Abstract
To investigate N metabolism of two contrasting Populus species in acclimation to low N availability, saplings of slow-growing species (Populus popularis, Pp) and a fast-growing species (Populus alba × Populus glandulosa, Pg) were exposed to 10, 100, or 1000 μM NH4NO3. Despite greater root biomass and fine root surface area in Pp, lower net influxes of NH4 + and NO3 – at the root surface were detected in Pp compared to those in Pg, corresponding well to lower NH4 + and NO3 – content and total N concentration in Pp roots. Meanwhile, higher stable N isotope composition (δ15N) in roots and stronger responsiveness of transcriptional regulation of 18 genes involved in N metabolism were found in roots and leaves of Pp compared to those of Pg. These results indicate that the N metabolism of Pp is more sensitive to decreasing N availability than that of Pg. In both species, low N treatments decreased net influxes of NH4 + and NO3 –, root NH4 + and foliar NO3 – content, root NR activities, total N concentration in roots and leaves, and transcript levels of most ammonium (AMTs) and nitrate (NRTs) transporter genes in leaves and genes involved in N assimilation in roots and leaves. Low N availability increased fine root surface area, foliar starch concentration, δ15N in roots and leaves, and transcript abundance of several AMTs (e.g. AMT1;2) and NRTs (e.g. NRT1;2 and NRT2;4B) in roots of both species. These data indicate that poplar species slow down processes of N acquisition and assimilation in acclimation to limiting N supply.
Key words: Gene expression, glutamate synthase, glutamine synthetase, net flux, nitrate reductase, nitrite reductase, plasma membrane H#x002B;-ATPase, poplar, stable carbon isotope.
Introduction
As woody crops, forest plantations hold a great potential for the pulp and paper industry, carbon mitigation, and biomass production for biofuels (Luo et al., 2006; Luo and Polle, 2009; Novaes et al., 2009; Studer et al., 2011). Plantations of some fast-growing tree species such as Populus spp. have been widely established in recent years (Weih, 2004; Polle and Douglas, 2010; Rennenberg et al., 2010). As a riparian species, Populus in its natural habitat is supplied with sufficient nitrogen (N) derived from intensive N-fertilization application in agriculture (Rennenberg et al., 2010; Koyama and Kielland, 2011). Due to the high demand of fertile soil for agriculture, however, poplar plantations have often been established on marginal lands where soil N is limiting (Rennenberg et al., 2010; Bilodeau-Gauthier et al., 2011). In this context, it is of particular importance to select poplar species with tolerance to low N availability.
The genus Populus contains about 30–40 species which may differ in N metabolism (Calfapietra et al., 2007; Finzi et al., 2007; Euring et al., 2012; Li et al., 2012). For instance, Populus tremula × Populus tremuloides is a species that often occurs on nutrient-poor soil. The growth and wood properties of this species are more responsive to different N levels than those of Populus trichocarpa which is adapted to fluctuating N supply (Euring et al., 2012). These results highlight that it is essential to better understand the distinctness of N metabolism in different poplar species in order to select poplars with tolerance to low N availability.
Although little information is available on responses of N metabolism in different woody plants to low N availability, distinct N metabolism has been reported in different herbaceous species or genotypes in response to N deficiency (Lawlor et al., 1987a , 1988, 1989; Lawlor, 2002; Hirel et al., 2007; Shen et al., 2013). For instance, some maize varieties displayed a higher capacity to absorb and utilize N than the others (Banziger et al., 1997; Toledo Marchado and Silvestre Fernandes, 2001). In cereal crops, earlier studies demonstrated that N deficiency in soil often led to altered root length and branching and decreased soluble protein concentration and photosynthetic activity (Maizlich et al., 1980; Lawlor et al., 1987b , 1989; Lawlor, 2002; Hirel et al., 2007). Plants with tolerance to low N are often associated with higher photosynthetic N use efficiency (PNUE), greater root length, and surface area per volume of soil (Lawlor, 2002; Hirel et al., 2007; Shen et al., 2013). However, the physiological and molecular mechanisms remain to be elucidated for plant species differing in tolerance to low N supply.
