Abstract
Inflammasomes are key signalling platforms that detect pathogenic microorganisms and sterile stressors, and that activate the highly pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-18. In this Review, we discuss the complex regulatory mechanisms that facilitate a balanced but effective inflammasome-mediated immune response, and we highlight the similarities to another molecular signalling platform — the apoptosome — that monitors cellular health. Extracellular regulatory mechanisms are discussed, as well as the intracellular control of inflammasome assembly, for example, via ion fluxes, free radicals and autophagy
A major role of the immune system is to maintain homeostatic tissue function. For example, sterile tissue damage that occurs after trauma needs to be detected and repaired. Pathogens that can invade and cause harm to tissues should be eliminated while our commensal microbiome must be tolerated, as it fulfils functions that are required for host survival. The innate immune system has a number of signalling receptors that recognize foreign molecular structures as well as self molecules that are altered, that have become too abundant or that emerge in areas normally devoid of these molecules1,2.
Innate immune signalling receptors monitor the extracellular space as well as many subcellular compartments for signs of infection, damage or other cellular stressors. The inflammasomes are a group of multimeric protein complexes that consist of an inflammasome sensor molecule, the adaptor protein ASC and caspase 1. Inflammasome formation is triggered by a range of substances that emerge during infections, tissue damage or metabolic imbalances. Once the protein complexes have formed, the inflammasomes activate caspase 1, which proteolytically activates the pro-inflammatory cytokines interleukin-1β (IL-1β)3 and IL-18. In addition, inflammasome activation causes a rapid, pro-inflammatory form of cell death called pyroptosis4.
With the discovery of pattern recognition receptors (PRRs), such as inflammasome sensor molecules, their signalling pathways and their ability to programme cellular immune responses, we are beginning to understand how the immune system protects the host at a molecular level. At the same time, we have learnt that immune dysregulation contributes to prevalent diseases in Western societies such as atherosclerosis, type 2 diabetes, cancer and neurodegenerative diseases. Thus, a fine balance must be maintained between the activation and inhibition of inflammation to allow the immune system to remove any sources of danger without causing harm to the host.
In this Review, we present an overview of the current understanding of inflammasome activation and regulation, and we discuss the recent findings about non- canonical processing of IL-1β. We also compare the structures and the regulation of inflammasomes with that of the apoptosome.
The inflammasomes and their co-receptors
Inflammasome components
At first glance, the inflammasomes are organized in a very simple manner: inflammasome sensor molecules (see below) connect to caspase 1 via ASC, which is an adaptor protein encoded by PYCARD that is common to all inflammasomes. ASC consists of two death-fold domains: one pyrin domain and one caspase activation and recruitment domain (CARD). ASC interacts with the upstream inflammasome sensor molecules via the pyrin domain5. This interaction triggers the assembly of ASC into a large protein speck consisting mainly of multimers of ASC dimers6,7. Using its CARD, ASC brings monomers of pro-caspase 1 into close proximity, which initiates caspase 1 self- cleavage and the formation of the active heterotetrameric caspase 1. Active caspase 1 proteolytically activates a number of proteins8, including pro-IL-1β and pro-IL-18 (REFS 9,10), and induces their release via a non-classical secretion pathway11.
The transcription of pro-IL-1β is induced by the activation of the transcription factor nuclear factor-κB (NF-κB), whereas pro-IL-18 is constitutively expressed and its expression is increased after cellular activation. Therefore, these potent pro-inflammatory cytokines are controlled by two checkpoints: transcription as well as maturation and release11. Caspase 1-mediated activation of members of the IL-1β cytokine family leads to the recruitment and the activation of other immune cells, such as neutrophils, at the site of infection and/or tissue damage.
Several inflammasome sensor molecules can trigger the formation of inflammasomes. Most of the inflammasomes that have been described to date contain a NOD-like receptor (NLR) sensor molecule, namely NLRP1 (NOD-, LRR- and pyrin domain-containing 1), NLRP3, NLRP6, NLRP7, NLRP12 or NLRC4 (NOD-, LRR- and CARD-containing 4; also known as IPAF). The NLR proteins, with the exception of NLRP1, have a tripartite domain organization; they contain an amino-terminal death-fold domain (NLRPs contain a pyrin domain, whereas NLRC4 contains a CARD), a central NACHT nucleotide-binding domain and carboxy-terminal leucine-rich repeats (LRRs)12. The NACHT domain has ATPase activity and is thought to have a role in the oligomerization of the proteins, whereas the LRRs have regulatory functions and might be involved in ligand interaction. The death-fold domains of the NLR proteins interact with those of ASC and/or caspase 1. In addition to these domains, human NLRP1 contains a function-to-find domain (FIIND) and a C-terminal CARD. In the original description of the inflammasome, human NLRP1 was shown to recruit and to activate an additional inflammatory caspase, namely caspase 5, via its CARD3.
NLRC4 and NLRP1 can both activate caspase 1 through their CARDs without recruiting ASC; however, the recruitment of ASC greatly enhances the formation of the complex and the processing of IL-1β7,13–16. Exactly how ASC is recruited to these inflammasomes remains unclear, as NLRC4 and mouse NLRP1B do not have pyrin domains. In a mammalian two-hybrid analysis, the CARD of NLRC4 was found to interact with the CARD of ASC17. We speculate that a CARD–CARD interaction between the NLR and ASC recruits a first layer of ASC, which in turn interacts with a second layer of ASC via pyrin–pyrin domain interactions.
Two other inflammasomes have been described that contain the PYHIN (pyrin and HIN domain-containing protein) family members absent in melanoma 2 (AIM2) and IFNγ-inducible protein 16 (IFI16) rather than an NLR18. AIM2 consists of a pyrin domain to recruit ASC and a DNA-binding HIN domain, whereas IFI16 has one pyrin domain and two HIN domains for DNA binding. Retinoic acid-inducible gene I (RIG-I) protein is also thought to assemble an inflammasome with ASC and caspase 1 (REF. 19), possibly via its CARDs. However, for some of the inflammasome sensors (including NLRP6, NLRP12, RIG-I and IFI16), the potential to form inflammasomes has not been well established and other functions have been described for these molecules. Indeed, NLRP12 can function as a positive regulator of dendritic cell migration or as a negative regulator of non-canonical NF-κB signalling20,21, and NLRP6 can negatively regulate innate immunity22. RIG-I is widely known as a PRR that senses RNA and that signals via mitochondrial antiviral signalling protein (MAVS) to induce an interferon (IFN) response2, and IFI16 has been suggested to be a DNA sensor that signals via the protein STING (stimulator of IFN genes; also known as TMEM173) to generate an IFN response23.
Numerous activators of the inflammasomes and several different activation pathways have been described (reviewed in REF. 24). The PYHIN proteins and RIG-I recognize nucleic acids18,19,25, whereas NLRC4 is activated by microbial proteinaceous ligands26. NLRP1 recognizes muramyl dipeptide, which is a bacterial peptidoglycan, and murine NLRP1B can also be activated by the lethal toxin from Bacillus anthracis13,27. Many triggers, including crystalline material, peptide aggregates and bacterial toxins, can stimulate NLRP3 (REF. 24). NLRP7, which is not expressed in mice, is activated by bacterial lipo-peptides28, and the triggers for NLRP6 and NLRP12 remain to be identified29–31.
In summary, the inflammasome sensor molecules can detect a broad range of molecular signatures to sense microorganisms and tissue stress. They are best known for triggering a robust inflammatory response via the activation of inflammatory caspases. However, it should also be noted that not all NLR molecules form inflammasomes and that other functions of NLRs, which are not as well understood, might be important contributors to an inflammatory response.
Cofactors for inflammasome activation
Some inflammasome sensor molecules require co-receptors to recognize their ligands or to be stabilized in their activated states. NLRP1 has been shown to bind directly to its ligand muramyl dipeptide in vitro and this was demonstrated to be sufficient to activate the assembly of an inflammasome. However, a requirement for the interaction of NLRP1 with nucleotide-binding oligomerization domain-containing protein 2 (NOD2), which is another receptor for muramyl dipeptide, has been described. This suggests that NOD2 is a vital component of the NLRP1 inflammasome32,33.
Recent studies by the groups of Vance34 and Shao35 have determined that the activation of NLRC4 also requires co- receptors34,35. In mice, NAIP (NLR family, apoptosis inhibitory protein) family proteins sense the proteinaceous NLRC4 activators and, in turn, activate the assembly of the NLRC4 inflammasome. NAIP5 and NAIP6 have been shown to detect flagellin, and NAIP2 has been shown to sense the type III secretion system component PrgJ34,35. Only one NAIP orthologue has been found in humans (known as NAIP) and this is necessary to sense a type III secretion needle protein35; however, it remains to be determined whether and how human NLRC4 senses flagellin without another NAIP homologue. Some human cell lines (for example, U937) do not respond to flagel-lin35. However, studies using primary human cells that were infected with Legionella pneumophilia have determined that small interfering RNA (siRNA)-mediated knockdown of NLRC4 results in enhanced bacterial growth only when L. pneumophilia expresses flagellin36, which indicates that human NLRC4 might be able to sense flagellin. It is possible that NAIP could have dual specificity in humans and that it could recognize both a type III secretion needle protein35 and flagellin36. Dual specificity could also be used by other inflammasome sensors that recognize a variety of structurally and chemically diverse activators.
Differential requirements for various NLRP3 activators
NLRP3 is unique among the NLRs in that its basal expression is not sufficient for inflammasome activation in resting cells37,38. Similarly to IL-1β generation, a two-checkpoint activation mechanism is in place for NLRP3 inflammasome activation; it requires a priming step (see below) and a second activation step that can be induced by various triggers. Given the broad range of NLRP3 activators24, direct binding of these activators to NLRP3 seems to be unlikely. Indeed, as for NLRC4, it is possible that ligand specificity can be achieved via recruitment of different cofactors. There is evidence to support this theory, as the formation of the NLRP3 inflammasome in response to non-crystalline activators can be promoted by guanylate-binding protein 5 (GBP5)39. In addition, oxidized mitochondrial DNA, which is released into the cytosol under cellular stress and is a possible NLRP3 inflammasome activator, was recently reported to interact with NLRP3 (REF. 40) (BOX 1). However, whether NLRP3 directly interacts with oxidized mitochondrial DNA or whether a cofactor is required for this interaction remains unknown.
Box 1|Sensing of mitochondrial stress.
