Abstract
Purpose.
The role of microRNA (miRNA) regulation in corneal wound healing and scar formation has yet to be elucidated. This study analyzed the miRNA expression pattern involved in corneal wound healing and focused on the effect of miR-133b on expression of several profibrotic genes.
Methods.
Laser-ablated mouse corneas were collected at 0 and 30 minutes and 2 days. Ribonucleic acid was collected from corneas and analyzed using cell differentiation and development miRNA PCR arrays. Luciferase assay was used to determine whether miR-133b targeted the 3′ untranslated region (UTR) of transforming growth factor β1 (TGFβ1) and connective tissue growth factor (CTGF) in rabbit corneal fibroblasts (RbCF). Quantitative real-time PCR (qRT-PCR) and Western blots were used to determine the effect of miR-133b on CTGF, smooth muscle actin (SMA), and collagen (COL1A1) in RbCF. Migration assay was used to determine the effect of miR-133b on RbCF migration.
Results.
At day 2, 37 of 86 miRNAs had substantial expression fold changes. miR-133b had the greatest fold decrease at −14.33. Pre-miR-133b targeted the 3′ UTR of CTGF and caused a significant decrease of 38% (P < 0.01). Transforming growth factor β1–treated RbCF had a significant decrease of miR-133b of 49% (P < 0.01), whereas CTGF, SMA, and COL1A1 had significant increases of 20%, 54%, and 37% (P < 0.01), respectively. The RbCF treated with TGFβ1 and pre-miR133b showed significant decreases in expression of CTGF, SMA, and COL1A1 of 30%, 37%, and 28% (P < 0.01), respectively. Finally, there was significant decrease in migration of miR-133b–treated RbCF.
Conclusions.
Significant changes occur in key miRNAs during early corneal wound healing, suggesting novel miRNA targets to reduce scar formation.
Keywords: CTGF, microRNA, corneal wound healing, gene expression
Substantial changes occurred in levels of 37 miRNAs in mouse corneas 2 days following excimer ablation, including a 14-fold decrease in miR-133b. Addition of pre-miR-133b suppressed expression of CTGF, COL1A1, and SMA in RbCF cultures.
Introduction
After corneal trauma, stromal wound healing is the result of a complex cascade of multiple factors including growth factors, cytokines, chemokines, proteases, and, most recently discovered, microRNAs (miRNAs). Directly after epithelial damage, the process of healing is initiated by multiple cytokines and growth factors released by the epithelial cells, keratocytes/corneal fibroblast, and/or the lacrimal gland.1–7 Keratocytes in normal, noninjured corneal stromal tissue are often described as being quiescent because synthesis of proteins, DNA, and RNA is very low. Following injury, quiescent keratocytes rapidly transform and become “activated keratocytes” (also termed activated fibroblasts or corneal fibroblasts), which increase DNA, RNA, and protein. Specifically, using microarrays to evaluate changes in expression levels of RNA, it was reported that expression levels of 5885 genes changed after the first 12 days of corneal wound healing in rabbits after photorefractive keratectomy (PRK).8
Activated keratocytes play major roles in repairing corneal tissue after injury. These activated keratocytes are identified approximately 6 hours postinjury by their increases of cell size and organelle content. Twelve to 24 hours after epithelial injury, activated keratocytes begin proliferating and migrating.9 The proliferating keratocytes give rise to activated keratocytes, fibroblasts, and myofibroblasts that repopulate the depleted stroma.10–14 One to 2 weeks after injury, keratocytes transform into myofibroblasts under the influence of transforming growth factor β (TGFβ) in the anterior stroma.15,16 Myofibroblasts are characterized by their high concentration of alpha smooth muscle actin (SMA). Myofibroblasts secrete many growth factors, including TGFβ, and have elevated expression of cadherins and TGFβ receptors.17
Corneal scarring, which is described clinically as corneal haze, is a major cause of impaired vision. Recently, the use of in vivo confocal microscopy revealed that the major reflective (light scattering) structures were actually activated fibroblasts and myofibroblasts in the wound area.18–22 Furthermore, the highly reflective property of the activated fibroblasts was shown to be due to the loss of corneal crystallin proteins that are present in high levels in the cytoplasm of quiescent, transparent keratocytes.20
The TGFβ system is an extremely powerful scar-promoting system in the cornea and other tissues. The fibrotic action of TGFβ is mediated by the upregulation of connective tissue growth factor (CTGF), which is a 38-kDa secreted, cysteine-rich peptide that stimulates the synthesis of collagen (COLA1A), SMA, and proliferation of fibroblasts.23–26 In pathological fibrosis, such as biliary fibrosis, sclerotic skin fibroblasts, corneal scar tissue, atherosclerotic blood vessels, and inflammatory bowel disease, there are increased levels of CTGF.27–30 Expression of CTGF and the CTGF-mediated effect of TGFβ induction of COLA1A synthesis by corneal fibroblasts increased significantly during corneal wound healing.31 Several experiments have demonstrated that TGFβ mediates CTGF; but the mechanism is not fully understood, and microRNAs may play an important role in the regulation of CTGF expression.
MicroRNAs are short RNA sequences, 20 to 23 nucleotides long, that have been identified as molecules participating in a novel mechanism that regulates gene expression at the translational and transcriptional levels.32,33 In most instances, a miRNA inhibits translation of the target gene by causing direct degradation of the target mRNA. Species encode several miRNAs with identical or similar mature sequences. Specifically, there are three known genes for miR-133 in mice: miR-133a-1, miR-133a-2, and miR-133b, found on chromosomes 2, 18, and 1, respectively.34 MicroRNAs involved in eye development of mice are widely documented (in the public domain) at www.mirneye.tigem.it, which is a database that shows the expression of over 200 miRNAs35; but miRNAs involved in corneal wound healing have not been explored.
