Abstract
Ongoing global genome characterization efforts are revolutionizing our knowledge of cancer genomics and tumor biology. In parallel, information gleaned from these studies on driver cancer gene alterations—mutations, copy number alterations, translocations, and/or chromosomal rearrangements—can be leveraged, in principle, to develop a cohesive framework for individualized cancer treatment. These possibilities have been enabled, to a large degree, by revolutionary advances in genomic technologies that facilitate systematic profiling for hallmark cancer genetic alterations at increasingly fine resolutions. Ongoing innovations in existing genomics technologies, as well as the many emerging technologies, will likely continue to advance translational cancer genomics and precision cancer medicine.
CASE FOR TUMOR GENOMIC PROFILING
Progress in cancer genomics research over the past few decades has reinforced the notion that cancer is driven by various types of genomic alterations. Although some cancers harbor frequently recurring alterations in one or a small number of genes (95% of chronic myelogenous leukemias harbor a reciprocal translocation between chromosomes 9 and 22 resulting in the BCR-ABL fusion gene1,2), other cancer types exhibit considerable heterogeneity in the constellation of alterations that drive the malignancy. Conversely, although many of these alterations show tumor type specificity (eg, BRAF mutations occur frequently in papillary thyroid carcinomas3 and cutaneous melanomas), they may also occur at lower frequencies across many other cancer types (eg, BRAF mutations are present in 2% to 20% of non–small-cell lung cancers [NSCLCs],4 colorectal adenocarcinomas,5 pediatric low-grade astrocytomas,5 and multiple myelomas6). This “long tail” of rare driver genetic events may pose particular technological and methodological demands in the molecular cancer diagnostics arena as more and more genetic alterations become clinically actionable.
Many genomic alterations create a dysregulated signaling cascade, and the derivative mutant proteins (or proteins up- or downstream in the same or related pathway) are thus potential (and sometimes potent) foci for targeted anticancer therapies. There are several clinical success stories of rational targeted therapies based on knowledge of the underlying genetics: activating mutations and small insertions/deletions in the epidermal growth factor receptor (EGFR) in NSCLC confer sensitivity to the EGFR tyrosine kinase inhibitors erlotinib and gefitinib,7–9 and BRAF mutations in melanoma (specifically at the V600 locus) are targets for BRAF inhibitors10; clinical trials have confirmed the utility of targeted therapies in these instances.11,12 Information on the mutational status of many known cancer genes can thus be used to design rational therapeutics for a given patient.13,14 Similarly, the concept of synthetic lethality15— identifying and targeting a secondary dependency of a cancer cell when the primary target is inhibited, exemplified by the sensitivity of BRCA1/2-deficient breast cancer cells to poly(ADP-ribose) polymerase inhibition—allows one to selectively target cancer-specific mutations effectively.
It seems clear, therefore, that knowledge of a spectrum of actionable genomic alterations within an individual tumor—whether mutations, chromosomal rearrangements, copy number changes, or epigenetic alterations—may ultimately facilitate individualized approaches for many patients with cancer. However, systematic and comprehensive profiling of cancers remains underdeveloped in many patient-oriented research or clinical settings. Disruptive advances in sequencing technologies over the past several years have rapidly advanced cancer research efforts and are poised to similarly transform the translational oncology landscape. As they accelerate toward the clinic, these technologies may enable robust readouts of the genetic content of a tumor, facilitate the deployment of clinical trials on targeted agents and, ultimately, inform more rational treatment of many patients with cancer.
FIRST-GENERATION SEQUENCING
The technological revolution in the field of genomics began more than 30 years ago with the discovery of methodologies that first enabled investigators to perform DNA sequencing.16,17 During the intervening years, major improvements in molecular biology, DNA separation and detection, process automation,18 and analytics facilitated the landmark sequencing of the first human genome in 2001.19,20 Among other things, this achievement established a baseline reference genome for subsequent resequencing efforts and instituted Sanger sequencing as the major technology in the first generation of genomic interrogation. Approaches for cancer gene sequencing at that time consisted of amplifying the exonic regions of specific gene(s) or gene sets by the polymerase chain reaction (PCR), followed by sequencing of PCR-amplified DNA products by using capillary-based instruments. Although this candidate gene sequencing approach was laborious, costly, and limited in scope, it nonetheless resulted in vitally important discoveries7–9,21,22 that laid a solid foundation for genome-scale cancer characterization efforts.
Limitations of Sanger Sequencing
As Sanger-based sequencing efforts moved increasingly into cancer research, it became clear that many tumor specimens and their derivative genomic DNA posed specific challenges that often confounded the detection of specific genomic alterations. For example, histologic variables such as the purity of a tumor specimen, the ploidy of the tumor cells, and the presence of subclonality or heterogeneity within a sample could significantly affect the sensitivity and specificity of DNA sequencing and other genomics technologies (reviewed in Meyerson et al23). In large part, these difficulties centered around the ability to distinguish the signal of a specific genomic alteration from a background of normal noise contributed by stromal admixture or “passenger” ploidy alterations within the tumor cell. Although Sanger sequencing is still considered the gold-standard molecular diagnostic technology for interrogation of mutations and insertions/deletions (indels) on a gene-by-gene basis (EGFR and KIT, among others), this technology has several limitations: it is insensitive to alterations that occur at an allele frequency lower than approximately 20%24 (a phenomenon that may reflect low tumor content in a specimen), it has limited clinical scalability beyond a few genes, and it is unable to detect structural rearrangements or DNA copy number changes. Newer sequencing technologies that have greater sensitivity (such as pyrosequencing) are also used in some clinical laboratories to detect small base-pair changes in genes, although this approach is also somewhat limited in scope compared with next-generation technologies.
