Abstract
Prostate cancer is the most common type of cancer in men and the second leading cause of cancer death in men in the United States. The recent surge of high-throughput sequencing of cancer genomes has supported an expanding molecular classification of prostate cancer. Translation of these basic science studies into clinically valuable biomarkers for diagnosis and prognosis and biomarkers that are predictive for therapy is critical to the development of precision medicine in prostate cancer. We review potential applications aimed at improving screening specificity in prostate cancer and differentiating aggressive versus indolent prostate cancers. Furthermore, we review predictive biomarker candidates involving ETS gene rearrangements, PTEN inactivation, and androgen receptor signaling. These and other putative biomarkers may signify aberrant oncogene pathway activation and provide a rationale for matching patients with molecularly targeted therapies in clinical trials. Lastly, we advocate innovations for clinical trial design to incorporate tumor biopsy and molecular characterization to develop biomarkers and understand mechanisms of resistance.
INTRODUCTION
Prostate cancer is the most common nonskin cancer and the second leading cause of cancer death in men in the United States.1,2 Although there has been significant progress in the treatment of prostate cancer, with the approval of three new therapies for metastatic prostate cancer3 this year, several challenges persist such as a means to match patients with targeted therapies and the implementation of rational combination therapies. The Institute of Medicine recently critiqued the cooperative clinical trial groups in oncology and recommended innovative trial design through the incorporation of predictive biomarker stratification for patient selection.4 A molecular classification of cancer has the potential benefits of improving response, minimizing the time and adverse effects of treating patients with ineffective therapies, and reducing the sample size needed to show efficacy. High-throughput sequencing technologies have accelerated the molecular characterization of prostate cancer and positioned opportunities for development of precision medicine for therapeutic decision making in this disease. Here we examine the current data on molecular alterations in prostate cancer, the progress in translating these findings into the clinic, and the challenges that lay ahead for translational genomics in prostate cancer.
Genomic results have the potential to be translated clinically as diagnostic, prognostic, or predictive biomarkers. Diagnostic biomarkers facilitate obtaining an accurate cancer diagnosis as part of screening or confirmatory testing. Prognostic biomarkers provide data on risk of disease progression or morbidity and thereby help determine which patients need additional treatment, such as Gleason score 6 (low risk) versus 8 (high risk) prostate cancer. Predictive biomarkers suggest a course of therapeutic action. Here we provide examples, including early potential of ETS gene rearrangements as a diagnostic biomarker, and comment on novel approaches to prognostic biomarker development. Germline line mutations have the potential to be diagnostic, prognostic, or predictive and are discussed in another review in Journal of Clinical Oncology. Finally, we focus our attention on an in-depth review of putative predictive biomarkers for molecularly targeted therapies in clinical trials.
ETS GENE FUSIONS AND URINE TESTING
Gene fusions in prostate cancer were first described in 2005 using a bioinformatics approach that detected outlier transcript expression of genes with microarrays.5,6 The most common chromosomal rearrangements involve the 5′ untranslated region of the androgen-regulated gene TMPRSS2 and members of the ETS transcription factor family, ERG or ETV1. The presence of these gene fusions is essentially 100% specific for prostate cancer. Subsequent studies have confirmed ETS-related gene fusions in approximately 50% of prostate-specific antigen (PSA) –screened and 15% to 35% of unscreened prostate cancers6 (Table 1). The most common gene fusion pair occurs with TMPRSS2 and ERG genes, accounting for approximately 90% of ETS gene fusions.7 Many reports have retrospectively examined the correlation of gene fusions to Gleason score, pathologic stage, and disease-specific survival, but thus far, the data have not been consistent.6 We suspect that some of these inconsistencies may be explained in part by the varied composition of the cohorts evaluated and differences in method of fusion detection. Nevertheless, the high specificity of fusions in prostate cancer has potential value in diagnostic testing by limiting false positives; this is reviewed elsewhere.19
Table 1.
Clinically Relevant Genomic Alterations in Prostate Cancer
| Gene | Alteration Type | Frequency (%) | Potential Treatment Hypotheses |
|---|---|---|---|
| ETS transcription factors | Rearrangement | 507 | Indirect targeting of ETS gene fusions through PARP or DNAPK inhibitors8 |
| Androgen receptor | Mutation | 509,10 | Androgen synthesis inhibitors, next-generation androgen receptor antagonists |
| Amplification | 509,10 | ||
| PTEN | Loss | 509 | PI3K pathway inhibitors11 |
| RB1 | Loss | 25 to 609 | Role in disease progression and castrate resistance12 with potential for targeting |
| PIK3CA | Amplification | 1010 | PI3K pathway inhibitors11 |
| Mutation | 510 | ||
| MYC | Amplification | 99 | Potential for targeting13 |
| 4010 | Neuroendocrine prostate cancer | ||
| AURKA | Amplification | 510 | Aurora kinase inhibitors14; co-occurs with MYC in 40% of neuroendocrine prostate cancers |
| AKT | Mutation | 1 to 215 | AKT inhibitors |
| RAF | Rearrangement | 1 to 216 | RAF inhibitors |
| Mutation | |||
| KRAS | Mutation | 19,18,18 | RAF, MEK, or PI3K inhibitors |
| Rearrangement |
NOTE. Summarizes alterations in prostate cancer that have a treatment hypothesis currently being explored preclinically or in clinical trials. The most common alterations with a treatment hypothesis involve ETS rearrangements, androgen receptor, and PTEN loss. With only a limited number of samples assessed, a majority of these alterations are not necessarily mutually exclusive.
