Abstract
Recent advances in proteomics have facilitated the analysis of the kinome ‘en masse’. What these studies have revealed is a surprisingly dynamic network of kinase responses to highly selective kinase inhibitors, thereby illustrating the complex biological responses to these small molecules. Moreover these studies have identified key transcription factors, such as c-Myc and FOXO (forkhead box O), that play pivotal roles in kinome reprogramming in cancer cells. Since many kinase inhibitors fail despite a high efficacy of blocking their intended targets, elucidating kinome changes at a more global level will be essential to understanding the mechanisms of kinase inhibitor pharmacology. The development of technologies to study the kinome, as well as examples of kinome resilience and reprogramming, will be discussed in the present review.
INTRODUCTION
“For every reaction there is an equal and opposite reaction”. The Third Law of Motion, as first proposed by Sir Isaac Newton in 1687, describes the nature of forces between two bodies. Quite possibly, this same principle may apply to the dynamic properties of the cell signalling networks mediated by the human kinome. The human kinome or collection of 518 protein kinases [1], is proving to be a highly dynamic network of signalling enzymes capable of rapidly responding and remodelling to selective pressures. There are now numerous examples of kinome adaptation circumventing the effects of selective kinase inhibitors, thereby facilitating the resilience of the kinome to inhibition [2,3]. Moreover, multiple mechanisms of feedback inhibition and kinase cross-talk have been identified as checkpoints in virtually every kinase signalling network. Not surprisingly, disruption of these feedback circuits or alterations in patterns of kinase cross-talk has significant effects on the kinome as a whole [4]. Specific kinase inhibitors have distinct effects on relieving feedback inhibition, depending on the kinases targeted. In several cases, including B-Raf and PKC (protein kinase C), kinase inhibitors cause paradoxical activation of kinases that they were originally designed to inhibit [5,6]. Importantly, adaptations of the kinome as such may ultimately determine the success or failure of these compounds as therapeutic agents. Hence a comprehensive understanding of the details of kinome adaptation or remodelling in response to activators and inhibitors is crucial to understanding the mechanisms of drug action and/or acquired drug resistance.
Kinomics can be described as the study of kinases ‘en masse’. The term kinomics was first published by Vieth et al. [7] in 2004 who proposed a chemogenetics approach to structurally characterize the kinome. While genomics-based analyses of the kinome are well established, only relatively recently have developments in proteomics technologies permitted the study of the kinome at the protein level [8]. Principal among these methods are new quantitative MS-based approaches that allow large-scale studies of kinase expression and activation. Improved methods of kinase enrichment now permit the study of the expressed and activated kinome, providing a significant advantage over that of genomics alone. Combined with advanced methods of phosphoproteomics, a highly sophisticated view of the kinome and its related network of phosphoproteins is emerging [9].
Dysregulation of kinase signalling is central to the development of oncogenesis and drug resistance. Hence protein kinases have been identified as some of the most attractive pharmaceutical targets for a variety of diseases, including cancer and diabetes [10–12]. However a major impediment to the successful application of single kinase inhibitors is the development of acquired drug resistance. This can occur through a multitude of mechanisms, including kinase mutations or overexpression of specific kinases. For an excellent up-to-date overview of this topic the readers are referred to the review by Barouch-Benton and Sauer [13]. Less well understood is how changes in the activity of any single kinase perturbs the kinome or how changes in the kinome can be specifically measured or quantified. Moreover changes in kinome activity are predicted to influence protein–protein interactions and function of kinase scaffolds [14,15]. The objective of the present review is to discuss recent methodological advances for studying the kinome, some recently discovered examples of kinome adaptations to targeted kinase inhibitors and, finally, to examine potential mechanisms involved in mediating kinome responses to selective therapeutic agents.
NEW PROTEOMICS STRATEGIES TO STUDY THE KINOME
The philosophy of studying the kinome as a network of kinases or ‘kinomics’ was first proposed by Johnson and Hunter in 2005 [16]. Initial efforts to study the kinome focused on identifying kinase substrates through large-scale phosphoproteomic analyses [17]. However, because of the massive complexity of the phosphoproteome, these studies have provided only a partial insight into the detailed regulation of the kinome [18]. Some studies have attempted to characterize the kinome through kinase activity assays [19]. However, a limitation to this approach is the considerable assay development required to profile large-scale changes in kinase activity. Recently, advances in proteomics and genomics technologies have revolutionized the analysis of the human kinome itself as an entity. RNA-seq analysis revealed that ~70% of the total kinome (370/518) is expressed in typical breast cancer cell lines [20,21]. Other studies performed extensive profiling of expressed kinases in a large numbers of tissues [22] or specific cancers [23]. Despite this information, important questions remain. For instance, how many of these kinases are active and how does the activity of the kinome change in response to different stimuli or inhibitors? Moreover, how does one study the untargeted or understudied members of the kinome where well-characterized reagents are lacking?