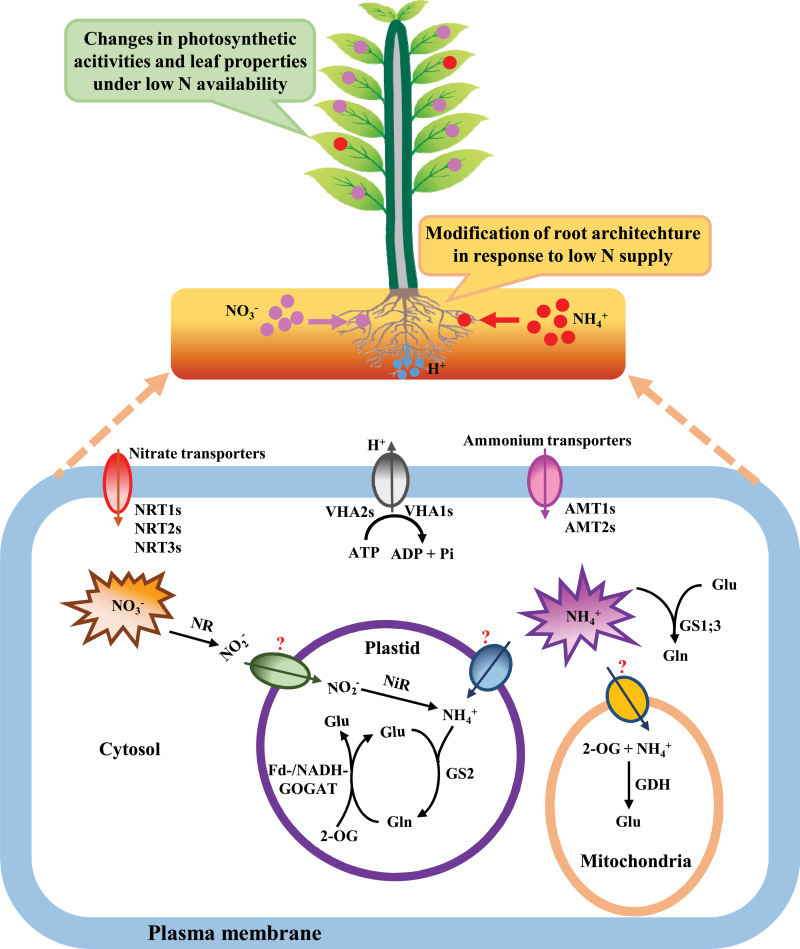
In plants, the N metabolism process involves uptake, transport, assimilation, and utilization for amino acid biosynthesis and ultimately for growth (Nunes-Nesi et al., 2010). Each of these steps may be regulated slightly different, leading to differences in N metabolism and performance of plants with distinct ecological requirements. In herbaceous plants, the N metabolism processes are well documented (Fig. 1), which may serve as a conceptual model to address N metabolism of different poplar species in response to low N availability.
Fig. 1.
A conceptual model of N metabolism in plants. In the uptake process, NH4 + and NO3 – enter the cytosol via ammonium (AMTs) and nitrate (NRTs) transporters, respectively, coupled with plasma membrane H+-ATPases (VHAs). After uptake in roots, NH4 + and NO3 – can be translocated to leaves or other parts of the plant. In the assimilation process, NO3 – is converted to NH4 + by the cytosolic nitrate reductase (NR) and the plastidic/chloroplastic nitrite reductase (NiR). Subsequently, NH4 + can be assimilated to glutamine (Gln) catalysed by glutamine synthetase (GS) isoenzymes either in the plastid or the cytosol. The Gln in the plastid with 2-oxoglutarate (2-OG) can be further converted to glutamate (Glu) by Fd- or NADH-dependent glutamate synthase (Fd/NADH-GOGAT). Additionally, in the mitochondrion, NH4 + can be assimilated to Glu with glutamate dehydrogenase (GDH). The synthesized N compounds provide precursors for amino acids, proteins, and other N-containing metabolites which can be utilized by plant growth. At the cellular level, N metabolism in plants can be affected by external low N availability. At the plant level, root characteristics, photosynthetic activity, and leaf properties can be altered in response to low N supply (this figure is available in colour at JXB online).