Abbreviation: PI3K, phosphatidylinositide 3-kinase.
PSA is widely offered for prostate cancer screening; however, it has limitations including false positives and the potential to result in overdiagnosis of indolent prostate cancers. To improve on PSA screening, Tomlins et al20 developed a multiplex assay combining PSA with urine testing for TMPRSS2–ERG fusion transcripts and PCA3 transcripts (noncoding RNA) to improve decision tools that predict the likelihood of cancer at time of biopsy. This diagnostic tool has the potential to help patients who may have benign prostate problems, such as benign prostate hypertrophy, avoid biopsies. Further prospective study is needed to demonstrate that this diagnostic tool selects for clinically significant or high-grade prostate cancer.
Prognostic molecular biomarkers are needed in prostate cancer to help differentiate aggressive from indolent disease. Several candidates have emerged, but clinical implementation has been limited by assay variation, small cohort size, and flaws in biomarker validation study design.19,21 In addition, prospective studies in prostate cancer are particularly challenging to complete because of the extended follow-up required to determine risk of relapse. Meanwhile, novel high-throughput approaches such as proteomics and genomics have the potential to systematically nominate candidates. One example is the emerging role of noncoding RNAs in cancer biology that may contain putative candidates for prognostic or predictive biomarkers.22
PUTATIVE PREDICTIVE BIOMARKERS IN PROSTATE CANCER
Over the past 5 years, rapidly evolving technology in nucleic acid sequencing has enabled large-scale sequencing projects of cancer genomes and transcriptomes22,23 with exhaustive identification of copy number alterations, somatic point mutations, structural rearrangements, and gene expression alterations.24,25 Before next-generation sequencing studies, Taylor et al17 published an integrative approach of the genomic and transcriptomic landscapes for prostate cancer. This work included arrays for copy number/gene expression and complete exon resequencing for mutations on a sample set of 218 prostate cancers comprising 181 primaries and 37 metastases. Since then, subsequent studies have employed next-generation sequencing strategies to further evaluate prostate cancer genomes. Kumar et al26 used exome capture and sequencing to characterize xenografts of prostate cancer metastases. Berger et al27 carried out comprehensive whole-genome sequencing on seven primary prostate cancers. This year, two additional studies performed exome capture and sequencing on 112 primary prostate cancers and 50 castrate-resistant prostate cancer (CRPC) metastases.9,28
Together, these and other studies have provided a snapshot of both primary and metastatic prostate cancers. To date, the most common molecular events in prostate cancer (Table 1) involve androgen receptor (AR) mutation or amplification,29–31 gene rearrangements of ETS transcription factors,5 and loss of phosphatase and tensin homolog (PTEN; phosphatidylinositide 3-kinase [PI3K] activation).32–34 Furthermore, there are aberrant mutually exclusive genes that occur at lower frequency (< 5%), including AKT,15 PIK3CA,17 and RAS mutations17 and BRAF mutations or fusions.16,35 The following is a discussion of these potentially actionable genomic alterations.
Common Genomic Events
ETS gene fusions.
Because of a prevalence of 50% of prostate cancers, a targeted therapy for ETS gene fusion products could affect thousands of patients with the disease. Currently, there are no therapies that directly target this gene fusion in clinical development. Like other gene fusions involving transcription factors, drug development has been hindered. Nonetheless, there are promising approaches emerging such as synthetic lethality screens and blocking specific protein-protein interactions of ETS transcription factors.36 Meanwhile, there are preclinical data that provide a rationale for targeting components of the transcription factor complex associated with ETS transcription factors through poly (ADP-ribose) polymerase (PARP) or DNAPK inhibition.8,37 Additionally, recent work by Knudsen et al38 has implicated PARP activity downstream of AR signaling, providing further rationale for dual AR and PARP inhibition. There are several clinical trials for metastatic CRPC evaluating PARP and DNAPK inhibitors and combined PARP and AR blockade, where knowledge of ETS fusion status will be a critical correlative element to the study (Table 2).
Table 2.
