No. of sequences | 211 | 105 | |
| No. (%) of pre-S2 start codon mutations | 0 | 8 (7.6) | <0.01 |
| No. (%) of pre-S2 deletion mutations | 0 | 8 (7.6) | <0.01 |
Percentage calculated as number of mutations/number of sequences. Data analysis was done by Fisher's exact test.
DISCUSSION
The long-term change of HBV LHBs variants has never been studied by large-scale community-based population surveys in China. Taking advantage of 1992 and 2005 serosurveys before and after universal immunization, we provided a meaningful comparison of LHBs variant HBsAg-seropositive populations within the same area. Convincing evidence for the influence of universal vaccination on the emergence of α determinant mutants comes from the following observations. (i) The prevalence of α determinant mutants in the child group increased from 6.5% in 1992 to 14.8% in 2005. The results are consistent with a previous study of vaccinated children in Taiwan (9). To further confirm the results, we also detected the mutations in the adult group within same area as a control. We assumed that massive immunization had little influence on their mutations. As expected, the prevalence of α determinant mutants in the adult groups showed little difference between the 1992 (9.4%) and 2005 (9.9%) surveys. Comparing the child groups with the adult groups, the difference in the mutation frequency provides novel evidence that massive immunization enhanced α determinant mutations. (ii) An increased number of mutated residues from the 2005 child group occurred at important residues that are neutralizing epitopes, such as G145 and T126. Although the G145R variant was reported to sparsely spread because of the lower serum HBV DNA level (25), the G145R mutation is still the most common mutation in the child group from the 2005 survey. The high prevalence is highly associated with vaccination selection (26, 27). On the contrary, in the adult group, the mutation profile is different, and the most frequently occurring mutations clustered at the first loop of the α determinant, such as M133 and T134. Such mutations are associated with virus secretion and disease development (28–30). Thus, the results showed that the child group mutations are under the pressure of vaccine immunization; however, adult group mutations are natural occurrences and disease related. In addition, the status of nucleoside analogue-resistant mutations was also determined. However, no drug-resistant mutants were discovered in either the child or adult groups (Table 7).
Table 7.
Prevalence of nucleotide analogue drug-resistant mutations
| Drugs | Resistance mutation | Mutation in groupa |
|||
|---|---|---|---|---|---|
| 1992 children | 2005 children | 1992 adults | 2005 adults | ||
| Lamivudine | V173L | V173A (HN) | N | N | N |
| I169T | N | N | N | N | |
| M204V/I | N | N | N | N | |
| L180M | N | N | N | N | |
| Famciclovir | L180M | N | N | N | N |
| Adefovir | A181V | N | N | N | N |
| I169 | N | N | N | N | |
| Entecavir | T184 | N | N | N | N |
| S202 | N | N | N | N | |
| Tenofovir | A194T | N | N | N | N |
N, no mutations were detected.
There have been limited numbers of epidemiology studies that investigated pre-S region substitution mutations. There is a high mutation frequency in the pre-S region relative to S region conservation. Some mutations are genotype related, particularly in genotype C, which has more subgenotypes in China (31). The HBV genotypes are highly endemic in China. Southern China has a higher genotype B prevalence, while genotype C is found mainly in the north (32). To avoid genotype-specific mutations, the sequences were aligned with strains of multiple genotypes. Following the statistical analysis, the following intriguing results were observed. (i) Children in the 2005 survey had a specifically high mutation frequency in the HBV S region. Results for genotypes B and C were consistent. This is the first demonstration that only S region and not pre-S region mutation frequencies have significantly increased (χ2 = 7.65; P < 0.01). Because children were universally vaccinated with the HBV vaccine, comprised of the HBV S protein, the specific increase in the S region mutation frequency in the child group of the 2005 survey is strong evidence for immunization enhancing mutation of the S region. In the adult group, the genotype C mutation frequency also increased between the 1992 and 2005 surveys; however, the increase is nonspecific, and the frequency of mutations in the pre-S region even had a slightly higher increase than in the S region. A possible reason for this is a difference in the prevalent HBV strains between the 1992 and 2005 surveys. (ii) HBV sequence mutation profiles between child and adult groups are different (Table 5). There were higher mutation frequencies in the adult group than in the child group. Because perinatal transmission is the primary way of infection in China (33), adult carriers always endure a longer time of infection. The results demonstrated that the HBV mutation frequency increased during the long course of infection. The results also suggested that the age of the individual upon infection is an important factor affecting the outcome of hepatitis B virus infection. The percentage of acquired sequences from HBsAg-positive sera was lower in the adult groups than in the child groups (Table 2). This is more evidence of the higher mutation frequency in adult groups.
