Abstract
The assembly of the proteasome — the cellular machine that eliminates unwanted proteins — is a carefully choreographed affair, involving a complex sequence of steps overseen by dedicated protein chaperones.
The proteasome degrades faulty or superfluous intracellular proteins, which are targeted to it for destruction by the attachment of the small ubiquitin protein. This multi-protein complex consists of a barrel-shaped core particle, which enzymatically degrades proteins, bound to one or two outer regulatory particles. The regulatory particle binds and unfolds ubiquitin-tagged proteins and transfers them to the catalytic chamber of the core particle. Whereas the assembly pathway of the core particle is quite well understood, little is known about how the regulatory particle is put together. That situation changes with the publication of two exciting papers in this issue, one by Roelofs et al.1 (page 861) and an accompanying paper by Park et al.2 (page 866), which report the details of regulatory-particle assembly in the yeast Saccharomyces cerevisiae.
The core particle of the proteasome consists of four heptameric rings: two inner rings of β-type subunits and two outer rings of α-type subunits. Although the structure of the regulatory particle has not been described in detail, it is known to consist of two parts — the base and the lid. The base is made up of six ATPase subunits (Rpt1–6) and three non-ATPase subunits (Rpn1, Rpn2 and Rpn13). The six ATPase subunits assemble into a ring, with the carboxy-terminal tails of the component Rpt proteins anchored in pockets in the α-subunits of the core particle3,4.
Until recently, it was believed that proteasome assembly was a relatively straightforward process, in which an intact core particle binds to two intact regulatory particles to generate the dumbbell-shaped proteasome. We now know that the assembly of the core particle involves multiple steps and protein-assembly factors (chaperones), which promote the correct arrangement of core-particle subunits and prevent inactive endpoints5.
Roelofs et al.1 and Park et al.2 reveal that, in the yeast proteasome, regulatory-particle assembly is also a complex process. They suggest that assembly of the hexameric ATPase ring of the base takes place on the pre-assembled core particle, and the carboxy-terminal domain of the base Rpt subunits is essential for docking with the core particle. Regulatory-particle assembly is facilitated by three chaperone proteins — Rpn14, Nas6 and Hsm3 — each of which binds to specific Rpt proteins1 and probably guides interaction between the Rpt proteins and the core-particle α-subunit1. The chaperone proteins also bind near the carboxy-terminal tail of the Rpt proteins, and the authors propose that they compete with the α-subunit pockets for binding the Rpt tails. In this way, the chaperone proteins may ensure that their partner Rpt binds to the correct α-subunit.
Mutations in the carboxy-terminal domain of the Rpt proteins that reduce their binding to the core particle result in increased amounts of free core particle, but, surprisingly, there is no evidence of free regulatory particle2. And although increased amounts of the lid sub-complex are detected, no free base is found2. These results suggest that the Rpt proteins are sequestered in intermediate assembly complexes, and Park et al.2 provide supporting evidence for this by identifying an intermediate subcomplex containing the chaperone Hsm3, the ATPases Rpt1, Rpt2 and Rpt5, and the non-ATPase subunit Rpn1, which they call base precursor 1 (BP1).
The order of addition of Rpt proteins to the core particle suggests an assembly line of sequential steps, in which the action of the Rpn14 chaperone precedes those of Hsm3 and Nas6. In this model (Fig. 1), formation of the hexameric ring of the regulatory-particle base is initiated by Rpn14, which anchors its binding partners Rpt4 and Rpt6 on the core particle. The non-ATPase subunit Rpn2 might perform a scaffolding role in stabilizing this complex, named the BP2–core particle (BP2-CP). The Rpn14-dependent step is followed by the delivery of additional ATPase subunits by BP1. The initial placement of Rpt4 and Rpt6 on the core particle might guide subsequent interactions between Rpt subunits and specific binding pockets on the core particle, ensuring that subunits in the emerging ring are correctly aligned, and locking the emerging ring firmly in place. These sequential steps yield an intact base–core-particle complex, which presumably sets the stage for the succeeding assembly of the lid to complete the regulatory particle. \
Figure 1. A model of proteasome regulatory-particle assembly1,2.
