Abstract
Cauliflower mosaic virus (CaMV) forms two types of inclusion bodies within infected plant cells: numerous virus factories, which are the sites for viral replication and virion assembly, and a single transmission body (TB), which is specialized for virus transmission by aphid vectors. The TB reacts within seconds to aphid feeding on the host plant by total disruption and redistribution of its principal component, the viral transmission helper protein P2, onto microtubules throughout the cell. At the same time, virions also associate with microtubules. This redistribution of P2 and virions facilitates transmission and is reversible; the TB reforms within minutes after vector departure. Although some virions are present in the TB before disruption, their subsequent massive accumulation on the microtubule network suggests that they also are released from virus factories. Using drug treatments, mutant viruses, and exogenous supply of viral components to infected protoplasts, we show that virions can rapidly exit virus factories and, once in the cytoplasm, accumulate together with the helper protein P2 on the microtubule network. Moreover, we show that during reversion of this phenomenon, virions from the microtubule network can either be incorporated into the reverted TB or return to the virus factories. Our results suggest that CaMV factories are dynamic structures that participate in vector transmission by controlled release and uptake of virions during TB reaction.
INTRODUCTION
During virus replication, assembly, and accumulation within the host cell, the viral products are most often confined within intracellular aggregates called “virus factories,” “viroplasms,” or “virus inclusions” (1). There is a large variety of viral inclusions, which differ by their localization, size, composition, and function. In most cases, viruses use these inclusions to concentrate cellular and viral proteins and thereby increase the efficiency of replication and assembly (2–4). These structures may also protect viral complexes from the cellular degradation machinery (2). Only in rare instances have other specific functions been attributed to inclusion bodies. For example, the inclusions formed during baculovirus infection, the polyhedra, are involved in virus transmission by protecting virions in the outside world until a new caterpillar host ingests them (5).
Cauliflower mosaic virus (CaMV), one of the top 10 viruses in plant pathology (6), forms numerous electron-dense inclusion bodies in the host cell cytoplasm that function early in infection as virus factories (VFs) (7, 8). CaMV VFs are nonmembranous structures that contain the multifunctional viral protein P6 as matrix, many virions, and the viral P3 protein, with the latter probably being associated with virions (8–10). Previous data suggest that P6 self-associates, which permits VF formation (11–13). The number and size (2 to 10 μm in diameter) of these inclusions depend on the stage of the viral cycle, CaMV isolate, and host plant (14). VFs play an important role early in the infection cycle as the site of viral protein synthesis, replication, and assembly, as well as for storage of newly formed virions (8, 15). However, later in infection when viral replication has ceased, VFs are assumed to be mere storage facilities for excess virions with no specific function.
A second type of CaMV inclusion is the transmission body (TB), named so because it is required for CaMV transmission by aphid vectors. The TB contains the totality of the viral protein P2, which is the aphid transmission factor or helper component (16–18). Besides P2, TBs also contain P3 and some scattered virions (19). CaMV aphid transmission is driven by a transmissible complex that forms either in the infected cells or in the vector. This complex is composed of the icosahedral virion (containing the viral genome enclosed in a shell of capsid protein P4), virus-associated protein P3, and helper protein P2 that mediates binding of P3 virions to the aphid vector (10, 20, 21) (Fig. 1). More precisely, the transmissible complex attaches to a dedicated structure in the aphid stylet tip, the acrostyle (22, 23). After acquisition from infected cells during vector feeding and attachment to the acrostyle, CaMV is transported to a new host plant and released by unknown mechanisms during probing punctures of aphids in epidermis and mesophyll cells (24).
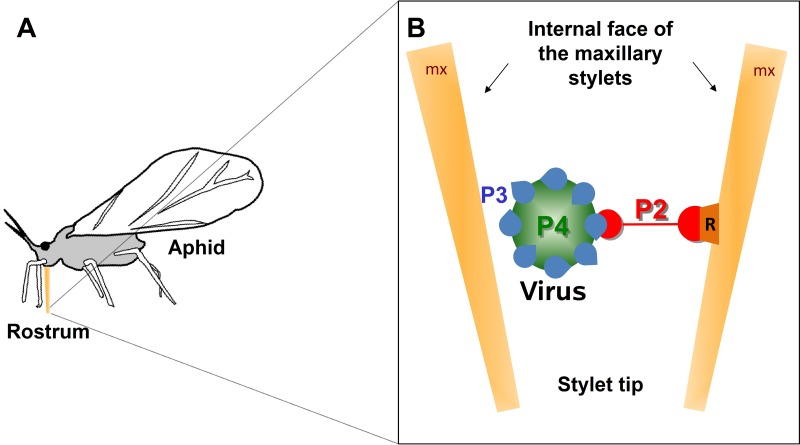
Fig 1.
CaMV transmissible complex. (A) The schematic drawing of an aphid shows the rostrum (orange) that harbors the needle-like stylet bundle when it is retracted and in resting position. During probing, the stylets glide through the rostrum and penetrate into the plant tissue. (B) The CaMV transmissible complex is composed of the viral helper protein P2 (red) that links the virus particle (P4; capsid protein in green) via the virion-associated protein P3 (blue) to a receptor (orange) at the tip of the aphid's maxillary stylets (mx).
It has long been suggested that TBs are involved in aphid transmission because they contain the totality of the protein P2 (16, 19, 25). Beyond their protein content, the TB was shown to be a specific structure enabling aphid transmission (26), adding this inclusion to the small list of viral inclusions that function in processes other than viral replication and assembly. However, the TB mode of action in relation to transmission has remained elusive. We showed previously that TB formation during the infection cycle requires an intact microtubule cytoskeleton (27), and that TBs are dynamic structures that respond rapidly to the presence and feeding of the aphid vector to facilitate virus acquisition (28). This study showed that TBs exist in a standby state, characterized by a spherical shape. However, when an aphid punctures infected tissues, the TBs in plant cells adjacent to the stylet path react instantly and undergo a spectacular transformation: tubulin suddenly enters into the TBs, and P2 relocalizes within seconds onto microtubules throughout the cell. At the same time and with the same kinetics, virions associate with the P2-decorated microtubules to form so-called mixed networks. The homogeneous redistribution of P2 and virions on the microtubules increases the efficiency of viral transmission, probably by increasing chances of stylet contact with transmissible complexes during aphid probing into cells. TB transformation is completely reversible: soon after removal of the stress inducing the formation of the mixed network, both P2 and virions are released from the microtubules and a standby TB reforms that is ready for another round of transmission.
A remaining question is the origin of virions that are recruited onto the mixed networks. While the source of microtubule-associated P2 is clearly the TB (it is the only P2 storage site within infected cells), the origin of virions aligning on the mixed networks is much less clear. Virions could originate from TBs or the cytosol, both of which contain some virions. Alternatively, they could stem from the numerous VFs scattered throughout the cytoplasm that contain the vast majority of virions (16, 19). As indicated above, thus far mature VFs have been regarded as a mere dead end for virions with no dedicated function, but if virions were recruited from VFs to mixed networks, they would turn out to be structures playing a significant role in vector transmission.
In this study, we show that there is a dynamic exchange of virions between VFs and TBs during TB transformation into mixed networks and its subsequent reversion. Thus, CaMV vector transmission not only is dependent on TB dynamics but also requires virion release from VFs. This relates another hitherto unsuspected CaMV inclusion to vector transmission and suggests an extensive reshuffling of the viral intracellular structures upon contact between infected plant cells and aphid stylets.