Advances in kinase enrichment have greatly facilitated the ability to study the kinome [24]. On the basis of the original method of Haystead et al. [25] using γ-phosphoryl-linked ATP-affinity columns to capture kinases, Knockaert et al. [26,27] were the first to demonstrate the utility of immobilized kinase inhibitors. Daub and co-workers [28] refined this approach by using a broad range of immobilized kinase inhibitors coupled to Sepharose beads. An inherent advantage of the latter approach is the nanomolar affinity binding of kinases to the inhibitor beads, thereby permitting efficient capture of a large fraction of the kinome. In addition, by increasing the diversity of the coupled inhibitors, a greater diversity of kinases could be captured [29]. Initially this technology was used to profile kinase inhibitor selectivity and identify new cellular targets for select kinase inhibitors such as SB203580, gefitinib, SU6668 and others [30–33]. Termed ‘Kino-Beads’ [34], this method was commercialized (Axxima, Cellzome, Ambit) and broadly applied to assess kinase inhibitor specificity across the kinome [29,34–37]. For example, Bantscheff et al. [34] performed quantitative analysis of the kinase targets for imatinib, dasatinib and bosutinib in K562 leukaemia cells. Similar studies have compared the selectivity of nilotinib and second-generation dual Abl/SFK (Src family kinase) inhibitors [38,39]. Others used this technology to compare the binding affinity of 38 kinase inhibitors across a panel of 317 kinases [36]. Importantly these papers marked some of the first studies to evaluate the specificity of structurally distinct inhibitors against large numbers of kinases in the kinome. The results of these and other studies demonstrated exquisite specificity for some inhibitors (lapatinib), whereas others [staurosporine, sunitinib, lestaurtanib (PKC412) and dasatinib] lacked specificity and bound many kinases from various kinase families [36,40].
More recent studies expanded this technology to profile changes in the kinome itself. Multiple investigators have used this strategy to demonstrate cell-type-specific sets of expressed kinomes using the immobilized inhibitors combined with LC (liquid chromatography)-MS/MS (tandem MS), phosphoproteomics analysis and quantitative MS [35,41,42]. In an interesting application, Daub et al. [43] used this approach to profile the change in the kinome from S- and M-phase-arrested cells. Specifically, the authors quantified over 219 kinases, identified over 1000 phosphorylation sites and identified multiple kinases not previously associated with mitotic progression.
Single kinase inhibitors have limited success as cancer therapies because tumours evade their action by activating alternative survival pathways, promoting acquired drug resistance [15]. As such, the requirement for effective combination therapies has become increasingly clear [44]. Until recently there has been no discovery mechanism to define the dynamic activity of the kinome in response to inhibitors. Typically studies of kinome adaptation have relied on immunoblotting of select kinases or the use of phosphoantibody arrays to assess kinome changes [45]. However, these methods are limited by phosphoantibody availability and specificity, and therefore limit the scale of analysis. An alternative approach is the chemoproteomics method described by Patricelli et al. [46,47]. This method involves covalently ‘tagging’ activated kinases with acyl phosphate derivatives and large-scale identification of activated kinases by a targeted LC-MS approach. An advantage of this approach is that inhibitor responses and inhibitor binding values can be established for kinases in the native cellular context [48]. However, the requirement for sophisticated methods of targeted MS limit the widespread application of this technology. Lastly, new technologies, such as mass cytometry, hold great promise for the highly sensitive detection of select members of the kinome and inhibitor responses in single cells [49,50]. Again, like the antibody array approach, these methods are dependent on highly specific antibodies to kinases and kinase phosphorylation sites.
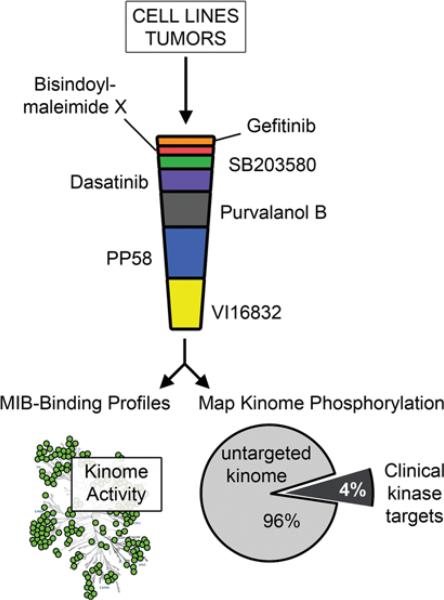
Recently, Duncan et al. [21] applied a chemical proteomics approach that couples kinase affinity capture with qMS (quantitative MS), providing a platform to profile global kinome activity in response to targeted therapies. Using a derivation of the original Daub approach [43], Duncan et al. [21] showed that changes in the expression or activation state of kinases could be selectively enriched using a cocktail of MIBs (multiplexed inhibitor beads) (Figure 1) [21]. Isolation of endogenous protein kinases from different cell types was carried out using MIBs consisting of multiple immobilized kinase inhibitors arranged in an inhibitor specificity gradient (specific→non-specific). Specific kinase inhibitors form the top layers of the MIBs, acting as a filter for abundant kinases. This critical pre-clearance step allows the subsequent retention of low-abundance kinases on the non-specific pan-kinase inhibitor beads. MIB-bound kinases are identified by off-line LC separation and high throughput MALDI (matrix-assisted laser-desorption ionization)–TOF (time-of-flight)/TOF-MS. An advantage of this approach is the large numbers of peptides quantified with high statistical confidence. Moreover, phosphorylation of MIB-enriched kinases can be simultaneously determined by titanium dioxide enrichment and LC-MS. Thus these methods combine kinase affinity capture with qMS and phosphoproteomics, providing a systems-wide approach to profile global kinome activity (Figure 1).
Figure 1. Monitoring kinome reprogramming in cells and tumours using MIBs in combination with MS.
Endogenous kinases are affinity-purified from cell lysates using MIB/MS and both MIB-binding ratios and phosphorylation of kinase activation loops were determined using qMS. Of the kinases identified in a recent study [21], only 4% are current targets for kinase inhibitor drugs, the remainder representing the untargeted cancer kinome [51].
