Abstract
In plants, numerous developmental processes are controlled by cytokinin (CK) levels and their ratios to levels of other hormones. While molecular mechanisms underlying the regulatory roles of CKs have been intensely researched, proteomic and metabolomic responses to CK deficiency are unknown. Transgenic Arabidopsis seedlings carrying inducible barley cytokinin oxidase/dehydrogenase (CaMV35S>GR>HvCKX2) and agrobacterial isopentenyl transferase (CaMV35S>GR>ipt) constructs were profiled to elucidate proteome- and metabolome-wide responses to down- and up-regulation of CK levels, respectively. Proteome profiling identified >1100 proteins, 155 of which responded to HvCKX2 and/or ipt activation, mostly involved in growth, development, and/or hormone and light signalling. The metabolome profiling covered 79 metabolites, 33 of which responded to HvCKX2 and/or ipt activation, mostly amino acids, carbohydrates, and organic acids. Comparison of the data sets obtained from activated CaMV35S>GR>HvCKX2 and CaMV35S>GR>ipt plants revealed unexpectedly extensive overlaps. Integration of the proteomic and metabolomic data sets revealed: (i) novel components of molecular circuits involved in CK action (e.g. ribosomal proteins); (ii) previously unrecognized links to redox regulation and stress hormone signalling networks; and (iii) CK content markers. The striking overlaps in profiles observed in CK-deficient and CK-overproducing seedlings might explain surprising previously reported similarities between plants with down- and up-regulated CK levels.
Key words: Arabidopsis thaliana, cytokinin, cytokinin oxidase/dehydrogenase, isopentenyl transferase, metabolome, proteome.
Introduction
Cytokinins (CKs) regulate diverse developmental processes in plants, including: the formation and activity of shoot meristems; apical dominance; leaf senescence; nutrient mobilization; seed germination; root, flower, and fruit development; plant–pathogen interactions; and stress responses. They also participate in various light-regulated processes, such as de-etiolation and chloroplast differentiation (e.g. Lochmanová et al., 2008; Argueso et al., 2009; Perlilli et al., 2010; Brenner et al., 2012; Novák et al., 2013).
Naturally occurring CKs are adenine derivatives substituted at the N 6-position with an isoprenoid or aromatic side chain. CK metabolism has been extensively reviewed (e.g. Strnad, 1997; Sakakibara, 2006; Kudo et al., 2010; Frébort et al., 2011). Briefly, the first step in isoprenoid CK biosynthesis, formation of isopentenyladenosine-5′-phosphates (iP-nucleotides) by transfer of an isopentenyl group to adenosine-5′-phosphates, is catalysed by adenosine phosphate-isopentenyl transferase (IPT; EC 2.5.1.27). The iP-nucleotides can be transformed to the corresponding zeatin-nucleotides, which are subsequently converted to active (free-base) CKs. CKs are inactivated either by conjugation, mainly reversible O-glucosylation and irreversible N 7- or N 9-glucosylation, or by degradation.
A key enzyme in CK inactivation is cytokinin oxidase/dehydrogenase (CKX; EC 1.5.99.12), which degrades CK bases, ribosides, nucleotides, and N 9 -glucosides by oxidative cleavage of the N 6 -unsaturated CK side chain, resulting in the formation of a side chain-derived aldehyde and adenine (or corresponding derivatives) (Galuszka et al., 2001, 2007; Kowalska et al., 2010). CKX activity was first found in crude tobacco culture in 1971 (Pačes et al., 1971) and it remains the only characterized enzymatic CK degradation mechanism in plants. Generally, only N 7-glucosides, O-glucosides, and dihydrozeatins are not susceptible to CKX degradation.
Constitutive ectopic overexpression in transgenic plants of both IPTs (e.g. Smigocki and Owens, 1988; Cline, 1994) and CKXs (e.g. Werner et al., 2001, 2003, 2010; Bartrina et al., 2011; Holst et al., 2011; Nishiyama et al., 2011) has dramatically contributed to knowledge of CK action. However, use of constitutive promoters has inherent limitations. For example, CKs are homeostastically regulated by a fine balance between synthesis, conjugation, and degradation. Synthesis was originally believed to occur in root apical meristems, but transcriptomic analysis has revealed that CKs are produced in discrete sites throughout the plant in diverse organs and tissues, including roots, shoots, and vascular tissues, at specific developmental stages (Sakakibara, 2006; Argueso et al., 2009; Perilli et al., 2010). CK receptors are also widely distributed both spatially and temporally in plants (Stolz et al., 2011). Thus, the constitutive overexpression of CK metabolism enzymes is clearly problematic for studying the fine tuning of CK signalling or elucidating the roles of CKs at different developmental stages. However, several systems are available for chemically inducing transgene expression in plants, thus offering possibilities for restricting transgene expression to a particular developmental stage or generation (e.g. Moore et al., 2006). One of these, a stringently regulated and highly responsive dexamethasone (DEX)-inducible gene expression system, has been used to study the effects of activating an agrobacterial isopentenyl transferase gene, ipt, in Arabidopsis (Craft et al., 2005; Hradilová et al., 2007; Lochmanová et al., 2008) and tobacco (Šámalová et al., 2005).
The molecular mechanisms underlying CK action have been intensively researched, but not the proteome- and metabolome-wide changes triggered by controlled reductions in bulk CK levels. Therefore, dynamic global responses in Arabidopsis seedlings to reductions and increases in the bulk CK pool, mediated by DEX-induced activation of a construct hosting HvCKX2, a barley CKX isoform 2 gene (Galuszka et al., 2004), have been examined. The proteomic and metabolomic responses to inducible HvCKX2 and ipt activation have also been compared. Here the results of these analyses are presented, and the implications of significant overlaps found in the responses, notably indications that homeostatic control mechanisms with opposite effects are triggered by CK depletion and overproduction, are discussed.
Materials and methods
Plant material, growth conditions, and dexamethasone treatment
The plants used in the experiments were transgenic CaMV35S>GR>HvCKX2 line 13, CaMV35S>GR>ipt line pOpBK-ipt 11 (Craft et al., 2005), and corresponding wild-type plants (Arabidopsis thaliana ecotype Col-0). Briefly, DEX-inducible CaMV35S>GR>HvCKX2 lines were prepared as follows. The genomic sequence of barley HvCKX2 (AF490591) was subcloned from the pDRIVE vector (Galuszka et al., 2004) into Gateway entry pENTR1A vector KpnI/NotI sites (Invitrogen) with Acc65I/NotI restriction enzymes. Subsequently, the gene was inserted into the pOpOn2.1 binary vector derived from the pOpOff2 vector (Wielkopolska et al., 2005; provided by Dr Ian Moore, Department of Plant Sciences, University of Oxford) downstream of the artificial pOp6 promoter (Supplementary Fig. S1 available at JXB online) via a Gateway LR clonase II recombination reaction (Invitrogen). Arabidopsis thaliana ecotype Columbia Col-0 plants were transformed by the flower-dip procedure (Bechtold et al., 1993). Based on screening for the DEX-induced cytokinin deficiency phenotype, a strongly responsive line 13 was selected out of 20 independent transformants and selfed to obtain homozygous offspring. A seed stock for large-scale experiments was harvested from T3 plants. Seeds were surface-sterilized and sown on Uhelon 120T (Silk & Progress, www.silkandprogress.cz) mesh placed on 1% (w/v) agar containing Murashige and Skoog (MS) medium (pH 5.7) supplemented with 5×10–4% (v/v) dimethylsulphoxide (DMSO), stratified at 4 °C for 3 d, and cultivated at 21 °C/19 °C day/night temperatures, with a 16h photoperiod (80 µmol m–2 s–1 light intensity) for 7 d in a growth chamber (AR36LX, Percival, http://www.percival-scientific.com/). Prior to the harvest, the Uhelon mesh with the seedlings was transferred onto fresh MS medium supplemented with 5×10–4% (v/v) DMSO (mock) or 20 µM DEX (Sigma-Aldrich, http://www.sigmaaldrich.com) in DMSO (final concentration, as for the mock) for: (i) 6, 12, 24, and 48h for analysis of endogenous CK levels following DEX induction; (ii) 12h and 48h for two-dimensional electrophoresis (2-DE) proteomic analysis; (iii) 48h for liquid chromatography–mass spectrometry (LC-MS) proteomic and metabolomic analysis, and quantitative reverse transcription–PCR (RT–qPCR) analysis. Seedlings were then harvested and frozen in liquid nitrogen.