MATERIALS AND METHODS
Plants, viruses, plasmids, and inoculation.
Two-week-old turnip plants (Brassica rapa cv. Just Right) were mechanically inoculated with wild-type CaMV strain Cabb B-JI (29), Cabb B-JI-ΔP2, or Cabb B-JI-P2Myc as described previously (27), and they were used for experiments at 14 days postinfection (dpi). Initial inoculation was with plasmid pCa24, encoding Cabb B-JI (29), pCa24-Δ2, encoding Cabb B-JI-Δ2 (27), or pCa24-P2Myc, encoding Cabb B-JI P2Myc (this publication). Subsequent inoculation was made with crude extracts prepared from infected plants (20). The stability of the mutants was verified by sequencing PCR-amplified CaMV DNA using infected plant extracts as the matrix. To obtain the P2Myc mutant, the myc tag (GAGCAGAAACTCATCTCAGAAGAGGATCTG) was introduced at position 1650 of the CaMV genome by site-directed mutagenesis of plasmid pCa24, using the QuikChange kit according to the manufacturer's instructions (Agilent Technologies). To obtain a P2 expression plasmid, the P2 sequence of CaMV Cabb S (30) was amplified by PCR and ligated using NcoI and BamHI sites between the modified 35S promoter (consisting of a duplicated 35S enhancer plus an additional tobacco etch virus [TEV] enhancer sequence) and the CaMV terminator sequences of plasmid pCK-GFP (31), replacing the green fluorescent protein (GFP) sequence with P2.
Aphids.
A nonviruliferous clonal Myzus persicae colony was reared under controlled conditions (22°C day/18°C night with a photoperiod of 14-h day/10-h night) on eggplant. The population was originally started from a single virginiparous female.
Isolation of protoplasts.
Protoplasts for all experiments except transfection were prepared by enzymatic digestion from healthy or infected (14 dpi) leaves of turnip plants as described previously (28). For experiments involving transfection, the protoplasts were prepared from infected (14 dpi) leaves of turnip plants as described previously (32). Leaves were cut into thin lamella and incubated in the enzymatic solution (1.5% cellulase Onozuka R10, 0.4% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 20 mM morpholineethanesulfonic acid [MES], pH 5.7, 10 mM CaCl2, 0.1% bovine serum albumin [BSA]) during 3.5 h at 28°C in the dark. Protoplasts were separated from undigested tissue by filtration through Miracloth (Merck Millipore), and washed 3 times with ice-cold W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7) by centrifugation at 72 × g for 5 min in a swing-out rotor. After 2 washes, protoplasts were placed for 30 min on ice before centrifugation. The final pellet was resuspended in MMg solution (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7) at a concentration of 0.8 × 106 to 1.5 × 106 cells/ml. Protoplasts were counted using a Malassez counting chamber (Preciss; Strasbourg, France). For each transfection, 100 μl of protoplasts was gently mixed with 20 μg of plasmid and 110 μl of polyethylene glycol (PEG) solution (40% PEG 3000, 0.2 M mannitol, 100 mM CaCl2) and incubated for 10 min at room temperature. After addition of 440 μl W5 solution, protoplasts were centrifuged at 72 × g for 2 min in a swing-out rotor. The pellet was resuspended in 100 μl of W5 solution. Protoplasts were cultured in 900 μl Gamborg solution in a 24-well cell culture plate at 20°C in the dark.
Labeling of virions with DAPI.
Forty μg of virions, purified as described previously (33), was incubated with 50 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) in 500 μl of H2O for 1 h at room temperature. The solution then was diluted in 10 ml of H2O and centrifuged at 40,000 rpm (rotor Ti70; Beckman Coulter) for 4 h at 4°C. The pellet was resuspended in 1 ml of 10 mM HEPES, pH 7.2. Twenty μl of DAPI-treated virions (approximately 2 μg) was used per transfection.
Drug and stress treatments.
To induce mixed networks, protoplasts contained in Eppendorf reaction tubes were exposed for 15 min to gaseous CO2 as described previously (28). For TB reversion, the CO2 atmosphere in the reaction tubes was replaced with fresh air by opening them for 15 min. Azide treatment of protoplasts was with 0.02% NaN3 during 40 min (28).
Electron microscopy.
Samples for electron microscopy were processed as described previously (28). Infected agarose-embedded protoplasts were fixed with 4% glutaraldehyde, postfixed with 2% OsO4, and embedded in Epon resin. After ultrathin sectioning, the grids bearing sections were observed in a Jeol JEM 100CX II electron microscope (Jeol) operated at 60 to 80 kV.
Antisera and immunofluorescence.
We used the antibodies and immunofluorescence technique described previously (28). For anti-P2 and anti-P4 coimmunolabeling, the anti-P2 antibody was directly conjugated with Alexa 488 fluorescent dye using a commercial kit (Life Technologies) after purification of IgG fractions from the crude antiserum by Pierce thiophilic adsorbant chromatography (Thermo Scientific).
Microscopy.
Slides were observed with a Zeiss LSM700 confocal microscope operated in sequential mode. Alexa Fluor 594 dye was excited with a 555-nm LED laser, and the mirror was set to record emission from 555 to 620 nm. The Alexa Fluor 488 fluorophore was excited with a 488-nm LED laser, and emission was collected from 490 to 530 nm. For collecting fluorescence of DAPI-colored nuclei, a 405-nm LED laser was used, and emission was captured from 405 to 530 nm. Images were captured at 0.3- to 0.5-μm intervals using a 63× oil immersion objective. Raw images were processed using LSM software.
Transmission tests.
Transmission tests were performed from protoplasts as described previously (34). Aphids were allowed to acquire virus from protoplast suspensions for 15 min (30 min for transfected protoplasts), then they were placed on 1-week-old turnip test plants (10 aphids per plant) for inoculation. Twenty to 24 plants were inoculated per replicate. Symptoms were scored 3 weeks later by visual infection. To preload aphids with recombinant P2, insects were allowed to feed for 10 min across stretched Parafilm membranes on crude extracts from Sf9 cells infected with a P2-expressing baculovirus as previously described (19).
Statistics.
The transmission tests using azide-treated cells (see Fig. 4H) were statistically evaluated. As the distribution was not normal, the nonparametric Mann-Whitney test was used. The VassarStats website (http://vassarstats.net/) was used to calculate P values.