Genotyping
A distinctive feature of several of the classical oncogenes (eg, RAS, BRAF, PIK3CA, and so on) is that interrogation of their entire coding sequence is not required to identify their most important activating mutations. Instead, a subset of critical oncogene point mutations (and small indels) affect so-called “hotspot” amino acid codons. For example, 80% to 99% of known mutations in EGFR, BRAF, and KRAS can be assayed by interrogating only 10 to 20 bases in each gene. This phenomenon lends itself well to platforms such as mass-spectrometric genotyping5,25 and allele-specific PCR.26 Accordingly, these technologies emerged as attractive alternatives to conventional Sanger sequencing—in terms of cost, throughput, and sensitivity—for the assessment of specific driver mutations in oncogenes. Indeed, multiple cancer centers worldwide have instituted such technologies to anchor their initial precision cancer medicine initiatives.5,26–32 Several of these programs have implemented enterprise-wide projects to systematically profile patient tumor samples for mutations in potentially actionable or informative genes and deposit these data into internal databases that may be used for research or mined to identify candidates for specific targeted clinical trials.5 Preliminary results from early clinical trials matching patients with targeted drugs on the basis of their genotype have yielded encouraging albeit preliminary results.33
Genotyping-based platforms are advantageous in terms of cost, throughput, scalability (dozens of genes can be interrogated), and their applicability to (relatively) poor-quality nucleic acid material, such as that derived from archival tumor specimens. However, such platforms have many drawbacks, including a limited sensitivity (a minimum of approximately 10% tumor allelic fraction), restricted breadth (number of genes and mutations), and an inability to detect multiple categories of genomic alterations (limited to an a priori set of highly recurrent hotspot mutations and indels). The deficiencies of these technologies are qualitatively similar to those that restrict Sanger sequencing and pose important limitations on the use of such platforms as clinical tools for comprehensive tumor genomic profiling.
It should be noted that other technologies aside from Sanger sequencing and allele-based mutation detection platforms have long been leveraged for the assessment of cancer driver events other than mutations—some of these are commonly used in clinical and molecular diagnostic laboratories today. Examples include array comparative genomic hybridization34 and fluorescent in situ hybridization, which allow for the detection of genomic imbalances including copy-number alterations and deletions. However, the allocation of tumor material, often of limited quantity and quality, to interrogate distinct types of genetic alterations using diverse profiling platforms presents several logistical and cost-related challenges.
NEXT-GENERATION SEQUENCING: A TECHNOLOGY REVOLUTION
By the second half of the last decade, it had become clear that although the number of plausibly actionable genetic alterations was expanding rapidly, no categorical genomics technology existed to identify them all at once. Indeed, as many as three distinct profiling platforms may have been necessary to detect clinically actionable base mutations, copy number alterations, and translocations (Fig 1) in a tumor specimen. Starting in 2005, however, several powerful DNA sequencing technologies emerged that were radically different from the capillary-based instruments used to analyze the initial reference human genome. These next-generation or massively parallel sequencing (MPS) technologies enabled an unprecedented depth and breadth of genomic interrogation that soon transformed the cancer genome discovery arena.35
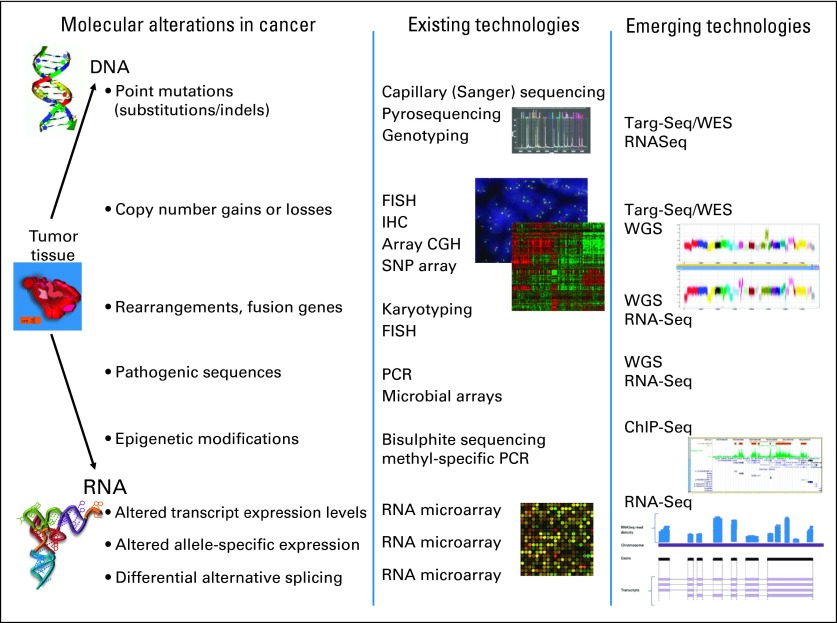
Fig 1.