A current challenge is to develop methods to study the untargeted or understudied kinome [51]. In this regard, the MIB strategy works remarkably well. Using MIBs, hundreds of kinases are enriched and quantified from across the spectrum of kinases, many of which have been barely studied due to a lack of reagents [21]. Kinases from all major kinome sub-families are captured, with a large percentage representing the understudied kinome. Characterization of the properties of kinase binding to MIBs further revealed that three factors determine binding: affinity, expression and kinase activity [21]. Thus MIB technology captures the majority of the expressed kinome, as estimated by RNA-seq, and can detect altered kinome activity profiles in response to stimuli or kinase inhibitors used to treat cancer. As such these methods provide a comprehensive unbiased method to profile changes in kinome remodelling or kinome adaptation to various cellular conditions or inhibitors. The application of these methods to study kinome reprogramming or remodelling in breast cancer is described below.
EXAMPLES OF KINOME RESILIENCE AND ADAPTATION TO INHIBITION
Raf kinase inhibitors and the activation of Raf/ERK (extracellular-signal-regulated kinase) signalling in cells
Oncogenic mutations in the kinase B-Raf are common in malignant melanomas [52]. The most frequent B-Raf mutation in melanoma is the V600E mutation and this, and other activating B-Raf mutations, result in chronic activation of the MEK [MAPK (mitogen-activated protein kinase)/ERK kinase]/ERK pathway, which is required for cell growth [53]. Consequently, small molecule inhibitors of Raf are attractive therapeutics for the treatment of melanoma and other Raf-driven cancers [54]. Interestingly, studies uncovered a surprising paradoxical resistance of Raf kinase signalling to select Raf inhibitors [55–57] (reviewed in [5]). Raf kinase inhibitors (PLX4720, sorafenib and 885-A), while highly effective at inhibiting mutant B-Raf, activated all three types of wild-type Raf (A, B and C) [55–57]. Using a chemical genetics approach, Poulikakos et al. [57] showed that inhibitor-mediated dimerization was responsible for the paradoxical activation of Raf by PLX4032. This compensatory activation is dependent on inhibitor binding to the ATP site of one Raf molecule, which stimulates dimerization and activation of the drug-free Raf protomer. This process requires Ras-GTP and stimulates growth through activation of the MAPK pathway [55–57]. Moreover, N-Ras mutation or up-regulated PDGFB (platelet-derived growth factor B) expression was shown to accompany the resistance of cells to PLX4032 [52]. Thus despite efficacy at inhibiting mutant B-Raf in tumours, Raf inhibitors pose a risk of enhancing Ras-dependent tumours through activation of wild-type Raf. In this regard, Raf inhibitors have been shown to increase cutaneous cell carcinomas [58]. While these studies provide an important cautionary example of the unexpected changes in the kinome in response to single kinase inhibitors, they also illustrate the importance of elucidating kinome responses to select inhibitors. As suggested from these findings, and observations that B-Raf mutations or amplification facilitate the development of MEK inhibitor resistance [59–62], strategies to co-target both Raf and MEK simultaneously have been proposed and tested [63,64]. In support of this idea, recent studies from Greger et al. [65] have demonstrated that co-targeting MEK and/or PI3K (phosphoinositide 3-kinase)/mTOR (mammalian target of rapamycin) prevented the acquired resistance to the B-Raf inhibitor GSK2118436 and suggest that co-targeting these two pathways is a viable strategy [65–67].
Paradoxical activation of Raf/ERK by nilotinib and Abl kinase inhibitors
CML (chronic myelogenous leukaemia) is a myeloid cell cancer commonly caused by the BCR-Abl fusion kinase oncogene [68]. While the majority of CML patients are successfully treated by small molecule kinase inhibitors such as imatinib, nilotinib or dasatinib [69,70], acquired drug resistance remains a problem, in particular mutation of the ‘gate-keeper’ residue (Thr315) of Abl (AblT315I), resulting in resistance to all three drugs [13,71]. Moreover, a recent study from Packer et al. [72] showed that these kinase inhibitors can exert a paradoxical activation of B-Raf and C-Raf kinase in drug-resistant CML, similar to that with the Raf kinase inhibitors described above. This response was also shown to require the activity of Ras and resulted in the activation of the MEK/ERK pathway, albeit weaker than that observed with the Raf kinase inhibitors [57]. The results of these studies demonstrated that these compounds are weak Raf inhibitors and thereby induced Raf dimerization as observed with PLX4032. In support of this, gatekeeper mutations in B-Raf (T529N) and C-Raf (T421N) that prevent inhibitor binding, showed a lack of Abl inhibitor- induced dimerization. Because of the observed paradoxical activation of Raf/MEK/ERK by these compounds, the authors tested the synthetic lethality of a MEK inhibitor. Co-treatment with the MEK inhibitor PD184352 blocked the nilotinib-induced activation of ERK and strongly induced apoptosis of these cells, whereas PD184352 alone was without effect [72]. Thus these studies reveal an important and unexpected ‘off-target’ effect of Abl kinase inhibitors on the MEK pathway and further illustrate the need to understand kinome responses to single kinase inhibitors.