Mitochondria are required to maintain cellular energy levels and so their health is crucial for cell viability. Mitochondrial dysfunction can trigger cell autonomous pathways that lead to apoptosis or to an inflammatory response. One of the molecules that signals that there is mitochondrial dysfunction is cytochrome c, which is an inner mitochondrial membrane protein and an essential component of the electron transport chain. After cytochrome c has leaked into the cytosol, it binds to apoptotic protease-activating factor 1 (APAF1), which is a scaffold protein for the activation of the initiator caspase 9, forming the apoptosome (REF. 137).
This intrinsic pathway of apoptosis is regulated by B cell lymphoma 2 (BCL-2) proteins138. Pro-apoptotic members of the family (such as BCL-2-associated X protein (BAX) and BCL-2 homologous antagonist/killer (BAK)) promote cytochrome c release from the mitochondria, whereas the anti-apoptotic family members (such as BCL-2 and BCL-XL) inhibit cytochrome c release138. Interestingly, these anti-apoptotic proteins also regulate the NLRP1 (NOD-, LRR- and pyrin domain-containing 1) inflammasome. BCL-2 and BCL-XL bind to NLRP1 via their loop domains, thereby suppressing the ability of NLRP1 to bind to ATP and to oligomerize139,140.
Mitochondrial stress can also be sensed by the NLRP3 inflammasome. Indeed, excessive production of reactive oxygen species (ROS)102 and release of oxidized mitochondrial DNA40 from stressed mitochondria have been suggested to activate the NLRP3 inflammasome. In addition, impaired mitochondrial homeostasis leads to changes in metabolites that are crucial for the function of the cell, such as the coenzyme NAD+ (REF 141). Low NAD+ levels inactivate the α-tubulin deacetylase sirtuin 2 (REF 142), which results in the accumulation of acetylated α-tubulin. Acetylated α-tubulin regulates the transport of mitochondria and helps the formation of an efficient interaction between the adaptor protein ASC and NLRP3 (REF 143). Furthermore, the RIG-I-like receptor (RLR) adaptor molecule mitochondrial antiviral signalling protein (MAVS), which is localized on the outer mitochondrial membrane, is required for optimal NLRP3 activation144.
Another intriguing mechanism by which lipopolysaccharide-activated macrophages can increase their output of interleukin-1β (IL-1β) is through a substantial increase in levels of the tricarboxylic acid cycle intermediate succinate145. This metabolite stabilizes the transcription factor hypoxia-inducible factor-1α, which leads to more sustained IL-1β transcription145. Therefore, monitoring of mitochondrial stress by apoptosomes and inflammasomes seems to directly link mitochondrial health and activity to cell death and inflammation.
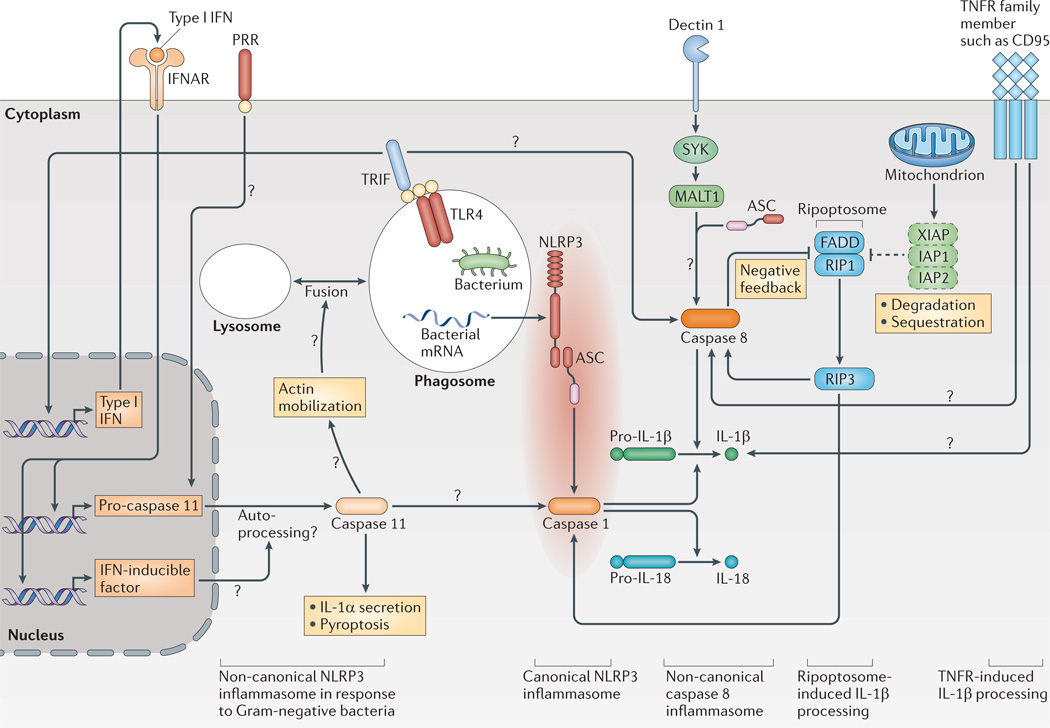
The activation of caspase 1 following the recognition of live Gram-negative bacteria depends on inflammatory pro-caspase 11 in mice (human orthologues are pro-caspase 4 and pro-caspase 5)41, which results in non-canonical NLRP3 inflammasome activation. Pyroptosis in response to live Gram-negative bacteria but not in response to other inflammasome triggers is entirely dependent on caspase 11, whereas caspase 1 is dispensable for this process41. Therefore, in contrast to the standard two-checkpoint model of priming and activation, three signals are required for full NLRP3 inflammasome activation in response to Gram-negative bacteria42,43. First, a bacterial Toll-like receptor (TLR) activator leads to cellular priming and upregulation of NLRP3 and pro-IL-1β expression (the priming checkpoint in the standard model)37,38. The second ‘activation checkpoint’ can be divided into two parts in response to bacteria. One checkpoint is that bacterial mRNA from live bacteria (also known as vita-PAMPs)44 activates NLRP3; the other checkpoint is that TLR4- and TRIF (TIR domain-containing adaptor protein inducing IFNβ)-dependent signalling — which is triggered by bacterial lipopolysaccharide (LPS) — mediate the secretion of type I IFNs, inducing pro-caspase 11 expression and activation by triggering the IFNα/β receptor (IFNAR) (FIG. 1).
Figure 1. Canonical and non-canonical activation of IL-1β.
NLRP3 (NOD-, LRR- and pyrin domain-containing 3) needs additional cofactors for the processing of interleukin-1β (IL-1β) in response to Gram-negative bacteria; this path way has been termed the non-canonical NLRP3 inflammasome. Toll-like receptor 4 (TLR4) signalling via TIR domain-containing adaptor protein inducing IFNβ (TRIF) induces the secretion of type I interferons (IFNs), which lead to the activation of caspase 11 via autocrine signalling through the IFNα/β receptor (IFNAR), possibly involving additional (not yet known) IFN-inducible factors. Caspase 11 is necessary for the activation of caspase 1 and has an independent role in IL-1α secretion and pyroptosis in response to Gram-negative bacteria. A possible mechanism for caspase 11 action might be its role in actin mobilization via cofilin, which might lead to increased fusion of lysosomes to phagosomes and, potentially, to increased leakage of bacterial mRNA (also termed vita-PAMPs) to the cytoplasm to activate NLRP3 (not shown). A platform consisting of mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), caspase 8 and the adaptor protein ASC (termed the non-canonical caspase 8 inflammasome) is formed in response to stimulation of dectin 1. Caspase 8 might also be activated by TLRs using the signalling adaptor TRIF in the presence of cycloheximide. In addition, the formation of the ripoptosome is triggered by the loss of inhibitor of apoptosis proteins (IAPs) with concurrent TLR stimulation. The ripoptosome — consisting of FAS-associated death domain protein (FADD) and receptor-interacting protein 1 (RIP1) — activates caspase 8 via RIP3. However, caspase 8 can also limit ripoptosome action on NLRP3. Members of the tumour necrosis factor receptor (TNFR) family might also induce pro-IL-1β cleavage, as has been shown for CD95 (also known as FAS). Question marks show pathways that are still speculative. PRR, pattern recognition receptor; XIAP, X-linked inhibitor of apoptosis protein.
The mechanisms by which caspase 11 is required to activate caspase 1 in response to bacteria are still under investigation and the exact timing of the different signals is not yet clear. In the context of NLRC4 activation, caspase 11 was shown to interact with cofilin. This interaction regulates actin polymerization and triggers the fusion of bacteria-containing phagosomes with lysosomes. These events facilitate the release of flagellin into the cytosol45, where NAIP family proteins can interact with flagellin and induce the activation of the NLRC4 inflammasome. It is possible that a similar caspase 11-dependent release of bacterial mRNA from the phagolysosomal compartment could mediate the activation of the NLRP3 inflammasome.
There is also a debate about how pro-caspase 11 expression is induced. One study found that the expression completely depends on IFN signalling42, whereas another study suggested that its expression could be induced independently of TRIF and IFN signalling46. Even if the expression of pro-caspase 11 could be induced independently of IFN signalling, caspase 11 activation in macrophages in response to Salmonella enterica subsp. enterica serovar Typhimurium infection is dependent on IFNAR1 as well as signal tranducer and activator of transcription 1 (STAT1)46. Therefore, an IFN-inducible factor might be required for the activation of caspase 11 during S. Typhimurium infection46.
Another recent study suggests a role for caspase 11 in defence against cytosolic bacteria that is independent of the known inflammasomes. These data have led to the development of a model in which a non-canonical caspase 11-containing inflammasome can be formed after cytoplasmic bacteria are sensed, which leads to pyroptosis but not to IL-1β release47. Further investigation is needed to determine when this complex is formed, under which circumstances caspase 11 activation will lead to NLRP3 activation and when it will cause cell death independently of the ‘canonical’ inflammasomes. A recent review discusses caspase 11 activation in more detail48.
Non-inflammasome processing of IL-1p
The inflammasomes and IL-1β processing in vitro and in vivo
Studies that have been carried out in vitro using cells that are deficient in inflammasome components have undoubtedly been instrumental in identifying several inflammatory triggers as activators of inflammasomes. However, the contribution of inflammasomes to IL-1β activation in vivo has not, at times, been nearly as convincing. The reduction in inflammation in mice that are deficient in the inflammasome components ASC or caspase 1 can be much less pronounced than that in mice lacking the IL-1 receptor (IL-1R) or IL-1β. It is probable that inflammasome-independent IL-1β activation mechanisms are involved in vivo that could account for the observed differences between the in vitro and in vivo studies. For example, the in vivo responses to particulates (diesel exhaust particles or silica) and to Mycobacterium tuberculosis have substantial caspase 1-independent but IL-1β-dependent components49–51. Indeed, a number of non-caspase proteases such as proteinase 3 have the ability to activate IL-1β in an inflammasome-independent manner11.
