This study developed a highly efficient serum-free pluripotent stem cell (PSC) neural induction medium that can induce human PSCs into primitive neural stem cells (NSCs) in 7 days, obviating the need for time-consuming, laborious embryoid body generation or rosette picking. This method of primitive NSC derivation sets the stage for the scalable production of clinically relevant neural cells for cell therapy applications in good manufacturing practice conditions.
Keywords: Astrocytes, Cell culture, Neural stem cell, Neural induction, Neural differentiation, Oligodendrocytes, Neuron, Nestin
Abstract
Human pluripotent stem cells (hPSCs), including human embryonic stem cells and human induced pluripotent stem cells, are unique cell sources for disease modeling, drug discovery screens, and cell therapy applications. The first step in producing neural lineages from hPSCs is the generation of neural stem cells (NSCs). Current methods of NSC derivation involve the time-consuming, labor-intensive steps of an embryoid body generation or coculture with stromal cell lines that result in low-efficiency derivation of NSCs. In this study, we report a highly efficient serum-free pluripotent stem cell neural induction medium that can induce hPSCs into primitive NSCs (pNSCs) in 7 days, obviating the need for time-consuming, laborious embryoid body generation or rosette picking. The pNSCs expressed the neural stem cell markers Pax6, Sox1, Sox2, and Nestin; were negative for Oct4; could be expanded for multiple passages; and could be differentiated into neurons, astrocytes, and oligodendrocytes, in addition to the brain region-specific neuronal subtypes GABAergic, dopaminergic, and motor neurons. Global gene expression of the transcripts of pNSCs was comparable to that of rosette-derived and human fetal-derived NSCs. This work demonstrates an efficient method to generate expandable pNSCs, which can be further differentiated into central nervous system neurons and glia with temporal, spatial, and positional cues of brain regional heterogeneity. This method of pNSC derivation sets the stage for the scalable production of clinically relevant neural cells for cell therapy applications in good manufacturing practice conditions.
Introduction
Human pluripotent stem cells (PSCs) can be differentiated into neural derivatives by a variety of different approaches [1]. The standard protocol, based on what had been developed for mouse embryonic stem cell cultures, requires a suspension culture to generate embryoid bodies, which can then be plated as an adherent culture to generate neural precursors that can be mechanically or enzymatically isolated. Several stages of neural stem cell (NSC) precursors have been identified. The earliest neuroepithelial precursor is the one described by Hitoshi et al. [2]. These investigators showed that a TRA-1-60−/SSEA4−/SOX1+ cell could be selected by fluorescence-activated cell sorting and clonally expanded under serum-free conditions to NSCs [2]. Human NSCs derived in this manner from PSC cultures appear largely similar to mouse NSCs but differ from them in the earlier expression of Pax6 and the absence of expression of epidermal growth factor (EGF) receptor [3]. Kalyani et al., using a different method, identified PSC-derived neuroepithelial precursor cells (NEPs) that were fibroblast growth factor (FGF)-dependent and appeared similar to the neuroepithelial precursors present in early fetal development [4]. NEPs could be propagated in culture, appeared to be positionally naïve, and could be induced to differentiate into more mature positionally determined EGF-dependent neural stem cells [5, 6]. These early precursors differ from their later-appearing counterparts in that they can be induced to differentiate into neural crest-like cells and generate peripheral nervous system derivatives. This ability is largely lost on further maturation, although fetal tissue-derived NSCs may still generate smooth muscle [7]. Even during fetal development, several stages of neural stem cell precursors can be identified. These can be distinguished based on their growth factor dependence or whether they reside in the ventricular zone, subventricular zone (SVZ), or the outer SVZ, and on their regional identity, which dictates the subtypes of neurons they will make [8–10].
What is clear from these studies is that although they retain multipotential differentiation, NSCs appear to acquire positional information that determines their differentiation potential soon after neural tube formation and that this information can also persist in neural cultures [10–12]. Further experiments have shown that positional information appears to be regulated by growth factors that mediate the expression of Hox and Hox-related family members, retinoic acid (RA), and FGF family members in caudalization [13] of the developing embryo and/or in culture to generate spinal derivatives, whereas bone morphogenetic proteins act to direct dorsal and Sonic hedgehog (SHH) ventral differentiation, respectively [14, 15]. The activity of these growth factors can be modulated by Wnt signaling, which often appears to regulate dorsal fate [16]. To obtain truly versatile NSC populations it is important to identify a process where NSC fate is specified from PSCs but the cells have not yet acquired a caudal phenotype. In this report we describe an approach that allows for the generation of positionally naïve NSCs that can readily differentiate into cranial and caudal derivatives. We show that these NSCs can be maintained and expanded in a good manufacturing practice (GMP)-manufactured culture medium for several passages while retaining their ability to form fore-, mid-, and hindbrain neurons.
Materials and Methods
Human Pluripotent Stem Cell Culture
H9 human embryonic stem cells (hESCs) and human induced pluripotent stem cell (iPSC) lines including episomal human induced pluripotent stem cells (hiPSCs) (Life Technologies, Carlsbad, CA, http://www.lifetech.com) and hiPSC clone III induced by Cytotune (Life Technologies) were maintained in either irradiated mouse embryonic fibroblast feeder layer or feeder-free conditions. For the feeder-containing conditions, the culture medium consisted of Dulbecco's Modified Eagle's Medium: Nutrient Mixture F-12 (DMEM/F12), 20% knockout serum replacement (KSR), nonessential amino acids, 0.1 mM β-mercaptoethanol, and 4 ng/ml basic fibroblast growth factor (bFGF) (all from Life Technologies). For feeder-free conditions, human pluripotent stem cells (hPSCs) were cultured in either StemPro hESC serum-free medium (SFM) (Life Technologies) containing DMEM/F12, StemPro hESC supplement, 1.8% bovine serum albumin, 0.1 mM β-mercaptoethanol, 8 ng/ml bFGF on Geltrex (Life Technologies)-coated culture dishes or Essential 8 medium (Life Technologies) containing DMEM/F12 and Essential 8 supplement on vitronectin (Life Technologies)-coated dishes. hPSCs were passaged every 3–5 days when they reached 70%–80% confluence.
