Abstract
Porphyromonas gingivalis is a Gram-negative anaerobic bacterium that colonizes the human oral cavity. It is implicated in the development of periodontitis, a chronic periodontal disease affecting half of the adult population in the USA. To survive in the oral cavity, these bacteria must colonize dental plaque biofilms in competition with other bacterial species. Long-term survival requires P. gingivalis to evade host immune responses, while simultaneously adapting to the changing physiology of the host and to alterations in the plaque biofilm. In reflection of this highly variable niche, P. gingivalis is a genetically diverse species and in this review the authors summarize genetic diversity as it relates to pathogenicity in P. gingivalis. Recent studies revealing a variety of mechanisms by which adaptive changes in genetic content can occur are also reviewed. Understanding the genetic plasticity of P. gingivalis will provide a better framework for understanding the host–microbe interactions associated with periodontal disease.
Keywords: adaptive virulence, genetic diversity, genetic variability, horizontal DNA transfer, pan-genome, Porphyromonas gingivalis
Porphyromonas gingivalis is a Gram-negative anaerobe that colonizes dental plaque biofilms in the human oral cavity, and is one of the few major pathogens responsible for the development of chronic periodontitis. Periodontitis is one of the most common bacterial infections of humans and 47% of the US adult population is affected by some form of this disease [1] . The pathological process in periodontitis destroys the structures supporting the tooth and is the leading cause of tooth loss in adults. The development of this disease is a multifactorial process involving interactions between the host and the microorganisms that colonize the oral cavity [2,3]. Persistent bacterial colonization by certain pathogenic microbes, coupled with a self-damaging host inflammatory response, results in destruction of soft tissues and loss of the bone surrounding the tooth. This disease process is chronic in nature and the inflammatory–infectious state can continue for years [4]. The natural outcome of this process is exfoliation of the tooth, which resolves the infection by removing the tooth-associated bacterial biofilm. Clinical intervention is primarily mechanical and involves scaling of the subgingival tooth surface to remove the plaque biofilm. In the event of significant loss of tooth-supporting tissues, surgical procedures are sometimes carried out to restore the periodontal architecture and stimulate regeneration of supporting tissues. There is a significant body of evidence linking the presence of active periodontitis to increased morbidity of certain systemic conditions including diabetes, cardiovascular disease, osteoporosis, preterm birth, respiratory diseases and rheumatoid arthritis [5]. While cause and effect relationships remain unclear, treatment of periodontal disease is beneficial in that it reduces chronic inflammation and the risks associated with the oral cavity as a focal point of infection [6].
P. gingivalis colonizes multispecies plaque biofilms at and below the gingival margin, and microbiome studies have also identified the deep crypts of the tongue as an additional habitat for these obligate anaerobes [7]. Residence under the gumline places P. gingivalis and other dental plaque microorganisms in contact with the host epithelium, and neutrophils and other innate immune effectors minimize the total number of bacteria present in the gingival sulcus (Figure 1A). This host immune response, supplemented by excellent oral hygiene, is sufficient to maintain healthy periodontal tissues. However, failures in immune defense or plaque control result in a higher bacterial burden and the subsequent development of gingival inflammation. Although present at low proportions in the oral microbial community, P. gingivalis is considered a ‘keystone’ pathogen, able to disrupt host–microbe homeostasis by molecular manipulation of select host protective mechanisms [8]. A multitude of studies have shown that P. gingivalis modulates innate host defense functions by subversion of IL-8 secretion, complement activity and TLR4 activation. This impairs the ability of the host to defend against the oral microbial community at large, resulting in an altered oral flora composition and subsequent inflammatory responses that contribute to the pathogenesis of periodontitis [9].
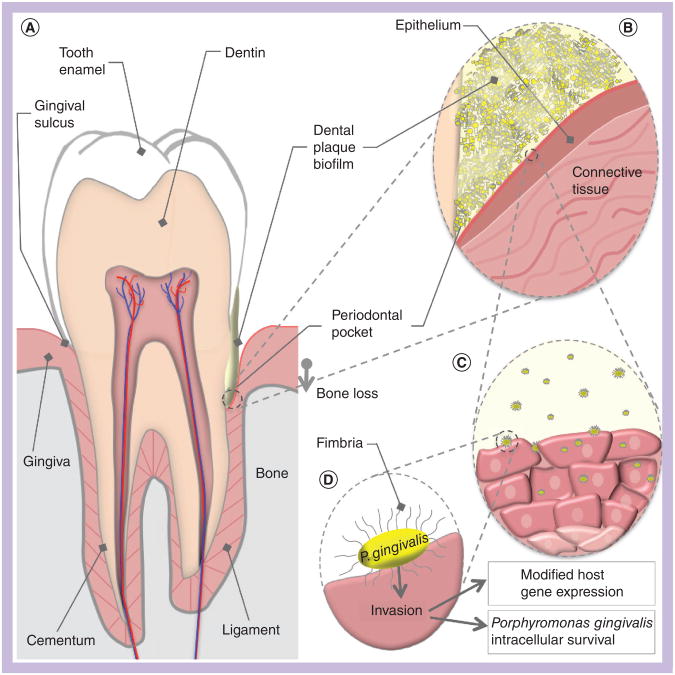
Figure 1. Porphyromonas gingivalis host environment and interactions.
(A) A cross-sectional schematic diagram of a human tooth. The periodontium is composed of the gingiva (pink) and the alveolar bone (grey). The left side represents periodontal health, while the right represents the changes that occur in periodontitis. Dental plaque is represented in yellow, in contact with the gingiva and tooth surfaces. Development of a chronic host inflammatory response ultimately results in a loss of connection between the gingiva and the root surface, creating a periodontal pocket. Bone is also resorbed as the bacteria advance down the surface of the tooth. (B) A magnified view of the periodontal pocket. The biofilm (yellow) is composed of many species of bacteria, of which Porphyromonas gingivalis is a small portion. P. gingivalis in the biofilm comes into direct contact with gingival epithelium. Secretion of P. gingivalis gingipains aids in the destruction of host antibodies and complement, and contributes to localized tissue destruction. (C) A number of P. gingivalis cells may leave the biofilm community and invade host cells. Internalized bacteria move from cell to cell into the deeper layers of the connective tissue. (D) Major fimbriae on the bacterial cell surface act as adhesins for host integrin receptors, and mediate internalization of the bacteria into the host cell cytoplasm. Postinvasion host cell gene expression is significantly modified; for example, reductions in cytokine production interferes with neutrophil recruitment to the periodontal pocket. The ability of P. gingivalis to modify local host responses to the biofilm flora results in the skewed immune response that drives periodontitis progression.
The collateral damage resulting from the battle between host and flora is loss of clinical attachment between tooth and bone, due to apical migration of the epithelial and connective tissue attachment together with alveolar bone loss [10]. In most untreated periodontal patients, the clinical attachment loss and ongoing periodontal inflammation generates a periodontal pocket, manifested as a deepened space beneath the gumline that exceeds 3 mm in depth, as measured from the margin of the gingiva to the bottom of the gingival sulcus (Figure 1A). This periodontal pocket can be very deep, reaching the apex of the root in some severe cases and provides a gradient of environmental conditions [11], with areas closest to the surface subjected to variable concentrations of oxygen, pH and temperature owing to local influence from the oral cavity. Deeper regions of the periodontal pocket are protected from disturbance and are more likely to be anaerobic [12,13], rich in blood and serum protein, and have a stable, slightly alkaline pH. Deep periodontal pockets evidently provide ideal conditions for P. gingivalis propagation, as cross-sectional surveys of periodontal pocket composition show the highest concentrations of P. gingivalis to be located in the deepest regions of the pocket [14]. Thus, the end-game strategy for this keystone pathogen is to manipulate host–flora interactions to create the ideal niche for survival. In this sense, P. gingivalis is considered a host-adapted pathogen, as chronic periodontal disease produces a slow-progressing morbidity with low risk of mortality.