In addition, IL-1α, which also activates IL-1R, could contribute to the inflammatory response in vivo. This could partly explain why IL-1R–deficient mice have a more severe phenotype than inflammasome component-deficient mice, in which only IL-1β production is affected. In contrast to IL-1β, IL-1α is constitutively expressed in many cell types and does not need to be processed for its biological activity; thus, biologically active IL-1α can be released during necrosis11. In addition, IL-1α can be found on the membrane of some cell types, where it is active11. IL-1α is also secreted in response to numerous NLRP3 inflammasome activators; however, its secretion is not strictly dependent on the inflammasome. Indeed, particulate activators of NLRP3, such as crystals, can induce IL-1α secretion in an NLRP3-independent manner52. Therefore, the discrepancies between the in vivo and in vitro data may be partially explained by inflammasome-independent processing of IL-1β and by the contribution of IL-1α, the secretion of which is regulated differently from that of IL-1β release.
The role of caspase 8 in IL-1β processing
One additional factor that can mediate IL-1β processing and activation is caspase 8 (FIG. 1). Recent studies indicate that caspase 8 might be able to cleave pro-IL-1β during immune responses. For example, following sensing of fungal components by the PRR dectin 1 that is expressed on human dendritic cells, signalling via the kinase SYK leads to the formation of a complex that is composed of caspase 8 and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1); this complex binds to ASC, which possibly recruits cleavage substrates53. The MALT1-ASC-caspase 8 complex directly mediates IL-1β maturation53. Thus, caspase 8 can be activated by a multiprotein complex that contains the canonical inflammasome adaptor ASC — this MALT1-ASC-caspase 8 inflammasome protein complex has therefore been termed a ‘non- canonical caspase 8 inflammasome’. However, exactly how MALT1, caspase 8 and ASC interact is not understood, as these proteins all have different death-fold domains. In addition, NLRP3 has been implicated in dectin 1-mediated IL-1β release in mice54 and it therefore remains to be established whether both of these mechanisms can function in parallel or whether they represent differences between the two species.
Other PRRs can also activate caspase 8. In the presence of the translation inhibitor cycloheximide, TRIF signalling that is downstream of TLR3 or TLR4 leads to pro-IL-1β processing by caspase 8 (REF. 55). Furthermore, in response to genotoxic stress, the ripoptosome assembles and activates caspase 8. The ripoptosome contains caspase 8, FAS-associated death domain protein (FADD) and receptor-interacting protein 1 (RIP1; also known as RIPK1), and forms spontaneously in response to loss or inhibition of the anti-apoptotic proteins X-linked inhibitor of apoptosis protein (XIAP), inhibitor of apoptosis protein 1 (IAP1; also known as BIRC3) and IAP2 (also known as BIRC2) (REF 56). The formation of this complex activates RIP3, which is necessary for the cleavage of pro-IL-1β by both the NLRP3-caspase 1 and the caspase 8 pathways56 (FIG. 1). However, two other studies contradict these findings. One study showed that the NLRP3 inflamma some is more active in caspase 8-deficient dendritic cells, which indicates that caspase 8 might limit ripoptosome signalling that would otherwise trigger NLRP3–caspase 1 activation57. Another study found that IAP1, IAP2 and TNF receptor-associated factor 2 (TRAF2) are important for the non-degradative polyubiquitylation of caspase 1, which enhances its activity58.
Other caspase 8-activating receptors of the tumour necrosis factor receptor (TNFR) family can also induce IL-1β activation. Indeed, activation of the TNFR family member CD95 (also known as FAS) can induce IL-1β processing that is independent of inflammasomes and of caspase 1 (REF. 59). In addition, it was recently shown that CD95 signalling mediates IL-1β and IL-18 processing through the activation of caspase 8 (REF. 60) (FIG. 1). Thus, more non-inflammasome pathways for the maturation of pro-IL-1β are emerging, and these pathways could contribute to the effects of IL-1β activation in vivo.
Cell-extrinsic inflammasome regulation
Cell-autonomous regulatory feedback loops, as well as complex networks of direct cell–cell signals and indirect signals via messenger substances, regulate cellular activation. Many important immune responses can only be efficiently induced when two or more signals act on the responding cell in a timely manner — for example, as has been described for IL-1β maturation and NLRP3 activation. One level at which inflammasomes are regulated is at the level of expression of the individual inflammasome components61.
Positive regulation and priming
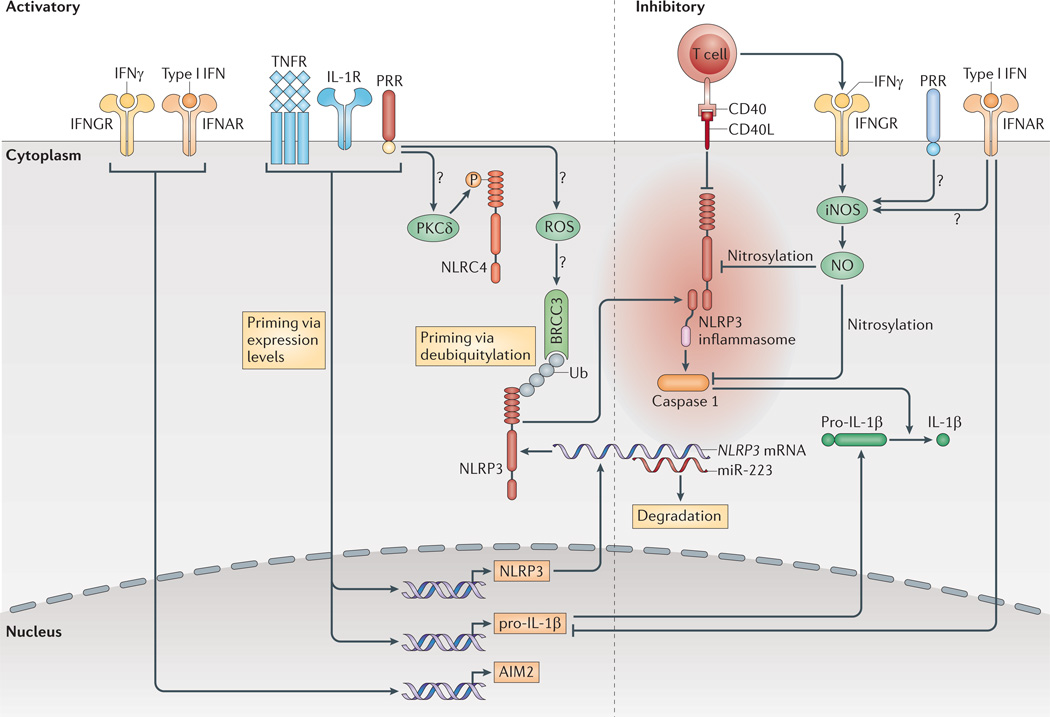
Many innate immune signalling or cytokine receptors, such as the TNFR, activate transcription of NLRP3 and thereby influence the susceptibility of immune cells to NLRP3 inflammasome triggers37,38 (FIG. 2). In addition to the transcriptional regulation of NLRP3, deubiquitylation of NLRP3 has been identified as an early priming event, which only occurs in response to PRR stimulation, possibly involving the production of reactive oxygen species (ROS)62. NLRP3 deubiquitylation is mediated by the K63-specific deubiquitinase BRCC3 (REF. 63). Therefore, the NLRP3 activation threshold is regulated by both a fast-acting post-translational mechanism and a slower-acting transcriptional activation of NLRP3 gene expression.
Figure 2. Extracellular signals regulate the inflammasomes.
Pro-interleukin-1β (pro-IL-1β) and NLRP3 (NOD-, LRR-and pyrin domain-containing 3) expression are induced by transcriptionally active pattern recognition receptors (PRRs) or by cytokine receptors. Furthermore, NLRP3 deubiquitylation by the K63-specific deubiquitinase BRCC3 is c rucial for its activation. Direct contact with mature or memory T cells inhibits the inflammasomes, probably via tumour necrosis factor receptor (TNFR) superfamily interactions. Type I interferons (IFNs) inhibit the transcription of pro-IL-1β , but also upregulate the expression of absent in melanoma 2 (AIM2). Both type I IFNs and IFNγ inhibit NLRP3 through the induction of nitric oxide (NO) via inducible nitric oxide synthase (iNOS), possibly with the requirement of concomitant priming by PRRs. Ub shows ubiquitylated proteins. Question marks show pathways that are still speculative. CD40L, CD40 ligand; IFNAR, interferon-α/β receptor; IFNGR, interferon-γ receptor; IL-1R, IL-1 receptor; miR-223, microRNA-223; NLRC4, NOD-, LRR- and CARD-containing 4; ROS, reactive oxygen species; PKCδ, protein kinase Cδ.
An early priming event has also been described for the AIM2 inflammasome, although the molecular mechanisms that are involved remain to be fully elucidated64. In addition, AIM2 expression can be induced by IFNβ and IFNγ65,66. NLRC4 can only be activated by S. Typhimurium after a specific serine residue has been phosphorylated by protein kinase Cδ (PKCδ; encoded by PRKCD), which depends on the recognition of both flagellin and the type III secretion system. This indicates that PKCδ might be activated by PRR signalling67. Therefore, at least some inflammasomes are only fully capable of responding to danger signals in situations in which either pro-inflammatory cytokines are present or innate signalling molecules sense noxious signals.
A few molecules, such as amyloid-β, can induce both NLRP3 priming through TLR activation and NLRP3 inflammasome activation68. However, the response to aggregated amyloid-β can be dramatically augmented when innate sensors or cytokine receptors become activated by additional stimuli. These priming mechanisms might be of great relevance in determining the magnitude of an inflammatory response to danger signals.
Conversely, genetic differences that influence the threshold of inflammasome activation might contribute to the development of chronic inflammatory or autoinflammatory diseases. It is known that about 60% of patients with cryopyrin-associated periodic syndrome (CAPS) carry activating mutations in the coding sequence of NLRP3 itself or in other inflammasome components69. This indicates that inflammasome hyperactivity may be influenced by additional mechanisms. Indeed, single nucleotide polymorphisms in the promoter region of NLRP3 were found in patients with CAPS69. In these patients, an increase in NLRP3 expression as opposed to the presence of a more active protein (as a result of mutations in the coding sequence) could conceivably result in a lower threshold of NLRP3 inflammasome activation in response to danger signals and this could trigger the development of CAPS. Therefore, several mechanisms ensure that inflammasomes are not accidentally triggered and that an appropriately balanced immune response to danger signals can occur in a regulated manner.
