Abstract
Chronic hepatitis B virus (HBV) infection remains an important global health problem. Stability of the episomal covalently closed circular HBV DNA (cccDNA) is largely responsible for the modest curative efficacy of available therapy. Since licensed anti-HBV drugs have a post-transcriptional mechanism of action, disabling cccDNA is potentially of therapeutic benefit. To develop this approach, we engineered mutagenic transcription activator-like effector nucleases (TALENs) that target four HBV-specific sites within the viral genome. TALENs with cognate sequences in the S or C open-reading frames (ORFs) efficiently disrupted sequences at the intended sites and suppressed markers of viral replication. Following triple transfection of cultured HepG2.2.15 cells under mildly hypothermic conditions, the S TALEN caused targeted mutation in ~35% of cccDNA molecules. Markers of viral replication were also inhibited in vivo in a murine hydrodynamic injection model of HBV replication. HBV target sites within S and C ORFs of the injected HBV DNA were mutated without evidence of toxicity. These findings are the first to demonstrate a targeted nuclease-mediated disruption of HBV cccDNA. Efficacy in vivo also indicates that these engineered nucleases have potential for use in treatment of chronic HBV infection.
Introduction
There are 387 million people who are chronically infected with hepatitis B virus (HBV) and at high risk for cirrhosis and hepatocellular carcinoma.1,2 Annually these complications cause ~600,000 deaths and HBV infection remains as an important global public health problem. The virion contains a relaxed circular DNA (rcDNA) genome that is formed following reverse transcription of HBV pregenomic RNA (pgRNA). After infection of hepatocytes, rcDNA is “repaired” to produce covalently closed circular DNA (cccDNA).3 This stable replication intermediate serves as the template for transcription of viral pgRNA and protein-coding mRNAs. The viral genome, which is remarkably compact, contains precore/core (C), polymerase (pol), surface (S) and X open-reading frames (ORFs) (Figure 1a).4 The pol ORF is the largest and encodes the enzyme responsible for priming and reverse transcription of pgRNA. Pre S1, pre S2 and S in-phase start codons of the S ORF initiate translation of the large, middle and major surface antigens (HBsAgs), respectively. The C ORF contains core and precore initiation codons that start translation of the overlapping nucleocapsid protein and secreted HBV e antigen. X encodes a protein that regulates viral gene transcription,5 is required for natural infection in vivo6,7 and is implicated in hepatocarcinogenesis.8 Each HBV ORF overlaps at least one other, and together they cover the entire genome. The regulatory cis-elements are therefore included within the protein-coding regions in a highly compact arrangement that restricts HBV sequence plasticity. As a result, emergence of escape mutants is limited and HBV is particularly susceptible to disabling effects of mutation.
Figure 1.
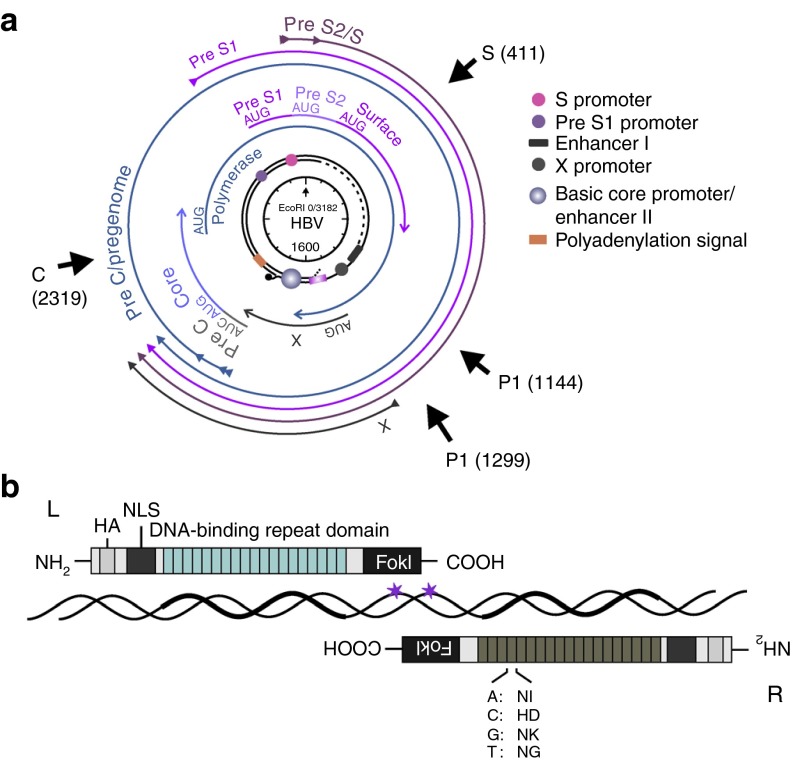
HBV target sequences and design of TALENs. (a) HB virion rcDNA, which is converted to cccDNA following hepatocyte infection, is schematically indicated at the center of the circular genome map. Nucleotide co-ordinates are calculated clockwise from the unique EcoRI site that occurs in HBV DNA. Approximate location of promoter, enhancer and cis regulatory elements are indicated as circular and rectangular symbols. The four viral ORFs, core, polymerase, surface and X, are shown as arrows immediately surrounding the genome. The four major viral transcripts are indicated as outermost arrows. Regions targeted by C, S, P1 and P2 TALENs are indicated as axial arrows. The HBV genome co-ordinates of the first nucleotide that is targeted by each of the TALENs is provided in parentheses. (b) Schematic representation of the left (L) and right (R) subunits comprising a TALEN dimer. The diagram shows N- and C- terminals, with hemaglutinin epitope (HA), nuclear localization signal (NLS), DNA binding domain and C-terminal FokI nuclease domain. Amino acids providing target base specificity of the repeat variable sequences are indicated below the R TALEN subunit.
Currently licensed HBV drugs include competitive reverse transcriptase inhibitors and immunomodulators.9 Availability of anti-HBV agents that counter the virus by disabling cccDNA would be useful to prevent reemergence of viral proliferation following treatment withdrawal. Nucleases that can be designed to cut cccDNA specifically have potential therapeutic utility. Repeated digestion of cccDNA by such endonucleases followed by nonhomologous end joining is mutagenic and therefore may inactivate the HBV replication intermediate. Engineered zinc finger nucleases (ZFNs) have been employed with moderate success to mutate HBV DNA.10 Co-transfection of cells in culture with ZFN pairs and plasmids containing HBV sequences resulted in cleavage of up to 26% of target sites. In a separate study, zinc finger proteins without nuclease domains were used to bind duck HBV regulatory elements.11 Viral gene expression was inhibited, but the suppression was transient.