In this study, the regulation of corneal wound healing was analyzed by examining the changes in expression of several miRNAs after excimer laser ablation of mouse corneas through use of a miRNA PCR Array. miR-133b had the greatest decrease in expression 2 days after ablation, and miR-133b was able to target CTGF and mediate the expression of the downstream mediators, SMA and COL1A1.
Materials and Methods
Animal Model
Adult male C57Black6 mice were used for this study, and the procedure was performed in accordance to the animal care guidelines published by the Institute for Laboratory Animal Research (Guide for the Care and Use of Laboratory Animals) and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Briefly, mice were anesthetized using avertin, and corneas were locally anesthetized using proparacaine eye drops. Both eyes of each mouse were ablated to a depth of 45 μm with a Nidek EC-5000 excimer laser (Nidek, Fremont, CA), creating a 2.0-mm-diameter central epithelium-to-stroma injury. The ablation conditions were specifically designed to remove all of the corneal epithelial cell layers and some of the stroma to simulate PRK. The animals were killed at 0 and 30 minutes and 2 days after excimer laser ablation. Three mice were ablated for a total of six corneas per time point.
Isolation of miRNAs and RT2 miRNA PCR Array
Whole corneas were collected at 0 and 30 minutes and 2 days postablation, and the six corneas were combined for each time point. Total RNA enriched with miRNA was extracted using MiRNeasy Mini Kit (Qiagen, Hilden, Germany). A NanoDrop spectrophotometer (ND-1000 Spectrophotometer; NanoDrop Technologies, Wilmington, DE) was used to quantify and assess the quality of the RNAs. First-strand reverse transcription was performed with the RT2 miRNA First Strand Kit (Qiagen) following the manufacturer's protocol. To examine the miRNA profile, cDNA was prepared according to the specifications of the cell differentiation and development RT2 miRNA PCR Array (Qiagen). This PCR array has 88 predefined miRNA genes that are reported to be involved in cell differentiation and development. RT2 miRNA PCR Array was performed using an Applied Biosystems 7300 Detection System (Applied Biosystems, Foster City, CA) per manufacturer instructions. Gene profiling and data analysis were performed using the mirScript miRNA PCR Array Data Analysis software provided by Qiagen.
The RT2 miRNA PCR Array was used to screen for miRNAs that appeared to have substantial changes in levels at 30 minutes or 2 days following excimer laser ablation. Four replicate samples of each of the three pooled samples were analyzed using the RT2 miRNA PCR Array; the average Ct values were calculated, and the miRNAs with levels that changed at least 2-fold at 30 minutes or 2 days compared to nonablated corneas were identified. The average Ct value for each miRNA probe can be considered the “biological average” for the six corneas in the pooled sample. Candidate miRNAs identified by this screening analysis were then selected for further assessment for effects on CTGF gene expression.
Isolation of Primary Rabbit Corneal Fibroblast
Cultures of rabbit corneal fibroblasts (RbCF) were established by outgrowth from corneal explants as described previously.36 Briefly, epithelial and endothelial cells were removed from corneas, and the stroma was cut into cubes of approximately 1 mm3. The isolated primary RbCF cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS until reaching confluence, with a change of media every 2 to 3 days. All supplies for isolation and culturing of these cells were purchased from Invitrogen (Carlsbad, CA) and Fisher Scientific (Atlanta, GA).
Gain or Loss of Function of miR-133b
Rabbit corneal fibroblast cells were seeded at a cell density of 3.5 × 104/well in six-well plates and at subconfluence were transfected with 50 nM pre-miR-133b, anti-miR-133b, pre-miR negative (pre-NC), or anti-miR negative control (anti-NC) (Applied Biosystems) for 72 to 96 hours using PureFection transfection reagent (System Biosciences, Inc., Mountain View, CA) according to the manufacturer's protocol.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from rat corneal tissues and cell cultures using Trizol (Invitrogen). The quantity and quality of the isolated RNAs were determined (ND-1000 Spectrophotometer; NanoDrop Technologies), and 10 ng (for miRNA) or 2 μg was reverse transcribed using specific stem-loop primer for miR-133b or random primers for CTGF and SMA according to the manufacturer's guidelines (Applied Biosystems). Quantitative RT-PCR was carried out using TaqMan or SYBR green expression master mix, TaqMan miRNA (for miR-133b and RNU6B), or TaqMan gene expression assays (for COL1A1) (Applied Biosystems). Following the manufacturer's instructions, reactions were incubated for 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C, and level of mRNA and miRNA expression was determined using Applied Biosystems 7300 Detection System with 18S and RNU6B used for normalization, respectively. Ct values of the target genes (CTGF, COL1A1, SMA) and mir-133b amplicon products were always less than 30 cycles, which indicates that the target nucleic acid sequences were abundant in the sample. All reactions were run in triplicate, and relative expression was analyzed with the comparative cycle threshold method (2−ΔΔCT) as described by the manufacturer (Applied Biosystems). The primer sequences used in the SYBR system for amplification of 18S were sense, 5′-GACGGACCAGAGCGAAAGC-3′, and antisense, 5′-CCTCCGACTTTCGTTCTTGATT-3′, respectively. The primers and probes used for amplification of CTGF and SMA in TaqMan system are as follows. For CTGF: sense, 5′-AGGAGTGGGTGTGTGATGAG-3′; antisense, 5′-CCAAATGTGTCTTCCAGTCG-3′, and probe, 5′-ACCACACCGTGGTTGGCCCT-3′. For SMA: sense, 5′-AGAGCGCAAATACTCCGTCT-3′; antisense, 5′-CCTGTTTGCTGATCCACATC-3′, and probe, 5′-CGGCTCCATCCTGGCCTCTC-3′.