Negative regulation of inflammasome activation
An inflammatory response should subside once it has carried out its function. For example, the release of proinflammatory cytokines such as IL-1β and IL-18 might not be required after an adaptive immune response has been initiated. Thus, it is not surprising that effector and memory CD4+ T cells have the capacity to inhibit the activation of the NLRP1 and NLRP3 inflammasomes in a contact-dependent manner, possibly via TNFR superfamily molecules such as CD40 ligand (CD40L)70. In addition, T cell-derived IFNγ has been shown to downregulate the activity of NLRP3 via activation of inducible nitric oxide synthase (iNOS) in a mouse model of tuberculosis71; nitric oxide (NO) induces NLRP3 nitrosylation and thereby inhibits NLRP3 activity. However, it seems to be crucial that TLR priming and IFNγ stimulation occur simultaneously, as IFNγ loses its inhibitory activity after sequential priming with IFNγ and a TLR stimulus70,72.
In addition, type I IFNs can reduce IL-1β and IL-18 release by functioning at two levels. First, type I IFNs inhibit the production of the pro-forms of these cytokines; second, they repress the cleavage of the cytokine proforms by the NLRP1 and NLRP3 inflammasomes72. For IFNβ, the mechanism of NLRP3 inhibition might also involve the induction of NO73. Timing is also important for the inhibitory effect of IFNβ; although LPS-induced IFNβ is necessary for caspase 11 expression and, thus, NLRP3 activation in response to bacteria42 (see above), IFNβ blocks NLRP3 inflammasome activation when the cells sense it before they have been primed, for example, by a TLR stimulus72. It is worth noting that IFNs can have seemingly opposing effects on different inflammasomes. IFNs upregulate AIM2 expression but they downregulate IL-1β expression and inhibit the NLRP3 inflammasome. These effects could be adapted to generate optimal antiviral responses, in which excessive responses to danger signals could be harmful for the host.
Furthermore, the amount of NLRP3 mRNA is tightly regulated by the microRNA miR-223, which leads to decreased NLRP3 protein levels and, thus, influences the threshold of NLRP3 activation74,75. Pro-inflammatory signals do not regulate miR-223; rather, miR-223 is expressed in a cell type-specific manner in innate immune cells — the lowest levels of expression are found in dendritic cells and the highest levels of expression are found in neutrophils. This might explain why dendritic cells have a comparatively low threshold for NLRP3 stimuli.
Taken together, these studies show that the inflammasome-mediated response can be adjusted by the activities of cell-extrinsic negative regulators and amplifiers. This ‘rheostat’-like fine-tuning facilitates the adjustment of inflammasome activation in response to danger signals and indicates that both systemic signals and the tissue context might influence the threshold of inflammasome activation (FIG. 2). Thus, priming and negative regulation might strengthen or limit the local response, respectively. In fact, it is conceivable that the effectiveness of certain anti-cytokine therapies that are used to treat chronic inflammatory diseases is partly mediated by alterations in the local responsiveness of inflammasomes to danger signals. Extrinsic regulators of inflammasome activity can also affect the overall responsiveness of the cell by influencing the cell-intrinsic regulatory mechanisms of inflammasome activation, which are described in the following section.
Cell-intrinsic inflammasome regulation
When the function of a cell is compromised or the survival of the cell poses a danger to the whole organism, it should be eliminated. As mentioned above, inflammasomes are localized in the cytosol and can be activated when microbial molecules are generated in this compartment. It is crucial that pathogens or protein aggregates that have been phagocytosed are subsequently neutralized in the phagolysosomal compartment. However, if the degradative function of lysosomes is defective or if the capacity of phagocytosis is overwhelmed, lysosomal damage occurs. This can activate the NLRP3 inflammasome, possibly involving the activity of lysosomal proteases76. In addition, the integrity of mitochondria is monitored by inflammasomes and apoptosomes, and mitochondrial stress can lead to inflammation and cell death (BOX 1). Indeed, inflammasomes (which induce pyroptosis through caspase 1 or caspase 11 activation) and apoptosomes (which activate caspase 9 in response to cytochrome c release from mitochondria) are two mechanisms by which compromised cells are eliminated. In order to be activated, the complexes need to oligomerize (BOX 2); the resulting different receptor complexes share many similarities in their regulation in response to cellular stress (discussed below).
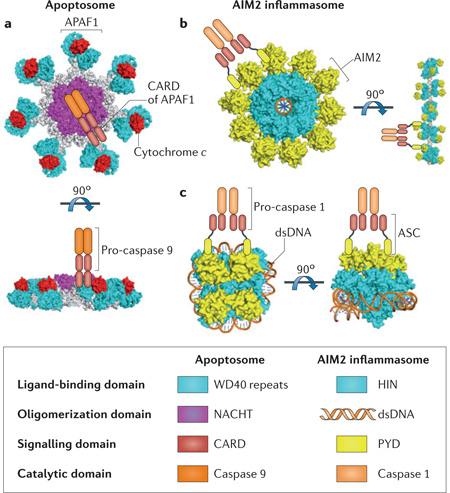
Box 2|Oligomerization of death-inducing molecular platforms.
Receptors of the death-inducing molecular platforms are activated by ligands such as cytochrome c for the apoptosome component apoptotic protease-activating factor 1 (APAF1), and double-stranded DNA (dsDNA) for the absent in melanoma 2 (AIM2) inflammasome. By contrast, the NLRP1 (NOD-, LRR- and pyrin domain-containing 1) inflammasome can be activated by self-proteolysis in its function-to-find (FIIND) domain131–133, which might be relevant to its proteolytic activation by the anthrax lethal toxin136. The cleaved fragments remain associated with each other but the proteolytic event enables NLRP1 to recruit the adaptor protein ASC and caspase 1. For the other receptors, ligand binding is thought to induce conformational changes, promoting oligomerization of the receptors into signalling platforms that recruit adaptors and activate effector caspases. Oligomerization of APAF1 and NOD-like receptors (NLRs) is mediated by their NACHT domains, which bind and hydrolyse deoxyadenosine triphosphate (dATP) or ATP146,147. However, AIM2 seems to use the multivalent ligand dsDNA as its oligomerization platform130. Both receptor oligomerization and ligand binding might be modulated by the regulatory domains of the receptors, including the WD40 repeats of APAF1, the HIN domain of AIM2 and the leucine-rich repeats (LRRs) of the NLRs. Two examples of the death-inducing platforms are shown in the box figure.
The apoptosome consists of seven APAF1 molecules that are activated by cytochrome c; this then recruits pro-caspase 9 molecules137,148,149 (see figure, part a). Six- or eight-fold symmetry of the apoptosomes has also been observed, which suggests that there are variable oligomerization states137,148,149. Two hypothetical models of the AIM2 inflammasome highlight the positioning of the activating DNA either at the centre (see figure, part b) or the periphery (see figure, part c) of the complex. It is possible that the physiological AIM2 inflammasome may contain characteristics of both models, and the oligomerization states can be variable owing to the sequence-independent nature of the DNA-binding. For clarity, only one caspase dimer is shown for each oligomeric platform. According to a recent model, the pyrin domain (PYD) of ASC can interact with itself in a helical manner to form filamentous structures5, which might mediate ASC pyroptosome formation6.
CARD, caspase activation and recruitment domain.
Regulation by ion fluxes, oxidative state and autophagy
Much of a cell’s energy is used to maintain ion gradients between the cytosol and the extracellular environment. Many enzymes and signalling pathways are regulated by dynamic changes in ion concentrations. In addition to its crucial role in cellular signalling, an intact ion gradient in healthy cells represents a mechanism that protects against cell death.
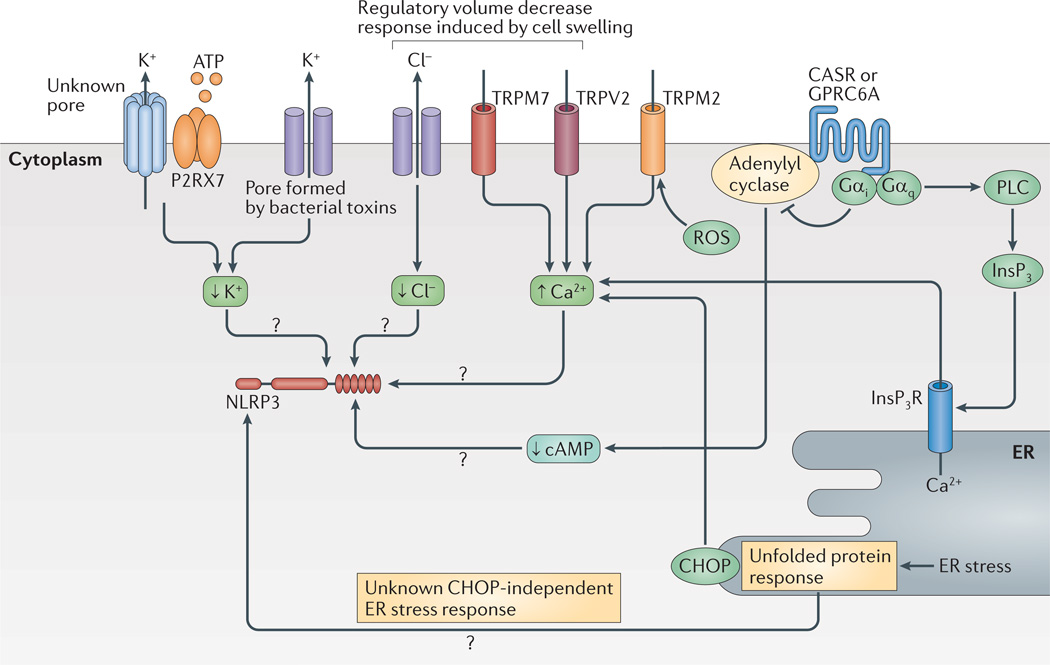
Indeed, the binding of cytochrome c that has been released from mitochondria to apoptotic protease-activating factor 1 (APAF1) and the subsequent assembly of the apoptosome only occurs when subphysiological K+ concentrations are reached in compromised cells77,78. Similarly, activation of the NLRP1B and NLRP3 inflammasomes depends on low K+ concentrations in intracellular compartments79, and a low K+ concentration promotes the assembly of the ASC speck6. In addition, high extracellular levels of K+ can block IL-1β release after NLRC4 and AIM2 inflammasome formation80,81, which indicates that low intracellular K+ levels might also be required for the activation of these inflammasomes. However, the concentrations of K+ that are necessary to block NLRC4 and AIM2 inflammasome formation are higher than those that are needed to block NLRP3 inflammasome formation. Interestingly, for NLRP7, a high extracellular K+ concentration only slightly reduces IL-1β release28. Why some inflammasomes are more sensitive to high extracellular concentrations of K+ than others is not understood. It was thought that the danger signal ATP and bacterial pore-forming toxins, which activate the NLRP3 inflammasome, could directly mediate K+ efflux through the hemichannel pannexin 1 (REFS 82–84); however, pannexin 1-deficient mice do not show diminished caspase 1 activation85. Thus, the mechanisms by which several inflammasome triggers can induce K+ efflux remain unclear.