Neural Induction of hPSCs, NSC Expansion, Cryopreservation, and Karyotyping
hPSCs cultured in either feeder-containing or feeder-free conditions were split as cell clumps into six-well plates at a density of 2–2.5 × 104 cells per cm2. Approximately 24 hours after splitting, culture medium was switched to Gibco PSC Neural Induction Medium (Life Technologies) containing Neurobasal medium and Gibco PSC neural induction supplement. Neural induction medium was changed every other day from day 0 to day 4 of neural induction. After day 4 of neural induction, neural induction medium was changed every day as cells reached confluence. At day 7 of neural induction, primitive NSCs (pNSCs) were dissociated with Accutase (Life Technologies) and plated on Geltrex-coated dishes at a density of 0.5–1 × 105 cells per cm2 in an NSC expansion medium containing 50% Neurobasal medium, 50% Advanced DMEM/F12, and neural induction supplement. NSC expansion medium was changed every other day until NSCs reached confluence at day 5 of pNSC plating. For pNSCs before passage 4, 5 μM ROCK inhibitor Y27632 (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) was added to NSC expansion medium at the time of NSC plating to treat cells overnight for the prevention of cell death. Dissociated pNSCs were cryopreserved in NSC expansion medium with 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich). Karyotyping of expanded pNSCs was performed by Cell Line Genetics (Madison, WI, http://www.clgenetics.com).
Differentiation of Expanded pNSCs to Neuronal, Astrocyte, and Oligodendrocyte Lineages
Expanded pNSCs at passage 6 (P6) were used to evaluate their differentiation potentials to triple lineages of neural cells. For neuron differentiation, Accutase-dissociated pNSCs were plated onto laminin (10 μg/ml; Life Technologies)-coated four-well chamber slides at a density of 5 × 104 cells per cm2 in a neuronal differentiation medium consisting of neurobasal medium, B-27, GlutaMAX (Life Technologies), nonessential animal acids, 20 ng/ml brain-derived neurotrophic factor (BDNF), 20 ng/ml glial cell-derived neurotrophic factor (GDNF) (all from Life Technologies), and 200 μM l-ascorbic acid (Sigma-Aldrich) for 14 days, and the culture medium was changed every 2–3 days. For astrocyte differentiation, dissociated pNSCs were plated onto Geltrex-coated four-well chamber slides at a density of 5 × 104 cells per cm2 in an astrocyte differentiation medium (DMEM supplemented with N2 and 1% fetal bovine serum; all from Life Technologies) for 21 days. Confluent cultures were passaged at a ratio of 1:4, and medium was changed every 2–3 days. For oligodendrocyte differentiation, dissociated pNSCs were plated onto Geltrex-coated dishes at a density of 5 × 104 cells per cm2 in a differentiation medium containing DMEM/F12 medium, N2, 10 ng/ml bFGF, and 20 ng/ml EGF (all from Life Technologies). After 6 days, medium was changed to include 10 ng/ml platelet-derived growth factor-AA (Life Technologies) for another 4 days. Following the removal of growth factors and N2, 30 ng/ml T3 (Sigma-Aldrich), 5 ng/ml NT3 (Life Technologies), 200 μM ascorbic acid (Sigma-Aldrich), and B27 were added for an additional 4 days in culture.
Differentiation to GABAergic Neurons
Dissociated pNSCs at P6 were plated onto polyornithine (20 μg/m; Sigma-Aldrich) and laminin (10 μg/ml; Life Technologies)-coated dishes at a density of 5 × 104 cells per cm2 and cultured in StemPro hESC SFM (Life Technologies) supplemented with 10 ng/ml activin (R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com), 10 ng/ml BDNF, and 10 ng/ml GDNF, but without FGF2. Culture medium was changed every other day. Subculture was performed when cells reached overconfluence during the differentiation, and GABAergic neurons were evaluated at day 15 after differentiation.
Differentiation to Dopaminergic Neurons
Dissociated P6 pNSCs were plated on polyornithine- and laminin-precoated 35-mm dishes at a density of 4 × 105 cells per cm2 in the NSC expansion medium. The next day, the spent medium was replaced with a dopaminergic induction medium consisting of Neurobasal medium, MEM nonessential amino acids, GlutaMAX, B27 supplement, 200 ng/ml SHH (Life Technologies), and 100 ng/ml FGF8 (Life Technologies) for 10 days, with medium changes every alternative day. On day 10, cells were dissociated with Accutase and plated onto laminin-coated four-well chamber slides at a density of 2 × 104 cells per cm2 in a dopaminergic maturation medium consisting of Neurobasal medium, MEM nonessential amino acids, GlutaMAX, B27 supplement, 200 μM ascorbic acid, 20 ng/ml BDNF, and 20 ng/ml GDNF to allow maturation of dopaminergic neurons for another 10 days with a medium change every other day.
Differentiation to Motor Neurons
For the specification of motor neurons, dissociated P6 pNSCs were plated at a density of 5 × 104 cells per cm2 on polyornithine- and laminin-coated plates in StemPro hESC SFM supplemented with 500 μM RA (Sigma-Aldrich), 200 ng/ml SHH, and 20 ng/ml activin (R&D Systems), with a medium change every other day for 12 days. Cells were passaged and replated between medium changes, as they reached confluence. For the maturation of motor neurons, cells were dissociated and plated onto polyornithine- and laminin-coated plates in StemPro hESC SFM with the reduction of FGF2 from 8 ng/ml to 4 ng/ml at day 13. Activin (20 ng/ml), BDNF (10 ng/ml), and GDNF (10 ng/ml) were added into medium for the maturation of motor neurons. The expression of Olig2 was analyzed at day 18 after differentiation. The maturation of motor neuron takes ∼3 weeks. During this time, medium was changed every 2 days followed by the expression analysis for HB9.