Clinical Trials for Targeted Therapies in Prostate Cancer
| Target | Ongoing Trials | Therapies | ClinicalTrials.gov Identifier | Genome-Based Enrichment or Randomization |
|---|---|---|---|---|
| ETS fusions | Study combining ABT-888, oral PARP inhibitor, with temozolomide in patients with metastatic prostate cancer | ABT-888, PARP inhibitor | NCT01085422 | ETS randomization |
| Study to assess safety and tolerability of oral CC-115 for patients with advanced solid tumors, non-Hodgkin lymphoma, or multiple myeloma | CC-115, DNAPK, and mTOR inhibitor | NCT01353625 | None | |
| Abiraterone acetate with or without veliparib in treating patients with metastatic prostate cancer | Veliparib, PARP inhibitor | NCT01576172 | None | |
| PI3K/mTOR/AKT pathway | Phase II study of BKM120 in men with metastatic CRPC | BKM120, PI3K inhibitor | NCT01385293 | None |
| Phase I/IIa, first time in human study of GSK2636771 in patients with advanced solid tumors with PTEN deficiency | GSK2636771, PI3K inhibitor | NCT01458067 | PTEN deficiency by immunohistochemistry | |
| Phase II study of the efficacy and safety of AP23573 in patients with taxane-resistant AIPC | Ridaforolimus, mTOR inhibitor | NCT00110188 | None | |
| Temsirolimus to reverse androgen insensitivity for CRPC | Temsirolimus, mTOR inhibitor | NCT01020305 | None | |
| Androgen signaling | Determine effect of MDV3100 on androgen signaling pathway in correlation with anti-tumor effects of MDV3100 to identify potential predictors of response or resistance to therapy | MDV3100, second-generation AR antagonist | NCT01091103 | None |
| Safety and efficacy study of MDV3100 in patients with CRPC who have been previously treated with docetaxel-based chemotherapy (AFFIRM)46 | MDV3100, second-generation AR antagonist | NCT00974311 | None | |
| Safety, pharmacokinetic and proof-of-concept study of ARN-509 in CRPC | ARN 509, second-generation AR antagonist | NCT01171898 | None | |
| Combinations | Bicalutamide with or without MK2206 in treating patients with previously treated prostate cancer | Bicalutamide plus MK2206, AKT inhibitor | NCT01251861 | None |
| Temsirolimus to reverse androgen insensitivity for CRPC | Temsirolimus (mTOR) | NCT01020305 | None | |
| Study of GDC-0068 or GDC-0980 with abiraterone acetate versus abiraterone acetate in patients with CRPC previously treated with docetaxel chemotherapy | Abiraterone plus GDC-0068 (AKT inhibitor) or GDC-0980 (PI3K/mTOR inhibitor) | NCT01485861 | None | |
| Private driver mutations | Abiraterone acetate with or without veliparib in treating patients with metastatic prostate cancer | Vemurafenib, RAF inhibitor | NCT01085422 | BRAF subset receives vemurafenib |
NOTE. There are multiple ongoing trials involving novel agents targeting ETS, PI3K pathway, and AR signaling in prostate cancer. Few studies are beginning to target multiple pathways through combination therapies. Although this table is not comprehensive, it is meant to provide an overview of directions for targeted therapies in prostate cancer.
Abbreviations: AFFIRM, A Study Evaluating the Efficacy and Safety of the Investigational Drug MDV3100; AIPC, androgen-independent prostate cancer; AR, androgen receptor; CRPC, castration-resistant prostate cancer; mTOR, mammalian target of rapamycin; PARP, poly (ADP-ribose) polymerase; PI3K, phosphatidylinositide 3-kinase; PTEN, phosphatase and tensin homolog.
AR.
The discovery that hormones regulate prostate cancer growth in the 1940s led to the first effective systemic therapies for prostate cancer and resulted in a Nobel Prize for Charles Huggins.39 In the 1990s, focused efforts on AR revealed escape mechanisms from androgen blockade through either mutation or amplification of AR.29,30 Additionally, these resistance mechanisms corroborate the mounting realization that androgen signaling remains important despite castrate levels of testosterone. Focus on persistent androgen signaling, including intratumoral testosterone production, has garnered support for the next generation of androgen synthesis inhibitors and AR antagonists.40–42 Abiraterone is a selective inhibitor of androgen synthesis through blockade of CYP17 used in the adrenal glands and testes, and within prostate tumors. In a randomized controlled trial against placebo, abiraterone resulted in improved overall and progression-free survival.43 Similarly, the next generation of anti-AR agents, including MDV310044–46 and ARN 509,47,48 has shown promising activity in men with metastatic CRPC.
Current data on prostate cancer genomics suggest more than 50% of metastatic CRPCs display AR amplification or mutation9 (Table 1). As the next generation of targeted therapies for androgen signaling are developed, genomic sequencing strategies may be useful in determining potential resistance mechanisms. Still unknown is the efficacy of combination therapy targeting both androgen synthesis and receptor.
In addition to mutations and amplifications involving AR, Dehm et al49 have described alternatively multiple spliced isoforms of AR that result in constitutively active androgen signaling. These variants generally function through loss of the C-terminal ligand-binding domain, preserving the N-terminal domain that contains the DNA binding and transactivation domains. Although multiple isoforms already have been revealed, unbiased next-generation sequencing approaches have identified additional isoforms and may yet be valuable for understanding other potential mechanisms of resistance.50,51 The frequency of AR splicing variants for patients with metastatic CRPC is not known. Thus, the relative significance of splicing and whether it occurs concurrently with other mechanisms of resistance, such as mutation or amplification, has not been clarified. This is complicated further by evidence suggesting genomic events such as cryptic splice sites or deletions may influence splicing patterns. Although there are currently no targeted therapies that prevent AR splicing, there are efforts to inhibit AR signaling by blocking the effects of the N-terminal domain that contains the DNA binding domain.52 The interplay of AR splice variants and next-generation antiandrogen therapies can be studied through comprehensive genomic and transcriptome data for patients with metastatic CRPC participating in clinical trials.
PTEN and PI3K pathway.