Engineered transcription activator-like effectors (TALEs) derived from the naturally occurring Xanthomonas plant pathogen have recently shown promise as alternative DNA-targeting proteins.12 Nuclease domains may be coupled to TALEs to form transcription activator-like effector nucleases (TALENs), which are capable of directed cleavage of specific DNA sequences. This site-specific cleavage has been reported to occur with greater efficiency, specificity and less toxicity than is achieved with ZFNs.13 Utility of TALEs and TALENs has been demonstrated in genetic studies,12 although to our knowledge none has reported potential therapeutic efficacy in vivo in disease models. To investigate the application of mutagenic nucleases to disabling HBV cccDNA, we have engineered TALENs that target four conserved and HBV-specific sites within the viral genome. We show that the TALENs with cognate sequences in the S or C ORFs efficiently introduce HBV-disabling mutations at the intended target sites in cell culture and in vivo.
Results
Propagation of HBV-targeting TALENs
To assess the utility of HBV cccDNA-targeting TALENs, we generated four TALENs that target sites within the S/pol, C/pol and pol ORFs of the HBV genome (Figure 1a). TALEN subunit pairs were derived from the AvrBs4 TALE protein scaffold (NH variant) as has previously been described.13 Left and right subunits were designed to bind two sequences of 19 nucleotides, each with a T residue at the 5′ end, on the sense and antisense strands of cccDNA. For optimal C-terminal FokI nuclease cleavage efficiency, the targets were separated by a spacer of 13 bp (Figure 1b) which has been reported to be optimal for the chosen TALEN architecture.13 The complete TALEN subunits each also included a nuclear localization signal and hemagglutinin epitope (HA). Conservation amongst viral isolates and absence of homologous sequences in mouse and human genomes were also used as criteria for selecting potential cccDNA targets. Comparison of HBV target sequences from representative genotypes of the World Health Organization (WHO) reference panel14 showed few mismatches at the intended TALEN subunit cognates (Supplementary Figures 1–4, online). An exception was found within the HBV A1, A2 and A3 subgenotype consensus sequences that were targeted by the R subunit of the C TALEN (Supplementary Figure 2, online). The target of these subgenotype viral consensus sequences contained an insertion of six base pairs. Detailed BLAST searching of human and murine genomes was carried out to identify potential off target binding of HBV TALENs. Sequences with 15 or more matches out of the 19 bases targeted by each TALEN subunit are provided in Supplementary Tables 1–4, online. A maximum sequence identity of 18 out of 19 bases was found, and none of the potential off target sites contained exact matches. Importantly, combinations of potential left and right TALEN cognates in human and murine DNA were positioned very far apart. Arrangement of the subunits on human or mouse DNA is therefore highly unlikely to favor mutagenic double stranded nuclease activity. Further analysis using TALENT 2.0 paired target finder software15 also did not identify potential human and mouse cognates for either the S or C TALENs or each of their dual left and dual right homodimers.
Anti-HBV efficacy of TALENs in cell culture
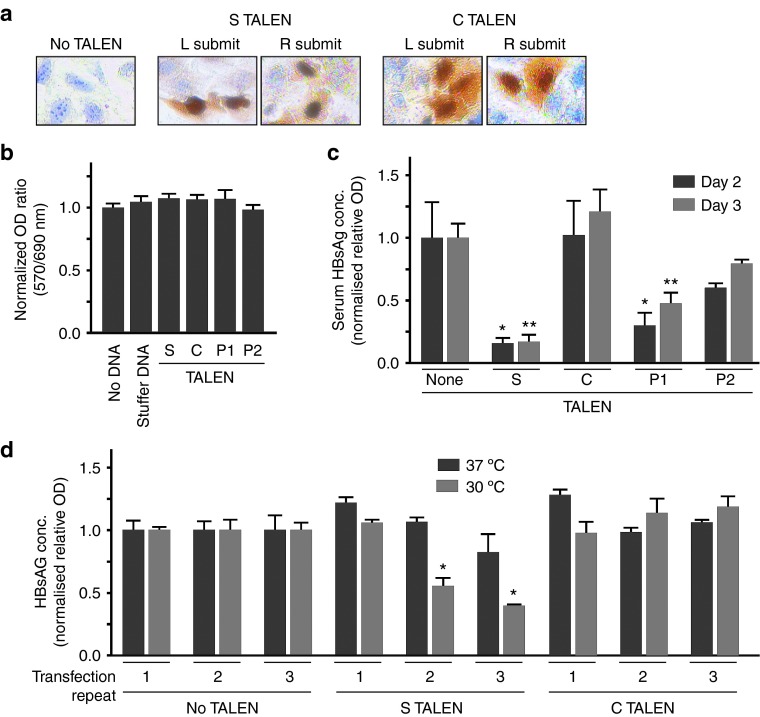
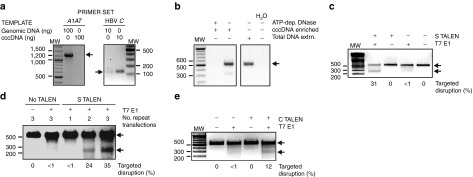
Transfection of cultured liver-derived Huh7 cells with TALEN-encoding plasmids, followed by immunodetection of the HA epitope, verified that nuclear expression occurs (Figure 2a) without evidence for cellular toxicity (Figure 2b and Supplementary Figure 5, online). Initial assessment of TALEN efficacy was determined after transient co-transfection of Huh7 cells with the pCH-9/3091 HBV replication-competent plasmid.16 Concentrations of HBsAg in the culture supernatants were significantly diminished in cells that had been transfected with the S TALEN and P1 TALEN (Figure 2c). Subsequent analysis indicated that inhibitory effects of the P1 TALEN might be through a transcriptional suppression mechanism rather than by direct cleavage of the target HBV DNA (see below). To assess efficacy in a more stringent model of HBV replication, the HepG2.2.15 cell line17 was transfected one- to three-times with S TALEN-expressing plasmids (Figure 2d). HBV replication in the HepG2.2.15 line occurs rapidly and each cell contains ~10 copies of cccDNA.17,18 This number is higher than the 1.5 copies per hepatocyte that has been estimated to be the average in liver cells of humans chronically infected with HBV.19 To enhance detection of TALEN-mediated HBV replication inhibition, cells were also cultured under mildly hypothermic conditions at 30 °C.20 Although not established conclusively, mildly hypothermic conditions are thought to slow DNA replication and cell division without significantly diminishing nuclease activity. As a result, mutagenic effects of nuclease digestion are more easily detectable when cells are cultured at the lower temperature. At both 37 and 30 °C, the initial transfection had no significant effect on HBsAg concentration in the culture supernatants. However, the second and third transfections had increasingly inhibitory effects on HBsAg secretion when cells were cultured under hypothermic conditions. As expected, transfection of the C TALEN, which targets a different part of the HBV genome, did not affect HBsAg secretion (Figure 2c,d). The P1 TALEN and P2 TALEN had modest or no inhibitory effect on HBsAg secretion from HepG2.2.15 cells respectively (Supplementary Figure 6, online).