Although P. gingivalis can be frequently detected in periodontal pockets, not all strains of P. gingivalis are equally pathogenic. Genetic heterogeneity within the species has been clearly linked to differences in outcomes related to host–pathogen interactions. There appears to be genetic adaptation occurring in this species as they transition from colonizers of healthy plaque to a major species in deep periodontal pockets, as species isolated from healthy sites have distinct genetic traits relative to those found in disease sites, consistent with a process of ‘inhost evolution’ or adaptive virulence [15] . In this review, the authors discuss the biological impact of genetic diversity on P. gingivalis pathogenicity and describe recent studies on gene transfer mechanisms underlying adaptive virulence.
P. gingivalis genetic diversity & virulence
P. gingivalis pathogenesis is well studied and these bacteria possess multiple virulence factors including gingipains (proteinases), major and minor fimbriae, hemagglutinins, hemolysin, iron uptake transporters, capsule and toxic outer membrane blebs [16,17]. These bacteria are also accomplished biofilm-formers, coaggregate with many other species found in dental plaque and contribute to the production of a protective biofilm exopolysaccharide matrix [3,18,19]. P. gingivalis directly binds to and invades gingival epithelial cells, and is able to spread from cell to cell into periodontal tissues (Figure 1C) [20,21]. Attachment of P. gingivalis to host cell surfaces is primarily mediated by the major fimbriae, which are long filamentous appendages on the surface of the bacterial cell (Figure 2). Interaction of the fimbriae with integrin host cell receptors mediates internalization of the bacteria and alters host cell signaling pathways (Figure 1D) [22–25]. Production of both gingipains and major fimbriae are mechanistically important for deregulation of immune function [2 6 –3 0], and expression of a capsule aids in evasion of immune effectors [31]. Many strains produce prolific membrane blebs, containing endotoxin from the outer membrane, which can stimulate innate immune responses (Figure 2). P. gingivalis is also capable of disseminating beyond the oral cavity via bacteremia and may contribute to distant sites of infection and aggravation of systemic diseases [5, 32–35] . In vitro and clinical studies have shown P. gingivalis to invade and survive in nonoral human cells, such as coronary arterial endothelial and placental cells, where they could potentially contribute to localized inflammatory responses [36–38].
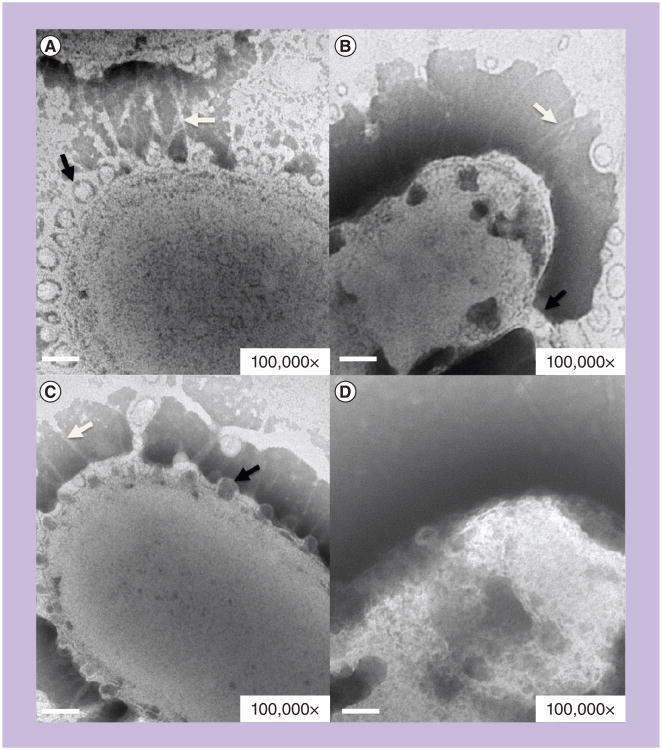
Figure 2. Transmission electron micrograph of Porphyromonas gingivalis strains 33277, 53977, 49417 and W83.
Porphyromonas gingivalis cells from 2 days' growth on blood agar stained with 2% phosphotungstic acid, shown at 100,000× magnification. Membrane blebs are indicated with black arrows, major fimbriae with white arrows. (A) Strain 33277 (fimA type I). (B) Strain 53977 (fimA type II). (C) Strain 49417 (fimA type III). (D) Strain W83 (fimA type IV) does not display any fimbriae nor any obvious evidence of membrane blebbing. Scale bar: 100 nm.
Sample preparation: Jennifer Kerr.
P. gingivalis can be detected in the gingival sulcus of healthy individuals and the periodontal pocket of periodontitis patients. As the conditions found in a healthy sulcus and a diseased pocket are quite different, these sites represent distinct environmental niches in the oral cavity. Furthermore, P. gingivalis may reside in a dental plaque biofilm or take up residence in host tissue. This ability to permanently colonize distinct niches implies that these bacteria have sophisticated mechanisms to adapt to the local environment. In vitro and animal model studies confirm the clinical observations: comparison of distinct P. gingivalis strains demonstrates extensive differences in behavior and pathogenic potential [28,30,31,39–52].
Underlying these behavioral differences, genetic variations exist for many of the major virulence factors of P. gingivalis (Table 1), and numerous studies have attempted to associate specific virulence alleles with initiation of disease. Most commonly, these studies have tracked a single genetic marker, the major fimbrial gene fimA. The fimA gene encodes the protein subunit that assembles to form the P. gingivalis surface fimbriae. The fimA gene has six alleles (types I, Ib, II, III, IV and V), as determined by DNA sequencing, and these alleles can easily be distinguished by PCR [53] . The genetic variability of fimA probably has a significant influence on host–pathogen interactions, as the fimbriae mediate several key aspects of virulence, including manipulation of host cell responses to bacteria. In studies of P. gingivalis isolates from healthy and diseased individuals, fimA type Ib, II and IV are more commonly associated with periodontal disease, whereas types I and III are found primarily in isolates from healthy individuals [45,46,52,54]. The human studies have been confirmed by animal studies, which found that strains containing fimA type II and IV are more cytotoxic and invasive, while strains with fimA type I and III are less inflammatory and more likely to form localized selflimiting infections [55]. The fimA type II allele is believed to contribute to improved virulence owing, in part, to its increased binding affinity for epithelial cell receptors compared with other fimA types [52,56].
Table 1.
Porphyromonas gingivalis genetic variations.
| Genetic variation | W83 | 33277 | 49417 | 53977 | TDC60 |
|---|---|---|---|---|---|
| fimA type (I, Ib and II–V) | IV | I | III | II | II |
| mfa (+/−) | − | + | Unknown | Unknown | + |
| ragAB type (1–4) | 1 | 4 | 1 | 3 | 4† |
| Capsule (K1–6 and K−) | K1 | K− | K4 | K3 | Unknown |
| kgp gingipain type (I and II) | I | II | Unknown | Unknown | II† |
| rgp gingipain type (A, B and C) | A | A | Unknown | Unknown | A |
| cTnPg1a (+/−) | − | + | − | + | + |
| ISPg4 copy number | 10 | 0 | 6 | 10 | 3‡ |
Table demonstrates known genetic variations between common laboratory strains of Porphyromonas gingivalis. fimA is the major protein subunit of the major fimbriae, Mfa is the presence or absence of minor fimbriae and RagAB is a surface lipoprotein associated with virulence. The capsule is characterized as one of six types or absent (K−), Kgp and Rgp are subtypes of the gingipain proteases, cTnPg1 is a conjugative transposon and ISPg4 is an example of an insertion sequence that varies in copy number between strains.