Categories of genomic alterations and technologies for detection. Many of the hallmark alterations in cancer are currently detected by using a multitude of existing technologies, often in a serial fashion, each using an appreciable amount of nucleic acid. Newer sequencing-based methodologies are capable of interrogating many types of cancer alterations in one composite, sensitive test. CGH, comparative genomic hybridization; ChIP-Seq, chromatin immunoprecipitation followed by massively parallel sequencing; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; PCR, polymerase chain reaction; RNA-Seq, RNA sequencing, also known as transcriptome sequencing; SNP, single nucleotide polymorphism; Targ-Seq, targeted sequencing; WES, whole-exome sequencing; WGS, whole-genome sequencing.
MPS technologies have three major advantages over conventional techniques. First, they brought forth an exponential decrease in the cost of sequencing. In short order, this economic disruption effectively democratized the genome sequencing arena, rendering this technology widely accessible to many investigators. For example, in 2007, a personal genome sequenced by using the Sanger method cost US$70 million36; by 2010, the cost for sequencing a genome by using MPS had dropped to approximately US$50,000, and in 2013, genome sequencing can be performed in a commercial or research setting for under US$5,000 (Fig 2). Second, the development of MPS conferred substantial increases in both the sensitivity (by sequencing to a high redundancy) and the scalability of sequencing, thus allowing a deep interrogation of thousands of genes—and eventually the entire exome—in a single sequencing lane. The first MPS technology reported a 100-fold improvement in throughput over Sanger sequencing37; in 2013, it is possible to sequence more than 10 human genomes in a single day by using an Illumina HiSeq sequencer. Third, these technologies provide the capability to detect multiple types of cancer genome alterations (base mutations, indels, copy number alterations, and rearrangements). MPS technologies (Table 1) vary in chemistries, enzymatic reactions, and signal detection methodologies, but they share common methodologic attributes that have produced the aforementioned groundbreaking improvements over capillary sequencing.
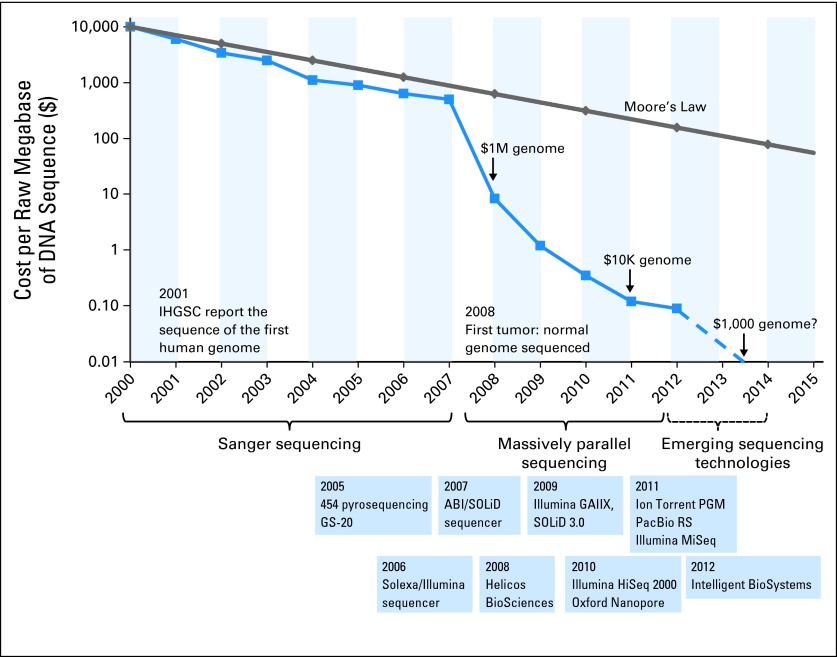
Fig 2.
The decrease in cost of genome sequencing facilitated by massively parallel sequencing technologies. The cost of sequencing has decreased at a rate faster than Moore's law in the past 10 years. The data from 2001 through 2007 represent the costs of generating DNA sequences by using Sanger-based chemistries and capillary-based instruments (first-generation sequencing platforms). Starting in 2008, the data represent the costs of generating DNA sequences by using second-generation sequencing technologies. The change in instruments represents the rapid evolution of DNA sequencing technologies that has occurred in recent years. Landmark events are also indicated on the timeline. The release of various second- and third-generation technologies is indicated in blue boxes. IHGSC, International Human Genome Sequencing Consortium. Data adapted from the National Human Genome Research Institute Web site.35
Table 1.