Pre-S deletion mutations are correlated with progressive liver disease development (15). Pre-S2 deletion and start codon mutations have particularly shown a high prevalence in occult carriers and cirrhosis and HCC patients (22, 23). It is a very interesting observation that pre-S2 start codon mutation and deletion frequencies were significantly higher in the adult groups than in the child groups. In fact, no pre-S2 mutation was observed in the child group. In the adult group, the pre-S2 deletion mutations were clustered at T cell-recognized epitopes (13). This phenomenon may demonstrate that most of the pre-S mutations occurred spontaneously and evolved during long-term coexistence with the host body but not through interpersonal infection. However, how HBV evolved with the host is an intriguing question, which may be related to liver cell maturity or host hormone level changes. Meanwhile, this result demonstrated that the LHBs mutation frequency increased in direct relation to the course of infection.
While some meaningful conclusions can be drawn from this comparison, there are some limitations of our study. First, only HBsAg-positive patients were enrolled in the study. Previous studies demonstrated that populations with HBsAg-negative but other positive marker profiles harbored even more mutations (9). Therefore, more studies will be conducted to investigate mutations in other serogroups in the future. Second, the vaccination backgrounds of the children were lacking in some areas, particularly in the 1992 survey. However, because massive vaccination in DSPs was initiated in 1986, the percentage of vaccinated children in 2005 was dramatically higher than that in 1992. According to the GX province record in the 2005 survey, in the population <20 years old, 85.9% of subjects were vaccinated for HBV, whereas only 3.8% of subjects aged >20 years had received HBV vaccination (16). Therefore, we investigated the mutation frequency of an adult group as control and showed that the mutation frequency was no different between the 1992 and 2005 surveys. Thus, we still can draw the conclusion that the massive vaccination program enhances mutations. Lastly, the PCR products were directly sequenced in the study. This method ignored quasispecies mutations; however, the method can determine that the sequence is the dominant strain.
In conclusion, our study provides information on a long-term basis for both child and adult populations and demonstrates some characteristics of virus evolution and mutation under massive vaccination. The mutation distribution differences were presented in LHBs, particularly in the pre-S region, between adult and child groups. The results showed that such mutations are due to long-term persistent infection, which can explain that the time of infection is correlated with disease prognosis.
ACKNOWLEDGMENTS
This work was supported by a grant from the Chinese Ministry of Science and Technology Program for Important Infectious Diseases Control and Prevention (2008ZX10002001).
We have no potential conflicts of interest in this study.
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Ganem D, Prince AM. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350:1118–1129 [DOI] [PubMed] [Google Scholar]
- 2.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45:529–538 [DOI] [PubMed] [Google Scholar]
- 3.Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y, Wang F, Zheng H, Guo J, Jia Z, Ma J, Wang H, Luo H, Li L, Jin S, Hadler SC, Wang Y. 2009. Evaluation of the impact of hepatitis B vaccination among children born during 1992-2005 in China. J. Infect. Dis. 200:39–47 [DOI] [PubMed] [Google Scholar]
- 4.Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y, Wang F, Zheng H, Guo J, Jia Z, Ma J, Wang H, Luo H, Li L, Jin S, Hadler SC, Wang Y. 2009. Epidemiological serosurvey of hepatitis B in China—declining HBV prevalence due to hepatitis B vaccination. Vaccine 27:6550–6557 [DOI] [PubMed] [Google Scholar]
- 5.Li RC, Yang JY, Gong J, Li YP, Huang ZN, Fang KX, Xu ZY, Liu CB, Zhao K, Zhuang H. 2004. Efficacy of hepatitis B vaccination on hepatitis B prevention and on hepatocellular carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi 25:385–387 (In Chinese.) [PubMed] [Google Scholar]
- 6.Oon CJ, Lim GK, Ye Z, Goh KT, Tan KL, Yo SL, Hopes E, Harrison TJ, Zuckerman AJ. 1995. Molecular epidemiology of hepatitis B virus vaccine variants in Singapore. Vaccine 13:699–702 [DOI] [PubMed] [Google Scholar]
- 7.Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. 2006. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol. Rev. 28:112–125 [DOI] [PubMed] [Google Scholar]
- 8.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325–329 [DOI] [PubMed] [Google Scholar]