Figure 2.
Expression and anti-HBV efficacy of TALENs in cultured cells. (a) Immunodetection of each of the L or R subunits of the S or C TALENs following plasmid transfection of cultured Huh7 cells. Representative fields under 400× magnification are shown. (b) Cell viability of liver-derived Huh7 cells following transfection of TALEN-encoding plasmids, measured using the MTT assay, was carried out 48 hours after transfection. (c) ELISA-based determination of concentrations of HBsAg in culture supernatants of Huh7 cells at time points of 2 and 3 days following transfection with pCH-9/3091 and each of the TALENs. (d) HBsAg concentrations in culture supernatants of HepG2.2.15 cells that were subjected to repeat transfections with plasmids encoding the S TALEN and C TALEN under hypothermic (30 °C) and normothermic (37 °C) temperatures. Data are represented as the means and the error bars indicate the SEM. Statistically significant differences are indicated by asterisks (*P < 0.05 and **P < 0.01).
TALEN-mediated targeted mutagenesis of cccDNA
To assess targeted TALEN-mediated mutagenesis of cccDNA, circular duplex DNA that had been subjected to ATP-dependent DNase treatment was isolated from HepG2.2.15 cells. PCR-based analyses were initially carried out to assess contamination of the cccDNA sample with cellular genomic DNA and HBV rcDNA (Figure 3a,b). To detect genomic DNA contamination, primer sets that amplify HBV C DNA or control cellular genomic sequences located in the A1AT gene were used. With the A1AT gene primers, DNA was efficiently amplified from cellular genomic DNA but not when using the cccDNA preparation as template (Figure 3a). Primers targeting the C sequence of HBV amplified DNA from both cccDNA and HepG2.2.15 cell genomic DNA preparations. Efficiency of amplification was however considerably higher when using the cccDNA preparation as template. Amplification of HBV C sequences from the genomic template was expected, and is likely to be derived from stable HBV integrants within HepG2.2.15 cells. To verify removal of rcDNA, DNA prepared from serum of HBV transgenic mice21 using different procedures was subjected to amplification with S gene primers (Figure 3b). Serum from these mice does not contain cccDNA. Each extracted serum sample contained approximately 5 × 106 copies of HBV rcDNA, which is a threefold excess of the estimated number of rcDNA copies in the starting material derived from the HepG2.2.15 cells. An S gene amplicon was not detectable when using the serum-derived sample that was subjected to the method used for cccDNA preparation from HepG2.2.15 cells. This procedure included cccDNA-enrichment and ATP-dependent DNase treatment. Collectively these data indicate that the cccDNA preparation used for the T7 endonuclease assays were not significantly contaminated with either cellular genomic DNA or HBV rcDNA.
Figure 3.
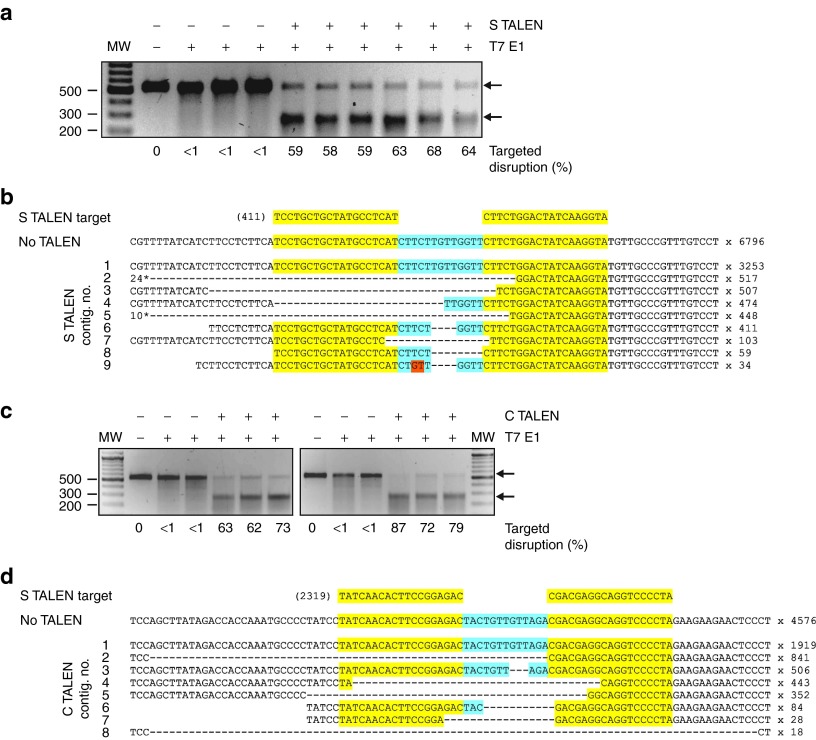
Targeted disruption of cccDNA extracted from HepG2.2.15 cells. (a) PCR analysis using A1AT or HBV C gene primer sets carried out on total genomic DNA or cccDNA isolated from HepG2.2.15 cells. Arrows indicate the A1AT and C gene amplicons of expected sizes of 1,224 and 125 bp, respectively. Sizes of DNA fragments (bp) in the molecular weight marker lane (MW) are shown on the left. (b) PCR amplification of DNA isolated from HBV transgenic mouse serum using the HBV S gene primer set. Template DNA preparation entailed methods using various combinations of total DNA extraction, cccDNA-enrichment and treatment with ATP-dependent DNase, or a no DNA template blank (H2O). The arrow indicates the amplicon of expected size of 520 bp. (c) T7E1 assay conducted on cccDNA isolated from HepG2.2.15 cells that had been subjected to three transfections with the S TALEN-encoding plasmids under hypothermic conditions. The arrows indicate the larger intact PCR product (520 bp) and smaller digested fragment (~260 bp). The measured target disruption is indicated below as a percentage. (d) Assessment of effects of each repeat transfection on targeted disruption of HBV sequences. Controls included DNA that had been isolated from cells that did not receive TALEN or amplicons that were not subjected to T7E1 digestion. (e) T7E1 assay carried out using similar procedures described in c, but after transfection of HepG2.2.15 cells with DNA encoding the C TALEN under mildly hypothermic conditions. In all panels, inverted images of ethidium bromide-stained gels are depicted.