Luciferase Reporter Assay
Rabbit corneal fibroblast cells were seeded at a cell density of 3.5 × 104/well in six-well plates and cultured until reaching subconfluence. The cells were then transiently cotransfected with pre-miR-133b or pre-NC at the above concentrations and luciferase reporter plasmid (1 μg/well) containing 3′ untranslated region (UTR) sequences for CTGF and TGFβ1, respectively (GeneCopoeia, Inc., Rockville, MD), using PureFection transfection reagent. Firefly and Renilla luciferase activities were measured after 48 hours of transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity, and the level of induction was reported as the mean ± SEM of three experiments performed in duplicates and compared with a ratio in cells transfected with pre-NC independently set at 1.
Immunoblotting
Total protein isolated from cells transfected with 50 nM pre-miR-133b, anti-miR-133b, pre-NC, or anti-NC for 96 hours was subjected to immunoblotting as previously described.37 Antibody against CTGF (Santa Cruz Biotechnology, Santa Cruz, CA) was used, and the membranes were also stripped and probed with α-tubulin antibody (Abcam, Inc., Cambridge, MA) serving as loading control. The band densities were determined using ImageJ (in the public domain at http://imagej.nih.gov/ij/) and normalized to α-tubulin accordingly, and their ratios were calculated by using the values in control groups as 1. At least five biological replicates of the experiment were performed to confirm consistency and statistical significance of the relative protein expression determined by the Western blots.
Cell Migration/Wound Healing Assay
The cell migration activity was determined using Radius 24-well assay kit (Cell Biolabs, San Diego, CA) consisting of a circular biocompatible gel in each well according to the manufacturer's instructions. Briefly, RbCF cells were seeded in the assay plates and cultured for 48 hours and then transfected with pre-miR-133b and pre-NC as described above. After 48 hours of incubation, the biocompatible gels were removed and the cells incubated for an additional 22 hours; the images of migratory cells were captured using an Olympus IX70 microscope equipped with digital camera (Olympus, Inc., Melville, NY).
Statistical Analysis
Whenever appropriate, the results were reported as mean ± SEM. Comparisons between two groups or among groups were made using unpaired nonparametric Student's t-test and analysis of variance (ANOVA) followed by Tukey's HSD post hoc multiple comparison, respectively. A P value < 0.05 was considered statistically significant.
Results
Signature of miRNAs Throughout Mouse Corneal Wound Healing at 0 and 30 Minutes and 2 Days
The miRNAs involved in mouse corneal wound healing after ablation were determined by using a miRNA PCR Array (Qiagen), which analyzes 86 different miRNAs at the same time. When compared to the 0-hour time point (no ablation), 30 minutes after ablation only 20 of 86 miRNAs had a substantial (greater than 2-fold) fold change (Table). At 30 minutes postablation, miR-106b had the greatest fold decrease at −6.34, and miR-22 had the greatest fold increase of 18.32. Two days after ablation, 37 of 86 miRNAs had a substantial (greater than 2-fold) fold change. At 2 days postablation, miR-133b had the greatest fold decrease at −14.33, and miR-22 had the greatest fold increase of 7.16.
Table. .
Relative Expression Levels of miRNAs in Ablated Mouse Corneas Compared to Normal Corneas
|
miRNA |
30 Minutes |
2 Days |
| mmu-miR-106b | −6.34 | 1.16 |
| mmu-miR-125b-5p | −1.25 | −2.14 |
| mmu-miR-122 | −1.08 | 4.26 |
| mmu-miR-126-3p | −1.26 | −3.6 |
| mmu-miR-22 | 18.32 | 7.16 |
| mmu-miR-92a | 2.31 | 3.15 |
| mmu-miR-10a | −2.63 | −2.12 |