In addition to the requirement for decreased K+ levels, osmotic pressure regulates NLRP3 inflammasome activation86. Cells that have been subjected to hypotonic solutions undergo cellular swelling concomitant with a decrease in intracellular K+ and Cl− concentrations. Interestingly, cell swelling induces a regulatory volume decrease response through transient receptor potential cation channels (TRPM7 and TRPV2) that triggers intracellular Ca2+ mobilization. The mobilized Ca2+ has many molecular targets, including TGFβ-activated kinase 1 (TAK1; also known as MAP3K7) (REF. 86). Although TAK1 might have a role in a PRR-dependent non-transcriptional priming pathway87 — that is, in the signalling pathway that leads to the deubiquitylation, for example, of NLRP3 — further investigation into its role is required. Another transient receptor potential channel — TRPM2 — has also been implicated in NLRP3 activation in response to crystalline substances88. This channel senses intracellular ROS and responds by opening itself to facilitate Ca2+ influx into the cell; this is intriguing considering that both ion fluxes and the oxidative state (see below) have important roles in NLRP3 inflammasome activation.
Another regulator of Ca2+, namely C/EPB-homologous protein (CHOP; also known as DDIT3), has been implicated in NLRP3 inflammasome activation89. However, the activation of CHOP following the induction of the unfolded protein response via inhibition of translation is insufficient for NLRP3 inflammasome activation90 and, therefore, the exact role of CHOP in NLRP3 inflammasome activation is unclear. Two recent studies have implicated calcium-sensing G-protein coupled receptors (GPCRs) in the activation of NLRP3: calcium-sensing receptor (CASR) and GPRC6A signal via Gαi and Gαq, which inhibit adenylyl cyclase and activate phosphorlipase C, respectively91,92. Thus, sensing of extracellular Ca2+ leads to reduced cyclic AMP (cAMP) levels through adenylyl cyclase inhibition and to increased cytoplasmic Ca2+ concentrations via the activation of phospholipase C. It is worth noting that the GPCR platelet activating factor receptor can also stimulate inflammasome formation92, which indicates that other GPCR pathways could control inflammasome activation. The role of cAMP in inflammasome activation remains unclear as one study has reported that cAMP can directly inhibit NLRP3 (REF. 91), whereas another study reported that cAMP levels had no direct influence on inflammasome activation92. Taken together, these studies show that NLRP3 activation is regulated by the ion content of the cell (FIG. 3); however, the molecular targets and the mechanisms by which ion fluxes regulate NLRP3 remain to be fully elucidated.
Figure 3. Ion fluxes and cell stress regulate the NLRP3 inflammasome.
K+ and Cl− efflux, as well as Ca2+ mobilization, have a role in the regulation of NLRP3 (NOD-, LRR- and pyrin domain-containing 3). K+ efflux is either achieved directly by pore-forming toxins such as nigericin or indirectly — for example, via the purinergic receptor for ATP P2RX7. Cl− and Ca2+ ion fluxes can be regulated during the regulatory volume decrease response by transient receptor potential receptors (TRPV2 or TRPM7). In addition, reactive oxygen species (ROS) can activate TRPM2 for Ca2+ influx. Ca2+ is also regulated by the unfolded protein response via C/EPB-homologous protein (CHOP). The unfolded protein response is also implicated in other pathways that regulate NLRP3. G-protein coupled receptors (GPCRs) such as calcium-sensing receptor (CASR) and GPRC6A regulate both Ca2+ levels and cyclic AMP (cAMP) levels via activation of phospholipase C (PLC) or inhibition of adenylyl cyclase, respectively. High cAMP levels might directly inhibit NLRP3 (not shown). Inositol-1,4,5-trisphosphate (InsP3) that is generated by PLC leads to release of Ca2+ from the endoplasmic reticulum (ER). How Ca2+ influences NLRP3 activation is not fully understood but it has many molecular targets. Question marks show pathways that are still speculative. InsP3R, InsP3 receptor.
The redox state of the cell is another important indicator of the viability of cells, and many signalling pathways are influenced by changes in the redox state. In particular, ROS facilitate the assembly of the apoptosome in several ways. For example, oxidative modifications to caspase 9 enable its recruitment to the apoptosome93. By contrast, nitrosylation of caspase 9 inhibits its function94. Similarly, caspase 1 can be inhibited by nitrosylation95, which suggests that modification by reactive molecules is a general mechanism for the regulation of caspase activity. The exact role of the redox state in NLRP3 inflammasome activation remains controversial. Originally, it was thought that ROS produced by NADPH oxidases following the phagocytosis of crystals activate the NLRP3 inflammasome96–98. However, studies that have been carried out either using cells that were deficient in NADPH protein components or using cells from patients with defective NADPH oxidase subunits have challenged this idea by showing that these patients have normal, or even increased, caspase 1 activity99–101. In addition, ROS are produced by stressed mitochondria and these mitochondria-derived ROS have been implicated in the activation of the NLRP3 inflammasome102. However, inhibitors of ROS that have been used in previous studies can inhibit NLRP3 priming rather than the actual activation of the inflammasome103. As ROS are involved in several signalling pathways, defining the exact roles of ROS in NLRP3 priming and activation remains challenging.
Furthermore, autophagy can regulate both apoptosis and inflammasome assembly, and, depending on the conditions, can be either pro- or anti-apoptotic104. For inflammasome activation and IL-1β release, autophagy is a negative regulator: mice that are deficient in autophagy-related protein 16-like 1 (ATG16L1) — an essential component of the autophagy machinery — show higher IL-1β levels in response to stimulation. This indicates that autophagy limits IL-1β activation or release105. The mechanisms by which autophagy regulates inflammasome activation are still under debate. One hypothesis suggests that autophagy is involved in the removal of ubiquitylated inflammasomes106 or pro-IL-1β molecules107. An alternative or additional mechanism could be that autophagy removes damaged mitochondria, which suggests that autophagy could prevent the release of mitochondrial ROS and DNA into the cytoplasm and thereby limit NLRP3 inflammasome formation102.
These studies show that inflammasomes are tightly regulated by intracellular ion concentrations (FIG. 3), by the redox state of the cell and by the nutritional situation of the cell. It seems that inflammasome activation is regulated at multiple levels and that several requirements need to be met for full inflammasome activation to occur.
Regulation by interacting host proteins
In addition to general cellular conditions such as ion concentrations and the nutritional state, specific regulatory proteins have evolved that regulate inflammasome formation (TABLE 1). The formation of the inflammasome can be controlled both at the level of inflammasome sensor molecules and, further downstream, at the level of ASC and caspase 1 interaction. As mentioned above, inflammasomes form large protein aggregates and there are two levels at which death-fold domains are used to recruit effector inflammasome components. First, ASC is recruited to the inflammasome receptors by homotypic pyrin domain interactions. Next, in a second death-fold domain interaction, ASC binds to and activates pro-caspase 1 via its CARD. Studies have elucidated inflammasome regulation at the level of death-fold domain interactions and regulatory proteins.
Table 1.
Regulatory proteins of the inflammasomes
| Regulatory protein |
Inflammasome component targeted |
Mechanism of action | Sources of experimental evidence | Refs |
|---|---|---|---|---|
| Accessory NLR proteins | ||||
| NOD2 | NLRP1 | Enhances NLRP1 inflammasome formation | Knockout mice and biochemical interaction |
32,33 |
| NAIP2* | NLRC4 | Necessary for recognition of the type III secretion system by the NLRC4 inflammasome |
RNAi and biochemical interaction | 34,35 |
| NAIP5 or NAIP6* |
NLRC4 | Necessary for recognition of flagellin by the NLRC4 inflammasome |
RNAi and biochemical interaction | 34,35 |
| NAIP‡ | NLRC4 | Necessary for recognition of flagellin and/or the type III secretion needle protein by the NLRC4 inflammasome |
RNAi, biochemical interaction and overexpression |
35,36 |
| CARD-containing regulatory proteins | ||||
| Caspase 12 and caspase 12L‡ |
Caspase 1 | Inhibits caspase 1 activity, probably by sequestration | Knockout mice and biochemical Interaction |
110,111 |
| CARD18‡ (also known as ICEBERG) |
Caspase 1 | Inhibits caspase 1 activity, probably by sequestration | Biochemical interaction and Overexpression |
113 |
| CARD16‡ (also known as COP1) | Caspase 1 | Inhibits caspase 1 activity, probably by sequestration | Biochemical interaction and Overexpression |
113 |
| CARD17‡ (also known as INCA) | Caspase 1 | Inhibits caspase 1 activity, probably by sequestration | Biochemical interaction and Overexpression |
114 |
| PYD-containing regulatory proteins | ||||
| POP1‡ | ASC | Inhibits inflammasome assembly, probably by sequestration |
Biochemical interaction and Overexpression |
115 |
| POP2‡ | ASC | Inhibits inflammasome assembly, probably by sequestration |
Biochemical interaction and Overexpression |
116,117 |
| ASC splice variant |
ASC§ or caspase 1§ |
Inhibits inflammasome assembly, probably by sequestration |
Biochemical interaction, co-localization and overexpression |
118 |
| NLRP10§ | ASC§ |
|
119–122 | |
| NLRP7‡§ | ASC§ |
|
28,123 | |
| Pyrin§ | ASC§ or IL-1β§ |
|
124,125 | |
| Other regulators | ||||
| GBP5 | NLRP3 | Enhances NLRP3 inflammasome formation in response to non-crystalline activators |
Knockout mice and biochemical evidence |
39 |
| BRCC3 | NLRP3 | Deubiquitylates NLRP3 as a priming step | RNAi and biochemical evidence | 63 |
| PKCδ | NLRC4 | Phosphorylates NLRC4 as a priming step | Knockout mice and biochemical evidence |
67 |
| BCL-2 | NLRP1 | Inhibits NLRP1 inflammasome activation by inhibiting nucleotide binding |
Knockout mice, biochemical evidence and overexpression |
139,140 |
| BCL-XL | NLRP1 | Inhibits NLRP1 inflammasome activation by inhibiting nucleotide binding |
Biochemical evidence and overexpression |
139,140 |
| IAPs§ | Caspase 1 | Increases the activity of caspase 1 by monoubiquitylation | Knockout mice (for IAP1 or IAP2) and biochemical evidence |
58 |
| IAPs§ | NLRP3 | Loss of IAPs leads to activation of the ripoptosome, which is necessary for caspase 1 activation |
IAP1, IAP2 and XIAP triple-knockout mouse |
56 |
| PKR | NLRP1, NLRC4, AIM2 and NLRP3 |
|
127, 128 | |
| Other regulators (cont.) | ||||
| HSP90 | NLRP3 | Necessary for activation, possibly owing to its chaperone function |
Biochemical evidence and inhibitor of HSP90 |
129 |
| SGT1 | NLRP3 | Necessary for activation, possibly owing to its chaperone function |
RNAi and biochemical evidence | 129 |
AIM2, absent in melanoma 2; BCL, B cell lymphoma; CARD, caspase activation and recruitment domain; FMF, familial Mediterranean fever; GBP5, guanylate-binding protein 5; HSP90, heat shock protein 90; IAP, inhibitor of apoptosis protein; IL-1β, interleukin-1β; NAIP, NLR family, apoptosis inhibitory protein; NLR, NOD-like receptor; NLRC4, NOD-, LRR- and CARD-containing 4; NLRP, NOD-, LRR- and pyrin domain-containing; NOD2, nucleotide-binding oligomerization domain-containing protein 2; PKCδ, protein kinase Cδ; PKR, protein kinase R; POP, pyrin domain-only protein; PYD, pyrin domain; RNAi, RNA interference; XIAP, X-linked IAP.