In the N uptake process, NH4 + and nitrate NO3 – in soil solution are the two major inorganic N forms for plant absorption. Although both ions can be used by plants, the energetic, biochemical, and molecular features of NH4 + and NO3 – are different for metabolism, leading to distinct net fluxes of both ions at the root surfaces and NH4 + or NO3 – preference of plants (Jackson et al., 2008; Patterson et al., 2010). Using 15N labelling, it was demonstrated that some woody plants prefer NH4 + (Rennenberg et al., 2009, 2010). Additionally, net fluxes of NH4 + and/or NO3 – at the root surfaces have been investigated using ion-selective microelectrodes in herbaceous and woody plants (Plassard et al., 2002; Gobert and Plassard, 2007; Hawkins et al., 2008; Hawkins and Robbins, 2010; Alber et al., 2012; Luo et al., 2013), providing a better understanding of electrophysiological processes of NH4 + and NO3 – acquisition. The net fluxes of both NH4 + and NO3 – are coupled with the activities of plasma membrane (PM) H+-ATPases in fine roots of Populus popularis (Luo et al., 2013). However, comparisons of fluxes of NH4 + and NO3 – at the root surface of poplar species with large differences in growth are missing.
The fluxes of NH4 + and NO3 – are mediated by various transporters for ammonium (AMTs) and nitrate (NRTs) (Rennenberg et al., 2010; Xu et al., 2012). Some AMTs and NRTs have been functionally elucidated in Arabidopsis thaliana (Wang et al., 2012; Xu et al., 2012). For instance, transcript abundance of AtAMT1;1 is strongly increased by N starvation and reduced upon NH4 + supply in Arabidopsis roots (Engelsberger and Schulze, 2012). Some NRT members such as AtNRT1;1, AtNRT1;2, AtNRT2;1, and AtNRT3;1 play pivotal roles in NO3 – uptake and signalling (Yong et al., 2010; Kotur et al., 2012; Wang et al., 2012). In the genome of P. trichocarpa, 14 putative AMTs have been documented (Tuskan et al., 2006; Couturier et al., 2007), whereas less is known about the NRT members (Plett et al., 2010; Rennenberg et al., 2010; Li et al., 2012). Several studies showed that transcripts of putative poplar transporters (e.g. AMT1;2, AMT1;6, AMT2;1, NRT1;1, NRT1;2, NRT2;4B, NRT2;4C, NRT3.1B, and NRT3.1C) were responsive to environmental fluctuations under non-limiting N conditions (Selle et al., 2005; Couturier et al., 2007; Dluzniewska et al., 2007; Ehlting et al., 2007; Plett et al., 2010; Li et al., 2012). However, it remains unknown how transcriptional regulation of these transporters responds to limiting N supply in different poplar species.
After uptake into the roots, a large amount of NH4 + can be assimilated locally and the remainder is translocated to leaves or other parts of the plant, whereas only a limited amount of NO3 – is assimilated in roots and most NO3 – is transported to leaves (Black et al., 2002; Xu et al., 2012). In the assimilation process, NO3 – is converted to NH4 + by nitrate reductase (NR) and nitrite reductase (NiR) (Xu et al., 2012). Subsequently, NH4 + can be assimilated to glutamine catalysed by glutamine synthetase (GS) (Castro-Rodriguez et al., 2011; Coleman et al., 2012). The formation of glutamate requires glutamine and 2-oxoglutarate in a reaction catalysed by glutamate synthase (GOGAT) (McAllister et al., 2012). Additionally, glutamate can be synthesized by glutamate dehydrogenase (GDH) under consumption of NH4 + and 2-oxoglutarate (McAllister et al., 2012). Very little is known about the response of these enzymes to low N supply in different poplar species.
Although forest plantations often grow on nutrient-poor soils (Johnson, 2006; Rennenberg et al., 2009), N-related studies in trees mainly addressed the effects of fertilization but less to uncover the responses to N-limitation (Cooke and Weih, 2005; Rennenberg et al., 2009, 2010; Lukac et al., 2010; Millard and Grelet, 2010). Recently, we found that growth, carbon, and N physiology, and wood properties of the fast-growing Populus alba × Populus glandulosa (Pg), which generally grows on relatively fertile soils, displays a stronger responsiveness to N-fertilization than the slow-growing P. popularis (Pp) which is often found on nutrient-deficient soils (Li et al., 2012). The differential responses are ascribed to prioritized resource allocation to the leaves and accelerated N physiological processes in the fast-growing Pg under higher N supply levels. However, it remains unknown how N metabolism processes and key components involved in these processes of Pp and Pg respond to external low N availability.