| mmu-miR-429 | 2.63 | 3.79 |
| mmu-miR-1a | −1.57 | −9.99 |
| mmu-miR-140 | −1.97 | −4.44 |
| mmu-miR-146b | 1.08 | 2.18 |
| mmu-miR-196a | 2.57 | −7.22 |
| mmu-miR-488 | −2.05 | −3.8 |
| mmu-miR-24 | 2.04 | −1.38 |
| mmu-miR-18a | −1.09 | 3.65 |
| mmu-miR-9 | 1.39 | −2.21 |
| mmu-miR-134 | −1.15 | −2.07 |
| mmu-let-7i | −2.23 | −2.28 |
| mmu-miR-222 | 1.47 | 2.29 |
| mmu-miR-99a | −1.38 | −2.12 |
| mmu-miR-21 | −1.37 | 3.36 |
| mmu-miR-451 | −1.98 | −6.08 |
| mmu-let-7c | −1.79 | −2.76 |
| mmu-miR-101b | −1.41 | −2.68 |
| mmu-miR-129-5p | 2.35 | −1.08 |
| mmu-miR-223 | 2.39 | 4.11 |
| mmu-miR-503 | −2.44 | 1.17 |
| mmu-miR-218 | 1.89 | 2.25 |
| mmu-let-7d | 1.1 | −2.02 |
| mmu-miR-10b | 2.04 | −2.19 |
| mmu-miR-133b | −1.61 | −14.33 |
| mmu-miR-144 | −1.92 | −4 |
| mmu-miR-33 | −1.72 | −3.03 |
| mmu-miR-124 | 1.31 | −2.69 |
| mmu-miR-150 | −1.13 | −3.83 |
| mmu-miR-130a | 1.02 | 2.01 |
| mmu-miR-17 | −2.11 | 1.1 |
| mmu-let-7b | −1.12 | −1.96 |
| mmu-miR-541 | −2.55 | −5 |
| mmu-let-7g | −2.79 | −3.64 |
| mmu-miR-375 | 2.7 | 5.58 |
| mmu-miR-142-3p | −2.36 | 4.91 |
Assessment of the Ability of miR-133b to Bind to the 3′ UTR of CTGF and TGFβ1
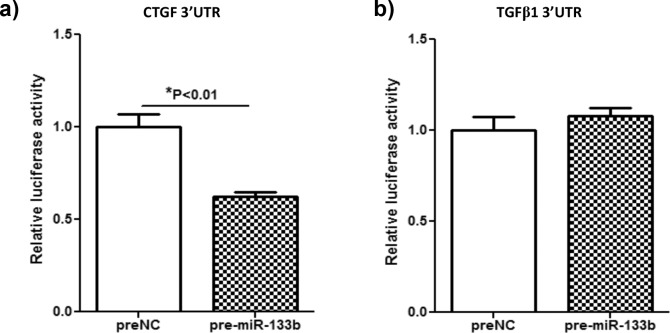
Since miR-133b had the greatest fold decrease at 2 days postablation, the ability of miR-133b to bind the target and inhibit translation of the profibrotic growth factors CTGF and TGFβ1 was predicted using TargetScan (in the public domain at www.targetscan.org). Shan et al.38 demonstrated that miR-133b can target TGFβ1 in atrial fibroblasts, although TargetScan predicted that miR-133b could target only CTGF. Therefore, the ability of miR-133b to target both CTGF and TGFβ1 was tested in RbCF. Briefly, RbCF were cotransfected with luciferase reporter plasmid including 3′ UTR of either CTGF or TGFβ1, pre-NC, or pre-miR-133b, and expression of the luciferase was determined (Fig. 1). As predicted by TargetScan, the 3′ UTR region of CTGF was targeted by pre-miR-133b as indicated by a significant decrease (P < 0.01) of approximately 38% in the luciferase activity (Fig. 1a). In contrast, there was no significant difference seen in the luciferase activity of the 3′ UTR region of TGFβ1 when transfected with the pre-NC compared to pre-miR-133b (Fig. 1b).
Figure 1. .
Determination of the ability of miR-133b to target the 3′ UTR of CTGF and TGFβ1. After dual transfection of a luciferase reporter plasmid that contained the 3′ UTR of either CTGF (a) or TGFβ1 (b) and either the pre-NC or the pre-miR-133b into RbCF, relative luciferase activity and significance were determined.
Relative Expression of miR-133b in RbCF Stimulated With TGFβ1
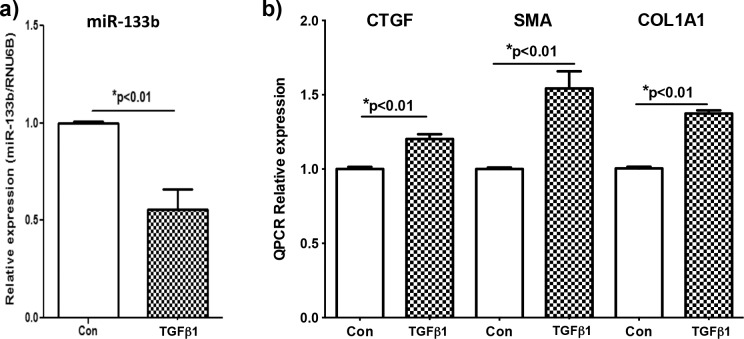
RbCF were stimulated with TGFβ1, and the expression of miR-133b was determined 12 hours after stimulation using qRT-PCR (Fig. 2a). Rabbit corneal fibroblasts stimulated with TGFβ1 had a significant reduction of 49% (P < 0.01) in miR-133b expression when compared to RbCF that were not stimulated with TGFβ1.
Figure 2. .
Endogenous expression levels of miR-133b, CTGF, SMA, and COL1A1 in RbCF. Using qRT-PCR, relative expression levels of miR-133b (a), CTGF (b), SMA (b), and COL1A1 were determined in unstimulated control (con) in RbCF or RbCF stimulated with TGFβ1 (2.5 ng/mL) for 12 (a) and 24 hours (b).
Relative Expression of CTGF, SMA, and COL1A1 in RbCF Stimulated With TGFβ1
Rabbit corneal fibroblasts were stimulated with TGFβ1, and the expression of CTGF, SMA, and COL1A1 was determined 24 hours after stimulation using RT-PCR (Fig. 2b). The relative expression levels of CTGF, SMA, and COL1A1 in RbCF stimulated by TGFβ1 were significantly greater, 20%, 54%, and 37% (P < 0.01), respectively, when compared to RbCF that were not stimulated with TGFβ1.