Comparison of Commercially Available Next-Generation Sequencing Technologies Detailing Similarities and Differences
| Technology | Library Construction | Sequencing Mechanism | Detection Mechanism | Maximum Read Length (bp) | Error Mode |
|---|---|---|---|---|---|
| First generation | |||||
| ABI Sanger | Bacterial cloning | Dideoxy chain termination | Fluorescence | 900 | End-of-read errors |
| Massively parallel sequencing, part 1 | |||||
| Roche 454 | Emulsion PCR on microbead surface | Polymerase-mediated incorporation of unlabelled nucleotides | Photon detection (light) | 700 | Errors in homopolymer runs |
| Illumina HiSeq | Amplification on glass surface | Polymerase-mediated incorporation of fluorescent nucleotides | Fluorescence | 250 | End-of-read errors |
| Life Technologies SOLiD | Emulsion PCR on microbead surface | Ligase-mediated addition of two base-encoded fluorescent oligonucleotides | Fluorescence | 75 | End-of-read errors |
| Massively parallel sequencing, part 2 | |||||
| Helicos | N/A (single molecule detection) | Polymerase-mediated incorporation of fluorescent nucleotides | Fluorescence | 32 | End-of-read errors |
| Life Technologies Ion Torrent | Emulsion PCR on microbead surface | Polymerase-mediated incorporation of unlabelled nucleotides | Ion sensing by semiconductors | 200 | Errors in homopolymer runs |
| Pacific Biosciences | N/A (single molecule detection) | Polymerase-mediated incorporation of fluorescent nucleotides | Fluorescence in real time | > 1,000 | Random errors |
| Oxford Nanopore | N/A (single molecule detection) | Depolymerization and cleavage of individual nucleotides | Ion sensing by nanopore in electrically resistant membrane bilayer | Approximately 50 | Errors generated by slipping or skipping of DNA |
NOTE. Massively parallel sequencing has evolved in two stages: part 1 includes technologies that were developed before 2008; part 2 indicates technologies that have one or more characteristics that define an emerging technology (ie, single-molecule sequencing or direct detection of nucleotide signal).
Abbreviations: N/A, not applicable; PCR, polymerase chain reaction.
Template Preparation and Library Construction
The initial preparatory steps for MPS are fewer and simpler than for Sanger sequencing: instead of stepwise PCR amplification and preparation of individual DNA fragments, the initial reaction produces a composite DNA library for sequencing that is formed by ligating synthetic adapters to the ends of a target (fragmented) DNA population. The library of fragments is subsequently PCR-amplified en masse to produce many copies of each fragment (this is one of the massively parallel steps). A common methodologic theme for MPS involves immobilization of the template DNA molecules onto a solid surface or support (either a bead or a glass slide). This allows spatial separation of molecules and enables simultaneous sequencing reactions (millions or billions) to be carried out in a highly parallel fashion (this is the definitive massively parallel step).
Sequencing Reactions
For most MPS technologies, sequencing reactions are executed as a series of repeating steps that are performed and detected sequentially (known as “sequencing-by-synthesis” [SBS]). Because these reactions are performed in pico- or nano-scale reactors in a massively parallel fashion, the products of many millions of reactions can be sequenced simultaneously, thus greatly reducing the cost of sequencing and increasing the speed at which data can be generated. In 2005, the first MPS technology—large-scale parallel pyrosequencing37— using SBS was reported; subsequent technologies also use SBS reactions coupled with different methodologies for signal detection38 (Table 1).
Paired-End Analysis
Several MPS technologies (Illumina and Life Technologies, among others) have the option of sequencing both ends of a DNA fragment in a sequencing library. Depending on the sequencing instrument and the methodology used for library construction, linear (paired-end) sequencing or circularized fragment (mate-pair) sequencing can be performed. The value of paired-end sequencing is an increased ability to map to a unique region of the genome and the ability to discover small- and large-scale structural variation in genomes.
The specifics of each step in the sequencing reaction differ for the various MPS platforms that have emerged in the past 7 years, which indicates the range of innovation in chemistry, molecular biology, and engineering required to produce sequence information in a massively parallel fashion (a comprehensive review can be found in Metzker et al36). Much of this innovation was enabled by funding from the National Human Genome Research Institute of the National Institutes of Health (NIH-NHGRI). Over the past 3 to 5 years, MPS technologies have advanced considerably to become faster and more accurate, modular, and cost-effective.39 This progress (coupled with competition among alternative platforms for leadership) has placed MPS in a seemingly permanent state of revolution. The extant MPS technologies vary not only in their mode of sequencing but also in accuracy, read length, throughput, coverage distribution and bias, variant detection, and cost40 and have been compared in several recent publications.41–43
APPLICATIONS OF MPS
The technological transformation in the field of DNA sequencing, coupled with substantial computational enhancements in the ability to align individual sequence reads against a reference human genome and identify variants, has both enlivened cancer genome discovery and enabled tumor genomic profiling at unprecedented scale, depth, and speed. It is now feasible to generate a fully comprehensive catalog of somatic mutations and to garner insights into altered biologic processes that contribute to the development and progression of human cancers. Importantly, MPS can interrogate different types of input material (DNA, RNA, or chromatin), varying proportions of the genome (whole genome, exome, transcriptome, or a subset of genomic regions) once several preprocessing modifications are incorporated (the various MPS applications are often termed DNA-Seq, RNA-Seq, ChIP-Seq, or methyl-Seq44). Together, these approaches yield comprehensive information on a markedly expanded constellation of (epi)genetic and gene expression-based alterations. Specific applications of MPS are outlined in more detail.