This study exposed Pp and Pg to low N levels. Measure ments of morphological (root characteristics), physiological (e.g. photosynthesis, net fluxes of NH4 +, NO3 –, and H+, accumulation of NH4 +, NO3 –, and NO2 –, total N concentration, and δ15N), and molecular (transcript levels of representative genes involved in N metabolism) parameters known to be important for acclimation to low N availability were conducted. Furthermore, multivariate analysis was applied to dissect the importance of parameters as contributors to the acclimation of N metabolism in both poplar species to low N supply. The following questions were specifically addressed: (i) do root morphology, photosynthesis, and N metabolism of Pp and Pg display different response patterns to limiting N supply? and (ii) what are the physiological and transcriptional regulation mechanisms of Pp and Pg in acclimation to low N availability?
Materials and methods
Plant cultivation and N treatment
Cuttings of the slow-growing P. popularis (Pp) and the fast-growing P. alba × P. glandulosa (Pg) were rooted as described previously (Li et al., 2012), and planted in pots (10 l) filled with fine sand. Plants were cultivated in a greenhouse (natural light, day/night 25/20 °C, 75% relative humidity) and provided with 50ml Long Ashton (LA) nutrient solution, which contains 1000 μM NH4NO3 (Dluzniewska et al., 2007) every other day. After 6 weeks, plants with similar height (c.60cm) were selected for further study. The root systems of selected plants were carefully washed with tap water. Plants of each species were divided into three groups with 18 plants for each group. Subsequently, plants of three groups from each species were cultivated in hydroponics with modified LA solution (0.5mM KCl, 0.9mM CaCl2, 0.3mM MgSO4, 0.6mM KH2PO4, 42 μM K2HPO4, 10 μM Fe-EDTA, 2 μM MnSO4, 10 μM H3BO3, 7 μM Na2MoO4, 0.05 μM CoSO4, 0.2 μM ZnSO4, and 0.2 μM CuSO4) containing 10, 100, or 1000 μM NH4NO3, respectively, and the nutrient solution was adjusted to pH 5.5. The LA solution was refreshed every 2 days. In the greenhouse, the position of each plant was randomly assigned and altered once a week. At the beginning of the N treatment, the apex of each plant was marked by using a laboratory marker to distinguish the shoots formed during the N treatment. After hydroponic cultivation with N treatments for 3 weeks from 20 June to 10 July, 12 plants of each group were used for gas exchange determination prior to harvest and the remaining six plants of each group were used for measurements of net fluxes of NH4 + and NO3 –.
Gas exchange and harvesting
The gas exchange of three mature leaves (leaf plastochron index = 8–10) formed during N treatment was determined for each plant. Net photosynthetic rates (A), stomatal conductance (g s), and transpiration rates were determined with a portable photosynthesis system (Li-Cor-6400, Li-Cor, Lincoln, NE, USA) and an attached LED light source (6400–02) as described by He et al. (2011). The instantaneous photosynthetic N use efficiency (PNUEi) was calculated based on A, foliar N concentration, and specific leaf area, as suggested by Li et al. (2012). As a stored photosynthate, starch concentration in harvested root and leaf tissues (see below) was analysed as suggested by He et al. (2013).
Since the daily rhythm of plants can affect physiological and molecular processes (Wilkins et al., 2009), the harvest was performed between 9:00 and 12:00. For harvest, the root system of each plant was well washed with corresponding LA solution containing 10, 100, or 1000 μM NH4NO3. Subsequently, the root system was wrapped in lab tissue paper to remove water on the root surface. Length of main root and fresh weight of roots were recorded. A part of roots (c.2g) of each plant was excised from the root system, scanned, and analysed by a WinRHIZO root analyser system (WinRHIZO version 2012b, Regent Instruments Canada, Montreal, Canada) as described by Luo et al. (2013). Leaves formed during the N treatment (above the mark on the stem) were harvested. Leaf discs for determination of specific leaf area were also collected and specific leaf area was calculated according to the method of Cao et al. (2012). Harvested roots or leaves were wrapped with tinfoil and immediately frozen in liquid N. Root or leaf samples were ground into fine powder in liquid N with a mortar and pestle and stored at –80 °C. Frozen powder (c.100mg) from roots or leaves of each plant was dried at 60 °C for 72h to determine the fresh-to-dry-mass ratio. For further biochemical analysis, equal weight of fine powder from roots or leaves of two plants within each group was combined to form a well-mixed sample.