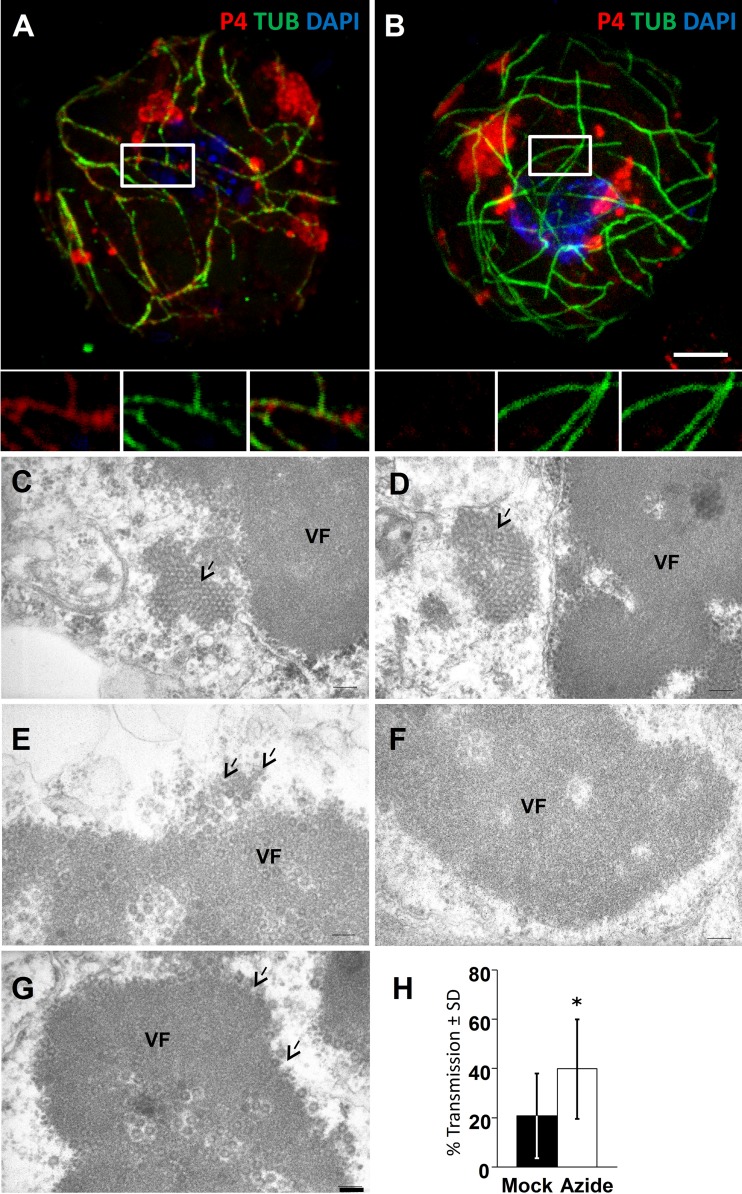
Fig 4.
Azide stress induces virus release from VFs. (A and B) Wild-type-infected (A) or CaMV-ΔP2-infected (B) cells were incubated for 40 min with 0.02% azide and then processed for immunofluorescence against P4 (red) and α-tubulin (green), and nuclei were counterstained with DAPI (blue). The images show that P4 relocalizes partially with microtubules after azide treatment in wild-type-infected cells but not in cells infected with the P2 deletion mutant. The main pictures show confocal projections, and the insets show single sections of the zones delineated in white. Scale bar, 5 μm for panels A and B. (C to G) Cells infected with CaMV-ΔP2 were either treated (C to E) for 40 min with 0.02% azide or were left untreated (F and G), and then they were processed for transmission electron microscopy. (C and D) Azide treatment induced appearance of paracrystalline virion arrays (arrows) close to VFs. (E) Azide-treated cells also displayed VFs with virions (arrows) decorating the VF-cytosol interface. (F) Control cells most often contained VFs with smooth virion-free borders; paracrystalline virion arrays were not observed. (G) Some VFs in unstressed cells had virions (arrows) seemingly budding at their surface. Note that VFs in both treated and untreated cells can also contain electron-transparent cavities filled with virions that have been reported before (35). Scale bar, 100 nm for panels C to G. (H) Azide-treated cells infected with CaMV-ΔP2 perform better in aphid transmission experiments than untreated controls. Aphids were preloaded by membrane feeding with recombinant P2 to compensate for the lack of P2 in CaMV-ΔP2-infected cells. They were then allowed to acquire virions from control (mock) or azide-treated (azide) infected cells before the aphids were transferred to healthy test plants for inoculation. Infected plants were scored 3 weeks later by visual inspection of CaMV symptoms. The asterisk indicates a significant effect of the treatment on transmission (P = 0.0014 by two-sided Mann-Whitney test; n = 18 from 3 independent experiments with 20 plants per replicate). SD, standard deviations.
RESULTS
Viral factories are connected to the microtubule network upon TB transformation.
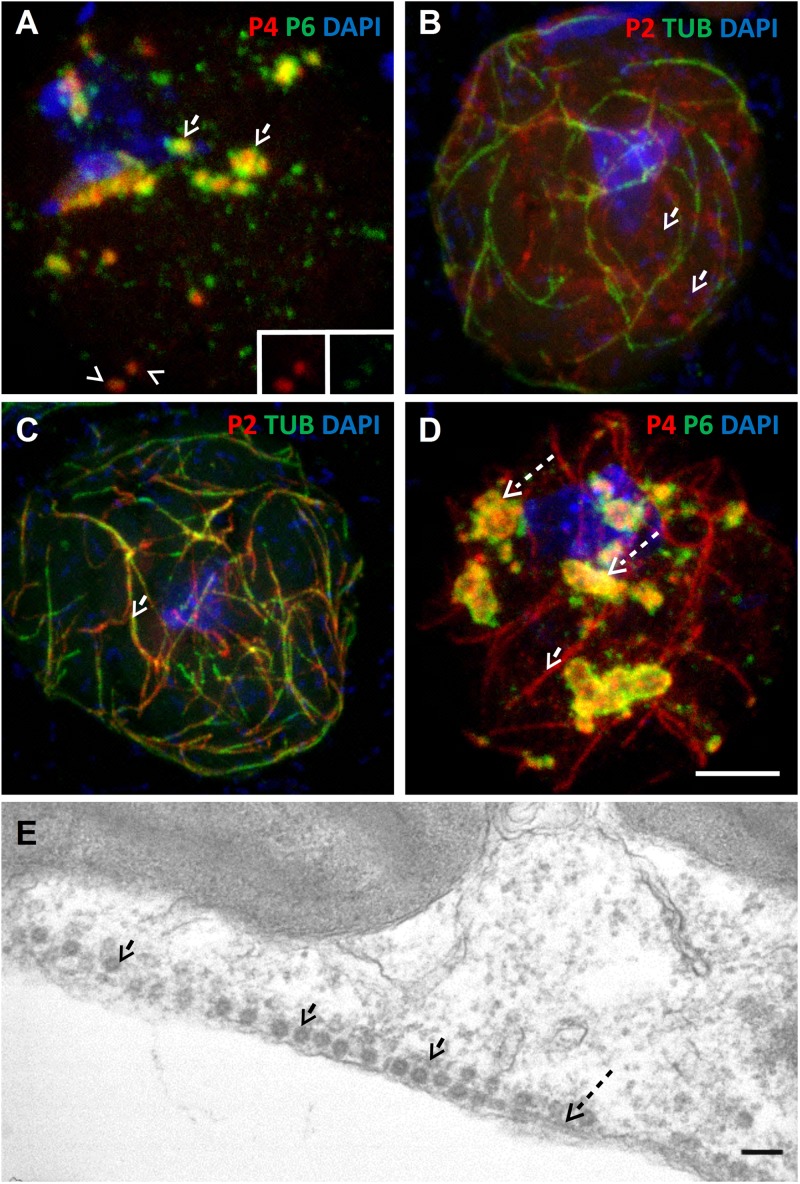
During TB transformation, induced naturally by aphid feeding or artificially by exposing cells to CO2 or to the chemical sodium azide, P2, and probably the virions contained within TBs, relocalize on microtubules and form mixed networks (28). Figure 2A illustrates that virions are localized mostly in numerous inclusions (likely VFs) in unstressed plant cells, whereas they also localize into mixed networks upon tissue exposure to CO2 (Fig. 2B). Transmission electron microscopy (Fig. 2C) shows that typical spherical virions are associated with microtubules that were identified by their appearance (rod-like structures of approximately 25-nm diameter displaying electron-dense rims). This confirms that the P4 label on the networks detected by immunofluorescence corresponds to virions and not to P4 aggregates. Thus, mixed networks are heavily charged with virions, as previously reported (28).Virions attached to the mixed networks are unlikely to originate exclusively from TBs, because these structures contain only about 5% of virions in a cell (19). Thus, we wanted to know whether the other 95% of virions contained within VFs represent virus sources that are alternative or complementary to mixed networks. For this, we used a protoplast system where TB dynamics are similar to those in intact tissue but are easier to manipulate (28, 34). As in infected tissue, mixed networks were induced in infected protoplasts by CO2 treatment. Figure 2D shows that CO2-stressed protoplasts displayed P4 foci, probably corresponding to VFs (see below), as well as P4 label on mixed networks, as previously reported (28). Close inspection indicated that the P4 label extended from VFs to mixed networks, suggesting a link between VFs and mixed networks. Indeed, optical single sectioning of the same protoplast showed continuation of the virus label from inclusions to microtubules with which the mixed networks are associated (Fig. 2D, insets). Approximately 90% of all cells (45 cells analyzed in 4 experiments with similar results) displayed these connections. In contrast, only about 10% (38 cells observed in 4 repetitions) of unstressed control protoplasts contained microtubules connected to VFs (Fig. 2E and insets). It is interesting that virions in CO2-stressed cells were always detected in VFs and on mixed networks but never on mixed networks alone. This suggests that if VFs can release virions, this phenomenon is only partial and VFs are not totally disrupted or emptied (Fig. 2D), a scenario complicating dissection of this phenomenon. To overcome this restriction in analysis of viral dynamics, we tried to obtain several independent and converging lines of evidence supporting the hypothesis that VFs contribute virions to the mixed networks.