2-DE MALDI TOF/TOF proteome profiling
Protein was extracted from frozen seedlings (180mg, ~250 seedlings), loaded onto Seppro IgY-Rubisco Spin Columns (Sigma-Aldrich), and processed according to the supplier’s manual. RuBisCO-depleted samples were pooled and extracted by acetone/trichloroacetric acid (TCA) extraction (Damerval et al., 1986). Dried protein was solubilized and separated, essentially as previously described (Lochmanová et al., 2008; Hradilová et al., 2010; Černý et al., 2011a , b ). Briefly, portions of extracts containing 150 µg of protein were applied to 7cm IPG strips with a non-linear pH gradient (3–10; Bio-Rad, http://www.bio-rad.com/), isoelectrically focused, then treated with buffers containing dithiothreitol (DTT) and iodoacetamide (Sigma-Aldrich) to reduce and alkylate the proteins, which were subsequently separated by SDS–PAGE. Gels were stained with colloidal Bio-Safe Coomassie G-250 (Bio-Rad), digitally imaged, and analysed using Decodon Delta 2D software (http://www.decodon.com). Two biological replicates with three technical replicates were used in the comparisons. Responses to DEX activation of proteins corresponding to detected spots were deemed significant if there was an absolute DEX/mock spot volume ratio ≥1.4, with t-test P-values <0.05 and similar profiles in two biological replicates. Spots with significant and reproducible changes were cut, digested with trypsin (Promega, http://www.promega.com/), desalted, and the resulting protein fragments were analysed using a 4800 Plus matrix-assisted laser desorption tandem time of flight (MALDI TOF/TOF) instrument (AB Sciex, http://www.absciex.com/). For details, see Supplementary Methods S1 at JXB online.
LC-MS proteome profiling
Further proteomic analyses were performed using a gel-free shotgun protocol based on nano-high-perfomance liquid chromatography (HPLC) and tandem mass spectrometry (MS/MS), as described elsewhere (e.g. Larrainzar et al., 2007). Briefly, two biological replicates, each consisting of ~300 Arabidopsis seedlings cultivated as described above, were pooled and analysed in three technical replicates. Proteins were extracted by combination of acetone/TCA and phenol extraction, and digested in solution with endoproteinase Lys-C and immobilized trypsin beads (Promega). The resulting peptides were desalted, dried, and dissolved in 0.5% (v/v) formic acid in 5% (v/v) acetonitrile, then analysed online by nanoflow C18 reverse-phase LC using a 15cm Ascentis Express Column (0.1mm inner diameter; Sigma-Aldrich) and an Eksigent ultra-high-performance liquid chromatography (UPLC) system (Eksigent, http://www.eksigent.com/) directly coupled to an electrospray ionization (ESI) source and an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, http://www.thermoscientific.com). Peptides were eluted with a 155min, 5–95% acetonitrile gradient. Dynamic exclusion settings were as described in Hoehenwarter and Wienkoop (2010). Raw files obtained from the MS analysis were searched against the TAIR10 Arabidopsis database using the Sequest algorithm. For identification and spectral count-based data matrix generation, Proteome Discoverer (v 1.3, Thermo Scientific) was used. Only high confidence peptides (false discovery rate <1%) with >7 ppm precursor mass accuracy and at least one distinct peptide per protein met identification criteria. Quantitative differences in protein abundance between DEX- and mock-treated samples were screened by spectral counting (Neilson et al., 2011) and were further manually validated by comparing respective peptide ion signal peak areas (Qual Browser 2.0.7, Thermo Scientific). Quantitative differences were deemed significant if there was an absolute DEX/mock ratio ≥1.5, with t-test P-values <0.05. For details, see Supplementary Methods S1 at JXB online.
GC-MS metabolome profiling
Polar metabolites were analysed essentially as described in Morgenthal et al. (2007). Briefly, two biological replicates, each consisting of ~100 Arabidopsis seedlings cultivated as described above, were pooled and analysed in three technical replicates. Metabolites were extracted with methanol/chloroform/distilled water [2.5:1:0.5 (v/v/v)], and clarified by centrifugation. The resulting polar phase was separated by adding 0.5ml of distilled water, dried, methoximated and silylated, then analysed using an Agilent 6890 gas chromatograph (Agilent, http://www.home.agilent.com) coupled to a Pegasus IV TOF mass spectrometer (LECO, http://www.leco.com/). The acquired data were analysed using ChromaTOF software (LECO). Quantitative differences in metabolite abundance were deemed significant if there was an absolute DEX/mock ratio ≥2.5, with t-test P-values <0.05. For details, see Supplementary Methods S1 at JXB online.
Quantification and identification of endogenous cytokinins
Endogenous CK contents of duplicate samples were analysed using the method described by Novák et al. (2003), with modifications described by Novák et al. (2008). Briefly, 150–200mg fresh weight of Arabidopsis seedlings were extracted in Bieleski buffer (Bieleski, 1964). A cocktail of stable isotope-labelled CKs (each at 1 pmol per sample) was added to the extracts to check purification recovery, and samples were purified using a combination of cation (SCX-cartridge) and anion (DEAE-Sephadex-C18-cartridge) exchange, followed by immunoaffinity chromatography, using a wide range of immobilized CK-specific monoclonal antibodies. Three resulting fractions were then analysed by UPLC–electrospray tandem mass spectrometry (using an UPLC Acquity System linked to a Xevo TQ MS spectrometer, Waters, http://www.waters.com). The concentrations of the various CKs were calculated using a standard isotope dilution method (Rittenberg and Foster, 1940).
RT–qPCR analysis
Total RNA was prepared from 50mg of seedlings that had been frozen in liquid nitrogen using TRIzol reagent (Invitrogen, http://www.invitrogen.com), and contaminating DNA was removed by DNase I. First-strand cDNA was prepared using SuperScript II reverse transcriptase (Invitrogen) and the oligo(dT) primer according to the manufacturer’s instructions. qPCR with specific UPL probes (Roche, http://www.roche.com) and primers designed by ProbeFinder Software was performed using a LightCycler 480 Instrument and LightCycler 480 Probes Master (Roche). The presented results are means obtained from three independent biological replicate experiments, each analysed in triplicate. Quantitative differences in transcript abundance were deemed significant if there was an absolute DEX/mock ratio ≥1.3, with t-test P-values <0.05. For details, see Supplementary Methods S1 at JXB online.
Results
Cytokinin pool dynamics following HvCKX2 activation in Arabidopsis seedlings
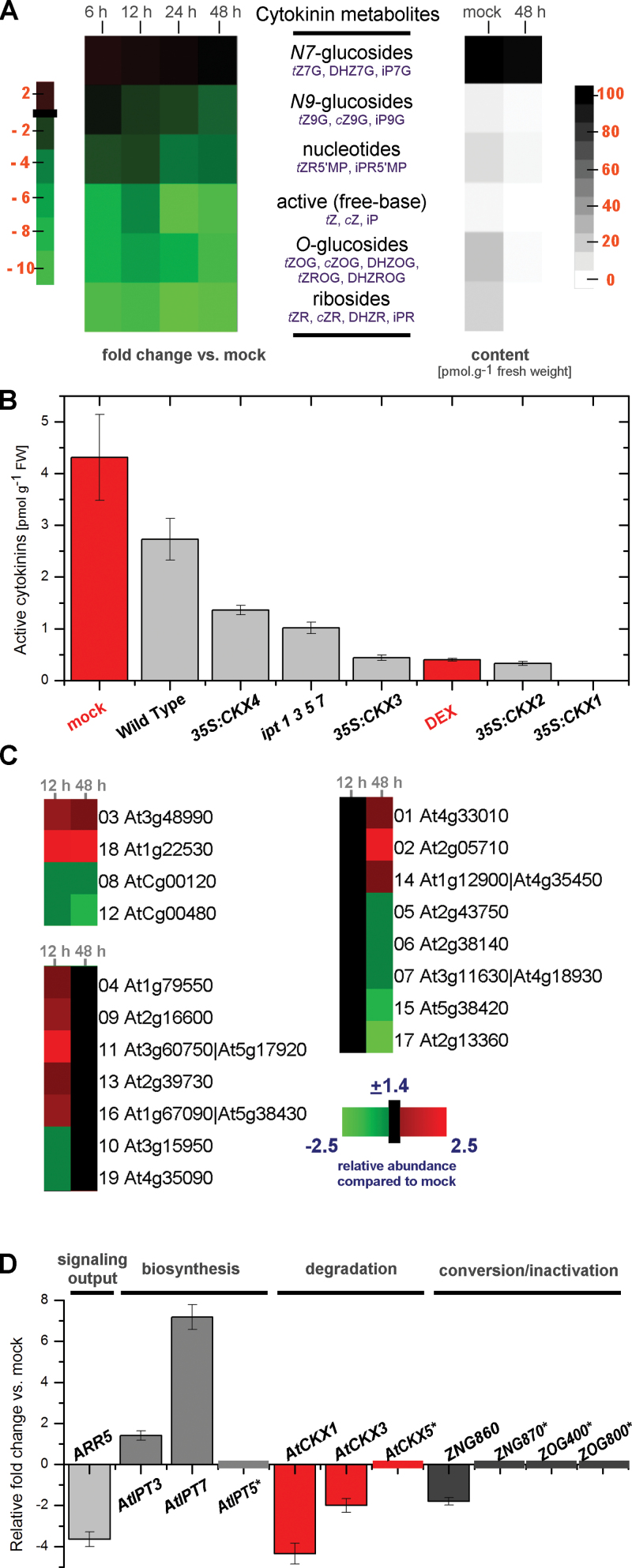
The binary pOp-HvCKX2/LhGR system of DEX-inducible HvCKX2 expression was used to decrease endogenous CK levels of light-grown Arabidopsis seedlings then associated proteomic and metabolomic changes were examined. To examine CK dynamics following HvCKX2 activation, 20 CK moieties were monitored in CaMV35S>GR>HvCKX2 line 13 seedlings cultivated for 7 d on mesh on solid MS medium and subsequently incubated on mesh on the surface of MS liquid medium without (mock) or with 20 µM DEX for 6, 12, 24, and 48h (Fig. 1A; Supplementary Fig. S2 at JXB online). Dramatic reductions were observed in tZ, cZ, and iP levels after 6h of DEX treatment, followed by oscillations in tZ levels, probably reflecting the activation of homeostatic mechanisms that stabilize hormonal ratios in seedlings. Similar, but less pronounced, tendencies were also apparent for cZ and iP. The varied reductions in the individual CK bases were reflected in a shift in the tZ:cZ:iP ratio from 32:1.3:1 at 6h to 4.2:0.5:1 after 48h. This shift is consistent with the higher substrate preference of HvCKX2 for tZ than for other CK bases (Galuszka et al., 2004) (for details, see Supplementary Fig. S2). Most of the CK pool consisted of biologically inactive CK metabolites, most prominently cytokinin-N 7-glucosides (CK7Gs). Unlike free CKs and most of their metabolites, the CK7G content increased by ~40% after just 6h of HvCKX2 activation. After 48h, the CK7G content had returned to levels comparable with those of mock-treated seedlings (Supplementary Fig. S3), which still represented ~90% of the CK pool. CK7Gs are reportedly not degraded either by HvCKX2 or by endogenous AtCKXs (Galuszka et al., 2007; Kowalska et al., 2010). Thus, their increase probably indicates an increased rate of CK biosynthesis—a mechanism whereby plants can counteract CK degradation by HvCKX2. The CK that increased to the highest level was iP7G. Accordingly, native plant IPTs produce iP-type CKs in vivo (e.g. Ueda et al., 2012), tZ is the preferred substrate for HvCKX2 (Galuzska et al., 2004), and CK degradation is a reaction concurrent with N 7-glucosylation in CK base inactivation (Brzobohatý et al., 1994; Frébort et al., 2011).