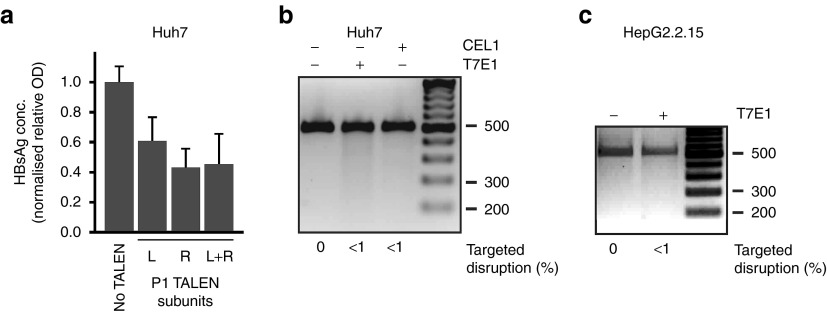
Assays using T7E1 (Figure 3c,d,e) and CEL1 (Supplementary Figure 7, online), were carried out on cccDNA preparations. According to the T7E1 assay, targeted disruption occurred in ~31% of cccDNA molecules after three transfections with the S TALEN under hypothermic conditions (Figure 3c). Moreover, increased targeted disruption of the cccDNA sequence following repeated transfection of HepG2.2.15 correlated with inhibition of HBsAg secretion (Figures 2d, 3d). After similar transfections of HepG2.2.15 cells, the C TALEN achieved disruption of cccDNA in 12% of C ORF targets (Figure 3e) but HBsAg secretion was not affected (Figure 2c,d). Interestingly, the P1 TALEN inhibited HBsAg secretion from transiently transfected Huh7 cells (Figure 2c) and HepG2.2.15 cells subjected to hypothermic triple transfection (Supplementary Figure 6, online). The effect in Huh7 cells was observed after transfection of the left and right P1 TALEN subunits individually (Figure 4a). However, targeted sequence disruption was not detectable (Figure 4b,c). Although the exact inhibitory mechanism is unclear, the P1 TALE elements may cause transcriptional repression through interaction with cognate sequences located within HBV enhancer I (Figure 1a).
Figure 4.
Inhibition of HBsAg secretion without targeted HBV DNA disruption following transfection by P1 TALEN-encoding plasmids. (a) ELISA-based determination of concentrations of HBsAg in culture supernatants of Huh7 cells at day 2 following transfection with plasmids encoding L, R and the combination of L and R P1 TALEN subunits. (b) T7E1 or CEL1 assays conducted on DNA isolated from Huh7 cells that had been subjected to transfection with each and a combination of L and R P1 TALEN-encoding plasmids. (c) T7E1 assay conducted on DNA isolated from HepG2.2.15 cells that had been subjected to three repeat transfections, under hypothermic conditions, with the combination of L and R P1 TALEN-encoding plasmids. In panels b and c, inverted images of ethidium bromide-stained gels are depicted.
Inhibition of HBV replication in vivo by TALENs
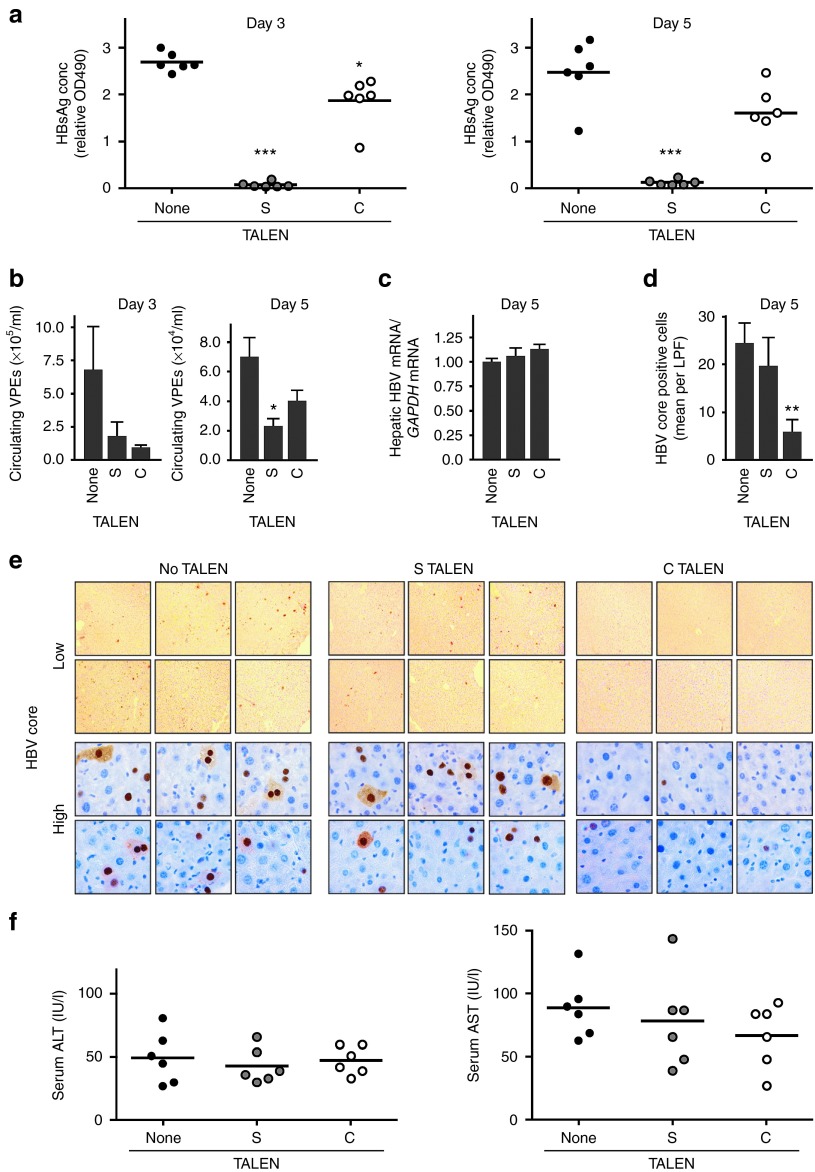
The hydrodynamic injection (HDI) method22 was employed to determine the effects of S and C TALENs on HBV replication in vivo. A plasmid that constitutively expresses Firefly luciferase was included to verify equivalent transfection efficiencies in vivo (Supplementary Figure 8, online). Anti-HBV effects of the TALENs were assessed by measuring serum HBsAg concentrations (Figure 5a) and quantitative PCR (qPCR) to determine viral particle equivalents (VPEs) (Figure 5b). The S TALEN effected knockdown of HBsAg by >90% on both days 3 and 5 after HDI. Circulating VPEs were decreased by ~70% after administration of either of the S or C TALENs. This result is in accordance with data from analysis in cultured cells (Figure 2c,d), and is likely to reflect specificity of the TALENs for their cognates. Intrahepatic mRNA concentrations were similar in control and TALEN-treated mice (Figure 5c). This observation may be expected as C and S TALENs do not have cognates within cccDNA transcription regulatory elements (Figure 1a) and are therefore unlikely to influence transcription activity. Since the S ORF is distinct from the C sequence, targeted disruption by the S TALEN should not affect HBV core antigen (HBcAg) expression. Indeed immunohistochemical detection of HBcAg in the liver of mice treated with the S TALEN was similar to that of the control animals. However, there was a decrease in measurable HBcAg in hepatocytes of mice that had received the C TALEN (Figure 5d,e). Histological assessment (Supplementary Figure 9, online) and measurement of serum transaminases (AST and ALT) (Figure 5f) indicated a minimal toxic effects in all groups, which is normal during recovery from the HDI procedure.
Figure 5.