Analysis of net fluxes of NH4 +, NO3 –, and H+
To analyse net fluxes of NH4 +, NO3 –, and H+ at the root surface, three white fine roots (c.1.5mm in diameter) were randomly selected from the root system of each plant. Net fluxes of these ions were measured non-invasively using scanning ion-selective electrode technique (SIET, SIET system BIO-003A, Younger USA Science and Technology, Falmouth, MA, USA) by Xuyue Science and Technology (Beijing, China). The SIET system and its application in net ion flux detection were described in detail (Li et al., 2010; He et al., 2011; Luo et al., 2013). The ion-selective microelectrode with 2–4 μm aperture was manufactured and silanized with a backfilling solution and an ion-selective liquid cocktail.
To find out the positions along the root where the maximal net fluxes of NH4 + and NO3 – take place, a preliminary experiment was performed using plants treated with 100 μM NH4NO3 by taking an initial measurement at the root apex, followed by measurements at 300 μm intervals (in the region of 0–2100 μm) or 5mm intervals (in the region of 5–30mm) along the root tip (Supplementary Fig. S1A, available at JXB online). Ion gradients near the root surface were measured by moving the ion-selective microelectrode between two positions (c.30 μm in distance) in a perpendicular direction to the root axis. The recording rate for these ion fluxes was one reading per 6 s and ion flux was recorded at each measurement point for 10min.
For the positions where the maximal net fluxes of NH4 + and NO3 – occur in roots, net fluxes of NH4 +, NO3 –, and H+ were further investigated in detail. A fine root was transferred to a Petri dish containing 10ml measuring solution (0.1mM KCl, 0.1mM CaCl2, pH 5.5) with 10, 100, or 1000 μM NH4NO3 according to the N treatment of the selected root, and equilibrated for 20min. Prior to the measurement, the root was transferred to a new Petri dish containing fresh measuring solution and net NH4 + fluxes were monitored for 10min. Afterwards, net NO3 – fluxes were measured for 10min in the same root and finally, net H+ fluxes were recorded for 10min in the root.
Isolation of the PM and measurement of PM H+-ATPase activity
PM vesicles of root cells were isolated according to the method of Sorgona et al. (2011) with minor modification. Briefly, fine powder of roots (c.2g) was homogenized with 3ml extraction solution containing 250mM sucrose, 10% (v/v) glycerol, 10mM glycerol-1-phosphate, 2mM MgSO4, 2mM EDTA, 2mM EGTA, 2mM ATP, 2mM DTT, 5.7% (w/v) choline chloride, 25mM 1,3-bis(tris (hydroxymethyl)-methyl-aminoethylether)propane (BTP, pH 7.6 with 2(N-morpholino)ethanesulphonic acid, MES), 1mM PMSF, and 20 μg ml–1 chimostatin at 4 °C. After centrifugation (12,700 g, 4 °C, 30min), the pellets were suspended over a 25/38% discontinuous sucrose gradient (5mM MES containing all protectants present in the extraction solution, pH 7.4). Afterwards, the gradient was centrifuged again (12,700 g, 4 °C, 60min). The pellets were resuspended in a medium containing 20% glycerol (v/v), 2mM EGTA, 2mM EDTA, 0.5mM ATP, 1mM PMSF, 2mM DTT, 20 μg ml–1 chimostatin, 5.7% choline chloride, and 5mM BTP buffered at pH 7.0 with MES and immediately frozen in liquid N and stored at –80 °C.
PM H+-ATPase activity was determined spectrophotometrically at 700nm as described by Sorgona et al. (2011) with minor modifications. In brief, assays were carried out at 30 °C in 0.5ml medium containing 30mM BTP/MES (pH 6.5), 5mM MgSO4, 50mM KCl, 4mM ATP, 0.6mM Na2MoO4, 100mM KNO3, 1.5mM NaN3, and 0.02% (w/v) polyxyethylene 20 cetyl ether, with or without 100 μM vanadate (an inhibitor of P-type H+-ATPase). The difference between these two activities was attributed to the PM H+-ATPase. Sodium azide and KNO3 were used as selective inhibitors of mitochondrial and tonoplast H+-ATPase, respectively (Zhu et al., 2009). The reaction was initiated by adding membrane vesicles (5–10 μg membrane protein) and stopped after 30min with a solution containing 2% (v/v) concentrated H2SO4, 5% (w/v) SDS, 0.7% Na2MoO4, and 10% ascorbic acid. After solubilizing the membrane vesicles with 0.5M NaOH (Gogstad and Krutnes, 1982), the total soluble protein was estimated according to Bradford (1976). PM H+-ATPase activity was expressed as that inhibited by 100 μM vanadate.