Fig. 1.
DEX-inducible HvCKX2 expression. (A) Time course of cytokinin depletion in CaMV35S>GR>HvCKX2 seedlings following HvCKX2 activation. Seven-day-old seedlings were transferred onto MS medium supplemented with DEX or mock and samples were collected for cytokinin determination 6, 12, 24, and 48h later. (B) Comparison of CK depletion in CaMV35S>GR>HvCKX2 and constitutive 35S:AtCKX transgenics, and the quadruple atipt1 3 5 7 mutant. Pool of active (free base) CKs; data for the CaMV35S>GR>HvCKX2 line (mock, DEX) are marked in red. Data for the 35S:CKX1-35S:CKX4 and ipt1 3 5 7 mutant used in this comparison are from 10-day-old Arabidopsis seedlings as reported by Nishiyama et al. (2011). (C) Proteins that responded to HvCKX2 activation identified by 2-DE analysis. MALDI TOF/TOF MS was used to identify proteins in spots representing differentially regulated proteins at each of the time points. Time courses of changes in their abundance are presented as a heatmap; accession numbers are as in the TAIR database. Numbers correspond to the spot number as given in Supplementary Table S1 at JXB online and in the 2-DE RuBisCO-depleted proteome map (Supplementary Fig. S5). See the Supplementary data for details. (D) Relative fold change in transcripts of 10 genes involved in CK metabolism and signalling by RT–qPCR 48h after HvCKX2 activation.
To obtain insights into the regulation of genes underlying homeostatic responses to CK degradation by HvCKX2, steady-state levels of transcripts of 10 genes involved in CK metabolism and signalling were examined by RT–qPCR 48h after HvCKX2 activation (Fig. 1D; Supplementary Table S9 at JXB online). The reduction in free CK bases correlated with reductions in transcripts of ARR5, a CK primary response gene (D’Agostino et al., 2000) that is frequently used to assess CK signalling. Similarly, genes involved in CK inactivation via either degradation (AtCKX1 and AtCKX3) or irreversible N-glucosylation (At5g05860 and At5g05870) were down-regulated, while expression of an O-glucosyltransferase gene (At1g22400 and At2g36800) was unaffected. AtIPT3 and AtIPT7 genes were up-regulated, in accordance with the presumed increase in the CK biosynthesis rate.
To assess the extent of CK depletion in the CaMV35S>GR>HvCKX2 seedlings, their measured CK contents after 48h of HvCKX2 activation were compared with previously reported contents of seedlings constitutively ectopically overexpressing AtCKX1, AtCKX2, AtCKX3, and AtCKX4 (encoding genuine Arabidopsis CKXs) and the CK biosynthesis-deficient atipt1 3 5 7 mutant (Fig. 1B; Supplementary Fig. S4 at JXB online). CKs were depleted more strongly in the CaMV35S>GR>HvCKX2 seedlings than in all the other transgenic and mutant lines except 35S:CKX1 plants.
Proteomic and metabolomic responses to cytokinin pool alterations
To obtain insights into molecular events triggered by the CK pool alterations, associated proteomic and metabolomic responses were examined. First, proteome changes caused by HvCKX2 activity were monitored by 2-DE of RuBisCO-depleted protein extracts, followed by image and MALDI TOF/TOF MS analyses of CaMV35S>GR>HvCKX2 seedlings, sampled 12h and 48h after HvCKX2 activation (corresponding to partial recovery from the initial shock and system stabilization, respectively, according to the observed changes in tZ contents). A total of 748 different protein spots were detected and quantified, of which 19 (~3%) showed significant changes in relative volume compared with mock-treated samples. Seven showed a significant transient response only at 12h, eight a delayed response at 48h, and four similar, early responses at 12h and 48h of DEX treatment (Fig. 1C; Supplementary Fig. S5, Table S1 at JXB online).
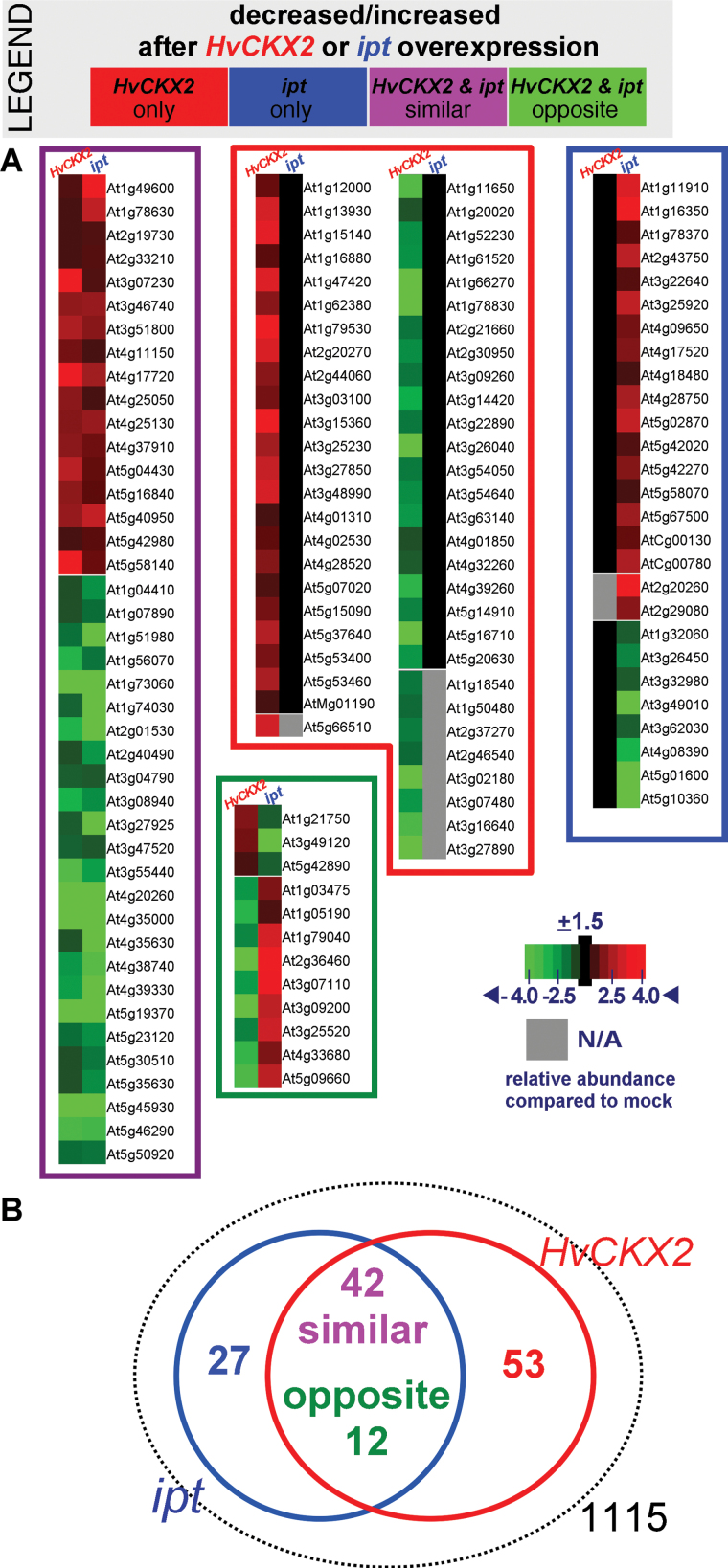
To increase proteome coverage, an LC-MS shotgun proteomic analysis of total soluble proteins was next conducted. As HvCKX2 activation probably results in at least local increases in CK levels due to AtIPT3 and AtIPT7 activation, transgenic CaMV35S>GR>ipt line pOpBK-ipt 11 (Craft et al., 2005), allowing DEX-inducible increases in CK levels, was analysed, in parallel to CaMV35S>GR>HvCKX2 line 13 at 48h after DEX treatment. In total, >1700 proteins were identified in at least one of three repeated LC-MS experiments. However, only 1115 were present in all three replicates (Fig. 2; Supplementary Table S3 at JXB online). The comparison of normalized spectral counts of DEX- and mock-treated samples revealed 153 potentially responsive proteins. Of these, 19 were removed after manual verification of the respective peptide ion signal peak areas, and 53 significantly responded only in CaMV35S>GR>HvCKX2 seedlings, about twice the number of proteins that exclusively responded to ipt activation. Large numbers of proteins responded to both HvCKX2 and ipt activation, 42 in similar and 12 in opposite directions. HvCKX2 was detected in CaMV35S>GR>HvCKX2 seedlings only upon DEX treatment (Supplementary Table S8, Fig. S9) confirming the tightness of the pOp-HvCKX2/LhGR system.
