Abstract
Here we report the de novo design and NMR structure of a four-helical bundle di-iron protein with phenol oxidase activity. The introduction of the cofactor-binding and phenol-binding sites required the incorporation of residues that were detrimental to the free energy of folding of the protein. Sufficient stability was, however, obtained by optimizing the sequence of a loop distant from the active site.
A major objective in chemical biology is the design of enzyme-like catalysts tailored to specific applications1,2. The design of an enzyme-like catalyst minimally requires the positioning of functional groups (for example, general acids, bases, metal ions) in an environment that promotes the desired reaction, ideally by stabilizing the transition state relative to the products and reactants. Computational methods have been used to redesign the binding sites of existing natural proteins3–6. An outstanding recent example of this approach combines computation and directed evolution to engineer a protein that positions a general base in a partially dehydrated pocket, thereby catalyzing the Kemp elimination reaction6. The value of kcat/Km is 2,600 M−1 s−1—only 102-fold slower than the corresponding second-order rate constant for the acetate-catalyzed reaction in acetonitrile, and 105-fold faster than the acetate-catalyzed reaction in water. De novo protein design is a more challenging approach in which the tertiary structure and catalytic site are both elaborated from first principles7–10. The ‘Due Ferri’ (two-iron; DF) family of proteins11 is a product of this approach. Although inspired by highly complex natural di-metal proteins, we designed the original DF1 protein, using a mathematical parameterization of the backbone sufficiently simple to be written on the back of an envelope. DF1 is a dimeric protein, each monomer consisting of a helix-loop-helix motif (Fig. 1a). The metal-binding site consists of four glutamate and two histidine residues as first-shell ligands, which are positioned in the core of the protein by hydrogen bonded interactions with two aspartate, tyrosine and lysine residues12.
Figure 1.
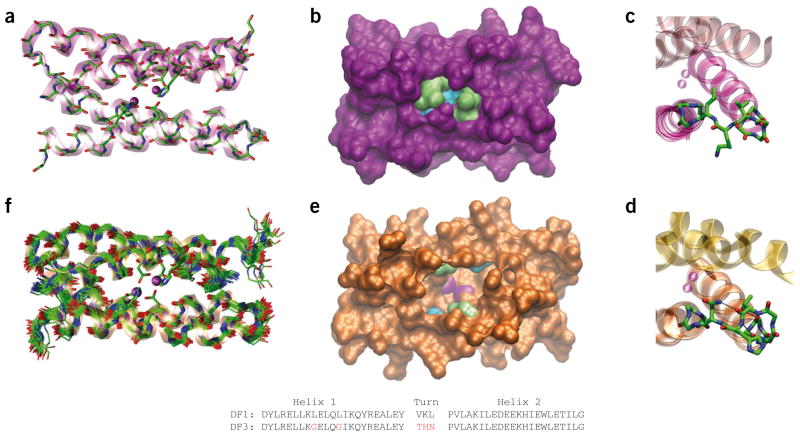
Comparison of DF1 and DF3 structures. (a) Di-Zn(II)-DF1 crystal structure. (b) Di-Zn(II)-DF1 surface representation, displaying the accessibility to the active site and highlighting the different residues in position 9 (lime) and 13 (cyan). (c) Di-Zn(II)-DF1 loop structure. (d) Di-Zn(II)-DF3 loop structure; N-capping and C-capping interactions are depicted. (e) Di-Zn(II)-DF3 surface representation, displaying the accessibility to the active site and highlighting the different residues in position 9 (lime) and 13 (cyan). The point mutations introduced in proximity of DF3 active site create a cavity large enough to allow substrates to approach the metal ions (depicted in magenta in e). In contrast, the active site is completely buried in DF1 (b). (f) NMR bundle of the best 30 minimized structures for di-Zn(II)-DF3. The structures were generated with Visual Molecular Dynamics (VMD; http://www.ks.uiuc.edu/Research/vmd/). DF1 and DF3 sequence comparison are also reported. Amino acid substitutions in the DF3 sequence are indicated in red. The atomic coordinates of di-Zn(II)-DF3 ensembles, chemical shifts and NMR constraints have been deposited in the Protein Data Bank under accession code 2KIK.
The structure of apo-DF1 (ref. 13) demonstrates that its six primary and four secondary ligands are preorganized with sub-Å accuracy within the core of the protein, even in the absence of bound metal ions. Thus, the protein scaffold dictates both the structure and the environment of the di-iron cofactor, which assembles spontaneously upon addition of Fe(II). The diferrous centers in DF proteins react rapidly with O2, with concomitant formation of an oxo-bridged di-Fe(III) species and reduction of molecular oxygen14,15.