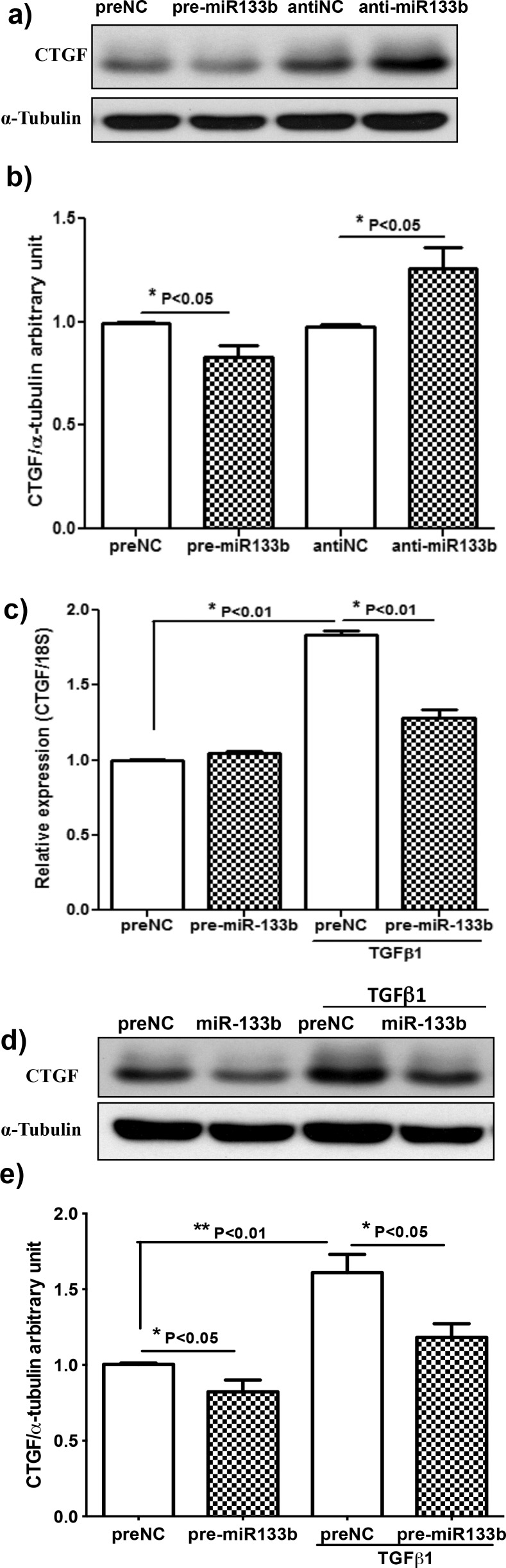
Effect of Pre-miR133b and Anti-miR-133b on CTGF mRNA and Protein Expression in RbCF
Rabbit corneal fibroblasts were treated with pre-miR-133b, scrambled pre-NC, anti-miR133b, or an anti-NC; and the relative levels of protein and mRNA of CTGF were analyzed using Western blots and qRT-PCR, respectively. Rabbit corneal fibroblasts treated with pre-miR-133b had a 16% (P < 0.01) reduction in CTGF protein levels, whereas RbCF treated with the anti-miR-133b had a significant increase of 29% (P < 0.05) (Figs. 3a, 3b). There was not a significant change in CTGF mRNA expression when RbCF treated with miR-133b were not stimulated with TGFβ1. In contrast, when RbCF were stimulated with TGFβ1 and treated with pre-miR-133b, there was a 30% reduction in CTGF mRNA expression (Fig. 3c). At the protein level, there was a 27% increase of CTGF when RbCF were stimulated with TGFβ1 and treated with pre-miR-133b (Figs. 3d, 3e).
Figure 3. .
Effect of anti-miR-133b and miR-133b on mRNA and protein levels of CTGF. (a) Using Western blot, relative expression levels of CTGF protein were determined after 96-hour transfection of prenegative control (NC), pre-miR-133b, anti-NC, and anti-miR-133b. (b) Protein expression levels were quantified by comparing to the α-tubulin control (n = 5). (c) Expression levels of CTGF mRNA from RbCF were quantified using qRT-PCR after 72-hour transfection of NC or pre-miR-133b and TGFβ1 (2.5 ng/mL) treatment for the last 24 hours. (d) Using Western blot, expression levels of CTGF protein were determined after 96-hour transfection of NC or pre-miR-133b and TGFβ1 (2.5 ng/mL) treatment for the last 48 hours. (e) Protein expression levels were quantified by comparing to the α-tubulin control (n = 8).
Effect of Pre-miR133b on SMA and COL1A1 Relative Expression in RbCF
The effects of pre-miR-133b or scrambled pre-NC on the relative expression levels of SMA and COL1A1 by RbCF that were stimulated or not stimulated with TGFβ1 are shown in Figure 4. As shown in Figure 4a, addition of TGFβ1 to cultures of RbCF treated with the scrambled pre-NC miRNA increased levels of SMA mRNA by 75% compared to cultures of RbCF not treated with TGFβ1. This demonstrates that transfection of the RbCF with a scrambled pre-NC did not produce a nonspecific effect in RbCF that reduced levels of SMA mRNA. Importantly, addition of pre-miR-133b to RbCF blocked 85% of the TGFβ1-stimulated induction of SMA. Similarly, as shown in Figure 4b, addition of TGFβ1 to cultures of RbCF treated with the scrambled pre-NC miRNA increased levels of COL1A1 mRNA by 33% compared to cultures of RbCF not treated with TGFβ1. Addition of pre-miR-133b to RbCF blocked 82% of the TGFβ1-stimulated induction of COL1A1. These results indicate that transfection of RbCF with pre-miR-133b blocks the TGFβ1-stimulated induction of SMA and COL1A1 mRNAs. This effect was not due to a nonspecific toxic effect of pre-miR-133b, since transduction with a scrambled pre-NC did not block the induction of the SMA and COL1A1 mRNAs by TGFβ1.
Figure 4. .
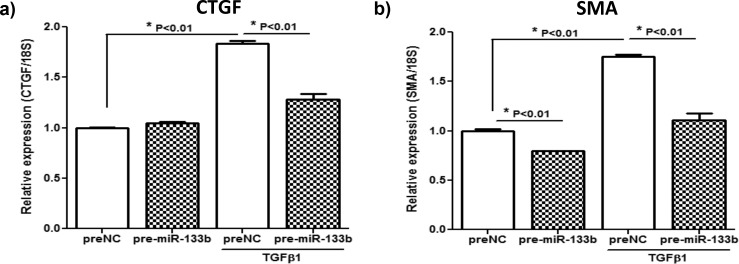
Effect of pre-miR-133b on relative expression levels of SMA and COL1A1 in RbCF. Using qRT-PCR, relative expression levels of SMA (a) and COLA1A (b) were determined after 72-hour transfection of NC or pre-miR-133b and TGFβ1 (2.5 ng/mL) treatment for the last 24 hours.