The PTEN gene is a tumor suppressor that encodes a protein phosphatase recurrently mutated in cancer. PTEN activity removes a phosphate from phosphoinositides at the plasma membrane (Fig 1) and negatively regulates the PI3K–AKT–mammalian target of rapamycin pathway. Thus, either loss or inactivation of PTEN genes leads to PI3K pathway activation. Somatic loss of PTEN occurs in multiple cancer types and provides a mechanistic rationale for PI3K pathway inhibitors.53 Through in vivo models, heterozygous loss of PTEN cooperates with other oncogenes to promote growth of prostate tumors.54
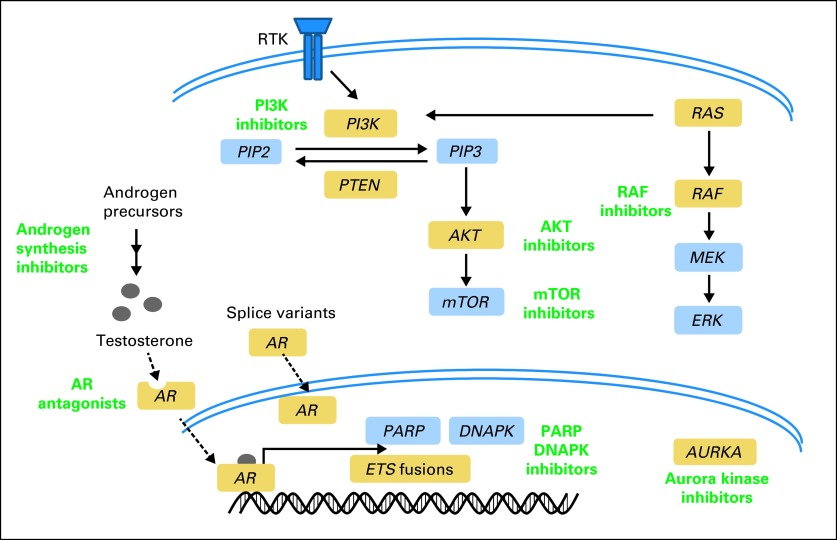
Fig 1.
Pathway-guided treatment in prostate cancer. This diagram highlights pathways for targeting in prostate cancer, including the phosphatidylinositide 3-kinase (PI3K) pathway, ETS rearrangements, and androgen signaling. Green indicates putative treatment hypotheses for relevant pathways. Each of these pathways can be driven by genomic aberrations, such as point mutations, copy number alterations, and rearrangements. The diagram also highlights less common driving mutations involving RAS, RAF, and AKT oncogenes. Furthermore, there is potential for cooperation between pathways because these are not necessarily mutually exclusive. Genes highlighted in gold have known genomic alterations. AR, androgen receptor; mTOR, mammalian target of rapamycin, PARP, poly (ADP-ribose) polymerase; RTK, receptor tyrosine kinase.
Genomic evidence of PTEN loss in prostate cancer through point mutation, deletion, or rearrangement has been observed in at least 50% of metastatic CRPCs9,27 (Table 1). However, this does not take into account other mechanisms of functional PTEN inactivation such as epigenetic or post-translational regulation. For example, Berger et al27 identified novel gene rearrangements involving the PTEN-interacting gene MAGI2, predicting that loss or mutation of MAGI2 would mimic functional PTEN inactivation. Certainly, there are other means to activate components of the PI3K-AKT pathway. Interestingly, recent studies have demonstrated a relationship for ERG fusions and androgen signaling with PTEN loss. A transgenic mouse model of ERG fusions was crossed into a heterozygous PTEN background and resulted in accelerated prostate tumor progression.34 This concurs with genomic data showing that PTEN loss and ETS fusions are not mutually exclusive events. With respect to androgen signaling, Carver et al34 demonstrated inhibition of either androgen or PI3K signaling resulted in reciprocal activation of the other pathway. This work has provided a rationale for dual inhibition of AR and PI3K signaling in clinical trials.
Retinoblastoma.
Retinoblastoma (RB1) is a tumor suppressor gene with somatic alterations in multiple cancers. RB1 has multiple cellular functions; with respect to cell cycle, it represses transcription of genes involved in DNA synthesis and mitosis. There are multiple means to somatically inactivate the RB1 pathway, including RB1 mutation or deletion, cyclinD1 expression, and p16INK4 mutation or loss. In prostate cancer, RB1 inactivation occurs through genomic deletion of RB1 in approximately up to 20% to 60% of cancers.9,55,55 Sharma et al12 demonstrated that RB1 inactivation resulted in enhanced AR expression dependent on the E2F1 transcription factor. Currently, researchers are actively exploring indirect targeting of RB1 inactivation through MDM2 and HDAC inhibitors.56
c-MYC.
The MYC oncogene is dysregulated in multiple cancers, including prostate cancer.57 When a MYC transgene was overexpressed in mice, all developed intraepithelial neoplasia and eventually invasive prostate cancer.58 Because of its expression in CRPC, researchers explored MYC as a potential mechanism of androgen independence, and in vitro MYC expression conferred AR-independent growth.59 Similarly, because of its co-occurrence with PI3K activation, Clegg et al60 generated mice with constitutive AKT1 and MYC activity and found accelerated development of invasive prostate cancer. Furthermore, although prostatic growth in AKT1 mice is sensitive to mammalian target of rapamycin inhibition, dual AKT1/MYC mice were less sensitive. Recent sequencing studies demonstrated that MYC is amplified in 10% of metastatic CRPCs.9 However, in a subset of prostate cancers with neuroendocrine differentiation, Beltran et al10 found that MYC amplification is more common (40%) and co-occurs with amplification of aurora kinase A (AURKA). AURKA is a kinase that regulates cell-cycle progression and is a putative target for cancer with several AURK inhibitors in clinical trials.14 Beltran et al demonstrated that MYC and AURKA–coamplified prostate cancer was sensitive to AURKA inhibitors in vitro and in vivo, thereby providing a rationale for trials in this molecular and histologic subset. Thus, targeting MYC is an active area of research and especially relevant to prostate cancer because of its cooperation with androgen independence, PI3K pathway, and aurora kinase activation.