For strain TDC60, allele types were derived based on genetic comparison to genome sequences for W83 and 33277.
ISPg4 exists as three truncated copies in TDC60.
Population structure
Numerous studies have measured genome-wide genetic variability in the P. gingivalis population using techniques such as random amplified polymorphic DNA analysis [57], restriction fragment length polymorphism [58], insertion sequence (IS) hybridization [59], cross-species microarray hybridization [41,60] and ribosomal intergenic spacer region analysis [61] . Several research groups have utilized multilocus sequence typing to measure the allele distribution of multiple housekeeping genes and, subsequently, utilize this measure of genetic variability to determine the population structure of these bacteria, defined as clonal or nonclonal [45,49,62,63]. In clonal populations, cells are descended from, and are genetically identical to, a single common ancestor; genetic diversity might arise by point mutation; however, there is little horizontal transfer of DNA. In nonclonal populations, cells are descended from multiple ancestors; therefore, some form of DNA exchange occurs between cells to generate diverse offspring. An Index of Association score is a quantitative measure of population structure: on one end of the spectrum, fully clonal populations score one; at the other end of the range, nonclonal populations are zero. According to the Multilocus Sequence Typing website for P. gingivalis [201], the P. gingivalis population structure falls in the nonclonal range with a score of 0.37 [64].
In nonclonal bacteria, the population displays a spectrum of phenotypes from commensal to pathogenic [65], and many different allele combinations can cause disease. Nonclonal population structures are common to human pathogens involved in chronic infections, resulting from horizontal DNA transfer between strains in vivo. This sexual behavior is thought to contribute to adaptation and persistence in the human host during changing niche conditions [65–67]. In well-characterized nonclonal pathogens such as Helicobacter pylori and Neisseria spp., natural competence is the mechanism that facilitates DNA exchange between strains that are coresidents in the host environment [15, 68 –70].
Regarding P. gingivalis, studies have attempted to more broadly link nonclonal population structure to pathogenic heterogeneity by analyzing the combinations of virulence alleles present in collections of P. gingivalis clinical isolates [31, 45] . In a study correlating fimA allele type to multilocus sequence profile, fimA type II was found to be the most common (34%) in a population of 82 strains of worldwide origin [45]. The fimA type II allele was found in seven distinct genetic backgrounds [61], suggesting that this allele confers a selective advantage during the disease process that ensures its distribution throughout the P. gingivalis population. Two other studies have attempted to correlate virulence in P. gingivalis strains to the presence of proteases, capsule production alleles and fimA alleles (Table 1). Inaba et al. found that strains containing the fimA type II allele were most virulent in mice if they possessed capsule types 4 or 5 (from seven types), and were more invasive if they had high levels of protease activity [44]. Yoshino et al. assessed the genotypes of 62 clinical isolates, determining the allele combinations for fimA (type I, Ib–V), gingpain kgp (I or II) and gingipain rgpA (A, B or C). Out of 36 possible allele combinations, 15 were found in the isolate collection. Of these 15 combinations, two allele profiles were found in 50% of the isolates. The two frequent allele combinations both contained the fimA type II allele [31] . Taken together, these results indicate that many different P. gingivalis strains have the potential to contribute to the pathogenesis of periodontitis; however, particular allele combinations are favored over others. This is consistent with the nonclonal population structure of P. gingivalis, in that many strains may cause disease, but some allele combinations are more virulent.
If certain allele combinations are favored in disease, why do other allele combinations persist in the P. gingivalis population? An important consideration in any population study is the presence of bias in the sample selection. The population structure and genetic diversity studies described above were done with clinical isolates from periodontal patients. The oral environment in these patients is distinct from individuals with healthy gingiva or with gingivitis. From an ecological perspective, these states of health or disease represent unique environmental niches, and certain alleles or allele combinations may favor survival in one niche over another. In the aforementioned studies, bacterial strains were selected mainly from periodontal patients, thus the diseased environmental niche is best represented. However, P. gingivalis is able to colonize both healthy and diseased gingival sulci. In the study comparing fimA alleles in diseased patients versus healthy individuals, healthy oral niches were more likely to be colonized by strains with fimA alleles I and III. P. gingivalis strain 33277 is less virulent in abscess models than isolates from diseased patients, has fimA allele I, produces no capsule and excretes low levels of gingipains (Table 1). Strain 33277 could therefore be considered a prototype for strains that successfully colonize the healthy oral cavity, and illustrative of strains that are under-represented in population genetics studies.
Whole-genome comparisons
Studies of population structure provide a wide-angle view of genetic diversity within a bacterial species, and alignment of whole genome sequences provides a complementary and a highly magnified assessment of variability. Whole-genome sequences are currently available from three strains of P. gingivalis: W83, 33277 and TDC60 [71–73]. These three strains are representative of the P. gingivalis nonclonal population structure in that they have diverse phenotypic behaviors. The W83 and 33277 isolates are well-studied laboratory strains. W83 has been demonstrated to be more cytotoxic and likely to disseminate in the host, while 33277 is better at biofilm formation and localized host cell invasion. The TDC60 strain is a strain recently isolated from a periodontal pocket and exhibits relatively greater pathogenicity in mice than strains W83 and 33277 [71] . The genome of strain 33277 contains 2090 protein-coding sequences (CDSs), while strain W83 contains 2023 CDSs and strain TDC60 has 2220 CDSs.
Computational alignment of these three genomes reveals that approximately 75% of the CDSs are conserved at greater than 90% identity. Strain 33277 has 461 CDSs, W83 has 415 CDSs and TDC60 has 382 CDSs that are designated as nonidentical or strain specific. Many of these strain-specific CDSs have functional homologs in the opposing strain sequence, implying that these CDSs represent allelic forms of the same gene. Others are unique to individual strains and many are hypothetical in function, thus providing an unknown benefit to the host strain. As shown in Figure 3, genetic variability between strains can be extensive and encompass entire genes or operons; these regions are considered strain-specific and compose approximately 20–25% of a given strains genome. Genetic variability can also be detected within open reading frames as point mutations or other genetic changes that are limited in size (Figure 3). Although the latter genetic changes are not dramatic, the resulting amino acid changes could result in significant functional alterations. Genetic differences can also be detected in intergenic regions and, therefore, have the potential to influence promoter or transcriptional terminator activity (Figure 3).
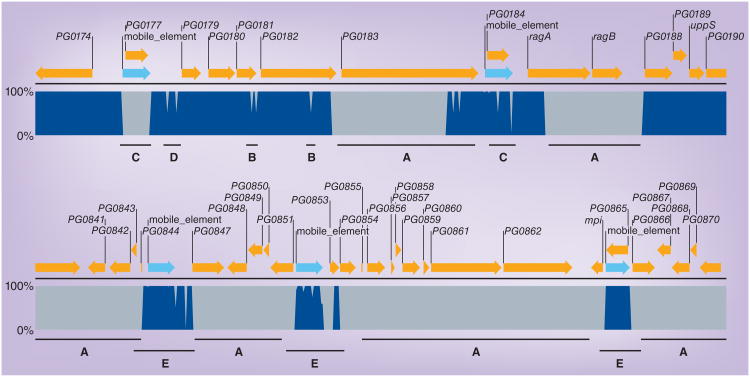
Figure 3. Examples of genomic differences between strains W83, 33277 and TDC60.