Anti-HBV efficacy of TALENs in vivo. (a) Groups of mice, each comprising six animals, were subjected to hydrodynamic injection with plasmids encoding S or C TALENs, HBV sequences and Firefly luciferase. Serum HBsAg concentrations were measured on days 3 (left panel) and 5 (right panel) in samples from each of the animals. (b) Circulating VPEs were determined using quantitative PCR on days 3 (left panel) and 5 (right panel) following HDI. (c) Using quantitative RT-PCR, intrahepatic HBV S mRNA concentrations were measured on day 5 following HDI. Results are presented as a ratio of HBV to GAPDH housekeeping mRNA concentrations. (d) HBV core antigen-positive cells per low power field (LPF) for each of the six mice of the three groups that had been subjected to HDI with or without TALEN-expressing plasmids. Data represent the mean numbers of positive cells and with error bars indicating the SEM. (e) Immunohistochemical detection of HBcAg in representative sections of animals that had been subjected to HDI with or without TALEN-expressing plasmids. Low (100×) and high (1,000×) power fields are shown. (f) Serum alanine transaminase (ALT, left) and aspartate transaminase (AST, right) activity in individual mice at day 5 following coadministration of plasmids encoding TALENs, HBV sequences and luciferase reporter. Data are represented as the means and the error bars indicate the SEM. Statistically significant differences are indicated by asterisks (*P < 0.05, **P < 0.01 and ***P < 0.001).
Targeted disruption of HBV sequences following HDI
The T7E1 assay demonstrated efficient target-specific mutagenesis of DNA extracted from livers of mice that had been treated with the S and C TALENs (Figure 6a,c). Detectable mutation in control mice was consistently <1%, while in the S and C TALEN-treated mice, 58–87% of amplified HBV sequences contained mutations. This proportion is significantly higher than what was observed in cultured HepG2.2.15 cells (Figure 3c,e). A higher ratio of TALEN-expressing plasmids to HBV target plasmid DNA following co-transfection in vivo using HDI is likely to be the primary reason for this. Moreover, the high cccDNA copy number in HepG2.2.15 cells makes targeted sequence disruption difficult to achieve in these cells. Since cccDNA is not formed when HBV replication-competent DNA is introduced into murine hepatocytes,23 structural variance of plasmid HBV DNA may also improve TALEN efficacy in vivo. Sequencing of HBV DNA amplified from hepatic extracts confirmed that intended target sites were mutated (Figure 6b,d). Wildtype sequences were exclusively detected in animals that did not receive TALENs, but a variety of target-specific sequence alterations was detected in DNA from animals receiving the S or C TALENs. Deletions within the target sites were the predominant mutation, which is in agreement with the recently reported analysis of TALEN-induced targeted disruptions in mammalian cells.24 Some mutations within the targets were unexpectedly particularly commonly represented. Although the exact reason for this is not clear, an unavoidable bias introduced during the two PCR amplifications of the deep sequencing protocol may be responsible. Nevertheless, collectively these results demonstrate that HBV-targeting TALENs are active in vivo and are capable of introducing disabling mutations at their intended HBV cognates.
Figure 6.
Targeted disruption of HBV DNA sequences in vivo. (a) T7E1 assay was carried out on hepatic DNA isolated from animals using similar procedures described in Figure 3. Each lane represents data obtained from an individual animal. Mice were subjected to HDI with solutions containing control (−) or S TALEN-encoding DNA (+). Heteroduplex DNA was treated in the presence (+) or absence (−) of T7E1. (b) Sequencing of S TALEN target regions following HDI. Amplified DNA was pooled from each of the groups of six mice and subjected to deep sequencing. TALEN targets are highlighted in yellow and the spacer sequence in blue. Commonly detected sequences are shown together with the number of sequence reads indicated on the right. Deletions are represented by dashed lines and substitutions are highlighted in orange. Contig. 2 and contig. 5 each had additional deletions of 24 and 10 bp respectively at their 5′ ends. (c) T7E1 assay carried out using procedures described in Figure 3 and a above, except that animals received control (−) or the C TALEN-encoding DNA(+). (d) Sequencing of C TALEN target regions following HDI. Data preparation and analysis was performed as described above in b.