Whole-Genome Sequencing
The first whole-genome sequencing (WGS) effort for a cancer—a cytogenetically normal acute myeloid leukemia—was reported in 200845 at an estimated cost of US$1.6 million. This ambitious effort required more than 100 sequencing runs using an Illumina machine. A year later,46 the cost had dropped to US$500,000.36 Since then, whole cancer genomes from breast,47–49 colorectal,50 liver,51 lung,52–54 medulloblastoma,55 melanoma,56 multiple myeloma,6 ovarian,57 and prostate58 cancers, among many others, have been reported. Large-scale cancer genome studies, such as those conducted by the International Cancer Genome Consortium (ICGC), are using MPS to comprehensively characterize tumors and generate genomes, transcriptomes, and epigenomes from more than 50 cancer types, thus providing a solid foundation for a complete catalog of oncogenic mutations.
WGS allows the identification of copy-number alterations and structural rearrangements at approximately 30- to 60-fold mean depth of coverage of both the tumor and its paired normal (germline) DNA49 (although accurate point mutation detection often requires deeper coverage, particularly if stromally admixed tumor material is queried).23 The paired-end nature of MPS59 can be leveraged effectively for the detection of DNA rearrangements, which underpin key translocation events in cancer (such as actionable gene fusions).60–64 WGS can also detect other types of genomic alterations, such as noncoding changes in unannotated regions of the genome21 as well as the presence of pathogenic, nonhuman sequences.65 Although the potential clinical utility of WGS has been demonstrated for a few tumor cases, the feasibility of performing WGS routinely has been questioned.66 For instance, WGS of archival (formalin-fixed paraffin-embedded) tumor material remains problematic (because of poor-quality DNA and resultant loss of genomic complexity), as does achieving a depth of coverage that is both sufficiently sensitive and cost-effective for clinical mutation detection (at least 100- to 200-fold sequencing coverage may be required for this application). Procuring server- or cloud-based storage for such large amounts of data also poses challenges. Finally, we do not yet understand the functional significance of many alterations and, thus, most WGS data are neither actionable nor informative at present. Nonetheless, additional advances in sequencing data generation and analysis may ultimately render WGS an appealing default approach for at least some clinical applications, since this can provide a permanent, patient-specific reference of tumor and germline genomic alterations that could be mined iteratively as new discoveries regarding clinical actionability are made.
Targeted Sequencing (whole-exome and cancer-specific panels)
An alternative approach to WGS involves the enrichment of a subset of the genome before MPS. By using this “target region” methodology, one can obtain much higher coverage with less raw sequence data generation and at a significantly lower cost than WGS. Several strategies have been effectively used for enrichment of so-called “target” cancer sequences of interest.4,67–70 One common approach uses a hybridization reaction between the genomic DNA and long, synthetic oligonucleotides. These DNA or RNA “baits” are either matrix-bound (array-based) or in solution (solution-phase), and a hybrid selection process is used to capture target DNA fragments from a “pond” of genomic DNA before sequencing.71 The performance of commercially available whole-exome sequencing (WES) bait sets (notably, Illumina, Agilent, and Nimblegen) or custom-designed bait sets can differ, and each platform has varying metrics in terms of input DNA required, bait length, specificity, target region coverage, genomic elements that can or cannot easily be targeted, and performance.72–75 Nonetheless, hybrid capture approaches represent a cost-effective means of selecting a subset of the cancer genome for sequencing,76 and thus have become a mainstay of MPS applications. This methodology underpins targeted MPS approaches ranging from a few hundred genes to the entire gene coding territory or exome.
WES and targeted sequencing, together with appropriate computational algorithms, allow robust detection of point mutations, amplifications, and deletion (as benchmarked against conventional techniques76,77), although detection of translocation events and rearrangements is more limited (this can be somewhat enhanced if hybrid capture baits targeting intronic regions are included). In principle, WES enables identification of a greater breadth of coding mutations and gene copy number events than targeted exome sequencing; however, targeted sequencing can achieve much greater depth of sequence coverage (and hence sensitivity) at a given cost. Moreover, inasmuch as targeted sequencing may focus on known cancer genes/mutations, their focused interrogation may obviate the need to sequence a matched normal sample from the same patient. Both approaches have been rendered successfully by using DNA from frozen and archival tumor material. Moreover, because the targeted region comprises only a small fraction (approximately 2%) of the genome, the vastly higher sequence coverage obtained per unit cost allows for the detection of lower-abundance somatic variants (thus revealing clinically useful information from stromally contaminated or heterogeneous tumor specimens). Targeted cancer sequencing is now available in the commercial setting as well as in Clinical Laboratory Improvement Amendments (CLIA) –certified hospital laboratories in several cancer centers, and initiatives devoted to precision cancer medicine enabled by MPS are also underway.