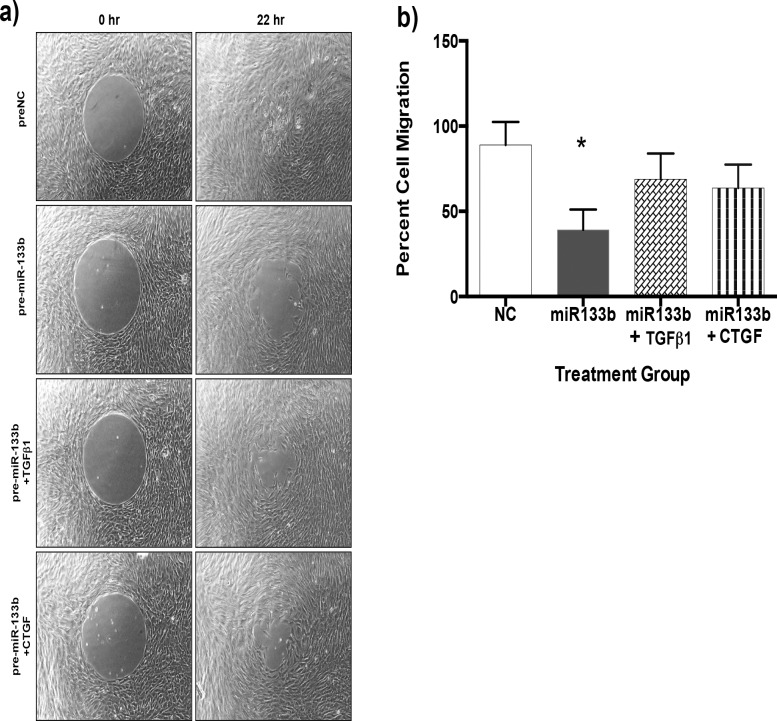
Effect of Pre-miR-133b on RbCF Migration
Rabbit corneal fibroblasts were transfected with either a scrambled pre-miR (pre-NC) or pre-miR-133b, and migration of the cells was quantified (Fig. 5). When the pre-NC was added, the cells fully migrated to close the gap after 22 hours. When compared to the pre-NC, the migration of the RbCF was significantly impeded by 55% (P < 0.05) when pre-miR-133b was added. When TGFβ1 and CTGF were added, the migration of RbCF was slightly impeded by 22% and 28%, respectively, but this was not significant when compared to the pre-NC (Fig. 5b). There was a significant (P < 0.05) increased migration due to the addition of TGFβ1 and CTGF when compared to the pre-miR-133b.
Figure 5. .
Effect of pre-miR-133b on migration of RbCF. A migration assay was performed using control RbCF or RbCF stimulated with TGFβ1 or CTGF and treated with either a NC or pre-miR-133b. (a) Cell migration assay of the different treatments. (b) Quantification of the cell migration due to the different treatments (*P < 0.05).
Discussion
While the role of miRNAs in corneal wound healing has not been investigated until this study, the expression of miRNAs in the whole eye and corneal development of mice was well documented by Karali et al.35 In fact, Karali et al. mapped the expression and changes in expression of over 250 miRNAs using microarrays during whole eye and corneal development in mice. Using RNA in situ hybridization (RNA ISH), they were able to locate the expression of the different miRNAs and created a comprehensive database showing the different developmental stages and the expression of the individual miRNA, which can be found (in the public domain) at www.mirneye.tigem.it as cited above. This database is an exciting and necessary step to understanding the role of miRNAs, but it does not address what miRNAs are integral in regulating corneal wound healing.
The present study is the first to determine the role of differentially expressed miRNA in regulating corneal wound healing and scar formation. The upregulation of CTGF by TGFβ causes corneal haze, which is characterized by light scattering due to the differentiation of fibroblast into myofibroblast and deposition of new irregular extracellular matrix.39 Moller-Pedersen20 has demonstrated that activated myofibroblasts identified by their increased expression of SMA in the cornea cause light scattering due to the lack of corneal crystallins. The mechanism that regulates the TGFβ/CTGF fibrosis systems has yet to be completely defined. This study demonstrates that miRNAs may play a key role in the regulation of the TGFβ/CTGF fibrosis pathway.
The relative changes in levels of 86 different miRNAs were assessed in mouse corneas at 30 minutes and at 2 days following excimer laser ablation. There were substantial changes (greater than 2-fold compared to values in normal unwounded corneas) in expression of 20 miRNAs at 30 minutes and in expression of 37 miRNAs at 2 days. Of particular interest was miR-133b, which had the most dramatic decrease in expression levels of all the miRNAs on day 2, decreasing 14.3-fold compared to values in normal corneas. In addition, miR-133b was predicted by TargetScan to target the CTGF mRNA, which was supported by the report of Duisters et al.40 that miR-133b regulates CTGF levels during myocardial matrix remodeling. Since CTGF is known to play major roles in generating corneal scarring and haze through inducing synthesis of SMA and COL1A1, investigating the effects of miR-133b on CTGF was warranted.31
An important general concept to recognize about miRNAs, including miR-133b, is that miRNAs are able to simultaneously influence the biological activity of multiple genes. For example, TargetScan predicts 536 conserved gene targets for miR-133b, including several additional growth factors and their receptors (fibroblast growth factor-1, fibroblast growth factor receptor, and epidermal growth factor), signal transduction pathway proteins (mitogen-activated protein kinase kinase kinase 3 and calmodulin 1), cell migration proteins (myosin IXb and myosin heavy chain 9), and 11 transcription factors. This can lead to more complex interpretation of results of experiments. For example, approximately 55% of the migration of RbCF in serum-containing medium was inhibited when pre-miR-133b was added to the culture medium. This indicates that 45% of the RbCF migration stimulated by serum-containing medium was due to factors that were not influenced by miR-133b. Addition of either TGFβ1 or CTGF to the pre-miR-133b–treated RbCF in serum-containing medium restored approximately 25% of the migration of the RbCF in serum-containing medium. These results indicate that only approximately 25% of the migration of RbCF in serum-containing medium is due to TGFβ or CTGF, and that other genes are also involved in stimulating migration of RbCF.