The goal of the current work was to introduce a phenol-binding site into DF1, converting it into a phenol oxidase, similar to the natural enzymes alternative oxidase (AOX) and plastid terminal oxidase (PTOX). These catalysts cycle between di-Fe(II) and di-Fe(III) states as they reduce O2 and then use the oxidizing equivalents to convert quinols to quinones16,17 (Supplementary Scheme 1). A successfully designed phenol oxidase should: (i) position the di-Fe(II) cofactor for facile oxidation with O2; (ii) stabilize the resulting di-Fe(III) cofactor, which otherwise would be highly insoluble in aqueous solution; (iii) bind phenols to allow two-electron oxidation of the substrate and regeneration of the diferrous cofactor; (iv) allow product release with entry to a new catalytic cycle; (v) minimize one-electron radical-forming processes that would give rise to Fenton chemistry or destruction of the catalyst. The elaboration of phenol oxidase activity in DF3 required expansion of the active site cleft to accommodate phenols. Previously, we explored possible solutions to this problem in a combinatorial manner through the design of DFtet, which is composed of four unconnected helical peptides18,19. Mixing different DFtet peptides produces catalysts for the oxidation of 4-aminophenol19 (1). However, the complex stoichiometry, marginal stability and tendency to undergo ligand-exchange reactions hamper attempts to fully characterize the structure and properties of the DFtet assemblies, which led us to introduce the corresponding mutations into the more well-characterized DF1 framework.
Access to the DF1 active site is severely limited by four bulky leucine side chains at positions 9 and 13 of each monomer (Fig. 1b), each of which is converted to glycine in the most active DFtet analog19. However, the introduction of helix-destabilizing glycine residues and loss of the hydrophobic driving force strongly destabilize the fold of the protein. Indeed, a single mutation of Leu13 to glycine destabilizes DF1 by 10.8 kcal mol−1 dimer−1, precluding the introduction of the second glycine residue13. To increase the conformational stability of the DF scaffold, we modified the sequence of the interhelical turn, which adopts an αR-αL-β conformation12. Previous attempts to redesign the protein to accommodate different turn types20,21 were not sufficiently successful to provide adequate stability to support the desired multiple glycine mutations. Thus, we used the Protein Data Bank database to derive statistical position-specific propensities for this solvent-exposed αR-αL-β turn21, leading us to change the original Val24-Lys25-Leu26 of DF1 to Thr24-His25-Asn26 (Fig. 1c,d). In models, His25 appeared capable of forming stabilizing hydrogen bonded C-capping interactions of helix 1, while Asn26 could either form N-capping interactions with helix 2 or with the carbonyl group of Thr24, depending on its rotamer. Introduction of this sequence, along with the two leucine-to-glycine mutations, resulted in a sequence designated DF3 (Fig. 1). Details of DF3 design are reported in the Supplementary Methods.
Addition of two equivalents of Fe(II) to DF3 in the presence of ambient oxygen led to rapid formation of a di-Fe(III) species with an absorbance spectrum characteristic of an oxo-bridged diferric cofactor (ε350nm 5,270 M−1 cm−1 per di-iron site)22. Spectroscopic titrations indicated that dithionite reduction and air oxidation of the cofactor are reversible over several cycles (Supplementary Fig. 1). Having established the ability of diferric DF3 to undergo reversible two-electron reduction/oxidation cycles, we evaluated its ability to catalyze the oxidation of 4-aminophenol (4-AP) to the corresponding benzoquinone mono-imine19 (2) (Supplementary Fig. 2a). In the presence of ambient oxygen, the protein followed Michaelis-Menten kinetics with values of 1.97 ± 0.27 mM and 2.72 ± 0.19 min−1, respectively, for Km and kcat (Table 1, kcat/Km = 1,380 M−1 min−1). Measurement of the reaction over the course of an hour indicated that the protein was capable of at least 50 turnovers.
Table 1.
Kinetics parameters obtained for the oxidation of the different substrates
| Protein | Substrate | Km (mM) | kcat (min−1) | kcat/Km (M−1 min−1) |
|---|---|---|---|---|
| di-Fe(III)-DF3 | 3,5-DTBCa | 2.09 ± 0.31 | 13.20 ± 1.21 | 6,315 |
| 4-APb | 1.97 ± 0.27 | 2.72 ± 0.19 | 1,380 | |
| PPDc | 8.87 ± 2.58 | 0.73 ± 0.03 | 83 | |
| OPDd | Not detected | |||
| G4-DFtet | 4-APb | 0.83 ± 0.06 | 1.30 ± 0.10 | 1,540 |
All experiments were performed in 100 mM HEPES/100 mM NaCl buffer (pH 7.0). Kinetic parameters represent mean values ± s.d., calculated by two independent measurements.
3,5-di-tert-butylcatechol.
4-aminophenol.
p-phenylendiamine.
o-phenylendiamine.