In the fullness of time, our understanding of cancer biology will likely be informed not only by genome sequencing (eg, chromosomal DNA-based) but also by identifying perturbations in transcriptomic, epigenetic, and protein space. Transcriptome sequencing (RNA-Seq)78 can capture the expressed genome of a tumor sample, thereby enabling robust detection of dysregulated genes,79 gene fusion events, and alternative splice isoforms.80,81–83 Chromatin immunoprecipitation and sequencing (ChIP-Seq)84 can be used to study interactions between proteins and DNA at a high resolution; analysis of the methylation status of particular CpG sites (methyl-Seq) may assist in interpreting the activation state of certain genes or regions. In the future, information from multiple datasets could be combined; toward this end, integration of sequencing methodologies has been shown to generate more informative mutational landscapes and highlight key cellular pathways and processes that become dysregulated.85,86 It should be emphasized that the clinical utility of these additional MPS applications remains to be determined; however, they will likely prove useful for near-term pilot studies of novel therapeutics as correlative science efforts. In this regard, a pilot study of integrative high-throughput sequencing (tumor WGS and RNA-Seq, tumor and matched normal WES) recently demonstrated promise in facilitating biomarker-driven clinical trials for oncology patients.14
LIMITATIONS OF MPS
The advantages of MPS (by now well recognized) are undergirded by its extremely high overall throughput and resultant low cost per sequenced base. For example, Illumina's HiSeq 2000 instrument can generate upwards of 600 Gb of sequence data per run. At the same time, there are several limitations to current MPS platforms. (1) When using a hybrid-capture approach, mission-critical exons may sometimes be missed (due to challenges with GC%, content, and uneven capture efficiency). (2) MPS technologies remain highly sophisticated. Although academic and commercial laboratories are trying to streamline processes, this approach still requires considerable expertise to routinely run MPS. (3) The turnaround time remains lengthy, driven by the stepwise process of fluorophore incorporation, detection, and washing on the sequencing instrument. Over the past year, three major MPS platforms released benchtop machines or reconfigured existing instruments to address the turnaround time; as a result, large amounts of MPS data may be generated after only about 24 hours on the sequencer. These advances suggest that clinical-grade testing—delivering results to a physician only a few days after receiving the sample—is now feasible.
EMERGING TECHNOLOGIES FOR GENOMIC PROFILING
Additional improvements (Fig 3) to MPS approaches hold further promise for increasing the power of MPS. Although these technologies remain in their infancy, further development could potentially herald the emergence of a new generation of sequencing technologies that evolve from amplified products to single molecules and from optical to electrical (or direct) detection.87–90 Although these approaches have the potential to increase throughput, generate longer sequence reads, and boost turnaround time, their performance with regard to sequence quality and output remains poorly characterized. Nonetheless, these ongoing efforts raise the possibility that WGS may someday become rapid and inexpensive enough to enable genome sequencing as part of routine care. A comprehensive review of these technologies is available in Schadt et al91; however, a high-level overview of exemplary topics follows.
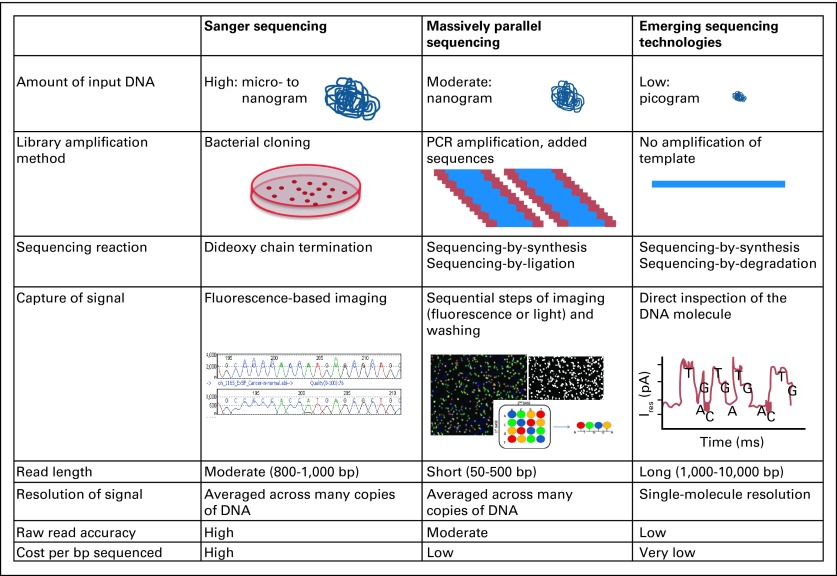
Fig 3.
Existing and emerging sequencing technologies. Comparison of the generations of sequencing technologies, illustrating the similarities and differences in methodologies and some sequencing characteristics. PCR, polymerase chain reaction.
Single-Molecule Sequencing
Single-molecule sequencing represents a logical destination for a technology in which the maximum amount of information is extracted from a minimum amount of nucleic acid. This is particularly true for cancer sequencing, in which primary specimens are often sparse, and the amount of DNA extracted from a specimen may limit the number and size of genomic studies that can be performed. Theoretically, direct interrogation of individual DNA molecules could obviate the need for PCR amplification procedures and lower the cost of sequencing to US$100 to US$1,000 per genome (at current target depths of sequence coverage).92
There are several categories of single-molecule sequencing technology.93 Nanopore sequencing is performed by threading individual DNA molecules through a pore formed by a single protein molecule.87,94–98 If successful, this approach may significantly increase read length, simplify sample preparation, and process low-input DNA amounts (picograms or less). Eventually, nanopore sequencing may also offer the potential to sequence RNA directly. However, its applicability to formalin-fixed paraffin-embedded–derived tumor DNA remains unproven.