COT (cancer Osaka thyroid oncogene) as an activator of resistance to Raf inhibitors
COT/TPL2 (MAP3K8) is a MAPKKK (MAPK kinase kinase) capable of functioning at the level of Raf [73]. To investigate the resistance to Raf kinase inhibitors in melanoma, Johannessen et al. [74] expressed ~600 kinase and kinase-related ORFs (open reading frames) and investigated resistance to Raf inhibitors (PLX4720) [74]. From these studies the authors identified COT/TPL2 as an activator of MEK, whose expression promoted acquired resistance to Raf inhibition. Furthermore, the authors demonstrated that COT/TPL2 expression facilitated the development of de novo Raf inhibitor resistance in B-Raf V600E cell lines and samples from patients with metastatic B-Raf V600E melanoma. In biopsies of relapsed tumours, COT/TPL2 (gene symbol MAP3K8) mRNA expression was increased, providing further evidence for overexpression of this kinase in resistant melanoma. Interestingly, COT/TPL2 expression also decreased sensitivity to MEK inhibitors (AZD6244 and CI-1040). Because of the potential effects of COT in Raf mutant cells, the authors suggest combinatorial treatments to target COT and MEK as a possible strategy to prevent drug resistance. These studies and those of others provide additional support for co-targeting Raf and MEK simultaneously [74,75].
PI3K inhibition activates ERK via HER (human epidermal growth factor receptor) 2 and/or HER3 in breast cancer: importance of FOXO (forkhead box O) signalling
A sophisticated view of kinome feedback regulation has been gleaned from studies on PI3K and Akt inhibitors. Of central importance is the role of feedback regulation of the mTOR pathway [76]. Rapamycin-dependent inhibition of mTORC (mTOR complex) 1 relieves feedback regulation of the IGF1R (insulin-like growth factor 1 receptor), thereby triggering a compensatory activation of IGF1R and Akt targets such as the transcription factor FOXO [77,78]. Because of the relationship between Akt and mTOR, it may not be surprising that inhibition of Akt results in similar mechanisms of kinase reprogramming. Specifically, Chandarlapaty et al. [79] showed that inhibition of PI3K/Akt relieved feedback inhibition and increased the expression of a unique set of receptor tyrosine kinases including HER3, IGF1R and the insulin receptor. In addition to expression, the phosphorylation of multiple RTKs (receptor tyrosine kinases) was also stimulated by Akt inhibition. Treatment of cells with Akti (Akt inhibitor) 1/2 or Akti 1/2/3, resulted in up-regulation of RTK transcripts in a FOXO-dependent manner. Given the significance of this RTK response, the authors tested the combination of HER kinase inhibitors (lapatinib and Iressa) with Aktis in xenograft models (BT-474 and NCI-H292) and showed that the combination of these inhibitors was highly effective at inhibiting tumour growth. Chakrabarty et al. [80] also showed that inhibition of PI3K with the small molecule inhibitor XL147 up-regulated the expression and activation of multiple RTKs, including HER3, in a manner dependent on HER2 [80]. Again, co-targeting the RTK response with trastuzumab or lapatinib was synergistic at inhibiting growth, compared with the action of these inhibitors alone. Similarly, Serra et al. [81] showed that the reactivation of HER signalling by PI3K/mTOR inhibition (BEZ235) resulted in activation of HER2-dependent ERK activation. Consequent inhibition of MEK/ERK with AZD6244 prevented this activation, and the combination of AZD6244 with BEZ235 was highly effective at inhibiting tumour cell growth [81]. Thus these studies exemplify the importance of elucidating the kinome feedback mechanisms and understanding their implications for successful therapeutic applications.
A further extension of these studies examined the effects of mTORC1/2 inhibition. While inhibition of mTORC1 with rapamycin leads to activation of Akt, ATP-competitive mTOR kinase inhibitors (AZD8055) do not [82]. However as shown by these authors, despite initial blockade of Akt phosphorylation (Thr308) through inhibition of mTORC2, the mTOR inhibitors caused a biphasic response that resulted in a sustained inhibition of mTOR1 despite activation of Akt Thr308 phosphorylation. Consequently, treatment with mTOR inhibitors reactivated Akt, again in a manner dependent on induced RTKs. Co-addition of RTK inhibitors with mTOR inhibitors resulted in the enhanced inhibition of tumour growth.
Limitation of EGFR [EGF (epidermal growth factor) receptor] inhibitors: cross-talk between EGFR and c-Met
Activation or mutation of the EGFR and downstream kinase pathways is common in GBM (glioblastoma multiforma), colorectal cancer, NSCLC (non-small-cell lung carcinoma) and TNBC (triple-negative breast cancer) [83]. This is often a result of EGFR mutations or overexpression, hence EGFR inhibitors have been investigated as possible therapeutics in these settings, with limited success [84,85]. Studies now demonstrate the difficulty of targeting this RTK is due to secondary gatekeeper mutations (T790M) that prevent EGFR inhibition or from activation of alternative RTKs following loss of EGFR activity. In particular, several studies in EGFR-mutant NSCLC identify Met amplification and activation as a mechanism for resistance to EGFR inhibitors, where Met amplification is observed in 15–22% of NSCLC patient's tumours that were resistant to EGFR inhibitors [86,87]. Multiple lines of evidence suggest that activated Met can compensate for EGFR inhibition by partially protecting against loss of phospho-EGFR and contributing to sustained activation of downstream growth signalling through ERK and Akt. A direct association between EGFR and Met has been observed, although it is unclear how this heterodimerization contributes to EGFR inhibitor resistance [88]. In such cases where Met is induced as a mechanism of EGFR inhibitor resistance, co-treatment with EGFR and Met inhibitors synergistically inhibited proliferation of cell lines, prevented tumour growth, and induced apoptosis in cell and tumour models. Similar studies demonstrate the remarkable plasticity of Met signalling in response to expression of EFGR mutations (de2-7 EGFR) in GBM [89].