Fig. 2.
Proteins responsive to HvCKX2 and ipt activation identified by gel-free LC-MS profiling. (A) Proteomes of 7-day-old CaMV35S>GR>HvCKX2 and CaMV35S>GR>ipt Arabidopsis seedlings treated with DEX or mock for 48h were resolved by C18 reverse-phase liquid chromatography and analysed using an LTQ-Orbitrap XL mass spectrometer. In total, 134 proteins (~12% of all proteins meeting identification criteria) that responded to 48h of HvCKX2 or ipt activation and were assembled into groups of proteins that were responsive (i) only to HvCKX2 (HvCKX2; red); (ii) only to ipt (ipt; blue); or to both HvCKX2 and ipt either (iii) similarly (purple) or (iv) oppositely (green). The heatmap represents fold changes in the abundance of proteins in seedlings after DEX treatment compared with the respective mock-treated control; accession numbers are as in the TAIR database. (B) Venn diagram illustrating the overlap in identified proteins. See Supplementary Table S2 at JXB online for detailed information on the identified proteins.
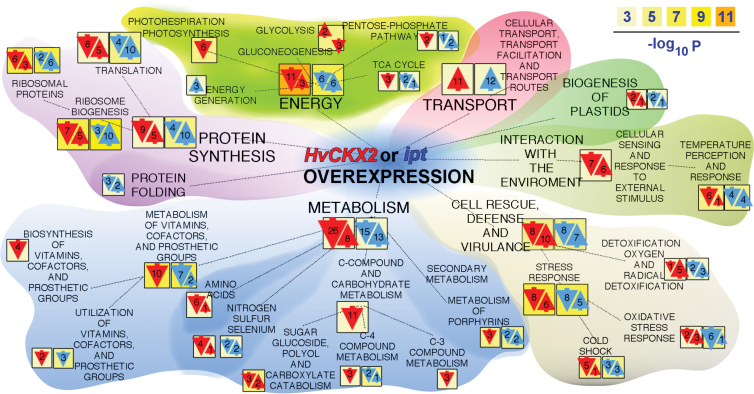
The subcellular location of each identified protein was determined using the SUBA database (http://suba.plantenergy.uwa.edu.au/; Heazlewood et al., 2007). The largest group of these proteins is located in chloroplasts (29.5%), followed by the plasma membrane (27%) and cytosol (11%) (Supplementary Fig. S6 at JXB online). Identified proteins were analysed by the BioMaps tool package (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/; Katari et al., 2010). Functional classification according to MIPS (Munich Information Center for Protein Sequences) revealed that the categories ‘energy’, ‘ribosome biogenesis’, ‘metabolism of vitamins, cofactors, and prosthetic groups’, and ‘stress response’ were most significantly enriched (Fig. 3). The general categories of enriched proteins were highly similar in samples from seedlings with both increases and decreases in bulk levels of endogenous CKs, but the overlap of individual proteins differed substantially. For example, only seven out of 20 proteins in the ‘stress response’ category were enriched in both sets of samples, six of which displayed similar responses to increases and decreases in CK levels (for details, see Supplementary Table S5).
Fig. 3.
Proteins that responded to HvCKX2 and ipt activation: functional classification according to the Munich Information Center for Protein Sequences (MIPS). The orientation and size of arrows indicate the direction of protein regulation (up or down) and number of proteins in a particular category, respectively. Red and blue: categories induced by HvCKX2 and ipt activation; the colour scale indicates the statistical significance of each category. The BioMap tool package (http://virtualplant.bio.nyu.edu/) was used for the analysis.
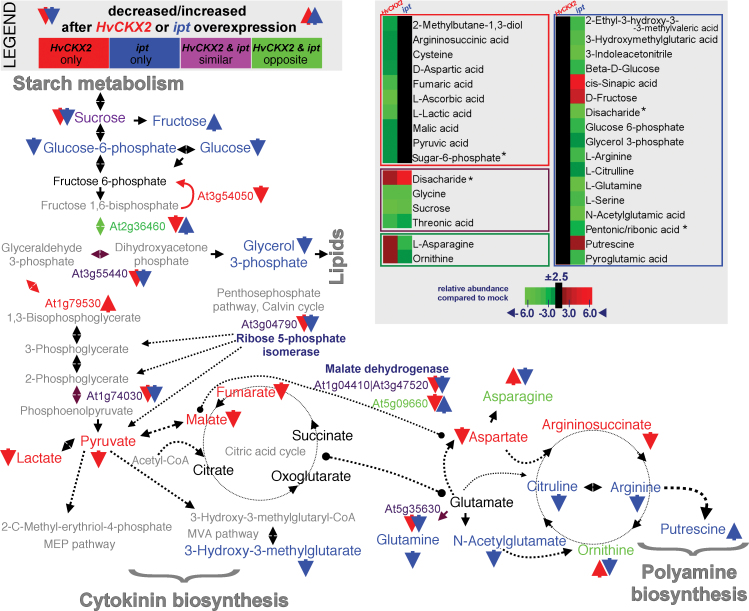
To assess the metabolomic impact of the proteome alterations, GC-TOF-MS was used to analyse changes in metabolite levels in both CaMV35S>GR>HvCKX2 and CaMV35S>GR>ipt lines following 48h of DEX treatment. The analysis resulted in a list of 79 annotated metabolites (Supplementary Table S4 at JXB online), including mostly amino acids, carbohydrates, and organic acids, 33 of which showed significant changes in relative abundance after DEX treatment (Fig. 4). Activation of HvCKX2 modulated levels of 16 metabolites. Of these, glycine, sucrose, threonic acid, and an unspecified disaccharide were similarly affected after ipt activation, while asparagine and ornithine showed opposite responses.
Fig. 4.
Integration of proteome and metabolome responses to HvCKX2 and ipt activation. Seven-day-old CaMV35S>GR>HvCKX2 (HvCKX2; red) and CaMV35S>GR>ipt (ipt, blue) Arabidopsis seedlings were treated with DEX or mock treated for 48h, then polar metabolites were analysed by GC-TOF-MS. In total, 33 metabolites (~41% of all annotated metabolites) significantly responded to HvCKX2 or ipt activation. Integration of metabolomic and proteomic (see Figs 1 and 2) data highlighted strong responses in carbohydrate metabolism and both the citric acid and urea cycles. Arrows represent the response direction (up or down); the colour coding is explained in the key; metabolites in black were detected, but their relative abundances were not affected more than ±2.5-fold. The heatmap represents the abundance of metabolites in seedlings after DEX treatment compared with their respective mock-treated controls; (*) metabolite annotated as several similar substances.
Discussion
Proteome profiling of cytokinin down- and up-regulation
The latest version of the Arabidopsis genome annotation lists >27 000 protein-coding genes, and alternative splicing and post-translational modifications (PTMs) may increase the number of different proteins in biological samples to at least 100 000. Both 2-DE- and LC-MS-based proteomic experiments do not usually cover more than several thousand. Thus, in terms of coverage, proteomic techniques lag far behind genomic techniques. Nevertheless, proteomic analysis is essential for elucidating biological processes as it provides information that cannot be deduced from genomic/transcriptomic data, ranging from data on protein abundance (Baginsky et al., 2010) to indications of post-translational control of protein activity (e.g. Černý et al., 2010). The 2-DE and LC-MS proteomic analyses of CaMV35S>GR>HvCKX2 and CaMV35S>GR>ipt seedlings covered ~6% of proteins currently deduced from the Arabidopsis genome. The former identified ~85% fewer proteins than the latter, which is not surprising as estimated amounts of the responsive proteins in the 2-DE Coomassie-stained spots ranged from 10ng to 600ng, far higher than the sensitivity limits of LC-MS-based analysis. Further, the overlap in responsive proteins was small; only three were detected in both data sets. Five proteins had shifted pI and/or molecular weight values, indicating the presence of PTM(s) not detected by MS. The other detected proteins did not meet criteria set for significant differences or were not found by LC-MS analysis (see 2-DE supplementary materials in Supplementary Table S1 at JXB online). These results confirm the benefits of 2-DE in qualitative analysis and screening for protein modifications, and imply a role for unspecified PTMs in response to CKX activity.