Expressed only in mice.
Expressed only in humans.
Two contradictory studies have been published about this interactor.
Caspase activity during cell death is regulated by CARD-containing proteins such as CARD8, which has also been shown to be a binding partner of NLRP3 (REF. 108). CARD8 has multiple functions in regulating apoptosis, one of which is to directly bind to pro- caspase 9 and to suppress its activation109. Similarly, mouse caspase 12, which is a paralogue of caspase 1, interacts with caspase 1 to reduce its activity110. A polymorphism in the caspase 12 (CASP12) gene in humans leads to either a truncated protein or to a full-length protein (caspase 12L)111. Similarly to mouse caspase 12, caspase 12L reduces cytokine production in response to LPS111, which — at high doses — can activate the NLRP3 inflammasome and thus caspase 1 without the need for a second signal. Therefore, caspase 12L and mouse caspase 12 probably function as decoy proteins for caspase 1, thereby limiting its activation. CARD-only proteins (COPs) can also inhibit caspase 1 activation112. CARD18 (also known as ICEBERG) and CARD16 (also known as pseudo-ICE and COP1) were the first ‘decoy’ COPs to be described113. Through their CARD–CARD interaction, these two inhibitors sequester pro-caspase 1 and inhibit its activation by the inflammasome. CARD17 (also known as INCA), which is another decoy protein, is upregulated by IFNγ to suppress IL-1β generation114. Thus, proteins containing a CARD can sequester caspase 1, thereby blocking the formation of a functional inflammasome complex.
An additional level of fine-tuning and regulation of the inflammasomes can be achieved by interfering with the pyrin–pyrin interaction in the inflammasomes. Several pyrin domain-only proteins (POPs) and other pyrin-containing proteins have been characterized. POP1 (also known as PYDC1 and ASC2) and POP2 (also known as PYDC2) inhibit the interactions between the inflammasome sensor molecules and ASC115–117. However, COPs and POPs have not been identified in the mouse genome, which indicates that a more complex regulatory system might be present in humans. The fact that COPs and POPs have not been identified in the mouse genome makes the study of these proteins difficult, and many of the COPs and POPs have only been investigated using overexpression studies and not using genetic deletion or RNA interference.
Alternative splicing of ASC can provide an additional level of regulation by giving rise to ASC variants that negatively regulate the assembly of the inflammasomes118. Furthermore, NLRP10 (also known as PYNOD) was suggested to negatively regulate inflammasomes by sequestering ASC119,120, but deletion of Nlrp10 in mice did not confirm this hypothesis121,122. NLRP7 has been suggested to be a negative regulator of inflammasomes; however, more recent findings indicate that NLRP7 might assemble an inflammasome in response to microbial acylated lipopeptides28,123.
The role of pyrin, which is a protein that is encoded by MEFV (Mediterranean fever gene), in inflammasome regulation also remains unclear. One study found that Mefv deletion in mice lead to increased IL-1β release without influencing caspase 1 activity or inflammasome assembly. These findings suggested that pyrin can inhibit IL-1β release downstream of the inflammasomes124. However, another study found that short-term IL-1β release was not impaired after Mefv gene deletion and that IL-1β was only inhibited after several days of stimulation125. In addition, mice carrying Mefv mutations that have been identified in patients with familial Mediterranean fever (FMF) showed ASC-dependent but NLRP3-independent release of IL-1β125. Therefore, whether pyrin is an inhibitor at the level of IL-1β release or whether it forms a ‘pyrin inflammasome’ remains to be determined.
In addition to death domain interactions targeting the formation of the inflammasome complex, regulation can occur at the level of the inflammasome sensor molecules. A recent report linked the inhibition of translation with NLRP3 inflammasome activation126. Protein synthesis could be inhibited by the phosphorylation of the initiation factor eukaryotic translation initiation factor 2 subunit-α (EIF2α; also known as EIF2S1). This phosphorylation event could be mediated by different kinases that sense endoplasmic reticulum (ER) stress, or by the presence of heavy metals or double-stranded RNA (dsRNA). Protein kinase R (PKR; also known as EIF2AK), which is activated by dsRNA and can phosphorylate EIF2α, has been suggested to be involved in NLRP3, NLRP1, NLRC4 and AIM2 inflammasome activation, and this was found to be dependent on its kinase activity127. However, another study concluded that PKR does not have any role in inflammasome activation128. The reasons for these seemingly contradictory results could be attributed to differences in mouse strain backgrounds and will need to be further evaluated. In another study, ER stress was shown to activate the NLRP3 inflammasome90 and, although the classical unfolded protein response was shown to activate EIF2α phosphorylation (consistent with the above observations), this pathway was not found to be necessary90. The authors proposed that another, not yet characterized, ER stress pathway activates the NLRP3 inflammasome90. Finally, heat shock proteins (HSPs) also have important roles in the regulation of cell death, and it has been shown that HSP90 and the co-chaperone ubiquitin ligase-associated protein SGT1 are required for NLRP3 activation (REF. 129).
Taken together, these studies show that the activation of inflammasomes can be regulated at different levels and that many proteins contribute to the overall response of these important signalling platforms. Elucidating the exact function of the different components seems to be challenging. Overexpression could considerably perturb the finely balanced thresholds of inflammasome activation, and it is possible that inflammasomes respond differently depending on culture conditions and/or genetic background.
Conclusion and future perspectives
In this Review, we summarize our rapidly evolving understanding of how inflammasomes are activated and we highlight the many mechanisms that are involved in their regulation. Although we have gained detailed knowledge about many aspects of inflammasome activation, several important questions remain unanswered or poorly understood.
One main conceptual question that remains unanswered is whether inflammasome sensors can directly interact with their triggers and, thus, whether they represent true receptor molecules. AIM2 can tightly interact with DNA and has recently been crystallized with its ligand; it therefore qualifies as a receptor molecule130. The NLR inflammasome sensors all have LRR domains that, for example, in TLRs, directly recognize lipids, nucleic acids or proteins. However, whether the NLR inflammasome sensor molecules can directly recognize their various triggers or whether they use accessory host proteins for this process remains a matter of debate. In the case of NLRC4, additional host molecules (that is, NAIPs) have been suggested to function as the direct receptors for the ligands34,35. Therefore, NLR inflammasome sensors could, in fact, function as adaptor molecules rather than receptors.
In addition, post-translational modifications of NLRs, such as phosphorylation67, ubiquitylation62,63 and even proteolysis131–133, have been suggested to be necessary for the activation of certain NLR sensors. Therefore, the modification of NLRs by host enzymes could be crucial for their activation and, at the same time, could represent a novel target for pharmacological intervention strategies. Future research should focus on the identification of host molecules that function upstream of the NLRs. In the case of NLRP3, the elucidation of the unknown factors that influence its activation represents a particular challenge, as these factors could influence both the priming and activation events. Therefore, combinatorial proteomic and genomic approaches, as well as overexpression systems in which priming is not required, could be instrumental.
Another important challenge will be to more precisely define the contribution of inflammasomes to the inflammatory response in vivo. The ability of a particular trigger to activate inflammasomes and IL-1β family cytokines in vitro does not necessarily indicate that inflammasomes fully control the inflammatory response to the respective trigger in vivo. It is becoming increasingly evident that the activation of inflammas-ome-independent IL-1β can substantially contribute to tissue inflammation. In acute inflammatory situations, in which neutrophils have important roles, inflammasomes can be functionally redundant in the activation of IL-1β and thus might only partially contribute to the inflammatory tissue response. Caspase 8, which has recently been implicated in IL-1β activation, could be one of the factors that contributes to inflammasome-independent responses in vivo. As many pathways can trigger caspase 8 activation, the relative contributions of caspase 1 and caspase 8 to IL-1β production and inflammation in vivo need to be carefully investigated in future studies. Furthermore, caspase 8 is known to mediate cell death; hence, it will be important to better understand the consequences of the different forms of cell death in the inflammatory tissue response.
Moreover, in this Review we compare the activation and regulation of inflammasomes to that of the apoptosome, which is another multimolecular caspase-activating platform. We note with interest that, in addition to the way in which the effector proteins of these platforms are tightly controlled, the activity of the triggers can be modulated; for example, cytochrome c can be modified to be more or less pro-apoptotic. Indeed, apoptosome activation by cytochrome c is only possible after cytochrome c has incorporated a haem group in the mitochondrion134. By contrast, nitrosylation of cytochrome c inhibits its ability to induce the formation of the apoptosome135. We predict that similar regulatory mechanisms are in place for the triggers of inflammasomes.