Uncommon and Private Genomic Events
PIK3CA.
Activating mutations in PIK3CA were described after exon resequencing an initial panel of colorectal cancer tissues and later were found across multiple cancer histologic subtypes, including breast and ovarian cancers.61 Functional characterization of PIK3CA hotspot mutations revealed downstream activation of PI3K pathway components, including AKT phosphorylation, and phenotypes, including cell proliferation.62 Its role as a driving oncogene and prevalence across multiple cancer subtypes53 have led to the development of PI3K inhibitors. Although some trials are focusing on disease groups, select trials are including molecular enrichment based on activating PIK3CA mutations for patients with any cancer histology.
In prostate cancer, PIK3CA mutations occur at a frequency of up to 5% and copy number gains of up to 10%10,63 (Fig 1). In contrast, as mentioned earlier, the loss of PTEN is more common in prostate cancer, reaching 50%. As PI3K inhibitors are developed in prostate cancer, an important question will be whether there is a clinical or functional difference between subsets defined by PIK3CA mutations versus PTEN loss. Furthermore, one can speculate that inhibition of PI3K in PIK3CA-mutant prostate cancer may drive reciprocal androgen signaling, and such molecular subsets could potentially benefit from dual blockade.
AKT.
AKT is a serine/threonine protein kinase and is a component of the PI3K pathway (Fig 1). Carpten et al64 discovered recurrent activating mutations in AKT1 after resequencing the coding regions for AKT1, AKT2, and AKT3 genes in breast, colorectal, and ovarian cancer tissues. Rather than mutations in the catalytic kinase domain, however, they identified a mutation, corresponding to an E17K amino acid substitution, in the pleckstrin homology domain affecting protein localization. The mutation resulted in constitutive AKT1 activation and in vitro and in vivo transforming properties. Boormans et al15 sequenced AKT1 in 188 prostate cancer specimens and identified the activating E17K mutation in 1.4% of cases. Because this is a rare occurrence in prostate cancer, recent genome and exome sequencing studies have not reported this mutation.
RAS-RAF pathway.
BRAF encodes a serine/threonine protein kinase involved in the RAS signaling cascade. Davies et al65 hypothesized that additional oncogenic kinase mutations might exist in downstream components of the RAS pathway. They sequenced exons of multiple RAS pathway genes in a panel of cancer cell lines and identified a recurrent mutation in the kinase domain of BRAF. An activating mutation, V600E, was recurrent in melanoma and other cancer subtypes and resulted in constitutive activation of the RAF kinase (Fig 1). In addition to the successful application of RAF and MEK inhibitors in BRAF-mutant melanoma,2,66 multiple inhibitors are in clinical development pipelines for other BRAF-mutant cancers.
With regard to prostate cancer, genomic studies of prostate cancer in North America have demonstrated canonical BRAF and KRAS mutations of approximately 1% to 2%.9,28 In addition to point mutations, activating gene fusions involving BRAF have been described in 1% to 2% of prostate cancers, as detected by transcriptome sequencing.16 These gene rearrangements result in the expression and constitutive activity of a RAF kinase that in vitro has transforming activity and is sensitive to RAF inhibitors. Similarly, gene fusions involving KRAS fusions were also identified in 1% of metastatic CRPCs and were sensitive to in vitro knockdown experiments.18 Thus, this represents a potential treatment hypothesis for patients with such gene fusions. Furthermore, it underscores the increasing complexity of potential driving mutations that may be relatively rare and the potential variety of mutation classes that may be possible (point mutations v rearrangements).
NOVEL GENOMIC ABERRATIONS IN PROSTATE CANCER
Recent sequencing studies in prostate cancer have uncovered a number of novel genomic alterations that have potential for clinical translation. In two studies of primary and metastatic prostate cancers, high-throughput sequencing revealed recurrent copy number alterations and point mutations.9,28 Here we highlight alterations in several genes: SPOP (13%), FOXA1 (3% to 4%), AURKA (40% of neuroendocrine prostate cancers), MED12 (4% to 5%), MAGI-2 (discussed previously), and CHD1; each is of particular biologic and potential clinical interest.
SPOP encodes a subunit of the ubiquitin ligase complex and was highlighted by Barbieri et al28 in their study of primary prostate cancers through exome sequencing. They observed a mutation frequency of 15%; other prostate cohorts ranged from 6% to 15%. Interestingly, SPOP mutations predominantly affected the substrate-binding region of the protein. Furthermore, they observed that SPOP mutations were mutually exclusive from ETS rearrangements, implying that these are separate events in prostate tumorigenesis. Although in vitro transfection of SPOP mutants did not affect cell growth or viability, cells displayed increased invasion. Taken together, SPOP-mutant prostate cancer may represent a novel molecular subset and has potential for therapeutic applications after further experimental biology.