Kinetic investigations using other di-amine and ortho-quinol substrates were consistent with the anticipated catalytic properties of di-Fe(III)-DF3 (Supplementary Methods). The active site cleft in the model of DF3 could also accommodate the widely studied substrate 3,5-ditert-butyl-catechol (3,5-DTBC)23 (3). Indeed, di-Fe(III)-DF3 catalyzed the oxidation of this catechol derivative to the corresponding quinone (3,5-DTBQ) (4) (Supplementary Fig. 2b), with a fivefold greater value of kcat/Km than that for 4-AP, reflecting an increase in kcat (Table 1). Amino-aniline substrates were expected to bind more weakly to the diferric center24. Indeed, kcat/Km for the two-electron oxidation of para-phenylenediamine (5) (Supplementary Fig. 2c) was 75-fold lower than the value for 3,5-DTBC, and no catalysis was observed for ortho-phenylenediamine (6).
To confirm the structural basis for DF3’s catalytic activity, its NMR solution structure was solved using the diamagnetic di-Zn(II) derivative as a surrogate for the diferrous form of the protein11 (Supplementary Methods and Supplementary Results). An ensemble of 30 refined structures (calculated using 15 restraints per residue, including 2 × 230 intraresidue, 2 × 185 sequential, 2 × 152 medium-range, 174 intermonomer and 2 × 47 intramonomer NOEs) was tightly clustered (Fig. 1f), with a root mean square deviation (r.m.s. deviation) of 0.68 Å for the main chain atoms and 1.39 Å for all heavy atoms (Supplementary Figs. 3 and 4; Supplementary Tables 1 and 2). The monomer consisted of two α-helices connected via an αR-αL-β turn spanning residues 24–26. As expected, the Nε proton of the His25 imidazole ring capped helix 1, forming a hydrogen bond to the amide carbonyl of residue 22. The side chain of Asn26 formed a hydrogen bond with the carbonyl oxygen of Thr24 in approximately 50% of the members of the ensemble (Fig. 1d). The solution structure of di-Zn(II)-DF3 was nearly identical to the guiding model, which was based on di-Mn(II)-L13G-DF1 (ref. 25). The backbone r.m.s. deviation for the bundle (residues 4–18, 29–41) was 1.2 Å; for comparison, the r.m.s. deviation between dimers of di-Mn(II)-L13G-DF1 in different crystalline environments ranges from approximately 0.6 to 1.2 Å25.
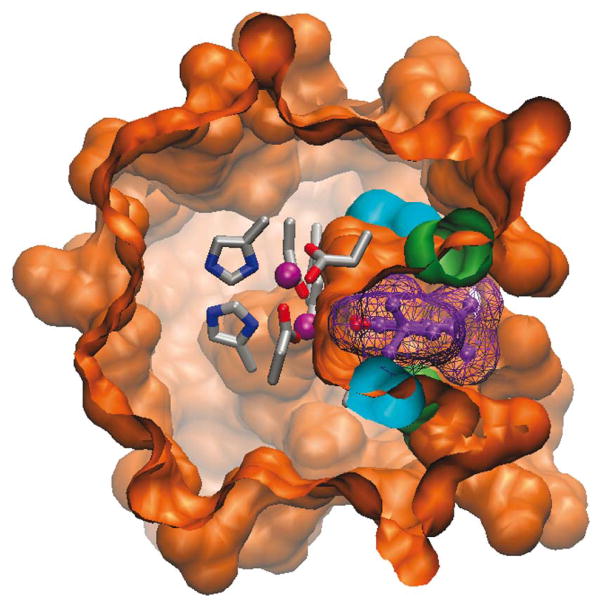
The di-metal cofactor, which lied at the bottom of an apolar pocket, had vacant ligation sites available to interact with incoming O2 and phenols (Supplementary Fig. 5). The 4-Glu, 2-His primary ligand field was stabilized by four second-shell interactions (Supplementary Fig. 6), resembling those in native proteins, including a proposed hydrogen bond between Tyr280 and the primary ligand Glu273 of AOX16,17. The L9G and L13G mutations created an active site cleft (Fig. 1e), which at its narrowest point matched the width of the substrate’s phenyl ring. Bulkier substituents at the 3 and 5 positions of a bound phenol could be accommodated in wider and more solvent-accessible regions of the cavity (Fig. 2).
Figure 2.
Model of substrate interaction with DF3 protein. Surface representation of the di-Zn(II)-DF3 NMR structure with a 3,5-DTBC molecule (violet) bound into the active site access channel. The surface associated with the bound 3,5-DTBC is also depicted. The model structure was generated with VMD.