Characterization of pNSCs and Differentiated Subtypes of Neural Cells
Immunocytochemical staining was used to determine the phenotypes of pNSCs and differentiated cells. pNSCs plated on Geltrex-coated four-well chamber slides or differentiated cells were fixed with 4% paraformaldehyde at room temperature for 15 minutes. After rinsing with Dulbecco's phosphate-buffered saline (DPBS) (Life Technologies), cells were permeabilized and blocked in a blocking buffer containing 0.1% Triton X-100 (Sigma-Aldrich) and 1% bovine serum albumin in DPBS for 30 minutes. After incubation with the primary antibodies at 4°C overnight in the blocking buffer and three rinses in DPBS, Alexa Fluor 488- and/or 594-conjugated secondary antibodies (1:1,000; Life Technologies) were used to visualize stained cells. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Fluorescent images were captured on an inverted fluorescence microscope (Nikon Instruments Inc., Melville, NY, http://www.nikoninstruments.com). The percentage of positive cells was counted with ImageJ software. Phase contrast images were taken with a Nikon inverted microscope.
The following primary antibodies were used: the pluripotent marker Oct4; NSC markers, including Sox1, Sox2, Pax6, and Nestin; neuronal markers, including βIII tubulin, TuJ1, and microtubule-associated protein 2; the astrocyte marker glial fibrillary acidic protein; the oligodendrocyte marker galactosylceramidase (GalC); the GABAergic marker γ-aminobutyric acid; the dopaminergic marker tyrosine hydroxylase (TH); the noradrenergic marker dopamine β-hydroxylase (DβH); and the motor neuron markers HB9 and Olig2. The sources of the antibodies and dilution ratios used are detailed in supplemental online Table 1.
Illumina Microarray Gene Expression Analysis
Fetal-derived NSCs were from Lonza (Walkersville, MD, http://www.lonza.com). Rosette-derived NSCs were from Life Technologies. Replicate total RNA samples were extracted from NSC populations using Trizol and the Purlink RNA isolation kit (Life Technologies). For microarray analysis, total RNA was processed and hybridized on Illumina Human ref-12 bead arrays following the manufacturer's specifications (Illumina, Inc., San Diego, CA, http://www.illumina.com). Sample amplification was performed using 100 ng of total RNA as input material using the Illumina RNA Amplification kit (Life Technologies) following the manufacturer's instructions. Labeling was achieved by use of the incorporation of biotin-16-UTP (PerkinElmer Life and Analytical Sciences, Boston, MA, http://www.perkinelmer.com) present at a ratio of 1:1 with unlabeled UTP. Labeled, amplified material (700 ng per array) was hybridized to Illumina Human ref-12 BeadChip. Arrays were scanned with an Illumina BeadArray Reader confocal scanner according to the manufacturer's instructions. The gene expression data were first normalized by using the robust multichip average approach [17]. The data were then analyzed by multidimensional scaling (MDS) [18], and the expression pattern was detected by clustering analysis and presented as dendrograms, scatter plots, and heat maps. The analyses were conducted using the R software package (http://www.r-project.org/) and the GenomeStudio software (Illumina).
Results
Derivation of Expandable Population of Primitive Neural Stem Cells
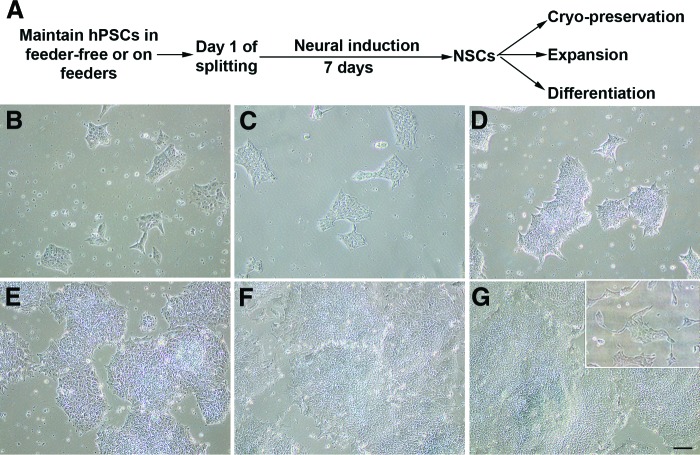
The work flow of neural induction medium is shown in Figure 1A. Human pluripotent stem cells can be efficiently differentiated to primitive NSCs without the need for rosette formation and rosette isolation using the neural stem cell induction medium. By starting with a million pluripotent stem cells, it is possible to get approximately 40–50 million NSCs with 40–50-fold increases in cell number in 7 days. The morphology of PSC transition to pNSC phenotypes is shown in Figure 1B through 1G. It is noteworthy that during the neural induction phase of PSCs, along with cell proliferation, a drastic change in cell morphology was observed along the duration of neural induction. At the initial stages of neural induction, cells expanded and displayed a small, round, globular morphology (Fig. 1C, 1D). As the duration of the culture progressed to day 4 (Fig. 1E), cells exhibited a more round morphology. By days 6–7 (Fig. 1F, 1G), cells continually propagated but tended to grow in a distinct uniform monolayer of cells or bundles of cells as compared with pluripotent stem cells.
Figure 1.
The morphology of cells during neural induction. (A): Workflow of NSC derivation from hPSCs. (B): hPSCs cultured in feeder-free conditions at day 1 of splitting with 10%–15% of confluence. (C–G): The morphology of cells at days 1 (C), 2 (D), 4 (E), 6 (F), and 7 (G) after neural induction. Inset in (G) shows passage 0 NSCs at day 1 of replating after dissociation. Scale bar = 100 μm. Abbreviations: hPSC, human pluripotent stem cell; NSC, neural stem cell.