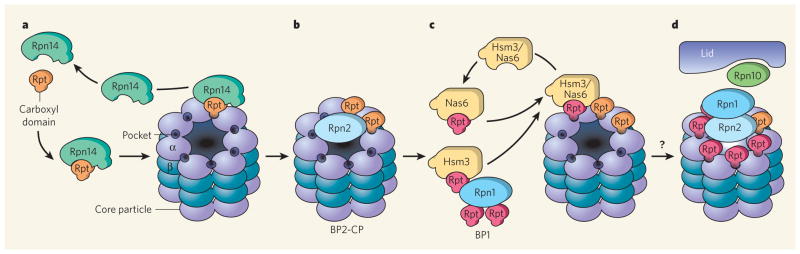
a, ‘Templating’ Rpt4 and Rpt6 proteins (orange) bind the Rpn14 chaperone and connect to the core particle. Rpn14 and the core particle α-subunit compete for binding to the carboxy-terminal domain of Rpt, and the chaperone is displaced following Rpt–α-subunit interaction. b, The non-ATPase subunit Rpn2 might stabilize this complex, termed BP2–CP. c, Other Rpt ATPases (red) are transferred to the core particle by chaperones Hsm3 and Nas6. Hsm3 is found in the intermediate complex BP1, which also contains Rpt1, 2 and 5 and Rpn1. The specific interactions among these proteins and their order of addition to the emerging Rpt ring are not known. For clarity, Rpn1 and Rpn2 are not shown on the core particle. d, Completion of the ATPase ring fully activates the proteasome. Formation of the base–core-particle complex is presumably followed by assembly of the intact proteasome. The interaction between the base and the lid is predicted to require Rpn10. Only one regulatory particle is shown.
Faulty interaction between the ‘templating’ ATPases (Rpt4 and Rpt6) and the core particle causes a more severe proteasome-assembly defect than mutations in the other ATPases (Rpt1, 2, 3 and 5)2. One interpretation of this result is that if Rpt4 or Rpt6 fail to bind to their partner α-subunits in the core particle, the pairwise interactions between the core particle and the ‘late-docking’ ATPases (Rpt1, 2, 3 and 5) could be adversely affected. By contrast, defects in the late-docking ATPases are expected to cause a milder defect in proteasome assembly because preceding core-particle interactions with Rpt4 and Rpt6 would steer them towards interacting with specific α-subunits. A previous report6 showed that the non-ATPase subunits Rpn1 and Rpn2 interact with the core particle and provide a platform for the ATPase ring, suggesting a crucial structural role for these non-ATPase subunits. However, as the carboxyl termini of Rpt subunits can interact directly with intra-subunit pockets in the α-subunit ring1–4, the specific role of Rpn1 and Rpn2 in base assembly is unclear.
These findings reiterate the complexity of proteasome assembly, which may allow the generation of proteasomes that vary in their composition and function to satisfy specific cellular needs. Moreover, as free base, intact regulatory particles and core particles can be detected in cell extracts, it is possible that alternative assembly mechanisms exist. Such inter actions may be governed by the same chaperone proteins, as these can be co-purified with the base and free regulatory particle1. One alternative model might predict base assembly occuring close to the α-subunits, with chaperone release synchronized to complete docking of the ring to the core particle.
As expected, the chaperone proteins are not detected in the mature proteasome — they are presumably released to continue their guiding role in assembly. However, Park et al.2 find that subtle alterations in the carboxy-terminal domain of the ATPase subunits allow their partner chaperones to remain bound to the core particle. Interestingly, one report7 has proposed that, in some cases, protein degradation by the core particle is followed by disassembly of the regulatory particle, although another study8 found that the proteasome remains intact. The role of the chaperone proteins during a cycle of protein degradation is unknown, and it remains to be tested whether the retention of regulatory-particle chaperones on proteasomes (using Rpt mutants) affects disassembly or reassembly. The interaction between regulatory-particle chaperones and the free regulatory particle shown by Roelofs et al.1 suggests that these chaperones are also required for promoting reassociation between the regulatory particle and the core particle.
Defects in the many human genes that encode proteins involved in the ubiquitin–proteasome pathway have been linked to cancers, such as breast cancer9, and neurodegenerative disorders, such as Parkinson’s disease10. Detailed structural characterization of the core particle led to the development of drugs that inhibit proteasome function, and these drugs have been beneficial in the treatment of a type of blood cancer called multiple myeloma11. However, a major limitation of such drugs is that the proteasome is essential in all cells, and altering its activity affects both normal and abnormal cells. The reports in this issue1,2, and other recent findings12–16, show that the requirement for regulatory-particle chaperones in proteasome assembly is evolutionarily conserved, revealing the chaperone proteins as promising targets for drug discovery.
References
- 1.Roelofs J, et al. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park S, et al. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DM, et al. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos CP, Dohmen RJ. Structure. 2008;16:1296–1304. doi: 10.1016/j.str.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. Nature Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babbitt SE, et al. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Kriegenburg F, et al. Cell. 2008;135:355–365. doi: 10.1016/j.cell.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 9.Downes M, Evans RM. Cancer Cell. 2008;13:184–185. doi: 10.1016/j.ccr.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Olanow CW, McNaught KS. Mov Disord. 2006;21:1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg AL. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 12.Le Tallec B, Barrault MB, Guérois R, Carré T, Peyroche A. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura Y, et al. Biochem Biophys Res Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 14.Funakoshi M, Tomko RJ, Kobayashi H, Hochstrasser M. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeki Y, Toh-e A, Kudo T, Kawamura H, Tanaka K. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, et al. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]