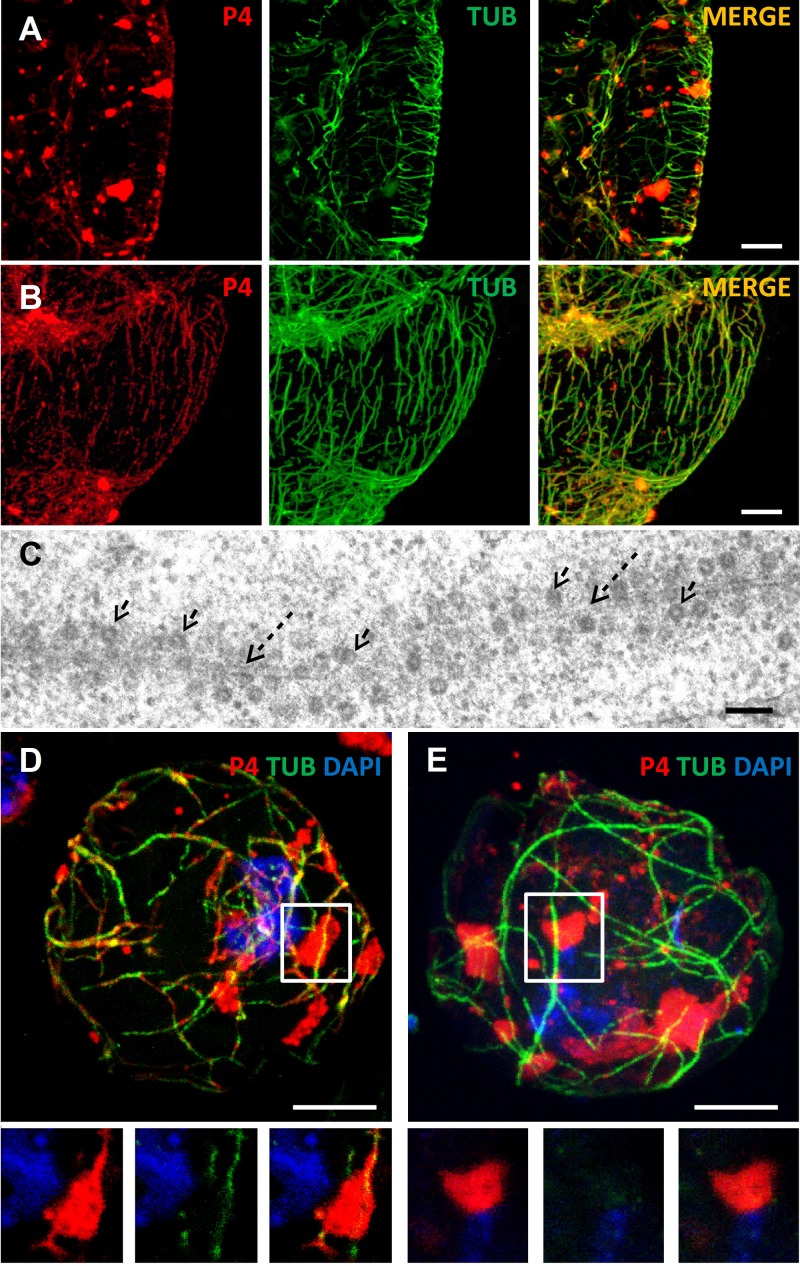
Fig 2.
Stress induces virion networks that extend from VFs. (A and B) Images show confocal projections of immunofluorescent coat protein (P4, red) and α-tubulin (TUB) (green) in unstressed leaf tissue (A) and in tissue exposed to CO2 stress that provokes TB disruption and mixed-network formation (B). While in unstressed cells P4 is located mainly in numerous inclusions, most of which probably correspond to VFs, P4 colocalizes with the microtubule network upon CO2 treatment (the merge of the two colors appears as yellow/orange in panel B). Scale bar, 5 μm for panels A and B. (C) Transmission electron microscopy of CO2-stressed infected leaves shows that virions (arrows) decorate microtubules (dashed arrows). Scale bar, 100 nm. (D and E) Images show confocal projections of CO2-stressed (D) and unstressed (E) infected protoplasts. Immunolabeling against P4 (red) and α-tubulin (green) in panel D reveals connections between virion networks (the mixed network) and VFs. The connections are visible in the insets in panel D that show a single optical section of the zone delineated in the projection. Such connections between VFs and the microtubule network were not observed in the projection of an unstressed cell (E) and in a single optical section of the boxed zone in panel E and shown in the insets. Nuclei were counterstained with DAPI (blue). Scale bar, 5 μm for panels D and E.
Ectopically induced TBs take up virions.
In contrast to TBs, which are dynamic structures throughout infection, mature VFs are believed to exchange none or little of their contents with the cytoplasm (35, 36). We wanted to know whether this is true or whether mature VFs can release significant amounts of virions, at least transiently. To test this, we used protoplasts infected with a CaMV P2 deletion mutant (denoted CaMV-ΔP2 in reference 27). These cells display wild-type-like VFs containing virions but no TBs (Fig. 3A). We transfected CaMV-ΔP2-infected protoplasts with a plasmid for ectopic expression of P2 to see whether a TB can form in a cell where the infection cycle of CaMV is completed and, most importantly, whether such de novo TBs would incorporate virions. If they did contain virions, they could only originate from the cytoplasm or from VFs. Sixteen hours after transfection with the P2 expression plasmid, virtually all transfected cells (200 cells counted, >20 repetitions) displayed inclusions with a typical TB phenotype, as judged by immunofluorescence (Fig. 3B). On one preparation, the TB phenotype was analyzed by electron microscopy, which revealed an electron-lucent inclusion with some dispersed virus particles that were virtually indiscernible from TBs in infected tissues (Fig. 3C). Moreover, the TB formed from ectopically expressed P2 had the same properties as wild-type TBs: it supported CaMV transmission by aphids from protoplast suspensions (Fig. 3D), and in about 70% of transfected cells (49 cells observed, 7 independent experiments), CO2 stress induced TB disruption and relocalization of P2 and virions on microtubules, i.e., formation of mixed networks (Fig. 3E).