FOXA1 encodes forkhead box A1, a transcription factor that has been shown to be a coregulator of AR.67 Identified point mutations involve the forkhead domain in DNA binding. FOXA1 mutants promoted tumor growth in xenografts while negatively affecting androgen signaling in vitro.
AURKA, as discussed earlier, encodes a kinase involved in cell-cycle regulation and is readily being targeted in clinical trials for cancer. Beltran et al10 identified co-occurring amplifications of MYC and AURKA in prostate cancer with neuroendocrine differentiation and demonstrated sensitivity to aurora kinase inhibition. These data provide a therapeutic rationale for exploring aurora kinase inhibitor in AURKA-amplified prostate cancer (Fig 2) and reflect an excellent example of the rapid translation of basic science discoveries through sequencing approaches into clinical trials.
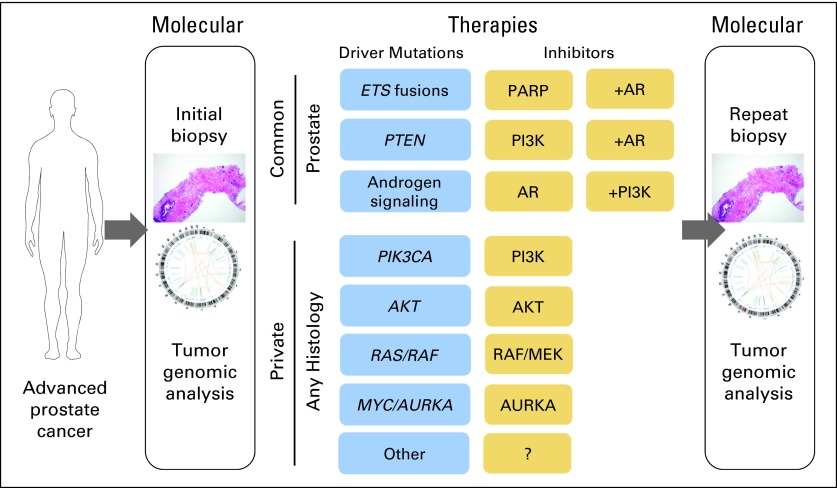
Fig 2.
Translating genomics for prostate cancer trials. This diagram outlines a path to genomics-driven trials in prostate cancer. Patients with advanced prostate cancer would undergo a fresh tumor biopsy for assessment of their current disease and molecular stratification to trials, including random assignment within those groups. Because of the prevalence of phosphatidylinositide 3-kinase (PI3K) pathway activation, ETS rearrangements, and androgen signaling, genomic enrichment of patients for these common disease subsets may follow traditional trial structures including combination treatments. However, for rare or private molecular disease subsets, these patients may be better suited to studies based on molecular aberrations and that include multiple histologies. For all studies, repeat biopsy for genomic assessment will be valuable for evaluating mechanisms of resistance, including tumor subclone selection. AR, androgen receptor; PARP, poly (ADP-ribose) polymerase.
MED12 is a subunit of a mediator complex and cyclin-dependent kinase 8, which is recurrently mutated in uterine leiomyomas. The mutation observed in prostate cancer was recurrent at one position in five patient cases and could conceivably affect cyclin-dependent kinase function.
CHD1 is a helicase DNA-binding protein involved in chromatin remodeling found to be homozygously deleted in 8% to 10% of prostate cancer array and next-generation sequencing studies.9,28,68,69 Knockdown experiments of CHD1 in nontumorigenic prostate epithelial cells resulted in increased invasiveness but were not transforming.68 Interestingly, CHD1 loss was associated with additional homozygous gene deletions. Furthermore, CHD1 loss seemed to be mutually exclusive from ETS rearrangements in the cohorts tested.
It is clear that additional experimental biology will be needed to delineate the roles of these novel findings. However, it is clear they represent an expanding molecular classification of prostate cancer through genomic technologies and promising opportunities for clinical translation.
CLINICAL DEVELOPMENT OF TARGETED THERAPIES
Clinical translation of prostate genomic data for patients is already under way, with clinical trials addressing androgen signaling, PI3K pathway activation, and ETS family rearrangements (Table 2). Trials for PARP inhibitors in prostate cancer are targeting sensitivity through ETS rearrangements and expression, and some trials are being enriched for patients with ETS rearrangements. With emerging data on effective second-generation AR antagonists, the question becomes whether androgen signaling has been fully shut down at time of disease progression, and what the mechanism of resistance is. In addition, promising preclinical work is ongoing for agents that target ETS fusions and the N terminus of AR. Because of the frequency of ETS family and AR aberrations in prostate cancer, these potential treatments could be widely applicable to a majority of patients with advanced prostate cancer.
Moving forward, the challenges for translating current achievements in prostate cancer genomics will be determining optimal combination treatments and addressing patients who have private or rare driving mutations. For example, the preclinical rationale for combined PI3K pathway and androgen signaling inhibition has spurred trials to address these hypotheses (Table 2). However, the majority of trials do not routinely incorporate molecular eligibility or stratification, such as identification of patients with PTEN or PI3K pathway aberrations. Integrating molecular analysis of tumor biopsies before and after therapy (Fig 2) will help develop and prove predictive biomarkers for those patients most likely to derive benefit. This can be done for all patients through retrospective analysis or prospectively for biomarker-enriched patients.70 Moreover, biopsy-based molecular analysis will provide the tissue data needed to understand emerging resistance to these investigational therapies and provide a rationale for the next therapy or combination approach.