The importance of self-proteolysis or proteolysis by the trigger anthrax lethal toxin in the activation of NLRP1 is still not fully understood131–133,136. It is possible that proteolysis of inflammasome sensor molecules could lead to multiple effects that have not yet been defined. The quest for the exact details of inflammasome activation is ongoing; it seems probable that signals emanating from mitochondria and lysosomes will provide important insights. In addition, we will probably learn from comparing these potential regulatory mechanisms with those that govern apoptosome and ripoptosome activation. The identification of upstream mechanisms and the discrimination between priming and activation could be instrumental in developing novel therapies on the basis of the specific inhibition of individual inflammasomes (BOX 3). Given their broad roles in mediating inflammatory pathologies, inflammasome inhibitors could become effective therapies many prevalent and emerging inflammatory diseases.
Box 3|Pharmacological interference of inflammasome activation.
The fact that the NLRP3 (NOD-, LRR- and pyrin domain-containing 3) inflammasome in particular is under the control of numerous regulatory mechanisms provides several opportunities for therapeutic intervention. It is conceivable that the biological effectiveness of certain anti-inflammatory agents is at least partly due to their ability to interfere with NLRP3 activation. Recently, the mechanism of action of certain anti-inflammatory therapies that have been in use for years has been partly ascribed to the inhibition of NLRP3. For example, the therapeutic application of interferon-β (IFNβ) is effective for most but not all patients with multiple sclerosis. IFN-mediated downregulation of NLRP3 inflammasome responses might be of particular relevance in this central nervous system pathology, which has been linked to inflammasome activation150. Interestingly, a recent study has documented that IFNβ therapy can only reduce the pathology in mouse experimental autoimmune encephalomyelitis models that are dependent on NLRP3, which indicates that IFNβ can actively reduce NLRP3-mediated brain inflammation in vivo151.
In addition, a number of small-molecule compounds have been shown to inhibit NLRP3 activation. For example, glyburide, which is a pharmaceutical compound that is used in the treatment of type 2 diabetes, has been shown to directly inhibit the NLRP3 inflammasome, albeit only at high doses152. Furthermore, the anti-inflammatory compound parthenolide, which is naturally present in the plant Tanacetum parthenium (also known as feverfew) that is used in herbal remedies, and the nuclear factor-κB (NF-κB) inhibitory compound Bay 11–7082 can both interfere with the ATPase activity of NLRP3, thereby hindering inflammasome activation153. The NLRP3 inflammasome is also inhibited by the TGFβ-activated kinase 1 (TAK1) inhibitor 5Z-7-oxozeaenol and its related compounds87. Future investments in the identification of small molecules that target these important intracellular signalling platforms or their upstream regulators could generate a novel class of highly effective anti-inflammatory therapeutics.
Acknowledgements
The authors would like to thank C. M. De Nardo and B. G. Monks for critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (NIH) and the Deutsche Forschungsgemeinschaft (DFG), Germany (to E.L.), and by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, USA (to T.S.X.). E.L. is a member of the excellence cluster ImmunoSensation in Bonn, Germany
Glossary
- ASC
An adaptor protein that was originally found to form protein precipitates in apoptotic cells that are termed protein specks
- Pyroptosis
A rapid form of cell death following caspase 1 activation which shares characteristics with both apoptosis (such as DNA fragmentation) and necrosis (such as cell swelling and rupture)
- Apoptosome
A large multimeric protein complex of apoptotic protease-activating factor 1 (APAF1) that recognizes cytochrome c release from damaged mitochondria and activates caspase 9
- Death-fold domains
Commonly found in proteins that are involved in cell death pathways and in inflammasomes. The four main death-fold domains — the pyrin domain caspase activation and recruitment domain (CARD), death domain and death effector domain — associate with each other through homotypic interactions
- NOD-like receptor
(NLR). A protein that contains amino-terminal pyrin caspase-recruitment domains or other signalling domains, followed by a NACHT domain and carboxy-terminal leucine-rich repeats. Some NLR proteins are involved in forming inflammasomes.
- Non-canonical NLRP3 inflammasome
An inflammasome-like complex containing NLRP3 (NOD-, LRR-and pyrin domain-containing 3), the adaptor protein ASC, caspase 1 and caspase 11. The term ‘non-canonical inflammasome’ is used loosely to describe an inflammasome-like complex that does not conform to the three ‘canonical’ components of a canonical inflammasome: an inflammasome sensor molecule ASC and caspase 1. Two other non-canonical inflammasomes have also been described so far: one containing dectin 1, MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1), ASC and caspase 8 (termed the non-canonical caspase 8 inflammasome), and a caspase 11-activating platform that has not yet been fully described
- Ripoptosome
A cytosolic multiprotein complex that induces cell death following genotoxic stress or depletion of inhibitor of apoptosis protein (IAP). The core ripoptosome contains receptor-interacting protein 1 (RIP1) FAS-associated death domain protein (FADD) and caspase 8, but it can also recruit other proteins such as RIP3
- Amyloid-β
An endogenous peptide that is generated by proteases in the brain. It is prone to , aggregation and plaque formation. Amyloid-β plaques are a hallmark of Alzheimer’s disease and can activate the NLRP3 (NOD- LRR- and pyrin domain-containing 3) inflammasome
- Cryopyrin-associated periodic syndrome
(CAPS). Characterized by NLRP3 (NOD-, LRR- and pyrin domain-containing 3) inflammasome hyperactivity and the excessive release of interleukin-1β, which leads to an autoinflammatory disease phenotype with periodic fever episodes, urticaria and often severe arthritis
- MicroRNA
Small endogenous RNA molecules that can recruit the RNA-induced silencing complex to an mRNA, leading to inhibition of translation or to degradation of the mRNA
- Autophagy
A homeostatic process during which cellular components are recycled through the lysosomal compartment. Its main triggers include nutrient starvation defective organelles and infection
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Eicke Latz’s homepage: http://www.iii.uni-bonn.de/
References
- 1.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 3.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 4.Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol. Rev. 2011;243:206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vajjhala PR, Mirams RE, Hill JM. Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J. Biol. Chem. 2012;287:41732–41743. doi: 10.1074/jbc.M112.381228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proell M, Gerlic M, Mace PD, Reed JC, Riedl SJ. The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem. J. 2013;449:613–621. doi: 10.1042/BJ20121198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 2007;282:36321–36329. doi: 10.1074/jbc.M708182200. [DOI] [PubMed] [Google Scholar]
- 9.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 10.Gu Y, et al. Activation of interferon-γ inducing factor mediated by interleukin-1β converting enzyme. Science. 1997;275:206–209. doi: 10.1126/science.275.5297.206. [DOI] [PubMed] [Google Scholar]
- 11.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 12.Ting JP-Y, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 15.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nature Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 16.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin-1β in Salmonella-infected macrophages. Nature Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 17.Geddes BJ, et al. Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem. Biophys. Res. Commun. 2001;284:77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]
- 18.Hornung V, Latz E. Intracellular DNA recognition. Nature Rev. Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 19.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nature Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 20.Arthur JC, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J. Immunol. 2010;185:4515–4519. doi: 10.4049/jimmunol.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand PK, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath GL, Schrum JE, De Nardo CM, Latz E. Intracellular sensing of microbes and danger signals by the inflammasomes. Immunol. Rev. 2011;243:119–135. doi: 10.1111/j.1600-065X.2011.01050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelaziz DH, Amr K, Amer AO. Nlrc4/Ipaf/ CLAN/CARD12: more than a flagellin sensor. Int. J. Biochem. Cell Biol. 2010;42:789–791. doi: 10.1016/j.biocel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nature Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 28.Khare S, et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–476. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vladimer GI, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu L-C, et al. A NOD2-NALP1 complex mediates caspase-1 -dependent IL-1β secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc. Natl Acad. Sci. USA. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferwerda G, et al. Engagement of NOD2 has a dual effect on proIL-1β mRNA transcription and secretion of bioactive IL-1β. Eur. J. Immunol. 2008;38:184–191. doi: 10.1002/eji.200737103. [DOI] [PubMed] [Google Scholar]
- 34.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510.References 34 and 35 demonstrate that NAIPs are crucial for ligand sensing and for the activation of the NLRC4 inflammasome.
- 36.Vinzing M, et al. NAIP and Ipaf control Legionella pneumophila replication in human cells. J. Immunol. 2008;180:6808–6815. doi: 10.4049/jimmunol.180.10.6808. [DOI] [PubMed] [Google Scholar]
- 37.Bauernfeind FG, et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-α mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141.This study is the first description of a regulator of NLRP3 that discriminates between crystalline and non-crystalline ligands.
- 40.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558.This reference shows the importance of caspase 11 as a cofactor of NLRP3 inflammasome activation in response to Gram-positive bacteria.
- 42.Rathinam VA, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurung P, et al. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J. Biol. Chem. 2012;287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akhter A, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity. 2012;37:35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broz P, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aachoui Y, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broz P, Monack DM. Noncanonical inflammasomes: caspase-11 activation and effector mechanisms. PLoS Pathog. 2013;9:e1003144. doi: 10.1371/journal.ppat.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer-Barber KD, et al. Caspase-1 independent IL-1β production is critical for host resistance to Mycobacterium tuberculosis and does not require TLR signaling in vivo. J. Immunol. 2010;184:3326–3330. doi: 10.4049/jimmunol.0904189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Provoost S, et al. NLRP3/caspase-1-independent IL-1β production mediates diesel exhaust particle-induced pulmonary inflammation. J. Immunol. 2011;187:3331–3337. doi: 10.4049/jimmunol.1004062. [DOI] [PubMed] [Google Scholar]
- 51.Kono H, Orlowski GM, Patel Z, Rock KL. The IL-1-dependent sterile inflammatory response has a substantial caspase-1-independent component that requires cathepsin C. J. Immunol. 2012;189:3734–3740. doi: 10.4049/jimmunol.1200136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross O, et al. Inflammasome activators induce interleukin-1α secretion via distinct pathways with differential requirement for the protease function of caspase-1. Immunity. 2012;36:388–400. doi: 10.1016/j.immuni.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 53.Gringhuis SI, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nature Immunol. 2012;13:246–254. doi: 10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- 54.Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 55.Maelfait J, et al. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1β maturation by caspase-8. J. Exp. Med. 2008;205:1967–1973. doi: 10.1084/jem.20071632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vince JE, et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity. 2012;36:215–227. doi: 10.1016/j.immuni.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 57.Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Labbe K, McIntire CR, Doiron K, Leblanc PM, Saleh M. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity. 2011;35:897–907. doi: 10.1016/j.immuni.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 59.Miwa K, et al. Caspase 1-independent IL-1β release and inflammation induced by the apoptosis inducer Fas ligand. Nature Med. 1998;4:1287–1292. doi: 10.1038/3276. [DOI] [PubMed] [Google Scholar]
- 60.Bossaller L, et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. J. Immunol. 2012;189:5508–5512. doi: 10.4049/jimmunol.1202121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lech M, Avila-Ferrufino A, Skuginna V, Susanti HE, Anders HJ. Quantitative expression of RIG-like helicase, NOD-like receptor and inflammasome-related mRNAs in humans and mice. Int. Immunol. 2010;22:717–728. doi: 10.1093/intimm/dxq058. [DOI] [PubMed] [Google Scholar]
- 62.Juliana C, et al. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Schroder K, et al. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 2012;217:1325–1329. doi: 10.1016/j.imbio.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 65.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 66.DeYoung KL, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 67. Qu Y, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012;490:539–542. doi: 10.1038/nature11429.This study decribes the importance of a pathogen-induced post-translational modification event for the activation of NLRC4.