The genome sequence of strain W83 was screened against strains 33277 and TDC60 to identify regions of conserved (dark blue) and divergent (grey) sequence. The upper alignment represents a region primarily encoding lipoproteins. The lower alignment demonstrates highly diverged regions flanked by conserved mobile elements. The genomic comparison was performed using CLC Genomics Workbench V4, using the internal BLAST function (Mask low-complexity; Expect 1; Match 1, Mismatch -2; Word size 30). The open reading frame annotations are from strain W83: yellow arrows represent coding sequence and light blue arrows represent mobile elements. ‘A’ denotes genes that are highly divergent between all three strains and are thus ‘strain-specific’. In the upper panel, ragAB encodes an outer membrane lipoprotein and PG0183 encodes a putative lipoprotein. ‘B’ denotes changes in DNA sequence in one of the three strains, TDC60, which result in limited changes in amino acid content. PG0181 and PG0182 encode putative lipoproteins; the genetic differences in strain TDC60 result in changes to 15 and 32 amino acids, respectively. ‘C’ denotes a mobile element that is found only in strain W83, ISPg4 to the left; and a mobile element found in all three strains, ISPg1 to the right. ‘D’ denotes genetic variation in the promoter region for PG0179. The promoter region is conserved between W83 and 33277, and divergent in TDC60. The two points of genetic variance begin 147 and 350 bp upstream of PG0179, and represent 28 and 25% genetic divergence over 127 and 385 bp, respectively. ‘E’ denotes conserved mobile elements flanking strain-specific genes. Two copies of the conserved ISPg1, left and middle, and one copy of ISPg2, right, flank regions containing W83-specifc genes encoding primarily hypothetical proteins. PG0861 is predicted to be a helicase and PG0862 a restriction endonuclease, suggesting that this region may encode functions associated with DNA transfer.
Beyond bioinformatic alignment of the three sequenced genomes, other multiple strain comparisons have been made using comparative genome hybridization microarrays. Brunner et al. compared seven P. gingivalis strains to a W83 microarray and identified genes that were: fully conserved in all strains; ‘aberrant’ or not identical between strains (allelic); or absent in the test strain. Similar to the genomic alignments, this study identified a common core genome comprised of approximately 80% of the genes in the W83 genome [74]. Interestingly, many of the genes that are divergent between strains are found in clusters, creating localized ‘hotspots’ of genetic diversity within the genome sequence [60,74]. One cluster of divergent genes is the capsule operon, which is composed of 14 open reading frames [72,75]. Other divergent regions contain open reading frames predicted to encode IS (Figure 3), transposases, CRISPR-associated gene sequences, restriction endonucleases or hypothetical proteins. The presence of transposases and CRISPR-associated genes implies that at least some of the P. gingivalis genome is ‘mobile’ and capable of rearrangement or horizontal transfer from one strain to another.
Mobile DNA & genetic diversity
Insertion sequences
ISs are the smallest forms of mobile DNA. They consist of one open reading frame encoding a transposase enzyme, surrounded by short repeated DNA sequences. If the transposase gene is expressed, the enzyme can bind to the DNA repeats, cut the DNA to remove the IS and reinsert the IS in a new location. At least seven distinct IS elements have been identified in strains of P. gingivalis and have been given a standardized nomenclature of ISPg (1–7) [76–80]. Most strains have multiple copies of more than one ISPg element and the IS element profile and copy number has been used to differentiate a collection of laboratory and clinical isolates (Table 1) [59]. The presence of IS elements in a bacterial genome can have multiple effects, including the inactivation of open reading frames, up- or down-regulation of gene expression via polarity effects, and can also result in wide scale genomic rearrangements. There is some evidence that IS elements in P. gingivalis are active. ISPg elements have been shown to influence levels of gingipain expression by insertion upstream in the promoter region [77, 81], and strain W83 lacks minor fimbriae owing to an ISPg1 insertion in the mfa gene [73,82]. These transposition events have a significant impact on genetic diversity and pathogenicity, as both gingipains and minor fimbriae are important for interactions with the host.
Whole genome alignments of the sequenced strains reveal a significant level of genomic rearrangements [71]. It is possible that transposonmediated recombination between repeated ISPg sequences dispersed on the genome might allow the rearrangement of the genomic structure (Figures 3 & 4C). Such genomic rearrangements can impact gene expression levels, as genes located in proximity to the chromosomal origin of replication may have a higher transcription level. Interestingly, a recent transcriptional mapping experiment demonstrated that during growth in standard laboratory conditions, antisense mRNA is expressed for ISPg elements. This would have the predicted effect of silencing expression of ISPg transposase genes, and preventing transposition activity [83]. I t will be interesting to discover if this transposition silencing i s released du r i ng cell stress, as low levels of ISPg activity could provide a mechanism of genetic adaptation to changing and stressful environmental conditions via random alteration of gene expression.
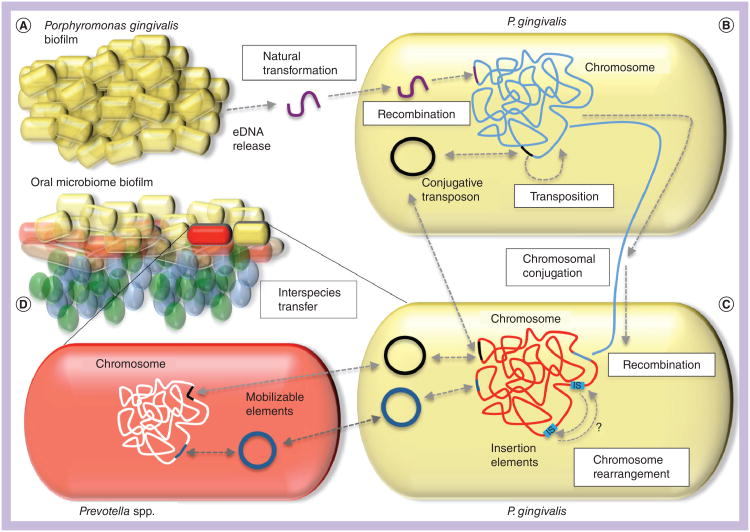
Figure 4. Mechanisms of DNA transfer in Porphyromonas gingivalis.
Porphyromonas gingivalis resides in dental plaque biofilms in the presence of diverse bacterial species and multiple strains of P. gingivalis may coexist within the same biofilm community. (A)P. gingivalis biofilms have been shown to release eDNA (purple) into the biofilm matrix. This DNA can be taken up by other strains of P. gingivalis by natural transformation. This process requires the comF gene in the recipient strain; comF proteins provide the molecular motor to bring DNA across the bacterial membrane. DNA that is homologous to the host cell genome may be integrated into the chromosome by homologous recombination. (B) Some strains of P. gingivalis contain conjugative transposons that are normally integrated into the chromosome (black line), but may excise and form a circular intermediate prior to conjugative transfer to a new host cell (black circle). Conjugative transposon cTnPg1 has been shown to transfer between P. gingivalis strains, is able to transfer into other closely related species such as Prevotella (D), and can also mobilize plasmids and other genetic elements between species (dark blue circles). (C) Conjugation can also mediate the transfer of chromosomal DNA between strains of P. gingivalis. This process is dependent on conjugative transposons or other traAQ-containing elements present in the donor strain and homologous recombination in the recipient. Allelic replacement of recipient DNA (red) with donor DNA (blue line) is mediated by homologous recombination. Finally, insertion elements (light blue boxes) are present in P. gingivalis and are capable of transposing into genes or their promoter regions, thus influencing expression. They are also likely responsible for the large scale genomic rearrangements that can be detected by comparison of multiple genome sequences. (D) cTnPg1 can be transferred to the closely related genus Prevotella. eDNA: Extracellular DNA.