Fig 3.
Ectopically expressed P2 forms a TB that recruits virions. (A) Confocal projection showing a protoplast infected with CaMV mutant CaMV-ΔP2 and immunolabeled against P4 (red) and VF matrix protein P6 (green). Virions and VFs colocalize (arrows). (B) CaMV-ΔP2-infected protoplasts were transfected with a P2 expression plasmid. In this case, immunolabeling against P4 (red) and the TB marker P2 (green) reveals P4 label in numerous VFs (arrows) and also in a rounded TB (dashed arrow). Scale bar, 5 μm for panels A and B. (C) Electron microscopy of a CaMV-ΔP2-infected protoplast transfected with a P2 expression plasmid shows a classical TB phenotype, where a few scattered CaMV virions (arrows) are distributed in an electron-lucent matrix. Scale bar, 100 nm. (D) De novo-formed TBs support aphid transmission. Aphids were allowed to feed for 30 min on CaMV-ΔP2-infected protoplasts transfected with P2 expression plasmid (Δ+P2) or on mock-transfected protoplasts infected with wild-type CaMV (BJI+PEG). They were then transferred to healthy turnip plants for inoculation, and infected plants were scored 3 weeks later for symptoms by visual inspection. Shown are results from 2 independent experiments using 24 test plants per condition. SD, standard deviations. (E) Confocal projection of a CO2-stressed, P2-transfected, CaMV-ΔP2-infected protoplast labeled for P2 (red) and α-tubulin (green). P2 and tubulin labels colocalize in mixed networks, with colabeling appearing as yellow/orange. Scale bar, 5 μm. Nuclei were counterstained with DAPI (blue) in panels A, B, and D.
Kinetics of ectopic TB formation showed that P2 label was not detected at any time point in VFs, indicating that P2 was translated free in the cytoplasm and assembled independently of VFs into TBs (not shown). This ruled out that virions were dragged along with P2 from VFs to TBs, as has been suggested to occur during initial TB formation early in the infection cycle (27). Thus, virions in this artificial TB could only have originated from mature VFs or from the cytoplasm.
Viral factories dispatch virions that support aphid transmission.
Our previous results showed that azide induces mixed networks where virions colocalize with P2 on microtubules (28). What might happen with virions if cells infected with CaMV-ΔP2 (i.e., containing no TBs and no P2) were incubated with azide? We performed this experiment. Figure 4A shows that azide-treated, wild-type-infected cells displayed P4 capsid protein labeling on microtubules, as reported previously (28). In CaMV-ΔP2-infected cells incubated with azide, however, no P4 label was detected on microtubules in 4 independent repetitions of the experiment, where about 100 cells were observed. This signified that either P2 is necessary for virion release from VFs, that VFs do not release virions, or that in the absence of P2, dispatched virions cannot associate with microtubules and accumulate somewhere else in cells. Because the immunofluorescence experiments did not resolve this question, we used electron microscopy with its greater optical resolution to visualize virions in protoplasts infected with CaMV-ΔP2 and treated with azide. These cells displayed virion aggregates or virion arrays entirely detached from, but in proximity to, VFs (Fig. 4C and D). We also often observed virions that seemed to be budding from the VFs' surfaces (Fig. 4E). In contrast, virion arrays could not be detected in the cytoplasm in unstressed cells infected by CaMV-ΔP2 (Fig. 4F), and virions budding from VFs were very rarely found (Fig. 4G).
We then assessed whether the VF-released virions can be efficiently acquired and transmitted by aphid vectors. Aphids were preloaded with recombinant P2 by membrane feeding as described previously (20). They were then allowed to feed on protoplasts infected by CaMV-ΔP2 before being transferred to healthy test plants for inoculation. Transmission after acquisition from azide-treated CaMV-ΔP2-infected protoplasts was significantly higher (P = 0.0014) (Fig. 4H). These results clearly show that transmission efficiency increased in the azide-treated cells. The only observable difference between these and control cells was the presence in the cytoplasm of free virions that seemingly stemmed from the VFs; thus, we conclude that these released virions are more accessible to the aphid vector than those enclosed in VFs.
Virions on microtubules originate from viral factories.
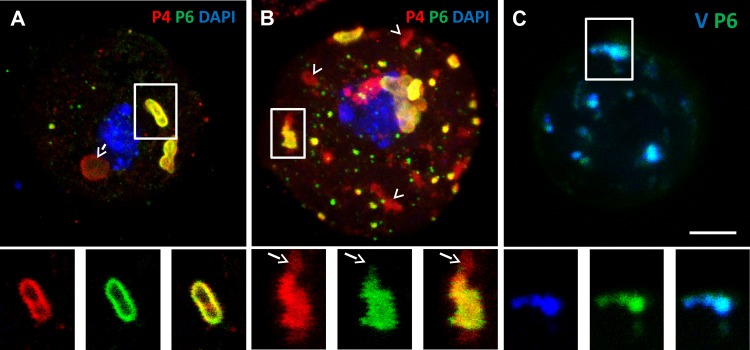
The results presented above show that VFs can release virions after stress treatment. However, it remained to be shown directly that these virions could associate with P2 on mixed networks in infected cells. For this, we used a CaMV mutant, CaMV-P2Myc, which contains a Myc tag sequence inserted at amino acid position 100 of P2. CaMV-P2Myc is infectious and forms normal-looking virion-containing VFs (Fig. 5A). Some VFs had a hardly visible P6 label, covered by the P4 label (Fig. 5A, arrowheads), but close inspection revealed that these inclusions contained some P6 nonetheless (Fig. 5A, insets). Interestingly, however, P2Myc did not assemble into a TB, and immunofluorescence microscopy revealed that it is distributed homogeneously in the cytoplasm (Fig. 5B). Despite the absence of TBs from CaMV-P2Myc-infected cells, CO2 induced relocalization of P2Myc onto microtubules (Fig. 5C), showing that its stress-induced redistribution is not inhibited.