Several papers have explored the processes that miRNAs regulate during wound healing, including cell proliferation, migration, angiogenesis, and tissue regeneration.41 In chronic nonhealing venous ulcers, there was increased expression of miR-21, -130a, and -203. Overexpression of miR-21 and miR-130a resulted in inhibition of re-epithelialization of human skin.42 In contrast, there was increased expression of miR-21 after 4 and 8 days in normal mice, and an increase in concentration of pre-miR-21 caused an increase in the migration of fibroblast.43 In mouse corneas after ablation, miR-22 had the greatest significant fold increase at both 30 minutes and 2 days. Similarly, Madhyastha et al.43 found that at both days 4 and 8 after full-thickness dermal wounding, miR-22 is upregulated in both normal and diabetic mice. Further analysis of the role of miR-22 is needed to fully understand how miR-22 contributes to corneal wound healing. Although this paper focused on the effect of miR-133b to influence profibrotic factors like CTGF, SMA, and COL1A1, the miRNAs listed in the Table could be used as a guide to further help define and understand the role of miRNAs in corneal wound healing and scar formation.
Acknowledgments
Supported by National Eye Institute Grants EY000587 (GSS), P30-EY021721(Vision Core Grant), and T32-EY07132 (training grant [PMR]), and the US Army Medical Research Acquisition Activity W81XWH-10-2-0917 (GSS).
Disclosure: P.M. Robinson, None; T.-D. Chuang, None; S. Sriram, None; L. Pi, None; X.P. Luo, None; B.E. Petersen, None; G.S. Schultz, None
References
- 1. Mohan RR, Liang Q, Kim WJ, Helena MC, Baerveldt F, Wilson SE. Apoptosis in the cornea: further characterization of Fas/Fas ligand system. Exp Eye Res. 1997; 65: 575–589 [DOI] [PubMed] [Google Scholar]
- 2. Mohan RR, Mohan RR, Kim WJ, Wilson SE. Modulation of TNF-alpha-induced apoptosis in corneal fibroblasts by transcription factor NF-kappaB. Invest Ophthalmol Vis Sci. 2000; 41: 1327–1336 [PubMed] [Google Scholar]
- 3. Mohan RR, Kim WJ, Mohan RR, Chen L, Wilson SE. Bone morphogenic proteins 2 and 4 and their receptors in the adult human cornea. Invest Ophthalmol Vis Sci. 1998; 39: 2626–2636 [PubMed] [Google Scholar]
- 4. Wilson SE, Liang Q, Kim WJ. Lacrimal gland HGF, KGF, and EGF mRNA levels increase after corneal epithelial wounding. Invest Ophthalmol Vis Sci. 1999; 40: 2185–2190 [PubMed] [Google Scholar]
- 5. Tuominen IS, Tervo TM, Teppo AM, Valle TU, Gronhagen-Riska C, Vesaluoma MH. Human tear fluid PDGF-BB, TNF-alpha and TGF-beta1 vs corneal haze and regeneration of corneal epithelium and subbasal nerve plexus after PRK. Exp Eye Res. 2001; 72: 631–641 [DOI] [PubMed] [Google Scholar]
- 6. Jester JV, Huang J, Petroll WM, Cavanagh HD. TGFbeta induced myofibroblast differentiation of rabbit keratocytes requires synergistic TGFbeta, PDGF and integrin signaling. Exp Eye Res. 2002; 75: 645–657 [DOI] [PubMed] [Google Scholar]
- 7. Wilson SE, Walker JW, Chwang EL, He YG. Hepatocyte growth factor, keratinocyte growth factor, their receptors, fibroblast growth factor receptor-2, and the cells of the cornea. Invest Ophthalmol Vis Sci. 1993; 34: 2544–2561 [PubMed] [Google Scholar]
- 8. Tuli S, Goldstein M, Schultz GS. Modulation of corneal wound healing. In: Krachmer JH, Mannis JM, Holland EJ. eds Cornea. 2nd ed. Philadelphia: Elsevier Mosby; 2005: 133–150 [Google Scholar]
- 9. Netto MV, Mohan RR, Ambrosio R Jr, Hutcheon AE, Zieske JD, Wilson SE. Wound healing in the cornea: a review of refractive surgery complications and new prospects for therapy. Cornea. 2005; 24: 509–522 [DOI] [PubMed] [Google Scholar]
- 10. Fini ME. Keratocyte and fibroblast phenotypes in the repairing cornea. Prog Retin Eye Res. 1999; 18: 529–551 [DOI] [PubMed] [Google Scholar]
- 11. Gan L, Hamberg-Nystrom H, Fagerholm P, Van SG. Cellular proliferation and leukocyte infiltration in the rabbit cornea after photorefractive keratectomy. Acta Ophthalmol Scand. 2001; 79: 488–492 [DOI] [PubMed] [Google Scholar]
- 12. Andresen JL, Ehlers N. Chemotaxis of human keratocytes is increased by platelet-derived growth factor-BB, epidermal growth factor, transforming growth factor-alpha, acidic fibroblast growth factor, insulin-like growth factor-I, and transforming growth factor-beta. Curr Eye Res. 1998; 17: 79–87 [DOI] [PubMed] [Google Scholar]
- 13. Denk PO, Knorr M. The in vitro effect of platelet-derived growth factor isoforms on the proliferation of bovine corneal stromal fibroblasts depends on cell density. Graefes Arch Clin Exp Ophthalmol. 1997; 235: 530–534 [DOI] [PubMed] [Google Scholar]
- 14. Musselmann K, Kane BP, Hassell JR. Isolation of a putative keratocyte activating factor from the corneal stroma. Exp Eye Res. 2003; 77: 273–279 [DOI] [PubMed] [Google Scholar]
- 15. Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993; 122: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson SE, Mohan RR, Hong JW, Lee JS, Choi R, Mohan RR. The wound healing response after laser in situ keratomileusis and photorefractive keratectomy: elusive control of biological variability and effect on custom laser vision correction. Arch Ophthalmol. 2001; 119: 889–896 [DOI] [PubMed] [Google Scholar]