Conjugative transposons
The genome sequences for P. gingivalis strains have regions that are similar to conjugative transposons (cTns) found in their close taxonomic relatives, the Bacteroides. Bacteroides species comprise the majority of the flora in the mammalian GI tract and are well-characterized genetically [84]. These bacteria have been shown to transfer DNA to other bacterial species in the intestine, and the primary mode of genetic exchange between Bacteroides spp. is mediated by cTns. The Bacteroides cTns are a diverse group o f 40–60 kb elements that encode multiple genes required for formation of a conjugal pore, with the genetic designation tra. The cTns do not have an origin of replication and are usually integrated in the chromosome of the host cell. In preparation for conjugation, these elements excise themselves from the chromosome, forming a circular intermediate (Figure 4b). This circular form is the substrate for conjugation and the conjugal pore transfers a single strand to the recipient cell. Once conjugation is complete, the single strands in both the donor and recipient are replicated, and the circular molecules integrate back into the chromosomal DNA. In addition to their own transfer, the cTns also mobilize Bacteroides plasmids and other transposons by providing the promiscuous conjugal pore encoded by the traAQ locus. This extrachromosomal transfer of plasmids and transposons by the traAQ locus has been implicated in the high rates of antibiotic resistance in Bacteroides species [85].
There are four putative cTns in strain 33277; these elements have been given the designation cTnPg. Three of the 33277 cTns encode a partial tra conjugation locus (cTnPg1-b, cTn Pg1-c and cTnPg2) as compared with functional tra gene regions encoded in the genome sequences of Bacteroides spp. One 33277 element, cTnPg1-a has a complete traAQ region, and has been shown to be a functional conjugative transposon, with behaviors similar to those of Bacteroides cTns [72,86]. As both oral and intestinal bacteria have similar cTns, it seems that these elements could play an important role in horizontal gene transfer between related strains or species of bacteria. cTnPg1 carries a gene that codes for a Na+-driven multidrug efflux pump; however, it was not determined if transfer of this element conferred antibiotic resistance. Naito et al. demonstrated that cTnPg1 could be transferred to the closely related genera Bacteroides and Prevotella (Figure 4D); therefore, this family of cTns could contribute to genetic diversity by mediating gene sharing within the Bacteroidetes phylum [86].
Plasmid mobilization
In the Bacteroides, the cTns can mobilize plasmids and other transposons by providing the promiscuous conjugal pore encoded by the traAQ locus. To date, no natural plasmids have been identified in P. gingivalis; however, plasmids introduced from close relatives, such as the Bacteroides, are functional and are frequently used for genetic manipulation of P. gingivalis[87]. Two Bacteroides- derived plasmids were used to test the mobilization functions of cTn-like elements in eight different strains of P. gingivalis[88]. Strain W83 and two others could not transfer plasmids to a recipient. By contrast, strain 33277 and four others could conjugate plasmids at frequencies between 10−5 and 10−7[88]. Therefore, the cTnPg1 found in 33277 is functionally similar to Bacteroides cTns in that, in addition to its ability to transfer itself, it can also mediate the transfer of other genetic elements. The other four strains capable of plasmid mobilization contain DNA sequences highly similar to cTn Pg1 [Tribble GD, Unpublished Data]. Strain W83 also contains a traAQ genomic locus that has a similar gene arrangement, but less than 36% homology at the DNA level, to cTnPg1 of 33277.
The 33277 cTnPg1-a traAQ locus contains a traP gene and in Bacteroides, the traP gene is required for plasmid conjugation; the W83 traAQ locus does not contain a traP homolog, which may explain the lack of plasmid transfer for W83 [88,89]. The locus of P. gingivalis W83 does not have genes for integration or excision, implying that this locus is not a conjugative transposon and is permanently anchored in the chromosome [88]. Based on these collective data, it appears that some strains of P. gingivalis have the ability to facilitate transfer of cTns and other accessory mobile elements, while others do not.
Chromosomal DNA transfer
While the presence of mobile DNA elements, such as ISs and cTns, can contribute to genetic diversity, these elements alone do not account for the extensive genetic variability and nonclonal population structure demonstrated by the P. gingivalis pan-genome. In other nonclonal bacterial species, mechanisms to exchange chromosomal DNA are present, most commonly via natural competence or by integrated plasmids, similar to the high frequency recombination systems found in Escherichia coli[90]. Recent investigations into P. gingivalis chromosomal transfer have identified both natural competence and conjugation mechanisms for DNA exchange between P. gingivalis strains [91]. Using antibiotic resistance markers engineered into the P. gingvalis genome, it was found that strains W83 and 33277 could exchange chromosomal DNA with each other. The resulting offspring were shown to be chimeric, as they contained DNA from both donor and recipient strains and the chimeras were demonstrated to have altered phenotypic properties as measured by biofilm assays [88]. It was also demonstrated, by whole genome footprinting with ISPG1, that widespread genome rearrangements did not occur during interstrain DNA transfer; however, exchange of regions large enough to swap entire genes did occur. This study illustrates that essential features of nonclonal population structure exist in P. gingivalis at the mechanistic level: transfer of chromosomal DNA between strains swaps alleles and generates diverse phenotypic behaviors.
In a series of experiments to better define the molecular events occurring during DNA exchange, it was determined that using a ‘dead donor’ assay was much more efficient at DNA transfer than mixing two living strains. In a dead donor assay, one strain is heat-killed before mixing with a live strain. In this scenario, the only transfer that can occur requires the live recipient strain to take up free DNA released by the heat-killed donor. The high transformation frequencies in dead donor assays implied that a natural transformation system is present in P. gingivalis strains. This was confirmed by mutating a predicted competence-associated gene comF in strain W83. The resulting comF mutant was unable to be transformed in a dead donor assay. These studies also documented the presence of extracellular DNA in P. gingivalis biofilms, which could provide a source of DNA for transformation and reassortment of alleles between strains (Figure 4A) [91]. Interestingly, two live comF mutants were still able to exchange chromosomal DNA at very low frequencies. A further series of experiments revealed that this residual DNA transfer was due to conjugation of chromosomal DNA mediated by the traAQ region (Figure 4b & C) [91]. In summary, overlapping mechanisms for chromosomal DNA exchange exist with P. gingivalis: natural competence occurs at the highest frequency and conjugation of chromosomal DNA is also possible, although this occurs at a lower frequency. As predicted by the nonclonal population structure of P. gingivalis, horizontal transfer of chromosomal DNA is possible, and as P. gingivalis has redundant mechanisms to ensure this process, it seems likely to be a critical component of its lifestyle.