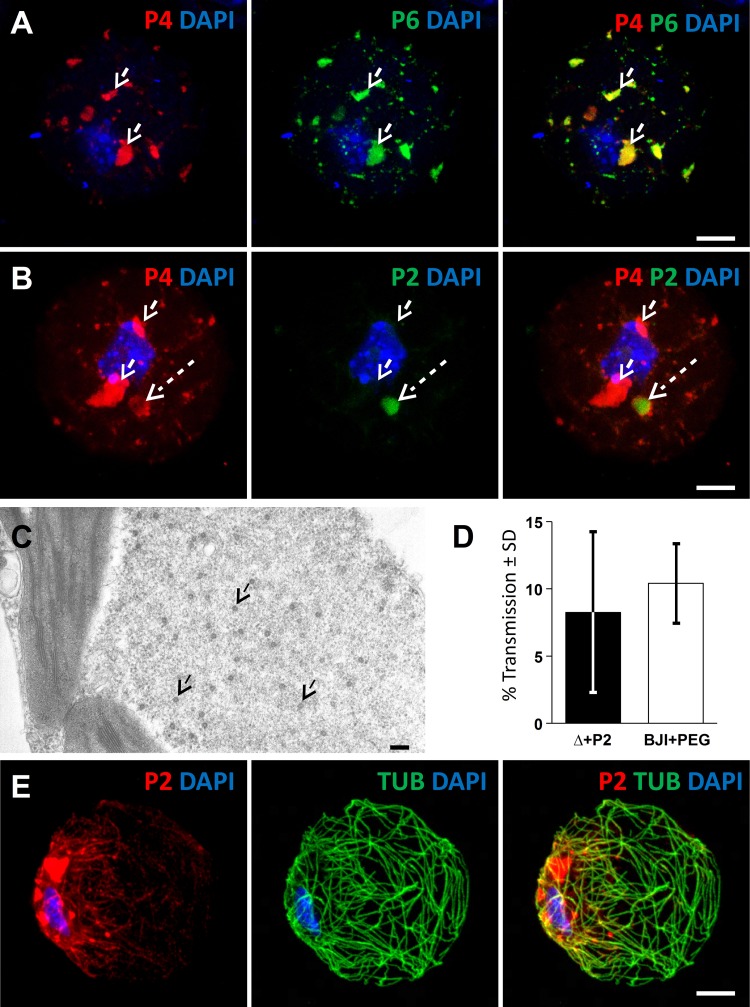
Fig 5.
CaMV-P2Myc mutant recruits virions from VFs. (A to D) Confocal projections of protoplasts infected with the CaMV-P2Myc mutant. (A) In unstressed cells, fluorescent label of P4 (red) and P6 (green) shows virions enclosed in VFs (arrows). The insets show single-channel presentations of the two VFs denoted by the arrowheads, where the P4 label covered the P6 signal. (B) The P2 label (red, arrow) and the α-tubulin label (green) reveal the presence of cytoplasmic P2Myc only, TBs are not present, and microtubule-associated P2Myc is barely detected. (C) Under conditions of CO2 stress, P2Myc (red) and α-tubulin (green) colocalize in mixed networks (arrow). (D) Under the same conditions, P4 (red) and P6 (green) colocalize in numerous VFs (dashed arrows), and P4 is also present as a series of networks (arrow). Nuclei were counterstained with DAPI (blue). Scale bar, 5 μm for all. (E) Transmission electron microscopy of CO2-stressed CaMV-P2Myc-infected protoplasts shows typical spherical CaMV virus particles (arrows) that decorate microtubules (dashed arrow). Scale bar, 100 nm.
The absence of TBs from cells infected with this mutant logically precluded any virions originating from TBs. This opened a nice opportunity to investigate, using induction of P2Myc networks by CO2, whether virions of another origin, namely, the VFs, can redistribute to mixed networks. Figure 5D and E demonstrates that CaMV-P2Myc-infected cells treated with CO2 accumulated a large quantity of virions on the mixed networks (75% of all cells contained mixed networks; 111 cells counted in 3 independent repetitions), suggesting that they were released from VFs and relocated on P2Myc-decorated microtubules.
Virions return to VFs after CO2 treatment.
An interesting property of the TB is that its transformation into a mixed network is reversible, and a new TB reforms after aphids withdraw their stylets or removal of CO2 or azide (28). As shown above, virions can be released from the VFs and accumulate on mixed networks. The next coherent question is whether virions can return to VFs upon reversion, when the mixed networks disappear. To investigate this possibility, we induced mixed networks in wild-type-infected CaMV protoplasts by CO2 treatment, allowed reversion and reformation of new TBs after removal of the gas, and localized virions by immunofluorescence microscopy during reversion. About 10% of unstressed control cells (Fig. 6A) contained some aggregation of virions against VFs. Other inclusions were either VFs displaying a perfect colocalization between virions and P6 or were TBs identified by the lack of P6 label. Cells fixed and examined during CO2 treatment, i.e., before reversion, displayed virus-containing VFs and virus-decorated mixed networks but rarely displayed virus aggregates next to VFs (Fig. 2D) (28). Reverting cells (Fig. 6B) displayed mostly normal VFs and reassembling TBs. However, 80% of the reverting cells displayed at least one VF with adjoining virion aggregates (3 independent experiments with a total of 300 cells analyzed for the 2 conditions). Thus, the accumulation of virions next to VFs seems to occur during the reversion process and might represent a congestion phenomenon, i.e., the uptake capacity of VFs for virions is temporally saturated. To provide direct evidence for the incorporation of virions from the cytoplasm into VFs, we labeled purified virions with the fluorescent nucleic acid staining dye DAPI. The fluorescent virions were then transfected into infected protoplasts, and the protoplasts were immunolabeled for the P6 protein to visualize VFs. Figure 6C shows VFs that are fluorescent due to the presence of DAPI-stained virions in them. This phenotype was observed in virtually all infected cells (100 cells analyzed in 3 independent repetitions), demonstrating that free exogenously supplied virions can migrate to VFs and be incorporated into them.
Fig 6.
Virions are packed against VFs during TB reversion. (A) The confocal projection shows that unstressed protoplasts contain mostly normal VFs (white frame) characterized by colocalization of the P4 and P6 label. The insets present single optical sections of the VF delineated by the white frame in the main picture. The arrow points to a TB, identified by its characteristic rounded shape with a cortex rich in P4 and by lack of P6 label. (B) Confocal projection showing that virions (red label) accumulate close to VFs (yellow merge of the P4 and P6 label) in reverting protoplasts. The insets present separated channels of a single optical section of the VF marked by the white frame in the confocal projection. The arrows in the insets point to P4 label adjoining a VF. The red-labeled inclusions devoid of adjacent P6 label (arrowheads in the main picture) probably correspond to reforming TBs that are characterized by the absence of P6 or to virion aggregates. (C) Confocal projection of a wild-type CaMV-infected protoplast transfected with DAPI-labeled virions in blue and P6 immunolabeled in green. The inset shows a single optical section of the VF delineated in the projection. It is apparent that the extrageneously supplied virions colocalize with VFs. Scale bar, 5 μm for all.
DISCUSSION
Taken together, our results confirm a dynamic exchange of virions between VFs, cytosol, TBs, and mixed networks both during initial TB transformation and during subsequent reversion. Therefore, both TBs and VFs are involved in vector transmission of CaMV: VFs provide virions for and retrieve them from the mixed networks at the right time, i.e., when TBs react to artificial stresses and, most likely, to aphid feeding on the host plant.
The presence of virions, although in widely varying amounts, in several compartments simultaneously (i.e., VFs, TBs, and the cytoplasm) made it difficult to determine the exact origin of the virions that were recruited onto microtubules. While it is very likely that all three sources provide virions for the mixed networks, our results suggest that VFs contribute the most. Indeed, after stress application, TB-less CaMV-P2Myc-infected cells displayed virion-decorated microtubules indiscernible from those in stressed wild-type-infected cells (compare Fig. 2C to 5E). This suggests that TBs allocate no or only a few virions for mixed networks. Theoretically, virions on P2Myc-decorated microtubules could have derived from preformed cytosolic P2Myc-virion complexes, but we think this is unlikely, because in these cells no above-background cytoplasmic P4 label, indicative of such complexes, was detected (Fig. 5A).