The de novo design of a catalytically active di-iron protein illustrates a well-known concept in natural proteins: “ Protein function and stability are often the result of opposite requirements” 13. This trade-off between conformational stability and function is particularly apparent in metalloproteins, which reflect a delicate interplay between opposing requirements for binding (favoring static, coordinately saturated geometries) versus function (often requiring coordinately unsaturated and sometimes unusual geometries). The requirement of forming a cavity to bind phenol provided yet another restraint opposing stability. Analysis of the thermodynamic stability of a series of DF1 mutants versus DF3 (Supplementary Results) reinforced the concept that the functional mutations were indeed thermodynamically destabilizing and that the improved turn-forming sequence helped improve the overall stability in the face of these mutations. The availability of the DF3 scaffold should now allow one to explore how changes to the first- and second-shell ligands as well as residues lining the substrate binding cleft affect the reactivity and catalytic properties of the protein.
Supplementary Material
Acknowledgments
The authors thank M. Trifuoggi for metal content analysis. This work was supported by the Italian Ministry of University and Scientific Research (PRIN 2007KAWXCL), the European Commission (Marie Curie Fellowship HPMD-GH-01-00113-03 to R.T.M.R.), the US National Institutes of Health (GM54616) and the US National Science Foundation (grant from the Materials Research Science and Engineering Center to the Laboratory for Research on the Structure of Matter at the University of Pennsylvania).
Footnotes
AUTHOR CONTRIBUTIONS
F.N., O.M. and A.L. designed experiments and analyzed results. R.T.M.R., M.F. and C.A. synthesized the protein and performed UV-vis and chemical denaturation studies. O.M. and M.F. determined the solution structure. F.N. and C.A. performed kinetic experiments. A.L and V.P. performed molecular modeling. W.F.D. and A.L. wrote the manuscript in consultation with the remaining authors.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
References
- 1.Bolon DN, Voigt CA, Mayo SL. Curr Opin Chem Biol. 2002;6:125–129. doi: 10.1016/s1367-5931(02)00303-4. [DOI] [PubMed] [Google Scholar]
- 2.Koder RL, Dutton PL. Dalton Trans. 2006:3045–3051. doi: 10.1039/b514972j. [DOI] [PubMed] [Google Scholar]
- 3.Lu Y, Berry SM, Pfister TD. Chem Rev. 2001;101:3047–3080. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]
- 4.Bolon DN, Mayo SL. Proc Natl Acad Sci USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, et al. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Röthlisberger D, et al. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 7.Bryson JW, et al. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 8.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 9.Razkin J, Lindgren J, Nilsson H, Baltzer L. Chem Bio Chem. 2008;9:1975–1984. doi: 10.1002/cbic.200800057. [DOI] [PubMed] [Google Scholar]
- 10.Johnsson K, Allemann RK, Widmer H, Benner SA, Rere Nature. 1993;365:530–532. doi: 10.1038/365530a0. [DOI] [PubMed] [Google Scholar]
- 11.Maglio O, et al. C R Chimie. 2007;10:703–720. [Google Scholar]
- 12.Lombardi A, et al. Proc Natl Acad Sci USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maglio O, Nastri F, Pavone V, Lombardi A, DeGrado WF. Proc Natl Acad Sci USA. 2003;100:3772–3777. doi: 10.1073/pnas.0730771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell CB, III, et al. Biochemistry. 2009;48:59–73. doi: 10.1021/bi8016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calhoun JR, et al. J Am Chem Soc. 2008;130:9188–9189. doi: 10.1021/ja801657y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berthold DA, Andersson ME, Nordlund P. Biochim Biophys Acta. 2000;1460:241–254. doi: 10.1016/s0005-2728(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 17.Berthold DA, Stenmark P. Annu Rev Plant Biol. 2003;54:497–517. doi: 10.1146/annurev.arplant.54.031902.134915. [DOI] [PubMed] [Google Scholar]
- 18.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. J Mol Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan J, DeGrado WF. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maglio O, et al. J Biol Inorg Chem. 2005;10:539–549. doi: 10.1007/s00775-005-0002-8. [DOI] [PubMed] [Google Scholar]
- 21.Lahr SJ, et al. J Mol Biol. 2005;346:1441–1454. doi: 10.1016/j.jmb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Vincent JB, Olivier-Lilley GL, Averill BA. Chem Rev. 1990;90:1447–1467. [Google Scholar]
- 23.Koval IA, Gamez P, Belle C, Selmeczi K, Reedijk J. Chem Soc Rev. 2006;35:814–840. doi: 10.1039/b516250p. [DOI] [PubMed] [Google Scholar]
- 24.Corbett JF, Gamson EP. J Chem Soc Perkin. 1972;2:1531–1537. [Google Scholar]
- 25.DeGrado WF, et al. Angew Chem Int Ed. 2003;42:417–420. doi: 10.1002/anie.200390127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.