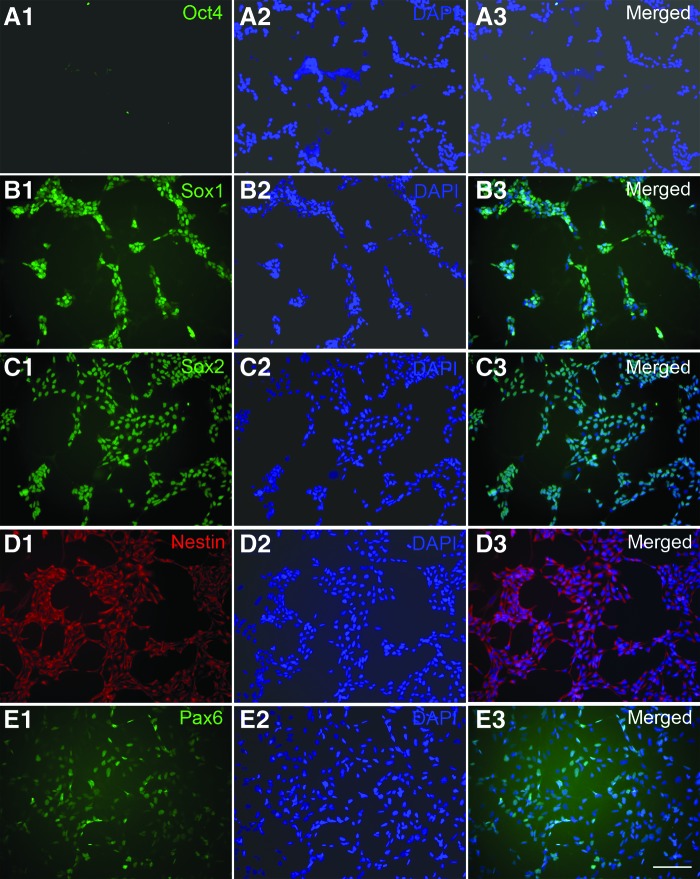
Cells at day 7 of neural induction were dissociated and plated for staining with pluripotent marker Oct4 and NSC markers. The cells showed distinct NSC morphology after the dissociation and replating at day 7 of neural induction (Fig. 1G, inset). Most of the cells were positive for NSC markers Sox1 (Fig. 2B1–2B3), Sox2 (Fig. 2C1–2C3), and Nestin (Fig. 2D1–2D3). A portion of cells also expressed Pax6 (Fig. 2E1–E3). Almost no Oct4+ cells were observed in the cultures (Fig. 2A1–2A3).The quantification data (Fig. 3A) showed that the percentage of Oct4+ cells was low (<1%) and that a high percentage of cells expressed Sox1 (∼90), Sox2 (>95%), and Nestin (>95%). The percentage of Pax6+ cells was ∼50%. These results suggest that cells derived from hPSCs by neural induction medium possess the NSC phenotype. To assess the expansion ability of NSCs induced by the induction medium, NSCs at P0 were propagated up to P5. When expanded in an NSC expansion medium prepared in Neurobasal medium and Advanced DMEM/F12 plus neural induction supplement, the cells exhibited clear morphology of NSCs with an 8–10-fold increase in cell number at each passage from P0 to P5 (Fig. 3B). The same morphological pattern could be observed at P10 (data not shown). When the NSCs were cryopreserved in NSC expansion medium with 10% DMSO, the cells exhibited good recovery, viability, and expansion (data not shown). Since this method of induction and derivation of pNSCs is different from a conventional rosette-based method, the potential effects on chromosome stability were examined by a karyotype analysis of metaphasic NSCs expanded to P5. The analysis revealed no gross chromosomal aberrations, thus suggesting that the cells preserved their normal karyotype (Fig. 3C).
Figure 2.
Expression of pluripotent and neural markers of neural stem cells (NSCs). (A–E): Passage 0 NSCs derived from H9 embryonic stem cells were dissociated and plated for staining with antibodies against the pluripotent marker Oct4 (A1–A3) and the neural markers Sox1 (B1–B3), Sox2 (C1–C3), Nestin (D1–D3), and Pax6 (E1–E3). Cell nuclei were stained with DAPI (blue). Scale bar = 100 μm. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
Figure 3.
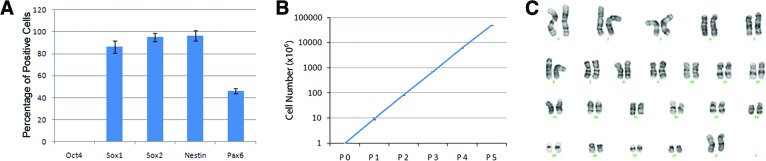
Quantification of neural stem cell (NSC) marker expression, expansion, and karyotyping. (A): The percentages of Oct4-, Sox1-, Sox2-, Nestin-, and Pax6-positive cells were counted over total cells in P0 H9 embryonic stem cell (ESC)-derived NSCs. (B): The dynamics of NSC expansion with an 8–10-fold increase in cell number of each passage. (C): No gross chromosomal aberrations were observed by karyotyping of P5 H9 embryonic stem cell-derived NSCs. Abbreviation: P, passage.
NSC generation and expansion by this culture medium was evaluated using three pluripotent stem cell lines: H9 hESCs, a Gibco episomal iPSC line, and a Sendai virus-derived iPSC line. Among these PSC lines, no significant difference was observed (data not shown). In addition, NSCs were derived from an additional two control iPSC lines and four Parkinson's disease-specific human iPSC lines in a reproducible manner, all of which yielded >70% SOX1 cells, indicating that disease lines also yield quality NSCs that were further used in downstream differentiation (data not shown). Although NSC derivation is highly reproducible, some iPSC lines of disease origin and aged samples might likely yield at various efficiencies. Analyses of expanded NSCs by immunocytochemistry showed consistent expression of NSC markers, including Sox1 (∼90%), Sox2 (>95%), Nestin (>95%), and Pax6 (∼50%), at passage 5. The few remaining Oct4+ cells in P0 NSCs were completely lost at P1 to P2 during NSC expansion. In addition, NSCs were derived using neural induction medium from a variety of PSC lines that were maintained in different pluripotent stem cell culture media, such as KSR on irradiated mouse feeders or feeder-free media, including StemPro hESC SFM, mTeSR1, and Essential 8 medium. As compared with NSC derivation with rosette formation (supplemental online Fig. 1), this method of pNSC derivation is fast and efficient, indicating that despite differences in the origin of PSC lines or differences in culture methods, such as feeder-based or feeder-free culture conditions, functional NSCs could be derived reproducibly and consistently.