Discussion
Targeted editing of genome sequences is now a rapidly expanding field. Recently reported successes of sequence-specific DNA modification indicate that the technology has considerable potential and may find use in treatment of a variety of diseases, including those caused by viral infections.25 To effect mutations, earlier studies employed homing endonucleases, such as I-SceI,26 chemical-based nucleases,27 mutagenic single stranded adeno-associated viruses28 and ZFNs.29 Although intended target sequence modifications were observed, considerable technical hurdles have impeded widespread use of these methods. Developments applying TALENs12 and the Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) associated (Cas)9-based RNA-guided nucleases21,30 to modifying genomic targets has been particularly impressive. However, recent evidence indicates that CRISPR-Cas nucleases are prone to off target mutagenesis.31 Among the designer nucleases, TALENs have been used successfully against a range of target sites and are currently considered as the most powerful platform for nuclease-based gene editing. Indeed, when compared with targeted HBV mutagenesis that has been reported when using ZFNs,10 the TALEN-mediated disruption to the viral sequences reported here is more efficient. Moreover, compared to ZFNs, TALENs have superior specificity, diminished toxicity and more predictable interaction with their targets.13
To date, TALENs have largely been used for modification of endogenous cellular genes. Efficient gene disruption has been observed in a variety of organisms.12 Most studies have been concerned with developing models for functional analysis, and little work has been carried out on using TALENs to disable pathology-causing genes, such as those encoded by viruses. The finding that TALENs are capable of efficiently disabling HBV targets therefore represents a substantial advance in therapeutic application of designer nucleases. Importantly, targeted disruption of cccDNA and concomitant decreases in markers of viral replication were demonstrated in HepG2.2.15 cells. Although cccDNA is not formed during HBV replication in the mice subjected to HDI, disruption of the intended HBV sequences without overt evidence of toxicity is a significant observation. Detailed analysis of the human or mouse genomes revealed that these organisms do not contain sites that would be predicted to be suitable S and C TALEN targets, which suggests that unintended double stranded breaks are unlikely to occur in normal mouse and human genomes. The highly economical use of HBV genetic material limits sequence plasticity and ability of the virus to escape the disabling effects of site-specific nucleases. Mutations, such as the deletions reported here, should drastically reduce viral fitness and render the virus replication defective. Efficient functioning in vivo, coupled with cccDNA disruption in HepG2.2.15 cells is, to the best of our knowledge, the first demonstration that shows potential antiviral therapeutic utility of engineered TALENs.
Despite encouraging results showing utility of TALENs against HBV, several challenges remain to be met before designer nucleases are used for HBV treatment. As with most gene therapies, achieving safe and efficient delivery of the therapeutic transgenes remains difficult. Engineering recombinant TALEN-expressing virus vectors and assessment in animal models where HBV cccDNA production occurs, such as xenografted uPA SCID mice32 and woodchucks infected with woodchuck hepatitis virus,33 will be important for preclinical evaluation. Information from these studies will also provide insights into the duration of TALEN expression that will be required to eliminate virus replication. It is likely that brief expression will be desirable to limit unintended effects and immune responses to the transgenes. An added consideration for the longer-term goals of using TALENs to treat HBV infection in humans, is that viral sequences are frequently integrated into the host hepatocyte genome.34 A deleterious effect, if any, of TALENs cleaving DNA at these sites of HBV integration is yet to be determined. Despite remaining hurdles, the efficiency and specificity of HBV DNA-targeting TALENs augurs well for use of these enzymes to inactivate expression of pathology-causing genes.
Materials and Methods
Propagating TALEN-expressing cassettes. TALEN subunit pairs, derived from the TALE protein AvrBs4 scaffold,13 were designed to target the S, C and pol ORFs of the HBV, subtype D (Figure 1a). TALE repeat arrays were assembled using Type IIS restriction enzyme cleavage and ligation as has been described.35 Repeat arrays comprising the left (L) and right (R) subunits of each complete TALEN were subsequently inserted into destination plasmid vectors encoding an upstream immediate early CMV promoter/enhancer, nuclear localization signal, HA and FokI nuclease domain (Figure 1b).13 Dimeric TALENs targeted sequences in the C (C TALEN), S (S TALEN) and pol (P1 and P2 TALENs) of HBV (Figure 1a).
Transfection of liver-derived cells and HBV knockdown analysis by ELISA. Two different liver-derived cell lines, Huh7 and HepG2.2.15,36 were used to determine TALEN efficacy in culture. An HBV replication-competent plasmid, pCH-9/3091,16 was used in transient cotransfections of Huh7 cells with plasmids expressing TALENs. Cells were cultured in DMEM (Lonza, Basel, Switzerland) supplemented with 10% FCS (Gibco BRL, UK), penicillin (100,000 U/ml), streptomycin (100,000 μg/ml), and maintained in a humidified incubator at 37 °C and 5% CO2.
Huh7 cells were seeded in 12-well plates at a density of 120,000 cells per well, one day prior to transfection. Polyethylenimine was used to transfect cells with 200 ng pCH-9/3091, 200 ng pCMV-GFP, and either 800 ng of plasmid expressing the left TALEN (SL, CL, P1L, P2L) and 800 ng of plasmid expressing the corresponding right TALEN (SR, CR, P1R, P2R), or 1,600 ng of pUC118. Fluorescence microscopy to detect GFP expression was used to confirm equivalent transfection efficiencies. HBsAg concentrations were measured using the Monolisa HBsAg ULTRA kit (Bio-Rad, CA, USA). Immunohistochemical detection of the HA epitope was performed using a primary mouse anti-HA monoclonal antibody (Sigma, MO, USA) in conjunction with the Ultra-Sensitive ABC Mouse IgG Staining Kit (Thermo Scientific, IL, USA). Cells were stained with 3,3′-diaminobenzidine (DAB) and counter-stained with hematoxylin.
HepG2.2.15 cells were seeded in six-well plates at a density of 140,000 cells per well, 1 day prior to transfection. Two hours before transfection, baseline HBsAg concentrations were measured. Polyethylenimine was used to transfect cells with 400 ng pCMV-GFP and either 2.3 µg of plasmid expressing the Left TALEN (SL, CL, P1L, P2L) and 2.3 µg of plasmid expressing the corresponding Right TALEN (SR, CR, P1R, P2R), or 4.6 µg of pUC118. One set of cells was incubated under standard growth conditions (37 °C and 5% CO2) while the other was maintained under mildly hypothermic conditions (30 °C and 5% CO2).20 Growth medium was replaced and HBsAg concentrations were measured on day 2 and day 3. On day 5, cells were harvested and a 1:5 dilution reseeded before repeat transfection.