Conclusion
Horizontal DNA transfer is well known for its spread of virulence traits such as antibiotic resistance genes and pathogenicity islands; however, its significance as a driving force in species diversity and niche adaptation is only recently becoming more obvious. The pan-genome concept describes a bacterial species as a pool of alleles that may be acquired, deleted or reassorted between coresident strains in response to pressures from the ecological niche [92]. For many bacteria, humans are their primary or only habitat, and adaptation and evolution is in response to selective pressures from the host environment. As each human host represents a unique habitat, bacterial genetic factors that contribute to colonization and persistence may need to be tailored to the specific host in order to maximize fitness of the microorganism. For potential pathogens, genetic adaptation to the host might contribute to the initiation of an infection after colonization, persistence and propagation in a hostile environment, evasion of the immune response and different disease outcomes in different hosts. In P. gingivalis, multiple forms of genetic transfer and recombination are occurring within the species by conjugation and transformation, and interspecies transfer of cTns and mobilizable elements can also occur. Therefore, mechanisms for maintaining genomic plasticity are well provided for and in the complex communities associated with oral biofilms, sources of exogenous DNA are plentiful [93,94].
In periodontal patients, elevated numbers of P. gingivalis are found in the periodontal pocket, and may compose up to 7% of the bacteria found in plaque samples. Individuals can be transiently coinfected by two, three or more strains simultaneously [95]. Theoretically, exchange of DNA between multiple strains occupying the same dental plaque biofilm could produce a pool of chimeric offspring with diverse phenotypes. Over a period of time, these offspring would compete with each other, with the most fit for the prevailing host conditions becoming the dominant strain within the plaque community. Over the lifetime of the host, one could predict multiple co-colonization events could occur, introducing new alleles to the local P. gingivalis gene pool. Assuming P. gingivalis is able to ‘dial-up’ the correct allele combinations, DNA exchange would allow continued fine-tuning of bacterial fitness and contribute to persistence in the ever-changing oral niche of the host. Beyond persistence, DNA exchange between strains could also be contributing to the clinical development of chronic periodontitis, by providing this organism with the correct ways in which to hack the host cellular signaling network.
In the complex lifestyle of P. gingivalis, initial colonization of plaque biofilms and survival in the nutrient deprived conditions of a healthy gingival sulcus will favor certain strains; whereas long-term survival and immune evasion in an inflamed periodontal pocket may favor different allelic combinations. Hence, selective pressure from the diverse oral environment maintains allelic diversity in P. gingivalis. The current authors predict that once a strain successfully colonizes the gingival sulcus, co-colonization with additional strains, even transiently, will allow genetic reassortment and the subsequent selection of new genetic variants with improved fitness for the changing environment [66]. At present, an interesting perspective is that strains that colonize the dental plaque of a healthy gingival sulcus are considered ‘less pathogenic’ than strains isolated from deep periodontal pockets. However, the true pathogenicity of P. gingivalis lies in its role as a keystone pathogen: defined by its ability to subvert early immune responses and send the host–pathogen interaction down a darker path. Therefore, one could redefine stages of pathogenicity for this bacterium and ‘healthy host’ colonizers involved in early subversion events are likely in ‘stealth’ mode, working to modify the host environment and continuing to persist in a shallow sulcus until the upper hand is gained [96]. Strains found in deeper periodontal pockets in the presence of a chronic immune response have likely shifted to a different mode, one that favors survival and transmission to a new sulcus or even a new host.
Future perspective
The study of P. gingivalis as a keystone pathogen in the polymicrobial development of periodontitis illustrates the difficult tasks faced by modern microbiologists and immunologists. With the advent of microbiome research, scientists and clinicians are challenged with dissecting the molecular interactions between the host and a highly complex flora. Changes in the composition of the human microbiome are being linked to a variety of chronic health conditions, including obesity [97], ulcerative colitis and irritable bowel syndrome [98,99], pulmonary exacerbations in cystic fibrosis [100] and even immunoregulatory dysfunction in Type 1 diabetes [101]. As with studies of periodontitis, determining if changes in the microbiome are a cause or effect of disease, identifying bacterial species that may act as a ‘keystone’ to disease progression and characterizing underlying molecular pathology will be key to linking microbiome data to other chronic health conditions.
Beyond the role of microbiome composition, understanding in vivo bacterial adaptations to the host is key to understanding fundamental questions about the role of microbial fitness, adaptation and evolution in chronic infections. Nonclonal bacterial species are a moving target, allowing them to persist within the host and potentially transform into a more pathogenic type. Our standard weapons against acute bacterial infection are ineffective at targeting chronic microbial disorders, but hopefully understanding the genetic plasticity of nonclonal bacteria such as P. gingivalis will provide a better framework for intervening in the host–microbe interactions associated with chronic disease.
Executive summary.
Porphyromonas gingivalis
Porphyromonas gingivalis is a Gram-negative, strictly anaerobic bacterium that colonizes the human oral cavity and is associated with the development of chronic periodontal disease.
P. gingivalis is found in anaerobic sites in the mouth such as the deep crypts of the tongue and in subgingival dental plaque.
Transmission occurs between household contacts, with elderly residents often transmitting to younger members.
P. gingivalis can be found in a healthy oral cavity in low numbers; in periodontitis the subgingival burden is much higher.
In the subgingival plaque biofilm, P. gingivalis comes into direct contact with host gingival tissues and circulating immune cells.
Host–pathogen interactions
P. gingivalis is defined as a keystone pathogen. Even in small numbers in the flora, the bacterium interacts with the host defense system to suppress cytokine expression, complement activity and Toll-like receptor activation.
Localized immune subversion ultimately leads to a pathologic relationship between host and oral flora, resulting in a chronic inflammatory response. Collateral damage to the periodontium results in the clinical signs of periodontal disease.
P. gingivalis possesses multiple virulence factors. For example, secreted protease enzymes called gingipains interfere with complement activity. Major fimbriae on the bacterial surface mediate adherence and internalization into host cells.
Genetic variability
The P. gingivalis population is composed of many different strains.
Pathogenic potential is variable between strains and most virulence factors have multiple alleles.
Particular combinations of virulence alleles are more common in active disease sites, while other allele combinations are present in healthy sites.
The nonclonal population structure predicts that new allele combinations are created by horizontal DNA transfer.
Gene-transfer mechanisms
P. gingivalis possesses multiple mechanisms for DNA transfer and recombination.
P. gingivalis insertion sequences can modify gene expression and mediate genomic rearrangements.
Conjugative transposons can transfer between strains and mobilize other elements and chromosomal DNA to recipients.
P. gingivalis secretes extracellular DNA into biofilms and displays natural competence; different strains occupying the same biofilm can therefore share DNA by transformation.
Gene transfer and recombination likely produce a diverse population of strains, which is then subject to fitness selection by the challenging conditions of the oral cavity.
Acknowledgments
Microscopist for Figure 2: K Dunner Jr funded by the Institutional Core Grant #CA16672 High Resolution Electron Microscopy Facility UTMDACC.