While it has been shown that EGFR inhibitor treatment can select for cells with pre-existing Met amplification, it is also evident that loss of EGFR activity can rapidly induce compensatory expression/activation of Met and other tyrosine kinases, including FGFR (fibroblast growth factor receptor) 2 and SFKs [90–93]. This induced kinase activity is independent on genetic mutation or amplification, and probably results, in part, from loss of negative-feedback regulation of suppressed kinases. Additionally, acute loss of upstream kinase signalling can trigger changes in transcription factor stability and/or activity that lead to induced expression of RTKs and their ligands. This was exemplified by Ware et al. [91], who defined loss of ERK activity downstream of EGFR inhibition as causal in de-repression of FGFR2/3 expression in NSCLC cell lines. These data demonstrate the diverse mechanisms of plasticity and resistance in the kinome of tumour cells under the selective pressure of targeted kinase inhibitors. With most oncogenic RTKs (such as EGFR) driving cell growth through conserved kinase signalling pathways, there is a high potential for cancer cells to circumvent targeted inhibition of any RTK by induced expression or activity of alternative kinases.
Cross-talk between Raf and LKB1
The LKB1/AMPK (AMP-activated protein kinase)/mTOR pathway is a major metabolic network that connects the cellular energy status to the regulation of protein synthesis and key metabolic events [94,95]. Activation of AMPK by LKB1 provides a braking mechanism to arrest protein synthesis through mTOR inactivation when cells are under energy stress or low energy. Hence studies have focused on the tumour suppressive activity of LKB1 [96]. Loss of this suppressive function in LKB1–/– cells or transformed cells results in an uncoupling of protein synthesis from growth signals [97]. Previous studies have shown that oncogenic B-Raf inhibits the tumour suppressor activity of LKB1 through phosphorylation [98]. In response to EGF or in B-Raf V600E melanoma cells, LKB1 is phosphorylated on Ser325 and Ser428 in an ERK- and RSK (ribosomal S6 kinase)-dependent manner respectively. Interestingly, this phosphorylation appears to disrupt the LKB1–AMPK complex and the ability of cells to respond to nutrient stress [97]. As such, B-Raf or Ras tumour cells may escape a major metabolic regulatory control mechanism. In support of this, expression of a phosphorylation-deficient LKB1 (S325A/S428A), inhibited the proliferation of melanoma cells [98]. Inhibition of MEK or RSK restored this regulatory switch by preventing the phosphorylation of LKB1 and maintaining AMPK regulation. Interestingly these studies suggest that an unexpected benefit of MEK inhibitors may be to restore this important metabolic checkpoint.
MEK inhibition, c-Myc stability and RTK reprogramming in TNBC
Kinase inhibitors often fail in cancer treatment because tumours evade their action by activating alternative survival pathways promoting drug resistance. This is a particular problem for TNBC which is among some of the most drug-resistant cancers. As such, a requirement for effective combination therapies has become increasingly evident; however, defining the response of the kinome ‘en masse’ to molecular therapy has historically been challenging. Recently, Duncan et al. [21] developed and utilized an innovative chemical proteomics approach (MIB/MS, described above), to define the dynamic reprogramming of the kinome in response to small-molecule inhibitors. Using MIB/MS chemical proteomics, Duncan et al. [21] evaluated kinome reprogramming of TNBC cells and tumours to MEK inhibitors, which are currently in Phase I/II clinical trials for a number of cancers, including TNBC. Interestingly, treatment of TNBC cell lines or a TNBC GEMM (genetically modified mouse model) with the highly specific allosteric MEK inhibitor AZD6244 resulted in rapid reprogramming of the kinome, with activation of multiple serine/threonine and tyrosine kinases, overcoming the initial MEK/ERK inhibition. Induced expression and activation of a number of pro-survival RTKs, including PDGFRβ (PDGF receptor β), AXL, VEGFR2 (vascular endothelial growth factor receptor 2) and DDR1 (discoidin domain receptor family, member 1) was observed within 24 h following MEK inhibitor treatment. Generation of AZD6244-resistant TNBC cell lines demonstrated that the inhibitor-induced RTK reprogramming was stable and accompanied by increased survival signalling through the PI3K/AKT, JAK (Janus kinase)/STAT (signal transducer and activator of transcription) and MEK/ERK pathways. Intriguingly, MIB/MS studies revealed that both MEK1 and MEK2 were initially inhibited by AZD6244; however, by 24 h MEK2 had escaped inhibition, reactivating ERK signalling. Knockdown of MEK2 in the presence of MEK inhibitor enhanced growth inhibition, suggesting that the TNBC cells acquired dependency on MEK2 signalling. Escape of MEK2 from AZD6244 inhibition is thought to occur due to two contributing factors: (i) MEK1, but not MEK2, is regulated by negative-feedback phosphorylation by ERK1/2, and (ii) MEK inhibitors have been shown to preferentially inhibit the inactive form of MEK1/2. Consequently, activation of upstream RTK signalling pathways following MEK inhibition lead to accumulation of activated MEK2, increasing the IC50 of AZD6244 for MEK2, and thus facilitating the reactivation of ERK signalling.