For patients with private or relatively rare aberrations (1% to 2%), completing clinical trials for these pathways will be severely limited in terms of geographic availability to patients and cost to cancer centers to maintain trials despite few patients enrolled. Additionally, trials will be problematic to complete within a timeframe acceptable to pharmaceutical companies if performed within a single-disease cohort. As a solution to challenges for these molecular subsets, we advocate approaching patients with pathway-based eligibility and open enrollment for any histology of metastatic cancer (Fig 2). Although such studies would have heterogeneous patient populations that could confound the trials, we consider these to be exploratory or compassionate studies.
To facilitate the molecular stratification of patients with these rare or private driving mutations, we and others have developed clinical tumor sequencing studies that include real-time next-generation sequencing from fresh biopsies of metastatic disease.71–73 Tumor characterization includes a combination of whole-genome or -exome and transcriptome sequencing, review of results through a multidisciplinary sequencing tumor board, and return of clinically actionable results. The Stand Up to Cancer program and the Prostate Cancer Foundation recently funded two multicenter studies to implement a clinical tumor sequencing program for patients with metastatic CRPC and melanoma. These studies will facilitate the concept of prediction medicine through molecular stratification in trials, identify novel molecular subsets, and facilitate understanding of resistance mechanisms to targeted therapies.
FUTURE DIRECTIONS AND CHALLENGES
Although application of prostate cancer genomic data is being translated into clinical trials, approaches employing high-throughput sequencing technologies have important limitations, including tissue biopsies, tumor microenvironment, epigenetics, and heterogeneity. Compared with other epithelial cancers, prostate cancer presents unique challenges for tumor biopsy assessments because a significant fraction of patients with metastatic prostate cancer have bone-only metastasis. This places limitations on tissue acquisition and handling for sequencing approaches. Up to half of bone biopsies yield poor or inadequate tumor tissue for sequencing evaluation. Although these sequencing strategies focus on cancer cell populations, they may miss significant interactions with the host, including tumor microenvironment and immune system.74 Similarly, prostate cancer, like other cancers, displays heterogeneity as has been seen in multifocal disease in primary prostate tissues. A key question will be whether multiple metastatic disease sites display such heterogeneity. Although they are likely to have dominant clones with driving mutations, it is unclear whether treatment decisions and responses will be affected by heterogeneity through emerging resistance. Thus, we anticipate that future advances in sequencing, such as single-cell sequencing, may facilitate microenvironment characterization. Whereas clinical implementation of high-throughput sequencing has focused on DNA and RNA level aberrations, little has been implemented with respect to epigenetic profiling strategies. However, the number of predictive epigenetic aberrations and matching therapies is limited. As the compendium of epigenetic predictive biomarkers and matching therapies grows and the cost of sequencing technologies continues to decrease, we anticipate incorporation of epigenetic profiling into both clinical application and basic research.
Despite these limitations, we look forward to the next decade as genomic technologies are clinically applied for advanced prostate cancer. The extensive molecular characterization of prostate cancer has positioned this disease for the development of clinical trials for personalized oncology. Translating genomics into trials will innovative trial design and facilitate early application of cutting-edge basic science for patients with prostate cancer.
Acknowledgment
We thank Karen Giles for assistance in manuscript preparation and submission.
Footnotes
Supported by American Cancer Society Mentored Scholar Research Grant No. MRSG-12-194-01, a Landon Foundation American Association for Cancer Research Innovator Award for Personalized Cancer Medicine, and a Prostate Cancer Foundation Young Investigator Award (S.R.) and by the Prostate Cancer Foundation, National Institutes of Health Early Detection Research Network Grants No. UO1CA111275 and R01CA132874-01A1, Prostate Specialized Program of Research Excellence Grant No. P50CA69568, and by a Doris Duke Charitable Foundation Clinical Scientist Award (A.M.C.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Arul M. Chinnaiyan, Gen-Probe/Hologic (C), Life Technologies (C), MolecularMD (C), Ventana (C), Wafergen (C), Metabolon (C) Stock Ownership: Arul M. Chinnaiyan, Life Technologies Honoraria: None Research Funding: None Expert Testimony: None Other Remuneration: Arul M. Chinnaiyan
AUTHOR CONTRIBUTIONS
Financial support: Arul M. Chinnaiyan
Administrative support: Arul M. Chinnaiyan
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 3.Attard G, de Bono JS. Translating scientific advancement into clinical benefit for castration-resistant prostate cancer patients. Clin Cancer Res. 2011;17:3867–3875. doi: 10.1158/1078-0432.CCR-11-0943. [DOI] [PubMed] [Google Scholar]
- 4.Young RC. Cancer clinical trials: A chronic but curable crisis. N Engl J Med. 2010;363:306–309. doi: 10.1056/NEJMp1005843. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly (ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltran H, Rickman DS, Park K, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A, Yeow WS, Ertel A, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 15.Boormans JL, Korsten H, Ziel-van der Made AC, et al. E17K substitution in AKT1 in prostate cancer. Br J Cancer. 2010;102:1491–1494. doi: 10.1038/sj.bjc.6605673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XS, Shankar S, Dhanasekaran SM, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 2011;1:35–43. doi: 10.1158/2159-8274.CD-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prensner JR, Rubin MA, Wei JT, et al. Beyond PSA: The next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhury AD, Eeles R, Freedland SJ, et al. The role of genetic markers in the management of prostate cancer. Eur Urol. 2012;62:577–587. doi: 10.1016/j.eururo.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 22.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A, White TA, MacKenzie AP, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taplin ME, Bubley GJ, Shuster TD, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 30.Visakorpi T, Hyytinen E, Koivisto P, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 31.Linja MJ, Savinainen KJ, Saramäki OR, et al. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–3555. [PubMed] [Google Scholar]