In vitro cell viability assay. The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (Sigma–Aldrich, MO, USA) colorimetric assay was used to determine TALEN-associated toxicity. Huh7 cells, 15,000 per well, were seeded in 96-well plates on the day prior to transfection. Groups of replicates included cells that were untransfected, mock transfected (25 ng pCH-9/3091, 25 ng pCMV-GFP, 200 ng pUC118) or transfected with TALENs (25 ng pCH-9/3091, 25 ng pCMV-GFP, 100 ng left TALEN plasmid and 100 ng right TALEN plasmid). Cell viability was assessed 3 days after transfection by adding 20 µl of 5 mg/ml MTT to each well, and incubating at 37 °C for one hour. Metabolism of the MTT to form the blue formazan was determined by measuring the ratio of optical density at a wavelength of 570 nm, to the background at 690 nm. In addition, a GFP survival assay was carried out according to a previously described protocol13 with minor modifications. Briefly, Huh7 liver cells were transfected, using procedures described above, with a four-fold excess of either S TALEN- or C TALEN-expressing plasmids relative to the GFP-encoding sequence. GFP-positive cells were counted after 48 hours and compared with the number detected in cells that had been transfected with control DNA.
Murine hydrodynamic injection of TALEN-expressing plasmids. Efficacy in vivo of TALENs was assessed using the HDI model of HBV replication.22 All experiments on animals were conducted according to protocols approved by the University of the Witwatersrand Animal Ethics Screening Committee. The bolus injectate was administered to 6-week-old NMRI mice as a saline solution comprising 10% of body weight. These solutions contained 8 µg HBV target DNA (pCH-9/3091), and either 32 µg of mock pUC118 or 16 µg of pairs of left and right TALEN-expressing plasmids. Additionally, 5 µg of reporter gene plasmid (pCMV-FLuc) was included as a control for delivery. To confirm equivalent hepatic delivery of plasmids, mice received an intraperitoneal injection of 150 mg/kg of d-luciferin (GoldBio, St. Louis, MO) on day 3, which was followed by bioluminescence imaging using a CaliperLS IVIS bioluminescence system (PerkinElmer, MA, USA). Blood was collected and serum diluted threefold in 1% PBS. Viral DNA was extracted using the QIAamp DNA Mini Kit blood spin protocol (QIAGEN, Hilden, Germany). AST and ALT activities were measured using an automated photometric analyzer (Roche Diagnostics, Switzerland). Mice were killed on day 5 and livers harvested. Total RNA was extracted from 100 mg of mouse liver using TRI Reagent (Sigma–Aldrich) followed by reverse transcription using the QuantiTect reverse transcription kit (QIAGEN, Germany). DNA extracted from circulating viral particle equivalents (VPEs) and reverse transcribed hepatic RNA was assayed using quantitative real-time PCR as previously described.37 Immunohistochemistry was used to determine intrahepatic HBcAg expression using Novocastra Lyophilized HBcAg mouse monoclonal antibody (Leica biosystems, Wetzlar, Germany).
T7 endonuclease 1 assay. To confirm that TALEN cleavage and targeted sequence disruption occurred at the intended target site, a mismatch-sensitive T7 endonuclease 1 (T7E1) (New England BioLabs, MA, USA)13 or CEL1 extract38 was used. HBV cccDNA was extracted from HepG2.2.15 cells as previously described39 with a minor modification employing EconoSpin plasmid binding resin (Epoch Life Sciences, Missouri, TX) followed by treatment with ATP-dependent DNase (Plasmid-safe, Epicenter Biotechnologies, Madison, WI) according to the manufacturer's instructions. Genomic DNA was extracted from HepG2.2.15 cells or mouse liver tissue using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). Total DNA was isolated from HBV serum of transgenic mice21 using procedures described above and employed previously.12 PCR analysis was used to confirm that cccDNA was successfully isolated from HepG2.2.15 cells. The alpha1 antitrypsin (A1AT) genomic DNA sequence and HBV C ORF respectively were amplified using standard conditions with the following primer sets: A1AT F 5′ TTCCCTGGTCTGAATGTGTG 3′ and A1AT R 5′ ACTGTCCCAGGTCAGTGGTG 3′ HBV C F 5′ ACCACCAAATGCCCCTAT 3′ and HBV C R 5′ TTCTGCGACGCGGCGA 3′. For A1AT amplification, 100 ng of either HepG2.2.15 genomic DNA or cccDNA preparation was used as template, whereas 10 ng of the similar DNA preparations were used as template for the HBV C ORF amplification. Sequences flanking the S and C TALEN binding sites were amplified under standard conditions using the following primer sets: S F 5′ CCTAGGACCCCTTCTCGTGT 3′ and S R 5′ ACTGAGCCAGGAGAAACGGG 3′ C F 5′ GAACTAATGACTCTAGCTACCT 3′ and C R 5′ CCTACAAACTGTTCACATTT 3′. PCR products were subjected to heteroduplex formation after denaturing 150 ng of amplified DNA at 95 °C for 5 minutes followed by slow cooling to 35 °C. Samples were treated with 3 U of T7E1 in 1× NEB buffer 2 (New England BioLabs, Ipswich, MA), incubated at 37 °C for 15 minutes then resolved electrophoretically. ImageJ software (version 1.46)40 was used to measure band intensities and targeted disruption was determined using the method described by Guschin et al.41
Sequencing. The GS Junior System (Roche 454 Life Sciences, Branford, CI) was used to perform ultra-deep sequencing of murine liver-derived DNA amplicons. Sequencing primers were designed to amplify a 450 bp region flanking the S and C TALEN binding sites. Multiplex identifiers (MIDs) were used in each primer set to distinguish between amplicons from the different groups of mice. Primers for mock and S TALEN HDI samples were: Mock F MID3 5′ CGTATCGCCTCCCT CGCGCCATCAGAGACGCACTCTCTCAATTTTCTAGGGGGAACTACCGTG 3′ Mock R MID3 5′ CTATGCGCCTTGCCAGCCCGCTCAGAGACGCACTCCGTCCGAAGGTT TGGTACAGCAACAG 3′ and S F MID4 5′CGTATCGCCTCCCTCGCGCCATCAGA GCACTGTAGTCTCAATTTTCTAGGGGGAACTACCGTG 3′ S R MID4 5′ CTATGC GCCTTGCCAGCCCGCTCAGAGCACTGTAGCGTCCGAAGGTTTGGTACAGCAACAG 3′. Primers for mock and C TALEN HDI samples were: Mock F MID7 5′ CGTATCGCCTCC CTCGCGCCATCAGCGTGTCTCTAGGGCCTAAAGTTCAGGCAACTCTTGTG 3′ Mock R MID7 5′ CTATGCGCCTTGCCAGCCCGCTCAGCGTGTCTCTAA GAATAAAGCCC AGTAAAGTTCCCCA 3′, and C F MID8 5′ CGTATCGCCTCCCTCGCGCCATCAGCT CGCGTGTCGGGCCTAAAGTTCAGGCAACTCTTGTG 3′ C R MID8 5′ CTATGCGC CTTGCCAGCCCGCTCAGCTCGCGTGTCAGAATAAAGCCCAGTAAAGTTCCCCA 3′. The initial PCR was performed using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Woburn, MA). All subsequent steps were completed according to the manufacturer's instructions.