Because in wild-type CaMV-infected cells no above-background virion signal was detected in the cytosol, we believe that this compartment does not constitute a major pool of virions. Consistent with this, electron microscopy observations of unstressed cells did not reveal significant amounts of cytosolic virus particles (Fig. 4F and G and Fig. 5E), as also reported earlier by others (35, 36). This suggests that virions do not diffuse freely in the cytoplasm in the absence of stress. However, while the cytosol is unlikely to be a virus stocking site, it might be used as the medium through which virions traffic from VFs to microtubules. The following observation provides evidence for this hypothesis: the TB-less CaMV-ΔP2 mutant released virions that were found as aggregates or paracrystalline arrays in the vicinity of VFs, a phenomenon never observed in cells infected with wild-type CaMV. A possible explanation is that in wild-type-infected cells, virions dispatched from VFs associate faster with P2-decorated microtubules to form mixed networks than they can accumulate in the cytoplasm. As CaMV-ΔP2 mutants cannot form mixed networks to absorb the virions, they accumulate by default in the cytoplasm. Interestingly, virion aggregates or arrays in the cytosol of infected cells have previously been reported for a CaMV mutant with a partially deleted P2 (37). In that work, the mutant phenotype was attributed to an increased instability of mutant virus VFs caused by deficient interaction between VFs (or its matrix protein, P6) and P2. Because we now know that P2 is absent from VFs, we offer another explanation: unintended stress during manipulations induced release of virions from VFs in this P2-defective mutant, which is visible as virion aggregates or arrays in the vicinity of VFs, just like we observed here with stressed CaMV-ΔP2-infected cells.
Some questions concerning the role and functioning of CaMV VFs in transmission remain. Do virions liberated from VFs attach directly to mixed networks as the observed connections between them and VFs suggest (Fig. 2D), do they cross the cytosol as the presence of virions close to VFs during TB recovery indicates (Fig. 6B), or both? At present, there is no clear answer for this question. A more intriguing issue is how the release and retrieval of virions is controlled so that VFs dispatch virions at the precise moment of TB dissociation and mixed network formation. The most likely hypothesis is that aphid feeding triggers a signal that is transduced simultaneously and independently to both TBs and VFs. This view is supported by the fact that VFs in cells infected by CaMV-ΔP2 liberate virions in the total absence of TBs. The nature of this signal remains unknown, but its characterization would be highly valuable for a better understanding of plant-aphid-virus interactions.
Virus particles have three principal functions during the CaMV life cycle: infection of the nucleus to ensure transcription, cell-to-cell and systemic movement to allow for intrahost spread, and vector transmission to ensure interhost propagation. These different functions must be coordinated so that free virions, disposed in the cytoplasm at the onset of infection by the vector or via a plasmodesm, can reach the nuclear pores and infect the nucleus due to nuclear localization signals that are accessible on the capsid (38). On the other hand, virions formed de novo might be detained in VFs to prevent reinfection of the nucleus, presumably because α-importin interacting with the capsid nuclear localization signal is excluded from VFs (38). The other two functions of virions, movement and transmission, depend on capsid-associated P3 protein that interacts directly either with the movement protein P1 (39) or with P2 (10, 19, 21). Here, aphid feeding activity might constitute a signal pattern eliciting controlled release of virions and concomitant TB transformation into mixed networks, enabling transmission. Another signal pattern might trigger release of virions from VFs without simultaneous mixed-network formation, allowing virions to interact with P1 and leading to cell-to-cell movement. The nature of the signals and whether they overlap remains unknown; we propose that the previously hypothesized redox switch involving the cysteine ring of P3 plays a role as a signal receptor (40). P3 is indeed a good candidate, because it is strategically well placed on the virus capsid surface (10), interacting with P1 and P2, and it is found in both VFs and TBs. P6 itself also might play a larger role than anticipated in these processes, because it has been shown to interact with P2 and P3 in yeast two-hybrid assays (41). Further, ectopically expressed P6 forms inclusions that associate with the endoplasmic reticulum and microtubules and move along actin filaments (42). Finally, P6 interaction with CHUP1, an actin-binding protein needed for chloroplast positioning (41), is required for infection. This indicates a role for intracellular movement of VFs in a yet-undefined step of the infection cycle (43).
In summary, here we define a new, entirely unanticipated function for CaMV VFs in virus transmission and suggest that CaMV VFs also contribute actively to other steps of the infection cycle (i.e., movement). A future challenge will be to attribute functions other than replication to viral inclusions from other viruses as well.
ACKNOWLEDGMENTS
We thank S. Le Blaye for plant care, S. Guttierez for creation of the CaMV-P2Myc mutant, Takii Seed Company for providing turnip seeds, and Rodrigo Piacentini Paes de Almeida for English language editing.
A.B. acknowledges a Ph.D. fellowship financed by the SPE Department of INRA and the Région Languedoc-Roussillon. Our work is financed by grants ANR12-BSV7-005-01 from ANR (Agence National de la Recherche, France) and RéVir from the SPE Department of INRA, both awarded to M.D.
Footnotes
Published ahead of print 4 September 2013
REFERENCES
- 1.Moshe A, Gorovits R. 2012. Virus-induced aggregates in infected cells. Viruses 4:2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heath CM, Windsor M, Wileman T. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, Lacomme C, Santa Cruz S, Oparka KJ. 2012. The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol. 158:1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salas ML, Andrés G. 2013. African swine fever virus morphogenesis. Virus Res. 173:29–41 [DOI] [PubMed] [Google Scholar]
- 5.Wood HA, Trotter KM, Davis TR, Hughes PR. 1993. Per os infectivity of preoccluded virions from polyhedrin-minus recombinant baculoviruses. J. Invertebr. Pathol. 62:64–67 [Google Scholar]
- 6.Scholthof K-BG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD. 2011. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12:938–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martelli GP, Castellano MA. 1971. Light and electron microscopy of the intracellular inclusions of Cauliflower mosaic virus. J. Gen. Virol. 13:133–140 [DOI] [PubMed] [Google Scholar]
- 8.Mazzolini L, Bonneville JM, Volovitch M, Magazin M, Yot P. 1985. Strand-specific viral DNA synthesis in purified viroplasms isolated from turnip leaves infected with Cauliflower mosaic virus. Virology 145:293–303 [DOI] [PubMed] [Google Scholar]
- 9.Covey SN, Hull R. 1981. Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology 111:463–474 [DOI] [PubMed] [Google Scholar]
- 10.Plisson C, Uzest M, Drucker M, Froissart R, Dumas C, Conway J, Thomas D, Blanc S, Bron P. 2005. Structure of the mature P3-virus particle complex of Cauliflower mosaic virus revealed by cryo-electron microscopy. J. Mol. Biol. 346:267–277 [DOI] [PubMed] [Google Scholar]