- 32.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 33.Vlietstra RJ, van Alewijk DC, Hermans KG, et al. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res. 1998;58:2720–2723. [PubMed] [Google Scholar]
- 34.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho NY, Choi M, Kim BH, et al. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006;119:1858–1862. doi: 10.1002/ijc.22071. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinopoulos PA, Papavassiliou AG. Seeing the future of cancer-associated transcription factor drug targets. JAMA. 2011;305:2349–2350. doi: 10.1001/jama.2011.727. [DOI] [PubMed] [Google Scholar]
- 37.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knudsen KE. Leveraging the dual functions of PARP1 in controlling AR and DNA repair to improve prostate cancer management. Presented at the American Association for Cancer Research Annual Meeting; March 31-April 4, 2012; Chicago, IL. [Google Scholar]
- 39.Huggins C. Prostatic cancer treated by orchiectomy; the five year results. J Am Med Assoc. 1946;131:576–581. doi: 10.1001/jama.1946.02870240008003. [DOI] [PubMed] [Google Scholar]
- 40.Mostaghel EA, Page ST, Lin DW, et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: Therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007;67:5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 41.Ryan CJ, Tindall DJ. Androgen receptor rediscovered: The new biology and targeting the androgen receptor therapeutically. J Clin Oncol. 2011;29:3651–3658. doi: 10.1200/JCO.2011.35.2005. [DOI] [PubMed] [Google Scholar]
- 42.Ang JE, Olmos D, de Bono JS. CYP17 blockade by abiraterone: Further evidence for frequent continued hormone-dependence in castration-resistant prostate cancer. Br J Cancer. 2009;100:671–675. doi: 10.1038/sj.bjc.6604904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scher HI, Beer TM, Higano CS, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: A phase 1-2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bono J, Fizazi K, Saad F, et al. Primary, secondary, and quality-of-life endpoint results from the phase III AFFIRM study of MDV3100, an androgen receptor signaling inhibitor. J Clin Oncol. 2012;30(suppl):281s. abstr 4519. [Google Scholar]
- 47.Clegg NJ, Wongvipat J, Joseph JD, et al. ARN-509: A novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72:1494–1503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rathkopf DE, Shore N, Antonarakis ES, et al. A phase II study of the androgen signaling inhibitor ARN-509 in patients with castration-resistant prostate cancer (CRPC) J Clin Oncol. 2012;30(suppl):325s. abstr TPS4697. [Google Scholar]
- 49.Dehm SM, Schmidt LJ, Heemers HV, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perry AS, Watson RW, Lawler M, et al. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 2010;7:668–680. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 52.Andersen RJ, Mawji NR, Wang J, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 53.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Cristofano A, De Acetis M, Koff A, et al. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 55.Aparicio A, Den RB, Knudsen KE. Time to stratify? The retinoblastoma protein in castrate-resistant prostate cancer. Nat Rev Urol. 2011;8:562–568. doi: 10.1038/nrurol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachdeva UM, O'Brien JM. Understanding pRb: Toward the necessary development of targeted treatments for retinoblastoma. J Clin Invest. 2012;122:425–434. doi: 10.1172/JCI57114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 58.Ellwood-Yen K, Graeber TG, Wongvipat J, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 59.Bernard D, Pourtier-Manzanedo A, Gil J, et al. Myc confers androgen-independent prostate cancer cell growth. J Clin Invest. 2003;112:1724–1731. doi: 10.1172/JCI19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clegg NJ, Couto SS, Wongvipat J, et al. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS One. 2011;6:e17449. doi: 10.1371/journal.pone.0017449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 62.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Sun X, Huang J, Homma T, et al. Genetic alterations in the PI3K pathway in prostate cancer. Anticancer Res. 2009;29:1739–1743. [PubMed] [Google Scholar]
- 64.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 65.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 66.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X, Gupta A, Wang Y, et al. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann N Y Acad Sci. 2005;1061:77–93. doi: 10.1196/annals.1336.009. [DOI] [PubMed] [Google Scholar]
- 68.Liu W, Lindberg J, Sui G, et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012;31:3939–3948. doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang S, Gulzar ZG, Salari K, et al. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2012;31:4164–4170. doi: 10.1038/onc.2011.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon R. The use of genomics in clinical trial design. Clin Cancer Res. 2008;14:5984–5993. doi: 10.1158/1078-0432.CCR-07-4531. [DOI] [PubMed] [Google Scholar]
- 71.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz C, Lenkiewicz E, Evers L, et al. Advancing a clinically relevant perspective of the clonal nature of cancer. Proc Natl Acad Sci U S A. 2011;108:12054–12059. doi: 10.1073/pnas.1104009108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanahan D, Coussens LM. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]