Chloroplasts host the highest proportion of CK-regulated proteins
Chloroplast proteins prevailed among proteins that responded to the altered CK levels. According to a prediction algorithm developed by Kaundal et al. (2010), chloroplast localization is expected for 33% of all 1115 proteins identified in the LC-MS profiling, and for 45% of the responsive proteins; a significant (1.4-fold) enrichment. Similar over-representation of chloroplast proteins was found in a previous study of early CK response proteins and phosphoproteins in Arabidopsis (Černý et al., 2011a ), and chloroplast proteins represented a major fraction of CK-induced proteins in dark-grown Arabidopsis seedlings identified by Lochmanová et al. (2008). Accordingly, chloroplasts have been linked to CK action on numerous levels. For example, a dynamic CK pool is reportedly present in chloroplasts (Benková et al., 1999) that is partly autonomous from the bulk cell CK pool (Polanská et al., 2007). Further, compartmentation into chloroplasts of some CK biosynthesis and metabolism pathways has been revealed (Brzobohatý et al., 1993; Kristoffersen et al., 2000; Takei et al., 2004; Kiran et al., 2006).
Previously reported and novel links in CK signalling
The LC-MS and 2-DE analyses identified 155 proteins that responded to the altered levels of endogenous CKs in the seedlings, 73 of which have been previously implicated in plant hormone, mostly CK (58), responses in large-scale transcriptomic and proteomic analyses (Rashotte et al., 2003; Brenner et al., 2005; Kiba et al., 2005; Nemhauser et al., 2006; Lochmanová et al., 2008; Chen et al., 2010; Černý et al., 2011a ; Brenner and Schmülling, 2012; Nishiyama et al., 2012). Direct comparison of data sets reported by different groups is not straightforward due to differences in experimental set-ups. Nevertheless, 34 proteins in the sets reported here showed similar responses to those observed in previous studies. In the set of responsive proteins yielded by LC-MS profiling, 12 displayed opposite responses to HvCKX2 and ipt activation, and thus may be indicative of bulk endogenous CK levels (Table 1). Of those 12 proteins, six are novel CK-response proteins: three ribosomal proteins, a key enzyme in haem biosynthesis (coproporphyrinogen oxidase, At1g03475), a peroxidase (At3g49120) reportedly involved in cell elongation (Passardi et al., 2005), and a sterol carrier protein (At5g42890) with a loss-of-function mutation phenotype correlating with known CK effects on root elongation, together with small cotyledon rosettes and short hypocotyls (Zheng et al., 2008). The other six proteins have already been linked to CK responses (Brenner et al., 2005; Černý et al., 2011a ; Brenner and Schmülling, 2012) and include, for example, an enzyme involved in lysine biosynthesis (At4g33680) and a peroxisomal malate dehydrogenase (At5g09660), mutation of which results, respectively, in aberrant growth and cell death (Song et al., 2004), and reduced growth rates in the dark (Pracharoenwattana et al., 2007). A recently assembled data set of genes with known loss-of-function mutant phenotypes in Arabidopsis (Lloyd and Meinke, 2012) provides information on biological functions of a further 41 proteins found here to respond to alterations in CK levels. Most of them (70%) are also essential for growth and development, including 15 for which loss of function is lethal or leads to embryo defects. Mutations in another 10 and eight of the proteins have been linked to defects in photosynthesis or biosynthesis of photosynthetic pigments and enhanced sensitivity to abiotic stresses, respectively (for details, see Supplementary Table S6 at JXB online).
Table 1.
Proteins and metabolites indicative of endogenous cytokinin levels
| ID (AGI/KEGG) | Recommended name | Correlation with cytokinin levels |
|---|---|---|
| At1g03475 | Coproporphyrinogen-III oxidase, chloroplastic | Positive |
| At1g05190 | 50S ribosomal protein L6, chloroplastic | Positive |
| At1g79040 | Photosystem II 10kDa polypeptide, chloroplastic | Positive |
| At2g36460 | Fructose-bisphosphate aldolase | Positive |
| At3g07110 | 60S ribosomal protein L13a-1 | Positive |
| At3g09200 | 60S acidic ribosomal protein P0-2 | Positive |
| At3g25520 | 60S ribosomal protein L5-1 | Positive |
| At4g33680 | LL-diaminopimelate aminotransferase, chloroplastic | Positive |
| At5g09660 | Malate dehydrogenase, glyoxysomal | Positive |
| At1g21750 | Protein disulphide isomerase-like 1-1 | Negative |
| At3g49120 | Peroxidase 34 | Negative |
| At5g42890 | Sterol carrier protein 2 | Negative |
| C00077 | Ornithine | Negative |
| C00152 | l-Asparagine | Negative |
ABA and CK cross-talk
Seventeen of the CK-responsive proteins identified here are abscisic acid (ABA) responsive, according to an analysis of overlaps in transcriptional responses to individual plant hormones by Nemhauser et al. (2006), who found 126 genes that are influenced by both CK and ABA (representing 46.8% and 4.3% of all identified CK- and ABA-modulated genes, respectively). A further 55 are ABA responsive according to data obtained from analyses of ABA-induced transcriptomic changes by Xin et al. (2005) and Matsui et al. (2008). These findings are consistent with the apparent involvement of ABA and CK cross-talk in several physiological processes, including stress responses (Tran et al., 2010; Ha et al., 2012) and seed germination (Wang et al., 2011). Furthermore, Nishiyama et al. (2011) found that CK deficiency decreases ABA content, but induces ABA hypersensitivity, which may be reflected in the up- or down-regulation of 36 proteins following HvCKX2 activation whose respective transcripts reportedly showed similar responses after plants were treated with exogenous ABA (Supplementary Table S7 at JXB online). In addition, seven proteins that showed no significant response to HvCKX2 overexpression are inversely up- or down-regulated in seedlings with increased levels of endogenous CK compared with ABA-treated plants (Supplementary Table S7). Proteins that were identified as CK responsive that are apparently involved in ABA–CK cross-talk include several well-known ABA response proteins, for example magnesium-chelatase subunit chlI (At4g18480; increased in response to ipt activation) and RNA-binding protein At2g21660 (decreased in response to HvCKX2 activation; mutant sensitive to ABA). They also included two proteins likely to be involved in ABA signalling (Yan et al., 2009): At3g26450 (decreased in response to ipt activation), which contains a ligand-binding domain similar to PYR/PYL/RCAR, and mitochondrial porin (At5g67500; increased in response to ipt activation).
Ethylene and CK cross-talk
Manipulation of endogenous CK levels resulted in increased abundance of two RNA-binding proteins (At4g17720 and At5g04430) previously identified in a phosphoproteomics study of ethylene responses (Li et al., 2009). HvCKX2 activity up-regulated a key enzyme in ethylene biosynthesis, ACC oxidase 2 (At1g62380), and affected a number of proteins previously found to be affected in ethylene-treated seedlings (Chen et al., 2011). In total, 18 proteins detected by Chen et al., were identified, 14 of which responded to HvCKX2 activation, five in the same direction as when seedlings were treated with ethylene. Ipt activation had milder effects, influencing three of 11 common proteins in the same manner as ethylene treatment (Supplementary Table S7 at JXB online). These findings indicate previously unrecognized links between ethylene and CK action or an alternative ethylene-independent regulation of the proteins originally recognized as ethylene responsive. The latter possibility is consistent with the finding that five of these proteins are also apparently ABA responsive.
Jasmonate signalling
Jasmonates are plant hormones that are mainly involved in plant responses to biotic or abiotic stresses. Jasmonate treatments reportedly have profound effects on plant growth, including reductions in the size of leaves and root systems (e.g. Zhang and Turner, 2008). Brenner et al. (2012) found that 20 genes involved in CK metabolism and signalling that were most strongly induced by CK treatment were rapidly down-regulated in response to methyl jasmonate (MeJA). However, this CK deficiency-like response to MeJA is not reflected in MeJA-responsive proteins in the present data set. HvCKX2 activity increased the amount of an uncharacterized protein, At1g13930, and thylakoid lumenal protein (At4g02530), while decreasing levels of β-glucosidase (At3g09260), ATP sulphurylase (At3g22890), tryptophan synthase (At4g01850), and phosphoserine aminotransferase (At4g35630). These are opposite effects compared with those of MeJA treatment (Nagano et al., 2005; Nemhauser et al., 2006; Jung et al., 2007a , b), indicating yet more complex interactions between the two plant hormones.
Redox-regulatory network responses to CK
Redox-regulatory components were over-represented in the present set of CK-responsive proteins. They included thioredoxin 3 (increased following activation of both ipt and HvCKX2), thioredoxin M and glutaredoxin (up-regulated in CaMV35S>GR>HvCKX2 seedlings), a ferredoxin superfamily protein (down-regulated in CaMV35S>GR>HvCKX2 seedlings), and 15 other proteins that are targets of thioredoxin regulation. Recently, a role for thioredoxin M in Mg chelatase regulation was indicated (Luo et al., 2012). Thus, thioredoxin signalling might provide a connection between CK action and ABA signalling. Further, CK-dependent regulation of the thioredoxin regulatory network could integrate CK and light signalling in chloroplasts. This is consistent with the known CK involvement in partial chloroplast biogenesis in dark-grown Arabidopsis (Chory et al., 1994; Lochmanová et al., 2008) and over-representation of chloroplast proteins among cytokinin-responsive proteins revealed in this study and previous studies by Černý et al. (2011a ) and Lochmanová et al. (2008).