Determination of NH4 +, NO3 –, and NO2 – concentration
NH4 + concentration in roots and leaves was determined based on the Berthelot reaction (Brautigam et al., 2007; Luo et al., 2013). In brief, fine power (c.100mg) was homogenized in an extraction solution (1ml 100mM HCl and 500 μl chloroform). The extraction solution was centrifuged (10,000 g, 4 °C, 10min) after shaking for 15min at 4 °C. The aqueous phase was transferred to a 2ml tube with 50mg activated charcoal, mixed well, and centrifuged (12,000 g, 4 °C, 5min) again. NH4 + concentration in the supernatant was determined spectrophotometrically at 620nm.
NO3 – concentration in samples was analysed as suggested by Patterson et al. (2010). Fine powder (c.100mg) was extracted in 1ml deionized water at 45 °C for 1h. After centrifugation (5000 g, 20 °C, 15min), the supernatant was used for nitrate quantification. The supernatant (0.2ml) was mixed thoroughly with 0.8ml of 5% (w/v) salicylic acid in concentrated H2SO4. After incubation at room temperature for 20min, 19ml of 2M NaOH was added to raise the pH to above 12. The solution was cooled to room temperature before NO3 – concentration was determined spectrophotometrically at 410nm.
NO2 – concentration in samples was quantified as described by Ogawa et al. (1999). Frozen fine powder (c.100mg) was extracted by an extraction buffer containing 50mM TRIS-HCl (pH 7.9), 5mM cysteine, and 2mM EDTA. After centrifugation (10,000 g, 20 °C, 20min), 500 μl supernatant was mixed with 250 μl 1% sulphanilamide and 250 μl 0.02% N-(1-naphtyl)-ethylene-diamine dihydrochloride in 3.0M HCl. NO2 – concentration was quantified spectrophotometrically at 540nm.
Determination of enzyme activities
Activities of NR (EC 1.7.99.4) and NiR (EC 1.7.2.1) were determined in roots and leaves according to the methods of Ehlting et al. (2007) and Ogawa et al. (1999), respectively. Briefly, frozen powder (c.200mg) was extracted in an extraction buffer (100mM HEPES-KOH (pH 7.5), 5mM Mg-acetate, 5mM DTT, 1mM EDTA, 0.5mM PMSF, 20mM FAD, 5mM Na2MoO4, 10% (v/v) glycerin, 1% (w/v) polyvinyl polypyrrolidone, 0.5% BSA, 0.1% (v/v) TritonX-100, and either 25mM leupeptine for leaves or 25mM chymostatine for roots). The crude extract was used for NR and NiR assays.
For NR, the extract was added to the reaction mixture (100mM HEPES-KOH (pH 7.5), 6.0mM KNO3, 6.0mM EDTA, 0.6mM NADH, 12mM FAD, 6mM Na2MoO4, 3mM DTT, and either 25mM leupeptine for leaves or 25mM chymostatine for roots) at 25 °C. The reaction was terminated after 20min by adding 0.6M Zn-acetate and 0.25mM phenazinemethosulphate. NO2 – formation in the solution was determined as in the assay of NO2 – concentration.
For NiR, 500 μl supernatant from NO2 – concentration assay was concentrated with a Amicon Ultra 10K filter (Millipore, Billerica, USA) to reduce nitrate ions. The concentrated supernatant was mixed with 500 μl solution containing 50mM TRIS-HCl (pH 7.5), 1mM cysteine, and 2mM EDTA. The NiR activity was determined by following the reduction of NO2 – in the assay. The assay solution contained 0.5mM NaNO2, 1mM methyl viologen, and the extract. The reaction was started by adding the reagent (0.12M Na2S2O4, 0.2M NaHCO3), incubated at 30 °C for 60min, and terminated by vigorous vortex until the colour of the methyl viologen disappeared completely. After adding 1M Zn-acetate, the mixture was centrifuged (10,000 g, 25 °C, 10min). The residual NO2 – in the reaction solution was determined as in the assay of NO2 – concentration.