- 17. Jester JV, Barry LP, Cavanagh HD, Petroll WM. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea. 1996; 15: 505–516 [PubMed] [Google Scholar]
- 18. Moller-Pedersen T, Li HF, Petroll WM, Cavanagh HD, Jester JV. Confocal microscopic characterization of wound repair after photorefractive keratectomy. Invest Ophthalmol Vis Sci. 1998; 39: 487–501 [PubMed] [Google Scholar]
- 19. Jester JV, Petroll WM, Cavanagh HD. Corneal stromal wound healing in refractive surgery: the role of myofibroblasts. Prog Retin Eye Res. 1999; 18: 311–356 [DOI] [PubMed] [Google Scholar]
- 20. Moller-Pedersen T. Keratocyte reflectivity and corneal haze. Exp Eye Res. 2004; 78: 553–560 [DOI] [PubMed] [Google Scholar]
- 21. Ivarsen A, Laurberg T, Moller-Pedersen T. Characterisation of corneal fibrotic wound repair at the LASIK flap margin. Br J Ophthalmol. 2003; 87: 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piatigorsky J. Review: a case for corneal crystallins. J Ocul Pharmacol Ther. 2000; 16: 173–180 [DOI] [PubMed] [Google Scholar]
- 23. Pelton RW, Moses HL. The beta-type transforming growth factor. Mediators of cell regulation in the lung. Am Rev Respir Dis. 1990; 142: S31–S35 [DOI] [PubMed] [Google Scholar]
- 24. Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell. 1993; 4: 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kikuchi K, Kadono T, Ihn H, et al. Growth regulation in scleroderma fibroblasts: increased response to transforming growth factor-beta 1. J Invest Dermatol. 1995; 105: 128–132 [DOI] [PubMed] [Google Scholar]
- 26. Grotendorst GR, Duncan MR. Individual domains of connective tissue growth factor regulate fibroblast proliferation and myofibroblast differentiation. FASEB J. 2005; 19: 729–738 [DOI] [PubMed] [Google Scholar]
- 27. Chen MM, Lam A, Abraham JA, Schreiner GF, Joly AH. CTGF expression is induced by TGF-beta in cardiac fibroblasts and cardiac myocytes: a potential role in heart fibrosis. J Mol Cell Cardiol. 2000; 32: 1805–1819 [DOI] [PubMed] [Google Scholar]
- 28. Igarashi A, Nashiro K, Kikuchi K, et al. Significant correlation between connective tissue growth factor gene expression and skin sclerosis in tissue sections from patients with systemic sclerosis. J Invest Dermatol. 1995; 105: 280–284 [DOI] [PubMed] [Google Scholar]
- 29. Dammeier J, Brauchle M, Falk W, Grotendorst GR, Werner S. Connective tissue growth factor: a novel regulator of mucosal repair and fibrosis in inflammatory bowel disease? Int J Biochem Cell Biol. 1998; 30: 909–922 [DOI] [PubMed] [Google Scholar]
- 30. Wunderlich K, Senn BC, Reiser P, Pech M, Flammer J, Meyer P. Connective tissue growth factor in retrocorneal membranes and corneal scars. Ophthalmologica. 2000; 214: 341–346 [DOI] [PubMed] [Google Scholar]
- 31. Blalock TD, Duncan MR, Varela JC, et al. Connective tissue growth factor expression and action in human corneal fibroblast cultures and rat corneas after photorefractive keratectomy. Invest Ophthalmol Vis Sci. 2003; 44: 1879–1887 [DOI] [PubMed] [Google Scholar]
- 32. Ambros V. The functions of animal microRNAs. Nature. 2004; 431: 350–355 [DOI] [PubMed] [Google Scholar]
- 33. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 34. Sokol NS. The role of microRNAs in muscle development. Curr Top Dev Biol. 2012; 99: 59–78 [DOI] [PubMed] [Google Scholar]
- 35. Karali M, Peluso I, Gennarino VA, et al. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010; 11: 715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Woost PG, Jumblatt MM, Eiferman RA, Schultz GS. Growth factors and corneal endothelial cells: I. Stimulation of bovine corneal endothelial cell DNA synthesis by defined growth factors. Cornea. 1992; 11: 1–10 [DOI] [PubMed] [Google Scholar]
- 37. Chuang TD, Luo X, Panda H. Chegini N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol Endocrinol. 2012; 26: 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shan H, Zhang Y, Lu Y, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009; 83: 465–472 [DOI] [PubMed] [Google Scholar]
- 39. SundarRaj N, Geiss MJ III, Fantes F, et al. Healing of excimer laser ablated monkey corneas. An immunohistochemical evaluation. Arch Ophthalmol. 1990; 108: 1604–1610 [DOI] [PubMed] [Google Scholar]
- 40. Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009; 104: 170–178 [DOI] [PubMed] [Google Scholar]
- 41. Banerjee J, Chan YC, Sen CK. MicroRNAs in skin and wound healing. Physiol Genomics. 2011; 43: 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pastar I, Khan AA, Stojadinovic O, et al. Induction of specific microRNAs inhibits cutaneous wound healing. J Biol Chem. 2012; 287: 29324–29335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madhyastha R, Madhyastha H, Nakajima Y, Omura S, Maruyama M. MicroRNA signature in diabetic wound healing: promotive role of miR-21 in fibroblast migration. Int Wound J. 2012; 9: 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]