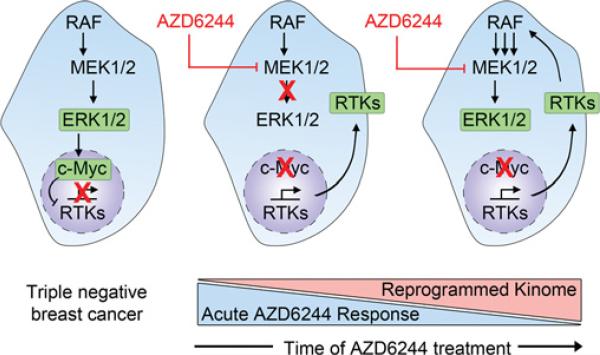
Defining how cancer cells rewire kinase networks in response to targeted inhibition of kinases presents a challenge that can be largely addressed by quantitative chemical proteomics, although elucidating the mechanisms of kinome reprogramming is far less straightforward. Directed investigation of the effects of disrupted feedback loops and/or signalling downstream of inhibited kinases must be implemented on a case-by-case basis. For example, Duncan et al. [21] found that treatment of both TNBC cells and the C3Tag TNBC GEMM with AZD6244 caused an acute loss of ERK activity, resulting in loss of phosphorylation of c-Myc at Ser62, rapid degradation of c-Myc protein and disruption of Myc-Max transcriptional activity (Figure 2). Destabilization of Myc-Max transcriptional repressor activity in response to MEK inhibition resulted in the induced expression and activity of a number of RTKs. Knockdown of ERK1/2 or c-Myc using siRNA (small interfering RNA) recapitulated the RTK activation by MEK inhibition, whereas stabilization of c-Myc protein levels using proteasome inhibitor treatment or expression of proteasome-resistant c-Myc mutants prevented activation of AZD6244-mediated RTKs. Collectively, Duncan et al. [21] discovered a unique mechanism of MEK inhibitor resistance, where MEK inhibition alleviates c-Myc-dependent repression of a number of pro-survival RTKs, leading to reactivation of MEK/ERK signalling and subsequent resistance. These studies were recapitulated with other MEK inhibitors (U0126 and GSK1120212) and demonstrate the pivotal role of Myc in kinome reprogramming ([21] and J.S. Duncan, unpublished work) (Figure 2).
Figure 2. Model describing the effects of targeted inhibition of MEK1/2 by AZD6244 in TNBC.
Shown is the proposed kinome reprogramming caused by loss of c-Myc protein and subsequent derepression of transcription of growth-promoting RTKs to reactivate MAPK signalling as described in Duncan et al. [21].
Identification of kinases activated in response to MEK inhibition using MIB/MS led Duncan et al. [21] to predict and test a novel combination therapy for treatment of TNBC. RNAi (RNA interference)-mediated knockdown of PDGFRβ, VEGFR2, DDR1 and AXL in TNBC cells in the presence of MEK inhibition enhanced growth inhibition, supporting RTK reprogramming driving MEK inhibitor resistance. Targeting the AZD6244-induced RTKs with the kinase inhibitor sorafenib synergistically enhanced growth inhibition and blocked the AZD6244-mediated reactivation of the ERK pathway in TNBC cells. Importantly, co-treatment of the TNBC GEMM with AZD6244 and sorafenib caused rapid tumour apoptosis and significant tumour regression, whereas single agents were ineffective. Taken together, Duncan et al. [21] used chemical proteomics to assess global kinome behaviour and its response to small-molecule inhibitors, and identified a new and effective combination therapy to treat TNBC.
SUMMARY
Recent methods have improved the ability to perform kinomics at a highly sophisticated and unprecedented level. The combination of deep sequencing and proteomics technologies will undoubtedly accelerate the analysis of kinome expression, activation and remodelling in different cell systems and in response to therapeutics and other stimuli. This unique opportunity will benefit clinicians and researchers alike who seek to evaluate the cellular responses to select kinase inhibitors. As such, examples of kinome reprogramming are now evident. Inhibition of the major signalling pathways, Raf/MEK and PI3K initiates profound kinome changes by altering the activity of transcription factors such as c-Myc and FOXO. Defining how the kinome responds, including many of the untargeted or understudied members, will be paramount to our understanding of the resilience of this important signalling network to select inhibitors.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant number DK037871 (to G.L.J. and L.M.G.)], the University Cancer Research Fund Team Science/Innovation Award (to G.L.J. and L.M.G.) and an NC-TraCS (North Carolina Translational and Clinical Sciences) Award (to L.M.G.).