Primitive NSCs Can Be Differentiated to Neuronal/Glial Lineages and Can Be Specified to Dorsal, Ventral, and Caudal Subneuronal Types
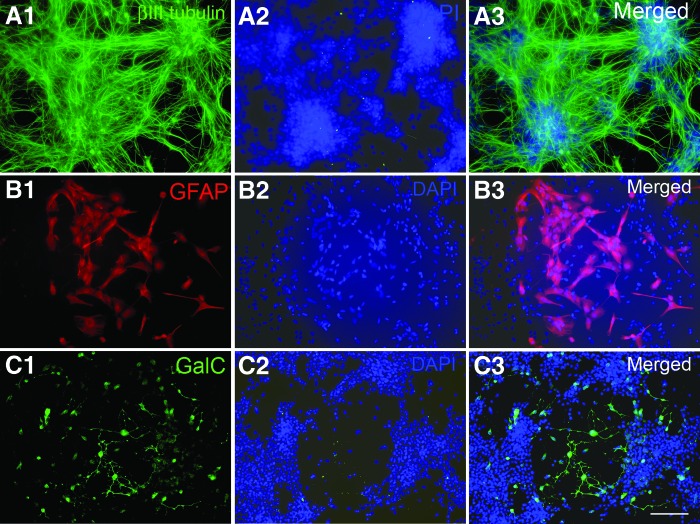
To test the differentiation potential of primitive NSCs derived by neural induction medium, we tested whether the NSCs could differentiate into generic neurons, astrocytes, and oligodendrocytes and further whether they retained the regional identity and ability to differentiate to different brain region-specific neurons, such as GABAergic neurons in the forebrain, dopaminergic neurons in the midbrain, and motor neurons in the hindbrain. As shown in Figure 4, NSCs expanded for six passages retained the ability to undergo neural, astrocytic, and oligodendroglial differentiation, as demonstrated by the expression of the neuronal marker βIII tubulin (Fig. 4A1–4A3), the astrocytic marker GFAP (Fig. 4B1–4B3), and the oligodendrocyte marker GalC (Fig. 4C1–4C3), respectively.
Figure 4.
Triple lineage differentiation of expanded neural stem cells derived from H9 embryonic stem cells. (A–C): Differentiated cells were stained with antibodies against the neuronal marker βIII tubulin (A1–A3), the astrocyte marker GFAP (B1–B3), and the oligodendrocyte marker GalC (C1–C3). Cell nuclei were stained with DAPI (blue). Scale bar = 100 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GalC, galactosylceramidase; GFAP, glial fibrillary acidic protein.
Furthermore, pNSCs were subjected to direct differentiation to brain region-specific neuronal subtypes. As shown in Figure 5, differentiated NSCs expressed the GABAergic marker GABA (Fig. 5A) and the dopaminergic marker TH (Fig. 5B). The TH-positive cells did not express the noradrenergic marker DβH (data not shown). Olig2 and HB9 are two critical transcription factors for motor neuron development. After the treatment with retinoic acid and SHH, differentiated cells expressed Olig2 (Fig. 5C) and HB9 (Fig. 5D). The analysis for the generation of subneuronal types of GABAergic, dopaminergic, and motor neurons demonstrated that region-specific neural types could be generated using a combination of specific growth factors such as activin and FGF2 for GABA neurons, SHH and FGF8 for dopaminergic neurons, and retinoic acid and SHH for motor neurons. Although the percentages of terminally differentiated lineages are different for each subtype, this analysis most importantly suggests that the primitive NSCs have retained the cues and ability to respond to regional specification signals. Furthermore, it is emphasized that although this is good morphological evidence of differentiation to neural, astrocyte, and oligodendrocyte lineages, the functional properties of each of these cell types further need detailed study.
Figure 5.
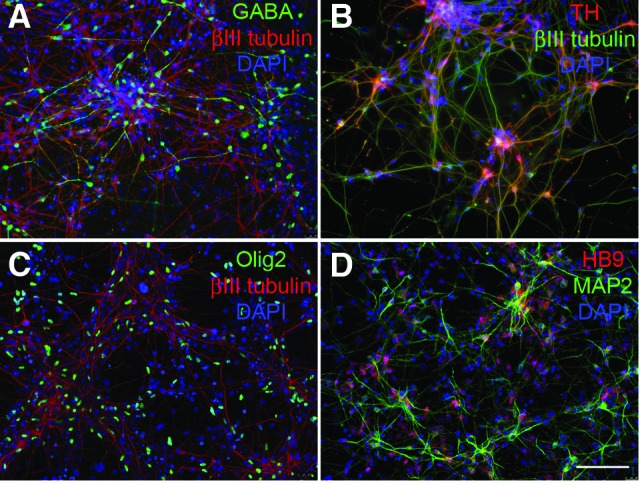
Subtypes of neuronal differentiation of expanded H9 embryonic stem cell-derived neural stem cells. Differentiated cells were stained with antibodies against the neuronal markers βIII tubulin (A–C) or MAP2 (D). Region-specific neuronal subtypes were evaluated by staining with antibodies against the GABAergic marker GABA (A), the dopaminergic marker TH (B), and the motor neuron markers Olig2 (C) and HB9 (D). Cell nuclei were stained with DAPI (blue). Scale bar = 100 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; MAP2, microtubule-associated protein 2; TH, tyrosine hydroxylase.