Ribosomal proteins: an information hub for cytokinin signalling?
Extensive regulation of transcripts encoding ribosomal proteins in response to CK was recently reported by Brenner and Schmülling (2012). A total of 76 ribosomal proteins were detected, 21% of which responded to altered CK levels (Supplementary Fig. S7 at JXB online). Functional classification ‘Ribosome biogenesis’ was also strongly represented, by 12 and 13 proteins that responded to HvCKX2 and ipt activation, respectively (Fig. 3). Four and four proteins in the two groups displayed identical and opposite responses, respectively. Phenotypically, CKs are known to alter leaf development and morphology (Werner et al., 2003; Hay and Tsiantis, 2010; Shani et al., 2010), and the differentially regulated ribosomal proteins are probably involved in the underlying molecular mechanism. In support of this hypothesis, Horiguchi et al. (2011) showed that ribosomal proteins significantly contribute to Arabidopsis leaf development. In addition, an L5e mutant (At3g25520; decreased and increased following HvCKX2 and ipt activation, respectively) has slightly pointed, serrated leaves, while an L4e mutant (At5g02870), which increased following ipt activation and is implicated in auxin responses (Rosado et al., 2010), has narrow, pointed first true leaves, short roots, retarded growth, and late flowering.
Urea cycle metabolites reflect likely cytokinin-dependent polyamine biosynthesis
The urea cycle is one of the few points unequivocally distinguishing ipt and HvCKX2 action. Decreases in arginine, citrulline and ornithine were observed following ipt activation, probably resulting from enhanced polyamine biosynthesis, which is reportedly triggered by increased CK levels (e.g. Walker et al., 1988) and is indicated in the present data set by an increase in the polyamine putrescine. The depletion in urea cycle metabolites provides an explanation for reductions observed in glutamine and other glutamate derivatives, which is also consistent with down-regulation of glutamine synthetase. Thus, glutamate is probably predominantly converted to citrulline directly or indirectly through its N-acetyl group to ornithine. HvCKX2 activation led to increased ornithine levels, possibly at the expense of argininosuccinate and aspartate pools. The ornithine accumulation indicates a decrease in cytokinin-dependent conversion of arginine to polyamines and that ornithine may be a possible marker of endogenous CK levels in the cell.
Pyruvate: a pivotal mediator of ipt and HvCKX2 action
Integration of the proteomic and metabolomic data highlighted complex responses of carbohydrate metabolism to the manipulations of endogenous CK levels. Demands on the sucrose pool clearly rose following activation of both ipt and HvCKX2, probably at the expense of starch biosynthesis via a mechanism that maintains the supply of pyruvate. IPT is reportedly specialized for plastid localization and preferential use of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBDP) as a prenyl donor in vivo (Ueda et al., 2012). In plastids, HMBDP is generated via the methylerythritol phosphate (MEP) pathway starting from pyruvate. It was previously found that pyruvate homeostatic mechanisms are sufficient to cope with increases in t-ZMP amounting to >10 nmol g–1 fresh weight in DEX-treated CaMV35S>GR>ipt seedlings (Hradilová et al., 2007). Increased pyruvate consumption is apparently compensated by increased glycolysis, as indicated by the observed depletion of sucrose, glucose, and glucose-6-phosphate. Interestingly, a >4-fold decrease was found in 3-hydroxy-3-methylglutarate, which is linked to a mevalonate pathway that generates prenyl donors in the cytosol, indicating that IPT may have unrecorded activity in the cytosol or that cytosolic isopentenyl pyrophosphate and plastid HMBDP levels may be connected by an unknown mechanism. Alternatively, CK might stimulate biosynthesis of an uncharacterized product originating from the terpenoid biosynthetic pathway. Pyruvate depletion following HvCKX2 activation could result from the increase in ornithine synthesis deduced from the metabolomic data set. An apparent local increase in CK biosynthesis in activated CaMV35S>GR>HvCKX2 seedlings (indicated by up-regulation of AtIPT3 and AtIPT7) would also increase demands on the pyruvate pool, but this would be unlikely to result in a detectable decrease in bulk pyruvate as pyruvate levels were not affected in activated CaMV35S>GR>ipt seedlings.
Cytokinin homeostatic mechanisms may account for significant similarities in phenotypes resulting from down- and up-regulation of bulk cytokinin levels
Up- and down-regulation of CK levels has been used to modulate a number of biological processes and engineer agriculturally important traits. However, comparison of effects of up- and down-regulation of CK levels on a particular biological process has occasionally resulted in seemingly conflicting conclusions. For example, up-regulation of CK production is reportedly an important factor in adaptation to salt stress in tomato and maize (Vyroubalová et al., 2009; Ghanem et al., 2011). However, cytokinin-deficient Arabidopsis plants are also reportedly strongly salt stress tolerant (Nishiyama et al., 2011). In the seedlings examined here, ipt and HvCKX2 activation yielded overlaps, of 31% and 12% of responsive proteins and metabolites, respectively, which cannot be explained solely as a general stress response. Fourteen of the overlapping proteins have been previously identified as cytokinin responsive, and the STRING (http://string-db.org; Szklarczyk et al., 2011) protein–protein interaction network analysis indicates that proteins in the overlapping set are associated with diverse processes, including chlorophyll biosynthesis, carbohydrate metabolism, thioredoxin regulation, protein transport, proteasome-mediated degradation, and ribosome biogenesis (Supplementary Fig. S8 at JXB online). A decrease in the bulk CK pool via HvCKX2 activity results in attenuation of cytokinin signalling (manifested here in down-regulation of ARR5 transcripts) which, in turn, induces de novo CK biosynthesis via a CK homeostatic mechanism, manifested by up-regulation of AtIPT3 and AtIPT7 transcripts (Fig. 5). This is likely to generate a local increase in CK contents at CK biosynthesis sites, as manifested by the increased bulk content of CK7Gs (non-degradable products of irreversible CK inactivation). Interestingly, increased CK contents have been found in Arabidopsis CK receptor mutants, indicating a homeostatic control of steady-state CK levels through signalling (Riefler et al., 2006).
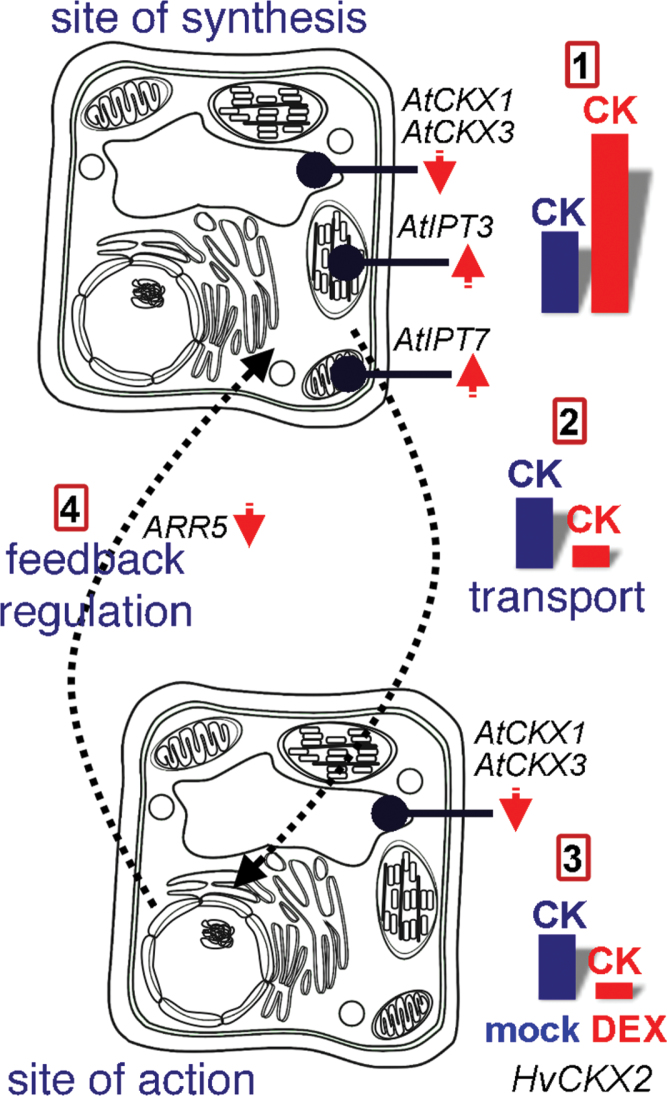
Fig. 5.
Hypothetical model of local CK increases in response to HvCKX2 activation. From a synthesis site (1), CKs are transported (2) to sites of action (3), and the synthesis rate is regulated by homeostasis-maintaining mechanisms (4). The closest HvCKX2 homologue in rice is apoplastic OsCKX7, and recent experiments have shown that HvCKX2 is predominantly localized in the leaf vasculature (P. Galuszka, unpublished results). Thus, HvCKX2 is a potent molecular tool for disrupting CK transport and (hence) decreasing cellular contents of active cytokinins, as evidenced by the down-regulation of a CK primary response gene ARR5, and AtCKX1 and AtCKX3 genes involved in CK down-regulation under standard conditions. In turn, CK biosynthesis is increased at synthesis sites via activation of AtIPT genes. Here, HvCKX2 activation resulted in up-regulation of AtIPT3 and AtIPT7, two of the three major AtIPT genes (Takei et al., 2004), of which AtIPT3 is the most prominent in shoots and the second most abundant in roots.