Abbreviations used
- Akti
Akt inhibitor
- AMPK
AMP-activated protein kinase
- CML
chronic myelogenous leukaemia
- COT
cancer Osaka thyroid oncogene
- DDR1
discoidin domain receptor family, member 1
- EGF
epidermal growth factor
- EGFR
EGF receptor
- ERK
extracellular-signal-regulated kinase
- FGFR
fibroblast growth factor receptor
- FOXO
forkhead box O
- GBM
glioblastoma multiforma
- GEMM
genetically modified mouse model
- HER
human epidermal growth factor receptor
- IGF1R
insulin-like growth factor 1 receptor
- LC
liquid chromatography
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- MIB
multiplexed inhibitor bead
- mTOR
mammalian target of rapamycin
- mTORC
mTOR complex
- NSCLC
non-small-cell lung carcinoma
- PDGF
platelet-derived growth factor
- PDGFRβ
PDGF receptor β
- PI3K
phosphoinositide 3-kinase
- qMS
quantitative MS
- RSK
ribosomal S6 kinase
- RTK
receptor tyrosine kinase
- SFK
Src family kinase
- TNBC
triple-negative breast cancer
- TOF
time-of-flight
- VEGFR2
vascular endothelial growth factor receptor 2
REFERENCES
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenzweig SA. Acquired resistance to drugs targeting receptor tyrosine kinases. Biochem. Pharmacol. 2012;83:1041–1048. doi: 10.1016/j.bcp.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gossage L, Eisen T. Targeting multiple kinase pathways: a change in paradigm. Clin. Cancer Res. 2010;16:1973–1978. doi: 10.1158/1078-0432.CCR-09-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye SV, Johnson GL. Inhibitors paradoxically prime kinases. Nat. Chem. Biol. 2009;5:448–449. doi: 10.1038/nchembio.f.11. [DOI] [PubMed] [Google Scholar]
- 6.Poulikakos PI, Rosen N. Mutant BRAF melanomas: dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Kinomics: structural biology and chemogenomics of kinase inhibitors and targets. Biochim. Biophys. Acta. 2004;1697:243–257. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 8.Workman P. Drugging the cancer kinome: progress and challenges in developing personalized molecular cancer therapeutics. Cold Spring Harbor Symp. Quant. Biol. 2005;70:499–515. doi: 10.1101/sqb.2005.70.020. [DOI] [PubMed] [Google Scholar]
- 9.Knight JD, Pawson T, Gingras AC. Profiling the kinome: current capabilities and future challenges. J. Proteomics. 2012;pii(12):S1874–S3919. 00710–5. doi: 10.1016/j.jprot.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Cohen P. Protein kinases: the major drug targets of the twenty-first century? Nat. Rev. Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 12.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu. Rev. Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 13.Barouch-Bentov R, Sauer K. Mechanisms of drug resistance in kinases. Expert Opin. Invest. Drugs. 2011;20:153–208. doi: 10.1517/13543784.2011.546344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr. Opin. Cell Biol. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26:641–650. doi: 10.1101/gad.186965.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat. Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr. Opin. Genet. Dev. 2010;20:23–30. doi: 10.1016/j.gde.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midland AA, Whittle MC, Duncan JS, Abell AN, Nakamura K, Zawistowski JS, Carey LA, Earp HS, 3rd, Graves LM, Gomez SM, Johnson GL. Defining the expressed breast cancer kinome. Cell Res. 2012;22:620–623. doi: 10.1038/cr.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilpinen S, Ojala K, Kallioniemi O. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS ONE. 2010;5:e15068. doi: 10.1371/journal.pone.0015068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zang ZJ, Ong CK, Cutcutache I, Yu W, Zhang SL, Huang D, Ler LD, Dykema K, Gan A, Tao J, et al. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res. 2011;71:29–39. doi: 10.1158/0008-5472.CAN-10-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutton CW. The role of targeted chemical proteomics in pharmacology. Br. J. Pharmacol. 2012;166:457–475. doi: 10.1111/j.1476-5381.2011.01778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haystead CM, Gregory P, Sturgill TW, Haystead TA. γ-Phosphate-linked ATP-sepharose for the affinity purification of protein kinases. Rapid purification to homogeneity of skeletal muscle mitogen-activated protein kinase kinase. Eur. J. Biochem. 1993;214:459–467. doi: 10.1111/j.1432-1033.1993.tb17942.x. [DOI] [PubMed] [Google Scholar]
- 26.Knockaert M, Gray N, Damiens E, Chang YT, Grellier P, Grant K, Fergusson D, Mottram J, Soete M, Dubremetz JF, et al. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chem. Biol. 2000;7:411–422. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 27.Knockaert M, Wieking K, Schmitt S, Leost M, Grant KM, Mottram JC, Kunick C, Meijer L. Intracellular targets of paullones. Identification following affinity purification on immobilized inhibitor. J. Biol. Chem. 2002;277:25493–25501. doi: 10.1074/jbc.M202651200. [DOI] [PubMed] [Google Scholar]
- 28.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 30.Wissing J, Godl K, Brehmer D, Blencke S, Weber M, Habenberger P, Stein-Gerlach M, Missio A, Cotten M, Muller S, Daub H. Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell. Proteomics. 2004;3:1181–1193. doi: 10.1074/mcp.M400124-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Brehmer D, Godl K, Zech B, Wissing J, Daub H. Proteome-wide identification of cellular targets affected by bisindolylmaleimide-type protein kinase C inhibitors. Mol. Cell. Proteomics. 2004;3:490–500. doi: 10.1074/mcp.M300139-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Godl K, Gruss OJ, Eickhoff J, Wissing J, Blencke S, Weber M, Degen H, Brehmer D, Orfi L, Horvath Z, et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 2005;65:6919–6926. doi: 10.1158/0008-5472.CAN-05-0574. [DOI] [PubMed] [Google Scholar]