Comparison of Gene Expression Profiles of Primitive NSCs With Those of Rosette-Derived NSCs and Human Fetal Cortex Isolated NSCs
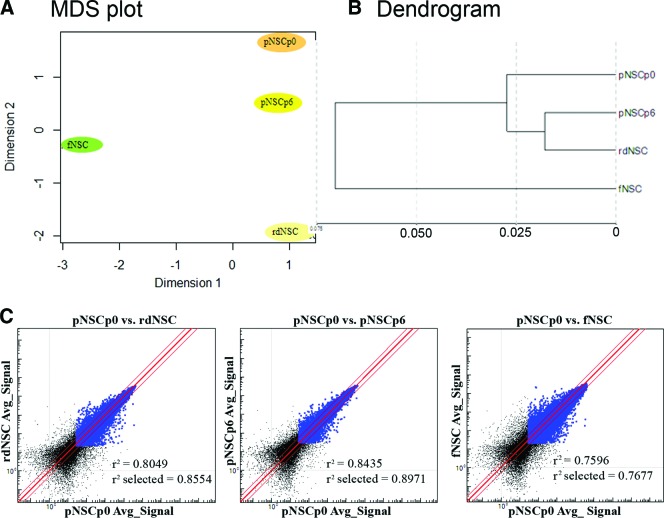
To evaluate a possible genotypic modulation under the derivation and expansion culture conditions, transcriptional analysis was performed. The transcriptomic analysis was carried out and compared with results from NSCs at day 7 in neural induction medium (pNSCp0), NSCs expanded by passaging six times (pNSCp6), rosette-derived NSCs (rdNSCs), and human fetal cortex-derived NSCs (fNSCs). A global gene-expression analysis was carried out using the Illumina bead array containing 46,000 full-length and splice-variant transcripts from the Human RefSeq database. Differential expression of genes detected at greater than a 0.99 confidence level was considered for analysis. The overall expression pattern is presented using an MDS plot and a dendrogram (Fig. 6A, 6B). Clustering among neural stem cell samples revealed that NSCs derived from PSCs (pNSCp0, pNSCp6, and rdNSCs) were clustered together compared with fNSCs, and this was further supported by scatter plots (Fig. 6C). The pairwise scatter plot comparisons revealed that the human fetal-derived NSCs exhibited greater variation than the NSCs derived at P0 or expanded to passage 6, suggesting that the greater variation might result from the origin of the NSC samples than from the derivation (P0) or expansion (P6) culture conditions. This is obvious from the comparison of fetal NSCs and NSCs derived from the current approach, where the correlation parameter r2 dropped to 0.77, whereas the r2 values across NSCs derived from PSCs were high: the r2 value for P0 NSCs versus P6 NSCs was 0.90, and that for P0 NSCs versus rdNSCs was 0.86.
Figure 6.
MDS dendrogram and scatter plot. (A, B): MDS plot and dendrogram to show clustering among examined neural stem cell samples. Pluripotent stem cell-derived neural stem cells (NSCs) were clustered together compared with fetal-derived NSCs and rdNSCs. (C): Scatter plots. The population similarity was shown in scatter plots. Correlation coefficients of whole genes (r2) and of genes expressed by both samples (r2 select) were calculated using GenomeStudio software. Abbreviations: Avg., average; fNSC, fetal cortex-derived neural stem cell; MDS, multidimensional scaling; pNSCp0, NSCs on differentiation day 7 in neural induction medium; pNSCp6, pNSCp0 passaged six times; rdNSC, rosette-derived neural stem cell, >28 passages.
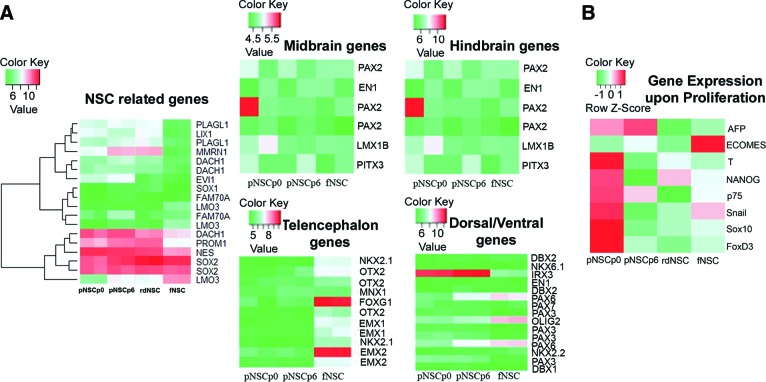
To demonstrate that the induction process is specific for the neural fate, we interrogated genes reported to be expressed by rosette-derived NSCs [19] and then examined expression on our NSC samples (Fig. 7A). The heat map indicates that most genes linked with neural stem cell fate were expressed. Both PSC-derived NSCs and fNSCs expressed widely accepted the neural stem cell genes NES, PROM, and SOX2. The expression value for SOX1 seems to be a false negative from probe mismatch, as the expression was confirmed by protein expression (Fig. 2). The expression of DACH1, NF2F1, and LIX1 was shared by all the tested NSC populations. However, differential expression was observed for MMRN1, PLAGL1, EVI1, LIX1, and RSPO3, whose expression was exempted from fNSCs but limited to PSC-derived NSCs. Furthermore, the expression profile was extended to examine the default regional propensity of the populations examined. fNSCs expressed genes related to the telencephalon (FOXG1, EMX1, EMX2, and OTX2), from which they were isolated. For pNSCp0 and pNSCp6, the genes related to dorsal hindbrain were highly expressed (IRX3, HOXB2, and HOXA2), suggesting the default identity of dorsal hindbrain. We next looked at the changes following derivation and expansion to passage 6 cells in culture in the expression of non-neural genes AFP, EOMES, Brachyury, Nanog, p75, Snail, Sox10, and FoxD3. We were able to show that upon expansion of P0 NSCs, most non-neural genes analyzed disappeared and the gene expression patterns returned to a fetal NSC gene expression pattern where contamination of non-neural population was minimized (Fig. 7B).
Figure 7.