- 68.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nature Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson JP, et al. Initial description of the human NLRP3 promoter. Genes Immun. 2008;9:721–726. doi: 10.1038/gene.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guarda G, et al. T cells dampen innate immune responses through inhibition of NLRP1 and NLRP3 inflammasomes. Nature. 2009;460:269–273. doi: 10.1038/nature08100. [DOI] [PubMed] [Google Scholar]
- 71.Mishra BB, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nature Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Hernandez-Cuellar E, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J. Immunol. 2012;189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 74.Haneklaus M, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 75.Bauernfeind F, et al. NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 76.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cain K, Langlais C, Sun XM, Brown DG, Cohen GM. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J. Biol. Chem. 2001;276:41985–41990. doi: 10.1074/jbc.M107419200. [DOI] [PubMed] [Google Scholar]
- 78.Purring-Koch C, McLendon G. Cytochrome c binding to Apaf-1: the effects of dATP and ionic strength. Proc. Natl Acad. Sci. USA. 2000;97:11928–11931. doi: 10.1073/pnas.220416197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pétrilli V, et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195.This study describes the role of K+ in NLRP1 and NLRP3 activation.
- 80.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arlehamn CS, Petrilli V, Gross O, Tschopp J, Evans TJ. The role of potassium in inflammasome activation by bacteria. J. Biol. Chem. 2010;285:10508–10518. doi: 10.1074/jbc.M109.067298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pelegrin P, Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1β release through a dye uptake-independent pathway. J. Biol. Chem. 2007;282:2386–2394. doi: 10.1074/jbc.M610351200. [DOI] [PubMed] [Google Scholar]
- 83.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1β release by the ATP-gated P2×7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Locovei S, Wang J, Dahl G. Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 2006;580:239–244. doi: 10.1016/j.febslet.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 85.Qu Y, et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J. Immunol. 2011;186:6553–6561. doi: 10.4049/jimmunol.1100478. [DOI] [PubMed] [Google Scholar]
- 86.Compan V, et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity. 2012;37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 87.Gong YN, et al. Chemical probing reveals insights into the signaling mechanism of inflammasome activation. Cell Res. 2010;20:1289–1305. doi: 10.1038/cr.2010.135. [DOI] [PubMed] [Google Scholar]
- 88. Zhong Z, et al. TRPM2 links oxidative stress to NLRP3 inflammasome activation. Nature Commun. 2013;4:1611. doi: 10.1038/ncomms2608.This study links Ca2+ mobilization to oxidative stress, both of which are important factors in NLRP3 activation.
- 89.Murakami T, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl Acad. Sci. USA. 2012;109:11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Menu P, et al. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis. 2012;3:e261. doi: 10.1038/cddis.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee GS, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rossol M, et al. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nature Commun. 2012;3:1329. doi: 10.1038/ncomms2339.References 91 and 92 describe that GPCRs, which sense extracellular Ca2+, activate the NLRP3 inflammasome by activating phospholipase C and by inhibiting adenylyl cyclase.
- 93.Zuo Y, et al. Oxidative modification of caspase-9 facilitates its activation via disulfide-mediated interaction with Apaf-1. Cell Res. 2009;19:449–457. doi: 10.1038/cr.2009.19. [DOI] [PubMed] [Google Scholar]
- 94.Zech B, Kohl R, von Knethen A, Brune B. Nitric oxide donors inhibit formation of the Apaf-1/ caspase-9 apoptosome and activation of caspases. Biochem. J. 2003;371:1055–1064. doi: 10.1042/BJ20021720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim YM, Talanian RV, Li J, Billiar TR. Nitric oxide prevents IL-1β and IFN-γ-inducing factor (IL-18) release from macrophages by inhibiting caspase-1 (IL-1β-converting enzyme) J. Immunol. 1998;161:4122–4128. [PubMed] [Google Scholar]
- 96.Cruz CM, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc. Natl Acad. Sci. USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van de Veerdonk FL, et al. Reactive oxygen species-independent activation of the IL-1β inflammasome in cells from patients with chronic granulomatous disease. Proc. Natl Acad. Sci. USA. 2010;107:3030–3033. doi: 10.1073/pnas.0914795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meissner F, et al. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–1573. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Bruggen R, et al. Human NLRP3 inflammasome activation is Nox1–4 independent. Blood. 2010;115:5398–5400. doi: 10.1182/blood-2009-10-250803. [DOI] [PubMed] [Google Scholar]
- 102.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 103.Bauernfeind F, et al. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 2011;187:613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383.This is the first study to describe that autophagy limits IL-1β production.
- 106.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nature Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harris J, et al. Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J. Biol. Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Agostini L, et al. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle–Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 109.Pathan N, et al. TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J. Biol. Chem. 2001;276:32220–32229. doi: 10.1074/jbc.M100433200. [DOI] [PubMed] [Google Scholar]
- 110.Saleh M, et al. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 2006;440:1064–1068. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- 111.Saleh M, et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 112.Stehlik C, Dorfleutner A. COPs and POPs: modulators of inflammasome activity. J. Immunol. 2007;179:7993–7998. doi: 10.4049/jimmunol.179.12.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1β generation by pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- 114.Lamkanfi M, et al. INCA, a novel human caspase recruitment domain protein that inhibits interleukin-1β generation. J. Biol. Chem. 2004;279:51729–51738. doi: 10.1074/jbc.M407891200. [DOI] [PubMed] [Google Scholar]
- 115.Stehlik C, et al. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-κB and pro-caspase-1 regulation. Biochem. J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dorfleutner A, et al. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect. Immun. 2007;75:1484–1492. doi: 10.1128/IAI.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-κB and disrupts ASC:CLR interactions. J. Immunol. 2007;178:3837–3845. doi: 10.4049/jimmunol.178.6.3837. [DOI] [PubMed] [Google Scholar]
- 118.Bryan NB, et al. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J. Inflamm. 2010;7:23. doi: 10.1186/1476-9255-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Imamura R, et al. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J. Immunol. 2010;184:5874–5884. doi: 10.4049/jimmunol.0900779. [DOI] [PubMed] [Google Scholar]
- 120.Wang Y, et al. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int. Immunol. 2004;16:777–786. doi: 10.1093/intimm/dxh081. [DOI] [PubMed] [Google Scholar]
- 121.Eisenbarth SC, et al. NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells. Nature. 2012;484:510–513. doi: 10.1038/nature11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Joly S, et al. Cutting edge: Nlrp10 is essential for protective antifungal adaptive immunity against Candida albicans. J. Immunol. 2012;189:4713–4717. doi: 10.4049/jimmunol.1201715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1β secretion. J. Biol. Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- 124.Hesker PR, Nguyen M, Kovarova M, Ting JP, Koller BH. Genetic loss of murine pyrin, the familial Mediterranean fever protein, increases interleukin-1β levels. PLoS ONE. 2012;7:e51105. doi: 10.1371/journal.pone.0051105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chae JJ, et al. Gain-of-function pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vyleta ML, Wong J, Magun BE. Suppression of ribosomal function triggers innate immune signaling through activation of the NLRP3 inflammasome. PLoS ONE. 2012;7:e36044. doi: 10.1371/journal.pone.0036044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He Y, Franchi L, Nunez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur. J. Immunol. 2013;43:1147–1152. doi: 10.1002/eji.201243187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nature Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- 130. Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014.This is the first crystal structure of an inflammasome sensor with its ligand.
- 131.D’Osualdo A, et al. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS ONE. 2011;6:e27396. doi: 10.1371/journal.pone.0027396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frew BC, Joag VR, Mogridge J. Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 2012;8:e1002659. doi: 10.1371/journal.ppat.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finger JN, et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 2012;287:25030–25037. doi: 10.1074/jbc.M112.378323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martin AG, Fearnhead HO. Apocytochrome c blocks caspase-9 activation and Bax-induced apoptosis. J. Biol. Chem. 2002;277:50834–50841. doi: 10.1074/jbc.M209369200. [DOI] [PubMed] [Google Scholar]
- 135.Nakagawa H, et al. Nitration of specific tyrosine residues of cytochrome c is associated with caspase-cascade inactivation. Biol. Pharm. Bull. 2007;30:15–20. doi: 10.1248/bpb.30.15. [DOI] [PubMed] [Google Scholar]
- 136.Levinsohn JL, et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Acehan D, et al. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 138.Llambi F, Green DR. Apoptosis and oncogenesis: give and take in the BCL-2 family. Curr. Opin. Genet. Dev. 2011;21:12–20. doi: 10.1016/j.gde.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bruey JM, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 140.Faustin B, et al. Mechanism of Bcl-2 and Bcl-XL inhibition of NLRP1 inflammasome: loop domain-dependent suppression of ATP binding and oligomerization. Proc. Natl Acad. Sci. USA. 2009;106:3935–3940. doi: 10.1073/pnas.0809414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. J. Biol. Chem. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Misawa T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nature Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 144.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl Acad. Sci. USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Duncan JA, et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl Acad. Sci. USA. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yuan S, et al. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qi S, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 150.Jha S, et al. The inflammasome sensor, NLRP3, regulates CNS inflammation and demyelination via caspase-1 and interleukin-18. J. Neurosci. 2010;30:15811–15820. doi: 10.1523/JNEUROSCI.4088-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Inoue M, et al. Interferon-β therapy against EAE is effective only when development of the disease depends on the NLRP3 inflammasome. Sci. Signal. 2012;5 doi: 10.1126/scisignal.2002767. ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lamkanfi M, et al. Glyburide inhibits the cryopyrin/ Nalp3 inflammasome. J. Cell Biol. 2009;187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Juliana C, et al. Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J. Biol. Chem. 2010;285:9792–9802. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]