- 33.Brehmer D, Greff Z, Godl K, Blencke S, Kurtenbach A, Weber M, Muller S, Klebl B, Cotten M, Keri G, et al. Cellular targets of gefitinib. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 34.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 35.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 37.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 38.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 39.Rix U, Remsing Rix LL, Terker AS, Fernbach NV, Hantschel O, Planyavsky M, Breitwieser FP, Herrmann H, Colinge J, Bennett KL, et al. A comprehensive target selectivity survey of the BCR-ABL kinase inhibitor INNO-406 by kinase profiling and chemical proteomics in chronic myeloid leukemia cells. Leukemia. 2010;24:44–50. doi: 10.1038/leu.2009.228. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, Bennett KL, Gao J, Song L, Eschrich S, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Tortora G, Bianco R, Daniele G, Ciardiello F, McCubrey JA, Ricciardi MR, Ciuffreda L, Cognetti F, Tafuri A, Milella M. Overcoming resistance to molecularly targeted anticancer therapies: rational drug combinations based on EGFR and MAPK inhibition for solid tumours and haematologic malignancies. Drug Resist. Updates. 2007;10:81–100. doi: 10.1016/j.drup.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 46.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 47.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graves LM, Litchfield DW. “Going KiNativ”: probing the native kinome. Chem. Biol. 2011;18:683–684. doi: 10.1016/j.chembiol.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 51.Fedorov O, Muller S, Knapp S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- 52.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osborne JK, Zaganjor E, Cobb MH. Signal control through Raf: in sickness and in health. Cell Res. 2012;22:14–22. doi: 10.1038/cr.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 57.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lacouture ME, O'Reilly K, Rosen N, Solit DB. Induction of cutaneous squamous cell carcinomas by RAF inhibitors: cause for concern? J. Clin. Oncol. 2012;30:329–330. doi: 10.1200/JCO.2011.38.2895. [DOI] [PubMed] [Google Scholar]
- 59.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, Edwards PA, Smith PD, Cook SJ. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci. Signaling. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 61.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signaling. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smalley KS, Flaherty KT. Integrating BRAF/MEK inhibitors into combination therapy for melanoma. Br. J. Cancer. 2009;100:431–435. doi: 10.1038/sj.bjc.6604891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poulikakos PI, Solit DB. Resistance to MEK inhibitors: should we co-target upstream? Sci. Signaling. 2011;4:pe16. doi: 10.1126/scisignal.2001948. [DOI] [PubMed] [Google Scholar]
- 65.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, Dickerson SH, Laquerre SG, Liu L, Gilmer TM. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol. Cancer Ther. 2012;11:909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 66.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 68.Larson RA, Golomb HM, Rowley JD. Chromosome changes in hematologic malignancies. Ca-Cancer J. Clin. 1981;31:222–238. doi: 10.3322/canjclin.31.4.222. [DOI] [PubMed] [Google Scholar]
- 69.Sawyers CL, Hochhaus A, Feldman E, Goldman JM, Miller CB, Ottmann OG, Schiffer CA, Talpaz M, Guilhot F, Deininger MW, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka MF, Kantarjian H, Cortes J, Ohanian M, Jabbour E. Treatment options for chronic myeloid leukemia. Expert Opin. Pharmacother. 2012;13:815–828. doi: 10.1517/14656566.2012.671296. [DOI] [PubMed] [Google Scholar]
- 71.Schenone S, Brullo C, Botta M. New opportunities to treat the T315I-Bcr-Abl mutant in chronic myeloid leukaemia: tyrosine kinase inhibitors and molecules that act by alternative mechanisms. Curr. Med. Chem. 2010;17:1220–1245. doi: 10.2174/092986710790936310. [DOI] [PubMed] [Google Scholar]
- 72.Packer LM, Rana S, Hayward R, O'Hare T, Eide CA, Rebocho A, Heidorn S, Zabriskie MS, Niculescu-Duvaz I, Druker BJ, et al. Nilotinib and MEK inhibitors induce synthetic lethality through paradoxical activation of RAF in drug-resistant chronic myeloid leukemia. Cancer Cell. 2011;20:715–727. doi: 10.1016/j.ccr.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vougioukalaki M, Kanellis DC, Gkouskou K, Eliopoulos AG. Tpl2 kinase signal transduction in inflammation and cancer. Cancer Lett. 2011;304:80–89. doi: 10.1016/j.canlet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi M. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 78.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc. Natl. Acad. Sci., U.S.A. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, Markman B, Rodriguez O, Guzman M, Rodriguez S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 85.Vivanco I, Mellinghoff IK. Epidermal growth factor receptor inhibitors in oncology. Curr. Opin. Oncol. 2010;22:573–578. doi: 10.1097/CCO.0b013e32833edbdf. [DOI] [PubMed] [Google Scholar]
- 86.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 87.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mueller KL, Yang ZQ, Haddad R, Ethier SP, Boerner JL. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J. Mol. Signaling. 2010;5:8. doi: 10.1186/1750-2187-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, Scott AM, Johns TG. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009;11:448–458. doi: 10.1593/neo.09230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, Bronson RT, Pfannl R, White F, Housman DE, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P, Helfrich BA, Doebele RC, Heasley LE. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS ONE. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wheeler DL, Iida M, Kruser TJ, Nechrebecki MM, Dunn EF, Armstrong EA, Huang S, Harari PM. Epidermal growth factor receptor cooperates with Src family kinases in acquired resistance to cetuximab. Cancer Biol. Ther. 2009;8:696–703. doi: 10.4161/cbt.8.8.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hardie DG. New roles for the LKB1→AMPK pathway. Curr. Opin. Cell Biol. 2005;17:167–173. doi: 10.1016/j.ceb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 95.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 96.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Esteve-Puig R, Canals F, Colome N, Merlino G, Recio JA. Uncoupling of the LKB1-AMPKα energy sensor pathway by growth factors and oncogenic BRAF. PLoS ONE. 2009;4:e4771. doi: 10.1371/journal.pone.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, Cantley LC. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]