Local increases in CK contents might at least partially explain the similarities in responses to HvCKX2 and ipt activation observed for 42 of the identified proteins. Similarly, both ipt activation and CK treatment can result in increases in AtCKX transcripts (Hoth et al., 2003; Rashotte et al., 2003; Brenner et al., 2005), which might lead to a local drop in CK content. Thus, local reductions in CK activity may follow ipt activation, which might, likewise, contribute to the similarity in observed responses to HvCKX2 and ipt activation. In this light, the apparent conflicts in the results of salinity stress experiments could be interpreted as follows. Salinity stress may be accompanied by an increase in CK biosynthesis. Rates of CK biosynthesis increase and CK signalling outputs are adjusted in plants with increased CKX and IPT activities, via mechanisms that are used to cope with environmental stresses. Such mechanisms would also explain the beneficial effects of exogenous CK application to wheat plants under high salinity conditions (Gadallah, 1999).
Conclusion
The comparative proteome- and metabolome-wide analysis of molecular events triggered by induced reductions and increases in bulk CK levels resulted in identification of numerous novel CK-responsive proteins and metabolites, including several that are potential reporters of up- and down-regulation of CK activity in planta. The data obtained fundamentally deepen our understanding of the roles CK in thioredoxin signalling and ribosome biogenesis, as well as cross-talk between CK and stress-related hormones. Further, the comparative analysis provided novel indications that CK homeostatic mechanisms may explain significant similarities in phenotypes resulting from reductions and increases in bulk CK levels, inter alia the highly similar responses to salinity stress of plants with both increased and decreased CK levels. The findings also highlight previously unrecognized challenges facing attempts to construct transgenic plants with improved temporal and spatial targeting of CK metabolism genes in order to enhance key agricultural traits including yield improvements and stress tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Schematic diagram of pOpOn2.1::gHvCKX2.
Figure S2. Time course of changes in active cytokinin contents in CaMV35S>GR>HvCKX2 seedlings following HvCKX2 activation.
Figure S3. Time course of change in CK conjugate contents in CaMV35S>GR>HvCKX2 seedlings following HvCKX2 activation.
Figure S4. Comparison of cytokinin depletion in CaMV35S>GR>HvCKX2 and constitutive 35S:AtCKX transgenics, and the quadruple atipt1 3 5 7 mutant.
Figure S5. Effects of HvCKX2 activation on the proteome of Arabidopsis seedlings.
Figure S6. Subcellular distributions of the differentially regulated proteins according to predictions and SUBA experimental data.
Figure S7. Ribosomal proteins that responded to HvCKX2 and ipt activation.
Figure S8. Protein–protein interaction network constructed using STRING.
Figure S9. Expression of ipt is evident, but not detectable at the protein level.
Table S1. 2-DE-based analysis.
Table S2. LC-MS proteome analysis: differentially regulated proteins.
Table S3. LC-MS proteome analysis: list of all identified proteins.
Table S4. GC-MS metabolome analysis.
Table S5. BioMaps results.
Table S6. Known loss-of-function mutant phenotypes for the differentially regulated proteins found by 2-DE and LC-MS profiling.
Table S7. Overlaps of cytokinin-responsive proteins and transcripts detected here and in previous proteomic and transcriptomic analyses of hormone action in Arabidopsis.
Table S8. Detectability of HvCKX2 and ipt in the LC-MS data.
Table S9. Steady-state levels of transcripts of 10 genes involved in CK metabolism and signalling by RT–qPCR 48h after HvCKX2 activation.
Methods S1. Overview of the experiments and supplementary Materials and methods
Acknowledgements
We thank Professor Petr Galuszka for critical comments on the manuscript, Dr Přemysl Souček for his help with selecting CaMV35S>GR>HvCKX2 line 13, and Dr Ian Moore for the CaMV35S>GR>ipt line pOpBK-ipt 11 seeds and pOpOn2.1 binary vector. This work was supported by grants 206/09/2062 and P305/12/2144 (Czech Science Foundation), MSM0021620858 (PJ; MEYS CR), ERA Net Patho Net (WH), and funds from the ERDF for ‘CEITEC–Central European Institute of Technology’ (CZ.1.05/1.1.00/02.0068). Access to the MetaCentrum computing facilities provided under the Projects of Large Infrastructure for Research, Development, and Innovations program (LM2010005), funded by the Ministry of Education, Youth, and Sports of the Czech Republic is highly appreciated. The Centre of the Region Haná for Biotechnological and Agricultural Research (grant no. ED0007/01/01) and the IGA (grant no. PrF_2013_023) are also gratefully acknowledged. M.S. was supported in part by the Operational Program Education for Competitiveness - European Social Fund (Project CZ.1.07/2.3.00/20.0165).
References
- Argueso CT, Ferreira FJ, Kieber JJ. 2009. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant, Cell and Environment 32, 1147–1160 [DOI] [PubMed] [Google Scholar]
- Baginsky S, Hennig L, Zimmermann P, Gruissem W. 2010. Gene expression analysis, proteomics, and network discovery. Plant Physiology 152, 402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. 2011. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana . The Plant Cell 23, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. 1993. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes Rendus de l Academie des Sciences Paris, Sciences de la vie/Life Sciences 316, 1194–1199 [Google Scholar]
- Benková E, Witters E, Van Dongen W, Kolář J, Motyka V, Brzobohatý B, Van Onckelen HA, Macháčková I. 1999. Cytokinins in tobacco and wheat chloroplasts: occurrence and changes due to light/dark treatment. Plant Physiology 121, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL. 1964. The problem of halting enzyme action when extracting plant tissues. Analytical Biochemistry 9, 431–442 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. 2012. Gene regulation by cytokinin in Arabidopsis. Frontiers in Plant Science 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. 2005. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. The Plant Journal 44, 314–333 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Schmulling T. 2012. Transcript profiling of cytokinin action in Arabidopsis roots and shoots discovers largely similar but also organ-specific responses. BMC Plant Biology 12, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohatý B, Morre I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. 1993. Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262, 1051–1054 [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Morre I, Palme K. 1994. Cytokinin metabolism: implications for regulation of plant growth and development. Plant Molecular Biology 26, 1483–1497 [DOI] [PubMed] [Google Scholar]
- Černý M, Doubnerová V, Müller K, Ryšlavá H. 2010. Characterization of phosphoenolpyruvate carboxylase from mature maize seeds: properties of phosphorylated and dephosphorylated forms. Biochimie 92, 1362–1370 [DOI] [PubMed] [Google Scholar]
- Černý M, Dyčka F, Bobál’ová J, Brzobohatý B. 2011. a Early cytokinin response proteins and phosphoproteins of Arabidopsis thaliana identified by proteome and phosphoproteome profiling. Journal of Experimental Botany 62, 921–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černý M, Skalák J, Kurková B, Babuliaková E, Brzobohatý B. 2011. b Using a commercial method for Rubisco immunodepletion in analysis of plant proteome. Chemické listy 105, 640–642 [Google Scholar]
- Chen R, Binder BM, Garrett WM, Tucker ML, Chang C, Cooper B. 2011. Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Molecular BioSystems 7, 2637–2650 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W, Weckwerth W. 2010. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. The Plant Journal 63, 1–17 [DOI] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M. 1994. A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiology 104, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG. 1994. The role of hormones in apical dominance—new approaches to an old problem in plant development. Physiologia Plantarum 90, 230–237 [Google Scholar]
- Craft J, Šámalová M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. 2005. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. The Plant Journal 41, 899–918 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. 2000. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiology 124, 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, De Vienne D, Zivy M, Thiellement H. 1986. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7, 52–54 [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P. 2011. Evolution of cytokinin biosynthesis and degradation. Journal of Experimental Botany 62, 2431–2452 [DOI] [PubMed] [Google Scholar]
- Gadallah M. 1999. Effects of kinetin on growth, grain yield and some mineral elements in wheat plants growing under excess salinity and oxygen deficiency. Plant Growth Regulation 27, 63–74 [Google Scholar]
- Galuszka P, Frébort I, Šebela M, Sauer P, Jacobsen S, Peč P. 2001. Cytokinin oxidase or dehydrogenase? Mechanism of cytokinin degradation in cereals. European Journal of Biochemistry 268, 450–461 [DOI] [PubMed] [Google Scholar]
- Galuszka P, Frébortová J, Werner T, Yamada M, Strnad M, Schmülling T, Frébort I. 2004. Cytokinin oxidase/dehydrogenase genes in barley and wheat: cloning and heterologous expression. European Journal of Biochemistry 271, 3990–4002 [DOI] [PubMed] [Google Scholar]
- Galuszka P, Popelková H, Werner T, Frébortová J, Pospíšilová H, Mik V, Köllmer I, Schmülling T, Frébort I. 2007. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. Journal of Plant Growth Regulation 26, 255–267 [Google Scholar]
- Ghanem ME, Albacete A, Smigocki AC, et al. 2011. Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) plants. Journal of Experimental Botany 62, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LP. 2012. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends in Plant Science 17, 172–179 [DOI] [PubMed] [Google Scholar]
- Hay S, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. 2007. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Research 35, D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W, Wienkoop S. 2010. Spectral counting robust on high mass accuracy mass spectrometers. Rapid Communications in Mass Spectrometry 24, 3609–3614 [DOI] [PubMed] [Google Scholar]