- 11.Cecchini E, Gong Z, Geri C, Covey SN, Milner JJ. 1997. Transgenic Arabidopsis lines expressing gene VI from Cauliflower mosaic virus variants exhibit a range of symptom-like phenotypes and accumulate inclusion bodies. Mol. Plant Microbe Interact. 10:1094–1101 [DOI] [PubMed] [Google Scholar]
- 12.Haas M, Bureau M, Geldreich A, Yot P, Keller M. 2002. Cauliflower mosaic virus: still in the news. Mol. Plant Pathol. 3:419–429 [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Leisner SM. 2002. Multiple domains within the Cauliflower mosaic virus gene VI product interact with the full-length protein. Mol. Plant Microbe Interact. 15:1050–1057 [DOI] [PubMed] [Google Scholar]
- 14.Shalla TA, Shepherd RJ, Peterson LJ. 1980. Comparative cytology of nine isolates of Cauliflower mosaic virus. Virology 102:381–388 [DOI] [PubMed] [Google Scholar]
- 15.Rothnie HM, Chapdelaine Y, Hohn T. 1994. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv. Virus Res. 44:1–67 [DOI] [PubMed] [Google Scholar]
- 16.Espinoza AM, Medina V, Hull R, Markham PG. 1991. Cauliflower mosaic virus gene II product forms distinct inclusion bodies in infected plant cells. Virology 185:337–344 [DOI] [PubMed] [Google Scholar]
- 17.Woolston C, Czaplewski L, Markham P, Goad A, Hull R, Davies J. 1987. Location and sequence of a region of Cauliflower mosaic-virus gene-2 responsible for aphid transmissibility. Virology 160:246–251 [Google Scholar]
- 18.Woolston CJ, Covey SN, Penswick JR, Davies JW. 1983. Aphid transmission and a polypeptide are specified by a defined region of the Cauliflower mosaic virus genome. Gene 23:15–23 [DOI] [PubMed] [Google Scholar]
- 19.Drucker M, Froissart R, Hébrard E, Uzest M, Ravallec M, Espérandieu P, Mani JC, Pugnière M, Roquet F, Fereres A, Blanc S. 2002. Intracellular distribution of viral gene products regulates a complex mechanism of Cauliflower mosaic virus acquisition by its aphid vector. Proc. Natl. Acad. Sci. U. S. A. 99:2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc S, Cerutti M, Usmany M, Vlak JM, Hull R. 1993. Biological activity of Cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology 192:643–650 [DOI] [PubMed] [Google Scholar]
- 21.Leh V, Jacquot E, Geldreich A, Hermann T, Leclerc D, Cerrutti M, Yot P, Keller M, Blanc S. 1999. Aphid transmission of Cauliflower mosaic virus requires the viral PIII protein. EMBO J. 18:7077–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzest M, Gargani D, Drucker M, Hébrard E, Garzo E, Candresse T, Fereres A, Blanc S. 2007. A protein key to plant virus transmission at the tip of the insect vector stylet. Proc. Natl. Acad. Sci. U. S. A. 104:17959–17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzest M, Gargani D, Dombrovsky A, Cazevieille C, Cot D, Blanc S. 2010. The “acrostyle”: a newly described anatomical structure in aphid stylets. Arthropod Struct. Dev. 39:221–229 [DOI] [PubMed] [Google Scholar]
- 24.Moreno A, Palacios I, Blanc S, Fereres A. 2005. Intracellular salivation is the mechanism involved in the inoculation of Cauliflower mosaic virus by its major vectors Brevicoryne brassicae and Myzus persicae. Ann. Entomol. Soc. Am. 98:763–769 [Google Scholar]
- 25.Nakayashiki H, Tsuge S, Kobayashi K, Okuno T, Furusawa I. 1993. Reasons for the low accumulation level of aphid transmission factor protein in infected leaves with an aphid-non-transmissible Cauliflower mosaic virus isolate, CM1841. J. Gen. Virol. 74:2469–2472 [DOI] [PubMed] [Google Scholar]
- 26.Khelifa M, Journou S, Krishnan K, Gargani D, Espérandieu P, Blanc S, Drucker M. 2007. Electron-lucent inclusion bodies are structures specialized for aphid transmission of Cauliflower mosaic virus. J. Gen. Virol. 88:2872–2880 [DOI] [PubMed] [Google Scholar]
- 27.Martinière A, Gargani D, Uzest M, Lautredou N, Blanc S, Drucker M. 2009. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus. Plant J. 58:135–146 [DOI] [PubMed] [Google Scholar]
- 28.Martinière A, Bak A, Macia J-L, Lautredou N, Gargani D, Doumayrou J, Garzo E, Moreno A, Fereres A, Blanc S, Drucker M. 2013. A virus responds instantly to the presence of the vector on the host and forms transmission morphs. eLife 2:e00183. 10.7554/eLife.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delseny M, Hull R. 1983. Isolation and characterization of faithful and altered clones of the genomes of Cauliflower mosaic virus isolates Cabb B-JI, CM4-184, and Bari I. Plasmid 9:31–41 [DOI] [PubMed] [Google Scholar]
- 30.Franck A, Guilley H, Jonard J, Richards K, Hirth L. 1980. Nucleotide sequence of Cauliflower mosaic virus DNA. Cell 21:285–294 [DOI] [PubMed] [Google Scholar]
- 31.Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. 1996. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc. Natl. Acad. Sci. U. S. A. 93:5888–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo S-D, Cho Y-H, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2:1565–1572 [DOI] [PubMed] [Google Scholar]
- 33.Hull R, Shepherd RJ, Harvey JD. 1976. Cauliflower mosaic virus: an improved purification procedure and some properties of the virus particles. J. Gen. Virol. 31:93–100 [Google Scholar]
- 34.Martinière A, Macia J-L, Bagnolini G, Jridi C, Bak A, Blanc S, Drucker M. 2011. VAPA, an innovative “virus-acquisition phenotyping assay” opens new horizons in research into the vector-transmission of plant viruses. PLoS One 6:e23241. 10.1371/journal.pone.0023241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitajima EW, Lauritis JA, Swift H. 1969. Fine structure of zinnial leaf tissues infected with Dahlia mosaic virus. Virology 39:240–249 [DOI] [PubMed] [Google Scholar]
- 36.Conti GG, Vegetti G, Bassi M, Favali MA. 1972. Some ultrastructural and cytochemical observations on Chinese cabbage leaves infected with Cauliflower mosaic virus. Virology 47:694–700 [DOI] [PubMed] [Google Scholar]
- 37.Givord L, Xiong C, Giband M, Koenig I, Hohn T, Lebeurier G, Hirth L. 1984. A second Cauliflower mosaic virus gene product influences the structure of the viral inclusion body. EMBO J. 3:1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsies A, Merkle T, Szurek B, Bonas U, Hohn T, Leclerc D. 2002. Regulated nuclear targeting of Cauliflower mosaic virus. J. Gen. Virol. 83:1783–1790 [DOI] [PubMed] [Google Scholar]
- 39.Stavolone L, Villani ME, Leclerc D, Hohn T. 2005. A coiled-coil interaction mediates Cauliflower mosaic virus cell-to-cell movement. Proc. Natl. Acad. Sci. U. S. A. 102:6219–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoh F, Uzest M, Drucker M, Plisson-Chastang C, Bron P, Blanc S, Dumas C. 2010. Structural insights into the molecular mechanisms of Cauliflower mosaic virus transmission by its insect vector. J. Virol. 84:4706–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz L, Raikhy G, Leisner SM. 2012. Cauliflower mosaic virus major inclusion body protein interacts with the aphid transmission factor, the virion-associated protein, and gene VII product. Virus Res. 170:150–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. 2003. Chloroplast unusual positioning 1 is essential for proper chloroplast positioning. Plant Cell 15:2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angel CA, Lutz L, Yang X, Rodriguez A, Adair A, Zhang Y, Leisner SM, Nelson RS, Schoelz JE. 2013. The P6 protein of Cauliflower mosaic virus interacts with CHUP1, a plant protein which moves chloroplasts on actin microfilaments. Virology 443:363–374 [DOI] [PubMed] [Google Scholar]