Direct Detection of Nucleotide Signal
Several approaches have queried the concept of direct imaging and detection of the nucleotide sequences comprising individual DNA molecules. An early approach involved semiconductor sequencing (Ion Torrent); others include transmission electron microscopy or transistor-based sequencing. Alternatively, direct observation of single DNA molecules in the context of DNA polymerase (in a highly parallel system) during DNA synthesis has been developed.92
ADDITIONAL APPLICATIONS OF EMERGING SEQUENCING TECHNOLOGIES
The sensitivity of many existing molecular diagnostics technologies is limited by stromal contamination and genetic heterogeneity within a cancer specimen. Moreover, the development of cancer drug resistance may often be attributed to (initially low-abundance) somatic mutations that undergo positive selection over the course of drug treatment.99–101 Thus, a potential advantage of emerging technologies involves the ability to query single cells (or a small number of cells). In the future, this approach could provide real-time information regarding genetic resistance mechanisms,102 demonstrate the evolution of tumor mutational landscapes, or monitor cancer mutations in circulating tumor cells.103 Profiling subclonal heterogeneity might also help predict patient response, thus informing the rational combination of targeted therapies to maximize efficacy13,104 and response and minimize resistance. Many emerging sequencing platforms may eventually have utility beyond DNA sequencing, including identification of patterns of methylation,105 comprehensive characterization of transcriptomes,78 and comprehensive characterization of translation.106
CHALLENGES IN CLINICAL APPLICATION OF MPS TECHNOLOGIES
Despite the optimism surrounding technologies for cancer genome characterization, several questions must be addressed to realize the potential of these technologies within the larger clinical context. The availability of biospecimens—whether resected tumor, biopsy, fine-needle aspirate, or other—and the purity of specimens (due to admixed cell populations, tumor ploidy, tumor heterogeneity, or other biologic confounders) are likely to remain limiting factors in obtaining sufficient tumor genomic material. Large sequencing data volumes may strain existing archival storage mechanisms, despite the development of resources107 and efficiencies that facilitate electronic data storage (such as compression108 or cloud-based systems). Several cancer centers and clinical and reference laboratories are implementing MPS approaches in a CLIA environment.109,110 From a clinical laboratory perspective, the challenges of regulatory issues, assay validation, proficiency testing, and quality control must be addressed (comprehensively reviewed in Schrijver et al111), and there is considerable uncertainty about the regulatory pathway for MPS testing, although efforts are underway to develop principles and guidelines.111
FUTURE DIRECTIONS
The acceleration in biologic and therapeutic understanding of cancer enabled by improvements in MPS technologies is well underway. Moreover, the sequencing technology arena seems poised for continued evolution. The resulting new platforms will have a continuing impact on cancer biology and biomedical research for years to come. In the near term, these technologies may also offer a tool that will provide physicians with relevant contextual genomic alterations for their patients with cancer. When paired with a robust interpretive framework to identify and interpret the underlying genomic information, this technology holds forth superb potential to accelerate precision cancer medicine.
Acknowledgment
We thank Chad Nusbaum (Broad Institute) for helpful discussions on technologies.
Glossary Terms
- DNA-Seq:
DNA sequencing (DNA-Seq) determines the nucleotide sequence of DNA. The amount of DNA sequenced can be the full complement of genetic material in a specimen (whole-genome), or a targeted portion of the genome (whole-exome, specific genes, targeted regions and so on).
- RNA-Seq:
RNA-Seq (also known as transcriptome sequencing) determines the nucleotide sequence of RNA as cDNA derived from (usually) mRNA, but also microRNAs or other RNAs. In tumor genomic profiling, RNA-Seq captures the expressed genome of a sample, and can enable robust detection of dysregulated genes, gene fusion events, and alternative splice isoforms.
- ChIP-Seq:
Chromatin immunoprecipitation followed by massively parallel sequencing (ChIPSeq) identifies the binding sites of DNA-associated proteins. In cancer genomics, this can be used (for example) to map global DNA binding sites of a transcription factor at a high resolution and determine how these interactions regulate gene expression.
- Methyl-Seq:
Methyl-Seq determines the methylation status of CpG islands (regulatory regions) across the genome, thereby assisting in the interpretation of the activation state of certain genes or regions.
- Paired-end reads:
Some sequencing technologies have the option of sequencing both ends of a DNA fragment in a library. Sequencing one end a linear fragment (representative of the original DNA) followed by sequencing the other end is termed “paired-end” sequencing. (This is different to mate-pair sequencing, where DNA fragments are circularized before sequencing). Paired-end sequencing allows more accurate mapping or placement of a DNA sequence on a reference genome. In cancer specimens, paired-end sequencing allows the detection of large- and small-scale structural genomic rearrangements.
Footnotes
Supported by the Dana-Farber Cancer Institute and by Grant No. R33 CA155554 from the National Institutes of Health.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Author's disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHOR'S DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author indicated no potential conflicts of interest.