- Holst K, Schmülling T, Werner T. 2011. Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. Journal of Plant Physiology 168, 1328–1334 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H. 2011. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. The Plant Journal 65, 724–736 [DOI] [PubMed] [Google Scholar]
- Hoth S, Ikeda Y, Morgante M, Wang X, Zuo J, Hanafey MK, Gaasterland T, Tingey SV, Chua N. 2003. Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Letters 554, 373–380 [DOI] [PubMed] [Google Scholar]
- Hradilová J, Malbeck J, Brzobohatý B. 2007. Cytokinin regulation of gene expression in the AHP gene family in Arabidopsis thaliana . Journal of Plant Growth Regulation 26, 229–244 [Google Scholar]
- Hradilová J, Rehulka P, Rehulková H, Vrbová M, Griga M, Brzobohatý B. 2010. Comparative analysis of proteomic changes in contrasting flax cultivars upon cadmium exposure. Electrophoresis 31, 421–431 [DOI] [PubMed] [Google Scholar]
- Jung C, Lyou SH, Yeu S, Kim MA, Rhee S, Kim M, Lee JS, Choi YD, Cheong J. 2007. Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana . Plant Cell Reports 26, 1053–1063 [DOI] [PubMed] [Google Scholar]
- Jung C, Yeu S, Koo Y, Kim M, Choi Y, Cheong J. 2007. Transcript profile of transgenic Arabidopsis constitutively producing methyl jasmonate. Journal of Plant Biology 50, 12–17 [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, et al. 2010. VirtualPlant: a software platform to support systems biology research. Plant Physiology 152, 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal R, Saini R, Zhao PX. 2010. Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiology 154, 36–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. 2005. Combinatorial microarray analysis revealing arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant and Cell Physiology 46, 339–355 [DOI] [PubMed] [Google Scholar]
- Kiran NS, Polanská L, Fohlerová R, et al. 2006. Ectopic over-expression of the maize β-glucosidase Zm-p60.1 perturbs cytokinin homeostasis in transgenic tobacco. Journal of Experimental Botany 57, 985–996 [DOI] [PubMed] [Google Scholar]
- Kowalska M, Galuszka P, Frébortová J, Šebela M, Béres T, Hluska T, Šmehilová M, Bilyeu KD, Frébort I. 2010. Vacuolar and cytosolic cytokinin dehydrogenases of Arabidopsis thaliana: heterologous expression, purification and properties. Phytochemistry 71, 1970–1978 [DOI] [PubMed] [Google Scholar]
- Kristoffersen P, Brzobohatý B, Höhfeld I, Bako L, Melkonian M, Palme K. 2000. Developmental regulation of the maize Zm-p60.1 gene encoding a β-glucosidase located to plastids. Planta 210, 407–415 [DOI] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. 2010. Metabolism and long-distance translocation of cytokinins. Journal of Integrative Plant Biology 52, 53–60 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladrera R, Arrese-Igor C, González EM. 2007. Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiology 144, 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wong WS, Zhu L, Guo HW, Ecker J, Li N. 2009. Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9, 1646–1661 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Meinke D. 2012. A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiology 158, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochmanová G, Zdráhal Z, Konečná H, Koukalová Š, Malbeck J, Souček P, Válková M, Kiran NS, Brzobohatý B. 2008. Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: a proteomic analysis. Journal of Experimental Botany 59, 3705–3719 [DOI] [PubMed] [Google Scholar]
- Luo T, Fan T, Liu Y, Rothbart M, Yu J, Zhou S, Grimm B, Luo M. 2012. Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiology 159, 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, et al. 2008. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant and Cell Physiology 49, 1135–1149 [DOI] [PubMed] [Google Scholar]
- Moore I, Samalova M, Kurup S. 2006. Transactivated and chemically inducible gene expression in plants. The Plant Journal 45, 651–683 [DOI] [PubMed] [Google Scholar]
- Morgenthal K, Wienkoop S, Wolschin F, Weckwerth W. 2007. Integrative profiling of metabolites and proteins: improving pattern recognition and biomarker selection for systems level approaches. Methods in Molecular Biology 358, 57–75 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Matsushima R, Hara-Nishimura I. 2005. Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana . Plant and Cell Physiology 46, 1140–1148 [DOI] [PubMed] [Google Scholar]
- Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. 2011. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. 2006. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475 [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Le DT, Watanabe Y, Matsui A, Tanaka M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K, Tran LP. 2012. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 7, e32124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, et al. 2011. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. The Plant Cell 23, 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Doležal K, Strnad M. 2008. Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry , 69, 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák J, Pavlů J, Novák O, Nožková V, Špundová M, Hlavinka J, Koukalová Š, Skalák J, Černý M, Brzobohatý B. 2013. High cytokinin levels induce a hypersensitive-like response in tobacco. Annals of Botany 112, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Tarkowski P, Tarkowská D, Doležal K, Lenobel R, Strnad M. 2003. Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass spectrometry. Analytica Chimica Acta 480, 207–218 [Google Scholar]
- Pačes V, Werstiuk E, Hall RH. 1971. Conversion of N-(Delta-isopentenyl)adenosine to adenosine by enzyme activity in tobacco tissue. Plant Physiology 48, 775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C. 2006. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223, 965–974 [DOI] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S. 2010. The molecular basis of cytokinin function. Current Opinion in Plant Biology 13, 21–26 [DOI] [PubMed] [Google Scholar]
- Polanská L, Vičánková A, Nováková M, Malbeck J, Dobrev PI, Brzobohatý B, Vaňková R, Macháčková I. 2007. Altered cytokinin metabolism affects cytokinin, auxin, and abscisic acid contents in leaves and chloroplasts, and chloroplast ultrastructure in transgenic tobacco. Journal of Experimental Botany 58, 637–649 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. 2007. Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. The Plant Journal 50, 381–390 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ. 2003. Expression profiling of cytokinin action in Arabidopsis. Plant Physiology 132, 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. 2006. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. The Plant Cell 18, 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg D, Foster GL. 1940. A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. Journal of Biological Chemistry 133, 737–744 [Google Scholar]
- Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, Kang B, Bressan RA, Raikhel NV. 2010. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. The Plant Cell 22, 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449 [DOI] [PubMed] [Google Scholar]
- Šámalová M, Brzobohatý B, Moore I. 2005. pOp6/LhGR: a stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. The Plant Journal 41, 919–935 [DOI] [PubMed] [Google Scholar]
- Shani E, Ben-Gera H, Shleizer-Burko S, Burko Y, Weiss D, Ori N. 2010. Cytokinin regulates compound leaf development in tomato. The Plant Cell 22, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigocki AC, Owens LD. 1988. Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proceedings of the National Academy of Sciences, USA 85, 5131–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Lu H, Greenberg JT. 2004. Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. The Plant Cell 16, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. 2011. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. The Plant Journal 67, 157–168 [DOI] [PubMed] [Google Scholar]
- Strnad M. 1997. The aromatic cytokinins. Physiologia Plantarum 101, 674–688 [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. 2004. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant and Cell Physiology 45, 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tran LP, Shinozaki K, Yamaguchi-Shinozaki K. 2010. Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signaling and Behavior 5, 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N, Kojima M, Suzuki K, Sakakibara H. 2012. Agrobacterium tumefaciens tumor morphology root plastid localization and preferential usage of hydroxylated prenyl donor is important for efficient gall formation. Plant Physiology 159, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyroubalová Š, Václavíková K, Turečková V, Novák O, Šmehilová M, Hluska T, Ohnoutková L, Frébort I, Galuszka P. 2009. Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiology 151, 433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MA, Roberts DR, Dumbroff EB. 1988. Effects of cytokinin and light on polyamines during the greening response of cucumber cotyledons. Plant and Cell Physiology 29, 201–205 [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng Y, Wu Y. 2011. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. The Plant Journal 68, 249–261 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell 15, 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. 2001. Regulation of plant growth by cytokinin. Proceedings of the National Academy of Sciences, USA 98, 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. 2010. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. The Plant Cell 22, 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C. 2005. A high-throughput inducible RNAi vector for plants. Plant Biotechnology Journal 3, 583–590 [DOI] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng Z. 2005. Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiology 139, 1350–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, He H, Tong S, Zhang W, Wang J, Li X, Yang Y. 2009. Voltage-dependent anion channel 2 of Arabidopsis thaliana (AtVDAC2) is involved in ABA-mediated early seedling development. International Journal of Molecular Sciences 10, 2476–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. 2008. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3, e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BS, Rönnberg E, Viitanen L, Salminen TA, Lundgren K, Moritz T, Edqvist J. 2008. Arabidopsis sterol carrier protein-2 is required for normal development of seeds and seedlings. Journal of Experimental Botany 59, 3485–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.