Heat map. (A): Expression of NSC-related genes and default regional identity of the sample population shown by a heat map generated by the R software. NSCs from pluripotent stem cell (PSC) origin expressed most of the NSC-related genes that are also expressed by fNSCs. Primitive NSCs expressed genes related to ventral hindbrain as in default. The gene expression data are log2 transformed. (B): Expression of select non-neural genes and identity of the sample population shown by heat map. NSCs from PSCs at passage 0 (P0) (pNSCp0) and expanded NSCs to P6 (pNSCp6), rdNSCs, and human fNSCs are included for comparison. The gene expression data are normalized (i.e., all genes have a mean of 0 and an SD of 1). Abbreviations: fNSC, fetal cortex-derived neural stem cell; NSC, neural stem cell; pNSCp0, NSCs on differentiation day 7 in neural induction medium; pNSCp6, pNSCp0 passaged six times; rdNSC, rosette-derived neural stem cell.
Discussion
To enable the transition of human pluripotent stem cells for cell therapy applications, it is an absolute necessity to develop efficient and reproducible differentiation methods for desired lineages such as neurons that are critical for cell therapy, drug discovery, and in vitro cellular/molecular modeling of neural development. One of the critical advances needed in this area is better methods to generate different neural subtypes with high efficiency. Although several protocols are available to generate neural stem cells from pluripotent cells, most of these methods involve an intermediate stage of an embryoid body formation and a manual rosette picking step. These methods suffer from several drawbacks, including variability in the quality of derived NSCs, labor intensiveness, and limited scalability [20–23]. Three research groups have reported that human neural stem cells can be generated and expanded short-term without the need of embryoid body formation in serum-free conditions by dual inhibition of SMAD signaling [1] or by synergistic inhibition of glycogen synthase kinase 3, transforming growth factor β, and Notch signaling pathways [24], or specifically toward anterior neural ectoderm cells using selective small molecules [25]. Although each of these methods contributed significantly in advancing the field, some of these methods are not easily reproducible because of variations in iPSC lines or scalable to the extent required for disease modeling or high throughput screens or for cell therapy, besides being limited to a specific brain regional fate. Here, we describe a method for the robust and reproducible induction to the neural stem cell fate starting with any pluripotent stem cell, using a commercially available current good manufacturing practice manufactured culture medium that eliminates variability, ensuring lot-to-lot consistency and scalable production of neural stem cells. Most importantly, this method enables the derivation of NSCs in a primitive state that retains the regional positional cues in brain development, making it easier for researchers to generate neural subtypes from all regions of the brain. The entire neural induction process involves a 7-day culture process initiated by plating specific cell densities of PSCs with culture medium changes every other day. On day 7 of the culture period, P0 NSCs can be harvested by simple enzymatic dissociation. The type of morphology transition to neural fate from PSCs allows culture of cells at much higher densities compared with cells grown as embryoid body followed by rosette formation and picking. Since this is a rosette-free culture system and the morphological changes caused in cultures of rosette versus rosette-free require different passaging frequencies, a direct comparison between the culture systems is difficult (supplemental online Fig. 1). The ability to expand human primitive NSCs at high densities while retaining multipotentiality provides a beneficial approach, since this study demonstrates as a proof of principle that a few subtypes of neurons can be generated, such as GABA neurons from forebrain, dopaminergic neurons from midbrain, and motor neurons from hindbrain. Additional studies are warranted to generate region-specific multiple neural types in human brain to fully explore function, genotype changes, and disease modeling of these neural cells.
Despite our nontraditional approach to neural induction, NSCs derived by this method still displayed multipotentiality with successful differentiation to generic neurons, GABA neurons, dopaminergic neurons, motor neurons, astrocytes, and oligodendrocytes. This differentiation capacity was retained even after expansion of NSCs, demonstrating the competence of the derived NSCs and the robustness of the neural induction method. Based on pairwise comparison of global gene expression, a close correlation between the expression patterns of NSCs grown as rosette-derived versus rosette-free-derived was observed. In addition, cells grown and expanded in neural induction media for longer passages continued to show a close correlation, suggesting minimal genotypic changes upon extended passaging.
Gene expression data comparing our NSCs with other NSCs, such as rosette-derived and human fetal brain-derived NSCs, is consistent with reported transcript profiles [26]. The global gene expression data show two main clusters, one with fetal NSCs and another containing rosette-derived and primitive NSCs, derived using this method. Importantly, pairwise comparison of pNSCs derived at P0 versus rosette-derived cells demonstrates fairly high correlation. The only noticeable change with global gene expression from P0 to P6 is the loss of expression of most non-neural genes. Although NSCs differentiated by our method are enriched for some dorsal hindbrain markers, they still retain the capacity to differentiate into neurons from all regions of the brain. Based on these results, it can be concluded that there is no significant drift in global gene expression with extended passaging in this medium system with scalable production of primitive NSCs that have the ability to both self-renew and differentiate into neural subtypes.
In addition to the data presented here, focusing on the expansion of human primitive NSCs and differentiation to multiple neural lineages, the medium culture system offers scope to generate exact neural cell types needed for central nervous system diseases, such as Parkinson's disease, amyotrophic lateral sclerosis, and Huntington's disease. A simplified platform such as common primitive NSCs to generate multiple neural types that exist in the brain would present a unique opportunity for researchers in academia, biotech, and cell therapy to model neurological diseases for drug discovery and translational cell therapy.
Conclusion
In summary, we have developed a culture medium that is manufactured in GMP conditions that enables the scalable production of neural stem cells from human pluripotent stem cells in a rapid and efficient way. This method and culture medium when used together yield NSCs (a) in highly reproducible quantities, bypassing the tedious rosette formation and picking steps eliminating variability; (b) from which neuronal, astrocyte, and oligodendrocyte cells can be differentiated; and (c) from which disease-relevant, region-specific neuronal cells such as forebrain GABAergic neurons, midbrain dopaminergic neurons, and hindbrain motor neurons can be generated. The culture system enables the creation of NSC banks in regulatory-friendly conditions suitable for drug screens and cell therapy. Maintenance of the NSC phenotype following expansion and normal karyotype is critical for clinical utility, and it is thus important that this medium culture system retains the properties critical for drug screening and cell therapy applications in a scalable, regulatory-compliant manner for meeting the needs of the regenerative medicine field.