REFERENCES
- 1.de Klein A, van Kessel AG, Grosveld G, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 4.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 12.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 16.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 17.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith LM, Sanders JZ, Kaiser RJ, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 19.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 21.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 23.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 24.Bar-Eli M, Ahuja H, Gonzalez-Cadavid N, et al. Analysis of N-RAS exon-1 mutations in myelodysplastic syndromes by polymerase chain reaction and direct sequencing. Blood. 1989;73:281–283. [PubMed] [Google Scholar]
- 25.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 26.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arcila M, Lau CY, Jhanwar SC, et al. Comprehensive analysis for clinically relevant oncogenic driver mutations in 1131 consecutive lung adenocarcinomas. Mod Pathol. 2011;24(suppl 1):404a. abstr 1721. [Google Scholar]
- 30.Arcila M, Lau C, Nafa K, et al. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beadling C, Heinrich MC, Warrick A, et al. Multiplex mutation screening by mass spectrometry evaluation of 820 cases from a personalized cancer medicine registry. J Mol Diagn. 2011;13:504–513. doi: 10.1016/j.jmoldx.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: A systems biology approach. J Clin Oncol. 2010;28:2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center Initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 35.National Human Genome Researce Institute. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2013. Feb 11, www.genome.gov/sequencingcosts.
- 36.Sudol M, Fritz JL, Tran M, et al. Evaluation of a system to screen for stimulators of non-specific DNA nicking by HIV-1 integrase: Application to a library of 50,000 compounds. Antivir Chem Chemother. 2011;22:67–74. doi: 10.3851/IMP1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quail MA, Smith M, Coupland P, et al. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Li Y, Li S, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam HY, Clark MJ, Chen R, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 44.Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 45.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 49.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 52.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 54.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pugh TJ, Weeraratne SD, Archer TC, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 61.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 64.Ju YS, Lee WC, Shin JY, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garraway LA, Baselga J. Whole-genome sequencing and cancer therapy: Is too much ever enough? Cancer Discov. 2012;2:766–768. doi: 10.1158/2159-8290.CD-12-0359. [DOI] [PubMed] [Google Scholar]
- 67.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porreca GJ, Zhang K, Li JB, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 69.Tewhey R, Warner JB, Nakano M, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner EH, Lee C, Ng SB, et al. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sulonen AM, Ellonen P, Almusa H, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark MJ, Chen R, Lam HY, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Asan, Xu Y, Jiang H, Tyler-Smith C, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parla JS, Iossifov I, Grabill I, et al. A comparative analysis of exome capture. Genome Biol. 2011;12:R97. doi: 10.1186/gb-2011-12-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lonigro RJ, Grasso CS, Robinson DR, et al. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia. 2011;13:1019–1025. doi: 10.1593/neo.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kinsella M, Harismendy O, Nakano M, et al. Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics. 2011;27:1068–1075. doi: 10.1093/bioinformatics/btr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maher CA, Palanisamy N, Brenner JC, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mardis ER. ChIP-seq: Welcome to the new frontier. Nat Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 85.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ohshiro T, Matsubara K, Tsutsui M, et al. Single-molecule electrical random resequencing of DNA and RNA. Sci Rep. 2012;2:501. doi: 10.1038/srep00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schloss JA. How to get genomes at one ten-thousandth the cost. Nat Biotechnol. 2008;26:1113–1115. doi: 10.1038/nbt1008-1113. [DOI] [PubMed] [Google Scholar]
- 90.Service RF. Gene sequencing: The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 91.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 92.Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 93.Braslavsky I, Hebert B, Kartalov E, et al. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci U S A. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zwolak M, Di Ventra M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 95.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lagerqvist J, Zwolak M, Di Ventra M. Influence of the environment and probes on rapid DNA sequencing via transverse electronic transport. Biophys J. 2007;93:2384–2390. doi: 10.1529/biophysj.106.102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajavelu A, Jurkowska RZ, Fritz J, et al. Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation. Nucleic Acids Res. 2012;40:569–580. doi: 10.1093/nar/gkr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scholz S, Liebler EK, Eickmann B, et al. Variation of the intercalating proline in artificial peptides mimicking the DNA binding and bending IHF protein. Amino Acids. 2012;43:289–298. doi: 10.1007/s00726-011-1073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 101.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 102.Smith CC, Wang Q, Chin CS, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 104.Gysin S, Salt M, Young A, et al. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flusberg BA, Webster DR, Lee JH, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ingolia NT, Ghaemmaghami S, Newman JR, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kodama Y, Shumway M, Leinonen R. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsi-Yang Fritz M, Leinonen R, Cochrane G, et al. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(suppl 5):S1–S32. doi: 10.6004/jnccn.2011.0137. [DOI] [PubMed] [Google Scholar]
- 110.Levy MA, Lovly CM, Pao W. Translating genomic information into clinical medicine: Lung cancer as a paradigm. Genome Res. 2012;22:2101–2108. doi: 10.1101/gr.131128.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schrijver I, Aziz N, Farkas DH, et al. Opportunities and challenges associated with clinical diagnostic genome sequencing: A report of the Association for Molecular Pathology. J Mol Diagn. 2012;14:525–540. doi: 10.1016/j.jmoldx.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]