This work was supported by Public Health Service grant DE-019634 from the National Institute for Dental and Craniofacial Research to GD Tribble, and by institutional financial support from the University of Texas Health Science Center at Houston School of Dentistry to GD Tribble and B-Y Wang.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. CDC periodontal disease surveillance workgroup Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91(10):914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 2.Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15(6):341–349. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- 3.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontology 2000. 2010;52(1):38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anerud A, Löe H, Boysen H, Smith M. The natural history of periodontal disease in man. Changes in gingival health and oral hygiene before 40 years of age. J Periodontol Res. 1979;14(6):526–540. doi: 10.1111/j.1600-0765.1979.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 5.Otomo-Corgel J, Pucher JJ, Rethman MP, Reynolds MA. State of the science: chronic periodontitis and systemic health. J Evid Based Dent Pract. 2012;12(Suppl. 3):S20–S28. doi: 10.1016/S1532-3382(12)70006-4. [DOI] [PubMed] [Google Scholar]
- 6.Parahitiyawa NB, Jin LJ, Leung WK, Yam WC, Samaranayake LP. Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. 2009;22(1):46–64. doi: 10.1128/CMR.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10(5):497–506. doi: 10.1016/j.chom.2011.10.006. Landmark publication on the host–pathogen interaction mechanisms underlying the Porphyromonas gingivalis‘keystone pathogen’ hypothesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91(9):816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontology 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 11.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38(Suppl. 11):S28–S35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 12.Mettraux GR, Gusberti FA, Graf H. Oxygen tension (pO2) in untreated human periodontal pockets. J Periodontol. 1984;55(9):516–521. doi: 10.1902/jop.1984.55.9.516. [DOI] [PubMed] [Google Scholar]
- 13.Loesche WJ, Gusberti F, Mettraux G, Higgins T, Syed S. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infect Immun. 1983;42(2):659–667. doi: 10.1128/iai.42.2.659-667.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilson DJ. Insights from genomics into bacterial pathogen populations. PLoS pathogen populations. 2012;8(9):e1002874. doi: 10.1371/journal.ppat.1002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James CE, Hasegawa Y, Park Y, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74(7):3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62(4):1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuboniwa M, Amano A, Hashino E, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 2009;9:105. doi: 10.1186/1471-2180-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda K, Tribble GD, Tucker CM, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69(5):1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontology 2000. 2010;52(1):68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154(Pt 10):2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handfield M, Baker H, Lamont R. Beyond good and evil in the oral cavity: insights into host–microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87(3):203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao S, Park Y, Hasegawa Y, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9(8):1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4(5):305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe K, Yilmaz O, Nakhjiri SF, Belton CM, Lamont RJ. Association of mitogenactivated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect Immun. 2001;69(11):6731–6737. doi: 10.1128/IAI.69.11.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets SM, Robles-Price AG, McKenzie RME, Casiano CA, Fletcher HM. Gingipaindependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci. 2008;13:3215–3238. doi: 10.2741/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagano K, Hasegawa Y, Abiko Y, Yoshida Y, Murakami Y, Yoshimura F. Porphyromonas gingivalis FimA fimbriae: fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA Genotypes. PLoS ONE. 2012;7(9):e43722. doi: 10.1371/journal.pone.0043722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Liang S, Hosur KB, et al. Differential virulence and innate immune interactions of type I and II fimbrial genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol. 2009;24(6):478–484. doi: 10.1111/j.1399-302X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce DL, Nishiyama SI, Liang S, et al. Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infect Immun. 2009;77(8):3294–3301. doi: 10.1128/IAI.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajishengallis G, Wang M, Liang S. Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J Immunol. 2009;182(11):6690–6696. doi: 10.4049/jimmunol.0900524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshino T, Laine ML, van Winkelhoff AJ, Dahlén G. Genotype variation and capsular serotypes of Porphyromonas gingivalis from chronic periodontitis and periodontal abscesses. FEMS Microbiol Lett. 2007;270(1):75–81. doi: 10.1111/j.1574-6968.2007.00651.x. [DOI] [PubMed] [Google Scholar]
- 32.Tonetti MS, D'Aiuto F, Nibali L, et al. Treatment of periodontitis and endothelial function. N Engl J Med. 2007;356(9):911–920. doi: 10.1056/NEJMoa063186. [DOI] [PubMed] [Google Scholar]
- 33.Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Natl Acad Sci USA. 2011;108(Suppl. 1):S4592–S4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skamagas M, Breen TL, LeRoith D. Update on diabetes mellitus: prevention, treatment, and association with oral diseases. Oral Dis. 2008;14(2):105–114. doi: 10.1111/j.1601-0825.2007.01425.x. [DOI] [PubMed] [Google Scholar]
- 35.Friedewald VE, Kornman KS, Beck JD, et al. The American journal of cardiology and journal of periodontology editors' consensus: periodontitis and atherosclerotic cardiovascular disease. Periodontol. 2009;80(7):1021–1032. doi: 10.1902/jop.2009.097001. [DOI] [PubMed] [Google Scholar]
- 36.Nakano K, Inaba H, Nomura R, et al. Distribution of Porphyromonas gingivalis fimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol. 2008;23(2):170–172. doi: 10.1111/j.1399-302X.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- 3.Saito A, Inagaki S, Kimizuka R, et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54(3):349–355. doi: 10.1111/j.1574-695X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 38.Inaba H, Kuboniwa M, Bainbridge B, et al. Porphyromonas gingivalis invades human trophoblasts and inhibits proliferation by inducing G1 arrest and apoptosis. Cell Microbiol. 2009;11(10):1517–1532. doi: 10.1111/j.1462-5822.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Marchesan JT, Morelli T, Lundy SK, et al. Divergence of the systemic immune response following oral infection with distinct strains of Porphyromonas gingivalis. Mol Oral Microbiol. 2012;27(6):483–495. doi: 10.1111/omi.12001. Describes the differences in downstream host responses associated with P. gingivalis genetic variability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biyikoglu B, Ricker A, Diaz PI. Strain-specific colonization patterns and serum modulation of multi-species oral biofilm development. Anaerobe. 2012;18(4):459–470. doi: 10.1016/j.anaerobe.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Igboin C, Griffen A, Leys E. Analysis of Porphyromonas gingivalis strain diversity. J Clin Microbiol. 2009;47(10):3073–3081. doi: 10.1128/JCM.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadkarni M, Chhour K, Browne G, Jacques N, Hunter N. Lysine gingipain (kgp) biovars of Porphyromonas gingivalis exhibit differential distribution on oral mucosal sites. J Clin Microbiol. 2009;47(10):3350–3352. doi: 10.1128/JCM.00753-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Enersen M, Olsen I, Caugant DA. Genetic diversity of Porphyromonas gingivalis isolates recovered from single ‘refractory’ periodontitis sites. Appl Environ Microbiol. 2008;74(18):5817–5821. doi: 10.1128/AEM.00225-08. Illustrates the number of diverse strains of P. gingivalis found in a single periodontal pocket. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inaba H, Nakano K, Kato T, et al. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol. 2008;23(1):29–35. doi: 10.1111/j.1399-302X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 45.Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46(1):31–42. doi: 10.1128/JCM.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodontol Res. 2007;42(6):511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 47.Kato T, Kawai S, Nakano K, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9(3):753–765. doi: 10.1111/j.1462-5822.2006.00825.x. [DOI] [PubMed] [Google Scholar]