Supplementary Material
Author Contributions
Y.Y.: neural induction medium conception, research and development work associated with medium development, data generation and analyses of neural induction, NSC expansion and differentiation into triple neural lineages, manuscript writing of methods and results; S.S., J.S., F.L., and M.Z.: global gene expression studies and analysis; B.S.J.: generation of GABA and motor neurons; Q.L. and X.Z.: dopaminergic differentiation generation and analysis; J.D. and K.B.: differentiation toward eye field cells; M.R. and N.M.: design review, data analysis and interpretation; M.C.V.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
Disclosure of Potential Conflicts of Interest
Y.Y., S.S., and M.C.V. have compensated employment at Life Technologies.
References
- 1.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–380. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hitoshi S, Seaberg RM, Koscik C, et al. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 2004;18:1806–1811. doi: 10.1101/gad.1208404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viti J, Feathers A, Phillips J, et al. Epidermal growth factor receptors control competence to interpret leukemia inhibitory factor as an astrocyte inducer in developing cortex. J Neurosci. 2003;23:3385–3393. doi: 10.1523/JNEUROSCI.23-08-03385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalyani AJ, Mujtaba T, Rao MS. Expression of EGF receptor and FGF receptor isoforms during neuroepithelial stem cell differentiation. J Neurobiol. 1999;38:207–224. [PubMed] [Google Scholar]
- 5.Kalyani A, Hobson K, Rao MS. Neuroepithelial stem cells from the embryonic spinal cord: Isolation, characterization, and clonal analysis. Dev Biol. 1997;186:202–223. doi: 10.1006/dbio.1997.8592. [DOI] [PubMed] [Google Scholar]
- 6.Rao MS. Multipotent and restricted precursors in the central nervous system. Anat Rec. 1999;257:137–148. doi: 10.1002/(SICI)1097-0185(19990815)257:4<137::AID-AR7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Denham M, Huynh T, Dottori M, et al. Neural stem cells express non-neural markers during embryoid body coculture. Stem Cells. 2006;24:918–927. doi: 10.1634/stemcells.2005-0151. [DOI] [PubMed] [Google Scholar]
- 8.Hsu YC, Lee DC, Chiu IM. Neural stem cells, neural progenitors, and neurotrophic factors. Cell Transplant. 2007;16:133–150. [PubMed] [Google Scholar]
- 9.Lim DA, Huang YC, Alvarez-Buylla A. The adult neural stem cell niche: Lessons for future neural cell replacement strategies. Neurosurg Clin N Am. 2007;18:81–92. ix. doi: 10.1016/j.nec.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Chojnacki AK, Mak GK, Weiss S. Identity crisis for adult periventricular neural stem cells: Subventricular zone astrocytes, ependymal cells or both? Nat Rev Neurosci. 2009;10:153–163. doi: 10.1038/nrn2571. [DOI] [PubMed] [Google Scholar]
- 11.Bauer S. Cytokine control of adult neural stem cells. Ann NY Acad Sci. 2009;1153:48–56. doi: 10.1111/j.1749-6632.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 12.Muguruma K, Sasai Y. In vitro recapitulation of neural development using embryonic stem cells: From neurogenesis to histogenesis. Dev Growth Differ. 2012;54:349–357. doi: 10.1111/j.1440-169X.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- 13.Irioka T, Watanabe K, Mizusawa H, et al. Distinct effects of caudalizing factors on regional specification of embryonic stem cell-derived neural precursors. Brain Res Dev Brain Res. 2005;154:63–70. doi: 10.1016/j.devbrainres.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Tozer S, Le Dréau G, Marti E, et al. Temporal control of BMP signalling determines neuronal subtype identity in the dorsal neural tube. Development. 2013;140:1467–1474. doi: 10.1242/dev.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes L, Ralls S, Wang H, et al. Duration of Shh signaling contributes to mDA neuron diversity. Dev Biol. 2013;374:115–126. doi: 10.1016/j.ydbio.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickx M, Van XH, Leyns L. Anterior-posterior patterning of neural differentiated embryonic stem cells by canonical Wnts, Fgfs, Bmp4 and their respective antagonists. Dev Growth Differ. 2009;51:687–698. doi: 10.1111/j.1440-169X.2009.01128.x. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 19.Koch P, Opitz T, Steinbeck JA, et al. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DS, Lee DR, Kim HS, et al. Highly pure and expandable PSA-NCAM-positive neural precursors from human ESC and iPSC-derived neural rosettes. PLoS One. 2012;7:e39715. doi: 10.1371/journal.pone.0039715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patani R, Compston A, Puddifoot CA, et al. Activin/Nodal inhibition alone accelerates highly efficient neural conversion from human embryonic stem cells and imposes a caudal positional identity. PLoS One. 2009;4:e7327. doi: 10.1371/journal.pone.0007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Hu J, Zhou L, et al. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J Cell Sci. 2011;124:1867–1877. doi: 10.1242/jcs.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou JM, Chu JX, Chen XJ. An improved protocol that induces human embryonic stem cells to differentiate into neural cells in vitro. Cell Biol Int. 2008;32:80–85. doi: 10.1016/j.cellbi.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Sun W, Zhang Y, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci USA. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surmacz B, Fox H, Gutteridge A, et al. Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem Cells. 2012;30:1875–1884. doi: 10.1002/stem.1166. [DOI] [PubMed] [Google Scholar]
- 26.Shin S, Sun Y, Liu Y, et al. Whole genome analysis of human neural stem cells derived from embryonic stem cells and stem and progenitor cells isolated from fetal tissue. Stem Cells. 2007;25:1298–1306. doi: 10.1634/stemcells.2006-0660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.