Statistical analysis. Mean and SEM were calculated for each data set. Two-tailed homoscedastic Student's t tests were performed using GraphPad Prism version 4.00 (GraphPad software, La Jolla, CA). P values of < 0.05 were regarded as statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Comparison of HBV genotype sequences in the region targeted by the S TALEN. Figure S2. Comparison of HBV genotype sequences in the region targeted by the C TALEN. Figure S3. Comparison of HBV genotype sequences in the region targeted by the P1 TALEN. Figure S4. Comparison of HBV genotype sequences in the region targeted by the P2 TALEN. Figure S5. Survival assay after co-transfection with GFP- and TALEN-expressing plasmids. Figure S6. HBsAg concentrations in culture supernatants of HepG2.2.15 cells that were subjected to repeat transfections with plasmids encoding each of the panel of 4 HBV-targeting TALENs under hypothermic (30 °C) temperatures. Figure S7. Detection of targeted HBV DNA disruption using CEL1 nuclease assay. Figure S8. Equivalent reporter gene expression in groups of mice that were subjected to HDI. Figure S9. Representative liver histology (hematoxylin and eosin staining) obtained from liver sections of each animal that had been subjected to HDI with or without TALEN-expressing plasmids. Low power fields (100 ×) are shown. Table S1. BLAST analysis of the Mus musculus genome to identify potential binding sites of combined and individual left and right subunits (SL and SR) of the S TALEN. Table S2. BLAST analysis of the human genome to identify potential binding sites of combined and individual left and right subunits (SL and SR) of the S TALEN. Table S3. BLAST analysis of the Mus musculus genome to identify potential binding sites of combined and individual left and right subunits (CL and CR) of the C TALEN. Table S4. BLAST analysis of the human genome to identify potential binding sites of combined and individual left and right subunits (CL and CR) of the C TALEN.
Acknowledgments
We are grateful to Mark Goosen and Adrian Puren (National Institute for Communicable Diseases, Johannesburg) for assistance with sequencing. Financial assistance from the South African National Research Foundation (NRF, GUNs 81768, 81692, 68339, 85981 & 77954), Medical Research Council, Poliomyelitis Research Foundation, Stella and Paul Loewenstein Charitable and Educational Trust, German Academic Exchange Service (DAAD) and the European Commission's 7th Framework Programme (PERSIST-222878) is thankfully acknowledged.
Supplementary Material
References
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, et al. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Chen HS, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, et al. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuthnot P, Capovilla A, Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol. 2000;15:357–368. doi: 10.1046/j.1440-1746.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- Ayoub WS, Keeffe EB. Review article: current antiviral therapy of chronic hepatitis B. Aliment Pharmacol Ther. 2011;34:1145–1158. doi: 10.1111/j.1365-2036.2011.04869.x. [DOI] [PubMed] [Google Scholar]
- Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82:8013–8021. doi: 10.1128/JVI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudy M, Hanschmann KM, Kress J, Nick S, Campos R, Wend U, et al. First WHO International Reference Panel containing hepatitis B virus genotypes A-G for assays of the viral DNA. J Clin Virol. 2012;55:303–309. doi: 10.1016/j.jcv.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40 Web Server issue:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B, Glebe D, Kann M. Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J Virol. 2006;80:5465–5473. doi: 10.1128/JVI.02303-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology. 2006;44:694–702. doi: 10.1002/hep.21299. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim JS. TALENs and ZFNs are associated with different mutation signatures. Nat Methods. 2013;10:185. doi: 10.1038/nmeth.2364. [DOI] [PubMed] [Google Scholar]
- Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR. Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol. 2012;86:8920–8936. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Stoddard BL. Homing endonucleases: structure, function and evolution. Cell Mol Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba Y, Sumaoka J, Komiyama M. Artificial DNA cutters for DNA manipulation and genome engineering. Chem Soc Rev. 2011;40:5657–5668. doi: 10.1039/c1cs15039a. [DOI] [PubMed] [Google Scholar]
- Hendrie PC, Hirata RK, Russell DW. Chromosomal integration and homologous gene targeting by replication-incompetent vectors based on the autonomous parvovirus minute virus of mice. J Virol. 2003;77:13136–13145. doi: 10.1128/JVI.77.24.13136-13145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. 2013High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnoldoi:10.1038/nbt.2623 (in press). [DOI] [PMC free article] [PubMed]
- Meuleman P, Libbrecht L, Wieland S, De Vos R, Habib N, Kramvis A, et al. Immune suppression uncovers endogenous cytopathic effects of the hepatitis B virus. J Virol. 2006;80:2797–2807. doi: 10.1128/JVI.80.6.2797-2807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Mason WS, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Lou X, Hua D, Yu W, Li L, Wang J, et al. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 2012;8:e1003065. doi: 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ely A, Bloom K, Weinberg MS, Arbuthnot P. Inhibition of hepatitis B virus replication with linear DNA sequences expressing antiviral micro-RNA shuttles. Biochem Biophys Res Commun. 2009;389:484–489. doi: 10.1016/j.bbrc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Till BJ, Burtner C, Comai L, Henikoff S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004;32:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK, Hall J, et al. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology. 2004;40:727–737. doi: 10.1002/hep.20353. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Tremblay JP, Xiao X, Aartsma-Rus A, Barbas C, Blau HM, Bogdanove AJ, et al. Translating the genomics revolution: the need for an international gene therapy consortium for monogenic diseases. Mol Ther. 2013;21:266–268. doi: 10.1038/mt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.