- 9.Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. 1999. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology 30:1312–1317 [DOI] [PubMed] [Google Scholar]
- 10.Chiou HL, Lee TS, Kuo J, Mau YC, Ho MS. 1997. Altered antigenicity of ‘a' determinant variants of hepatitis B virus. J. Gen. Virol. 78(Part 10):2639–2645 [DOI] [PubMed] [Google Scholar]
- 11.Coleman PF. 2006. Detecting hepatitis B surface antigen mutants. Emerg. Infect. Dis. 12:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuckerman JN, Zuckerman AJ. 2003. Mutations of the surface protein of hepatitis B virus. Antiviral Res. 60:75–78 [DOI] [PubMed] [Google Scholar]
- 13.Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. 2006. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology 130:1153–1168 [DOI] [PubMed] [Google Scholar]
- 14.Choi MS, Kim DY, Lee DH, Lee JH, Koh KC, Paik SW, Rhee JC, Yoo BC. 2007. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J. Viral Hepat. 14:161–168 [DOI] [PubMed] [Google Scholar]
- 15.Wang HC, Huang W, Lai MD, Su IJ. 2006. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 97:683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen LP, Zhang Y, Wang F, Zhang S, Yang JY, Fang KX, Yu T, Wang XY, Zhang WY, Bi SL. 2011. Epidemiological changes in hepatitis B prevalence in an entire population after 20 years of the universal HBV vaccination programme. Epidemiol. Infect. 139:1159–1165 [DOI] [PubMed] [Google Scholar]
- 17.Bian T, Zhang Y, Cao YQ, Wang JJ, Shen LP, Wang F, Wang Y, Bi SL. 2008. The analysis of hepatitis B virus genetic characterization and ratio of mutation in low-age group of HuNan. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 22:205–207 (In Chinese.) [PubMed] [Google Scholar]
- 18.Magnius LO, Norder H. 1995. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology 38:24–34 [DOI] [PubMed] [Google Scholar]
- 19.Kajiwara E, Tanaka Y, Ohashi T, Uchimura K, Sadoshima S, Kinjo M, Mizokami M. 2008. Hepatitis B caused by a hepatitis B surface antigen escape mutant. J. Gastroenterol. 43:243–247 [DOI] [PubMed] [Google Scholar]
- 20.Purdy MA. 2007. Hepatitis B virus S gene escape mutants. Asian J. Transfus. Sci. 1:62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oon CJ, Chen WN. 1998. Current aspects of hepatitis B surface antigen mutants in Singapore. J. Viral Hepat. 5(Suppl 2):17–23 [DOI] [PubMed] [Google Scholar]
- 22.Khan N, Guarnieri M, Ahn SH, Li J, Zhou Y, Bang G, Kim KH, Wands JR, Tong S. 2004. Modulation of hepatitis B virus secretion by naturally occurring mutations in the S gene. J. Virol. 78:3262–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, Zhou F, Waters J, Karayiannis P, Luo K. 2001. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology 34:1027–1034 [DOI] [PubMed] [Google Scholar]
- 24.Pollicino T, Zanetti AR, Cacciola I, Petit MA, Smedile A, Campo S, Sagliocca L, Pasquali M, Tanzi E, Longo G, Raimondo G. 1997. Pre-S2 defective hepatitis B virus infection in patients with fulminant hepatitis. Hepatology 26:495–499 [DOI] [PubMed] [Google Scholar]
- 25.Hsu HY, Chang MH, Ni YH, Chiang CL, Chen HL, Wu JF, Chen PJ. 2010. No increase in prevalence of hepatitis B surface antigen mutant in a population of children and adolescents who were fully covered by universal infant immunization. J. Infect. Dis. 201:1192–1200 [DOI] [PubMed] [Google Scholar]
- 26.Cooreman MP, Leroux-Roels G, Paulij WP. 2001. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J. Biomed. Sci. 8:237–247 [DOI] [PubMed] [Google Scholar]
- 27.Lada O, Benhamou Y, Poynard T, Thibault V. 2006. Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J. Virol. 80:2968–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. 2013. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One 8:e54486. 10.1371/journal.pone.0054486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S. 2010. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J. Virol. 84:12850–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathet VL, Feld M, Espinola L, Sanchez DO, Ruiz V, Mando O, Carballal G, Quarleri JF, D'Mello F, Howard CR, Oubina JR. 2003. Hepatitis B virus S gene mutants in a patient with chronic active hepatitis with circulating anti-HBs antibodies. J. Med. Virol. 69:18–26 [DOI] [PubMed] [Google Scholar]
- 31.Bian T, Shen LP, Wang F, Wang Y, Zhang LW, Zhang Y, Bi SL. 2008. Cloning of the full-length genome of A recombinant hepatitis B virus strain and phylogenetic analysis. Bing Du Xue Bao 24:255–259 (In Chinese.) [PubMed] [Google Scholar]
- 32.Zeng G, Wang Z, Wen S, Jiang J, Wang L, Cheng J, Tan D, Xiao F, Ma S, Li W, Luo K, Naoumov NV, Hou J. 2005. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J. Viral Hepat. 12:609–617 [DOI] [PubMed] [Google Scholar]
- 33.Yao JL. 1996. Perinatal transmission of hepatitis B virus infection and vaccination in China. Gut 38(Suppl 2):S37–S38 [DOI] [PMC free article] [PubMed] [Google Scholar]