- 48.Davila-Perez C, Amano A, Alpuche-Solis AG, et al. Distribution of genotypes of Porphyromonas gingivalis in Type 2 diabetic patients with periodontitis in Mexico. J Clin Periodontol. 2007;34(1):25–30. doi: 10.1111/j.1600-051X.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- 49.Enersen M, Olsen I, van Winkelhoff AJ, Caugant DA. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J Clin Microbiol. 2006;44(1):35–41. doi: 10.1128/JCM.44.1.35-41.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall LMC, Fawell SC, Shi X, et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect Immun. 2005;73(7):4253–4262. doi: 10.1128/IAI.73.7.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40(2):147–152. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 52.Zheng C, Wu J, Xie H. Differential expression and adherence of Porphyromonas gingivalis fimA genotypes. Mol Oral Microbiol. 2011;26(6):388–395. doi: 10.1111/j.2041-1014.2011.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis. J Periodontal Res. 2009;44(1):1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 54.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MPA. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19(4):224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 55.Nakano K, Kuboniwa M, Nakagawa I, et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004;19(3):205–209. doi: 10.1111/j.0902-0055.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 56.Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. Functional differences among fimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70(1):277–285. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ménard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63(7):2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittstock M, Schmidt H, Flemmig TF, Karch H. Heterogeneity of the prtC gene of Porphyromonas gingivalis. Oral Microbiol Immunol. 2001;15(1):33–39. doi: 10.1034/j.1399-302x.2000.150106.x. [DOI] [PubMed] [Google Scholar]
- 59.Califano JV, Arimoto T, Kitten T. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J Periodontal Res. 2003;38(4):411–416. doi: 10.1034/j.1600-0765.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- 60.Chen T, Hosogi Y, Nishikawa K, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186(16):5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rumpf RW, Griffen AL, Leys EJ. Phylogeny of Porphyromonas gingivalis by ribosomal intergenic spacer region analysis. J Clin Microbiol. 2000;38(5):1807–1810. doi: 10.1128/jcm.38.5.1807-1810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koehler A, Karch H, Beikler T, Flemmig TF, Suerbaum S, Schmidt H. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology. 2003;149(Pt 9):2407–2415. doi: 10.1099/mic.0.26267-0. [DOI] [PubMed] [Google Scholar]
- 63.Frandsen EV, Poulsen K, Curtis MA, Kilian M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;69(7):4479–4485. doi: 10.1128/IAI.69.7.4479-4485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enersen M. Porphyromonas gingivalis: a clonal pathogen?: Diversities in housekeeping genes and the major fimbriae gene. J Oral Microbiol. 2011 doi: 10.3402/jom.v3i0.8487. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spratt BG, Maiden MC. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci. 1999;354(1384):701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Ellegren H, Sheldon BC. Genetic basis of fitness differences in natural populations. Nature. 2008;452(7184):169–175. doi: 10.1038/nature06737. Outlines the impact of the genomics revolution on ecological studies of fitness. [DOI] [PubMed] [Google Scholar]
- 67••.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4(8):577–587. doi: 10.1038/nrmicro1460. Focuses on genomics and bacterial pathogen studies. [DOI] [PubMed] [Google Scholar]
- 68.Baltrus DA, Guillemin K, Phillips PC. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution. 2008;62(1):39–49. doi: 10.1111/j.1558-5646.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- 69.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5(6):441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 70.Kang J, Blaser MJ. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat Rev Microbiol. 2006;4(11):826–836. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe T, Maruyama F, Nozawa T, et al. Complete genome sequence of the bacterium Porphyromonas gingivalis TDC60, which causes periodontal disease. J Bacteriol. 2011;193(16):4259–4260. doi: 10.1128/JB.05269-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naito M, Hirakawa H, Yamashita A, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15(4):215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185(18):5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brunner J, Wittink FRA, Jonker MJ, et al. The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis. BMC Microbiol. 2010;10:252. doi: 10.1186/1471-2180-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aduse-Opoku J, Slaney JM, Hashim A, et al. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun. 2006;74(1):449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong H, Chen T, Dewhirst FE, Fleischmann RD, Fraser CM, Duncan MJ. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect Immun. 1999;67(7):3416–3423. doi: 10.1128/iai.67.7.3416-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewis JP, Macrina FL. IS195, an insertion sequence-like element associated with protease genes in Porphyromonas gingivalis. Infect Immun. 1998;66(7):3035–3042. doi: 10.1128/iai.66.7.3035-3042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang CY, Bond VC, Genco CA. Identification of a second endogenous Porphyromonas gingivalis insertion element. J Bacteriol. 1997;179(11):3808–3812. doi: 10.1128/jb.179.11.3808-3812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maley J, Roberts IS. Characterisation of IS1126 from Porphyromonas gingivalis W83: a new member of the IS4 family of insertion sequence elements. FEMS Microbiol Lett. 1994;123(1–2):219–224. doi: 10.1111/j.1574-6968.1994.tb07225.x. [DOI] [PubMed] [Google Scholar]
- 80.Maley J, Shoemaker NB, Roberts IS. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P gingivalis insertion-sequence element. FEMS Microbiol Lett. 1992;72(1):75–81. doi: 10.1016/0378-1097(92)90492-7. [DOI] [PubMed] [Google Scholar]
- 81.Simpson W, Wang CY, Mikolajczyk-Pawlinska J, et al. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67(10):5012–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sawada K, Kokeguchi S, Hongyo H, et al. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect Immun. 1999;67(11):5621–5625. doi: 10.1128/iai.67.11.5621-5625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Høvik H, Yu WH, Olsen I, Chen T. Comprehensive transcriptome analysis of the periodontopathogenic bacterium Porphyromonas gingivalis W83. J Bacteriol. 2012;194(1):100–114. doi: 10.1128/JB.06385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith CJ, Tribble GD, Bayley DP. Genetic elements of Bacteroides species: a moving story. Plasmid. 1998;40(1):12–29. doi: 10.1006/plas.1998.1347. [DOI] [PubMed] [Google Scholar]
- 86.Naito M, Sato K, Shoji M, et al. Characterization of the Porphyromonas gingivalis conjugative transposon C TnPg1: determination of the integration site and the genes essential for conjugal transfer. Microbiology. 2011;157(7):2022–2032. doi: 10.1099/mic.0.047803-0. [DOI] [PubMed] [Google Scholar]
- 87.Bélanger M, Rodrigues P, Progulske-Fox A. Genetic manipulation of Porphyromonas gingivalis. Curr Proto Microbiol. 2007;Chapter 13 doi: 10.1002/9780471729259.mc13c02s05. Unit13C.2. [DOI] [PubMed] [Google Scholar]
- 88.Tribble GD, Lamont GJ, Progulske-Fox A, Lamont RJ. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J Bacteriol. 2007;189(17):6382–6388. doi: 10.1128/JB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bacic M, Parker AC, Stagg J, et al. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J Bacteriol. 2005;187(8):2858–2869. doi: 10.1128/JB.187.8.2858-2869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roberts AP, Mullany P. Genetic basis of horizontal gene transfer among oral bacteria. Periodontology 2000. 2006;42:36–46. doi: 10.1111/j.1600-0757.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 91.Tribble GD, Rigney TW, Dao DHV, et al. Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis. MBio. 2012;3(1):e00231–11. doi: 10.1128/mBio.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011;365(4):347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu L, Chen X, Skogerbø G, et al. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics. 2012;100(5):265–270. doi: 10.1016/j.ygeno.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 94•.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–244. doi: 10.1038/nature10571. Identified genes transferred between species and identified the oral cavity as a site of frequent gene sharing. [DOI] [PubMed] [Google Scholar]
- 95.Leys EJ, Smith JH, Lyons SR, Griffen AL. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J Clin Microbiol. 1999;37(12):3906–3911. doi: 10.1128/jcm.37.12.3906-3911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hajishengallis G. Porphyromonas gingivalis–host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11(6–7):637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 98.Noor SO, Ridgway K, Scovell L, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99••.Pédron T, Sansonetti P. Commensals, bacterial pathogens and intestinal inflammation: an intriguing ménage à trois. Cell Host Microbe. 2008;3(6):344–347. doi: 10.1016/j.chom.2008.05.010. Reviews the potential interactions between the host flora and intestinal inflammation. [DOI] [PubMed] [Google Scholar]
- 100.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105(39):15070, 15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vaarala O, Atkinson MA, Neu J. The ‘perfect storm’ for Type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.Porphyromonas gingivalis MLST database. http://pubmlst.org/pgingivalis.