GS (EC 6.3.1.2) activity was analysed spectrophotometrically as proposed by Wang et al. (2008). Frozen fine powder was homogenized at 4 °C in 50mM TRIS-HCl extraction solution (pH 8.0) containing 2mM MgCl2, 2mM DTT, and 0.4M sucrose. After centrifugation (15,000 g, 4 °C, 20min), the supernatant was used for GS activity assay. The assay solution contained 0.35ml of 40mM ATP and 0.8ml of 0.1M TRIS-HCl buffer (pH 7.4) with 20mM Na-glutamate, 80mM MgSO4, 20mM cysteine, 2mM EGTA, and 80mM NH2OH. After adding the enzyme extract to the assay solution, the mixture was incubated (37 °C, 30min) and the incubation was stopped by addition of the reagent (0.37M FeCl3, 0.2M trichloroacetic acid, 0.6M HCl). After centrifugation (5000 g, 4 °C, 15min), the absorbance of the supernatant was recorded at 540nm. GS activity was expressed as 1 μmol γ-glutamyl hydroxamate formed per min.
Activities of GOGAT (EC 1.4.7.1) and GDH (EC 1.4.1.2) were assayed in roots and leaves based on the method of Lin and Kao (1996). Fine powder (c.100mg) was extracted with 10mM TRIS-HCl buffer (pH 7.6), 1mM MgC12, 1mM EDTA, and 1mM β-mercaptoethanol at 4 °C. After centrifugation (15,000 g, 4 °C, 30min), the supernatant was used for determination of enzyme activities. The GOGAT assay solution contained 0.2ml of 20mM l-glutamine, 25 μl of 0.1M 2-oxoglutarate, 50 μl of 10mM KCI, 0.1ml of 3mM NADH, and 0.25ml enzyme extract in a final volume of 1.5ml made up with 25mM TRIS-HCl buffer (pH 7.6). After addition of l-glutamine, the decrease in absorbance was recorded spectrophotometrically at 340nm. The GDH assay mixture contained 0.15ml of 0.1M 2-oxoglutarate, 0.15ml of 1M NH4Cl, 0.1ml of 3mM NADH, and 0.5ml of the enzyme extract in a final volume of 1.5ml made up with 0.2M TRIS-HCl buffer (pH 8.0). After addition of enzyme extract, the decrease in absorbance was monitored spectrophotometrically at 340nm.
Determination of total carbon and nitrogen and stable isotopes
Root samples and mature leaves used for gas exchange measurements were harvested for total C and N and stable isotope (13C and 15N) analysis. Total C and N concentration was determined according to the method of Luo and Polle (2009). The stable C isotope (13C) was analysed based on the protocol of Cao et al. (2012). Fine powder (c.50mg) was dried in an oven at 80 °C. The dried powder (c.0.8mg) was sealed under vacuum in a quartz tube with copper oxide and silver foil and combusted for at least 4h at 800–850 °C. The CO2 from the combustion tube was extracted and purified cryogenically. The isotopic ratio of the extracted CO2 was determined by an elemental analyser (NA 1110, CE Instruments, Rodano, Italy) and a mass spectrometer (Delta Plus, Finnigan MAT, Bremen, Germany) with an interface (Conflo III, Finnigan MAT, Bremen, Germany) according to the method of Werner et al. (1999). The 13C/12C ratio is expressed as parts per thousand deviation from the Pee Dee Belemnite standard. Carbon isotope composition (%) was calculated as δ13C = (Rsa – Rsd)/Rsd × 1000, where Rsa and Rsd are the ratios of 13C to 12C of the sample and the standard, respectively. The standard was referred to CO2 in air.
Stable N isotope composition (δ15N) was analysed similarly to 13C with minor modifications, according to the method of Yousfi et al. (2012). The standard was referred to N2 in air.
Determination of mineral nutrients, soluble sugars, soluble protein, and phenolics
Mineral elements in roots and leaves were analysed by an inductively coupled plasma-atomic emission spectrometer (Spectroflame, Spectro Analytical Instruments) based on the protocol of Heinrichs et al. (1986).
Concentrations of total soluble sugar in root and leaf tissues were analysed by the anthrone method of Yemm and Willis (1954) with minor modifications (He et al., 2013). The standard curve was established by using a serial of diluted solutions of glucose. The final absorbance of total soluble sugar and starch (expressed as glucose equivalent) in samples was determined at 620nm.
Soluble protein in plant materials was extracted and used for quantification (Bradford, 1976). Soluble phenolics were determined according to the method of Swain and Goldstein (1964) with themodification by Luo et al. (2008).
Analysis of transcript levels of genes involved in N uptake and assimilation
The transcriptional changes of genes implicated in N uptake and assimilation were analysed by reverse-transcription quantitative PCR (qPCR) based on the method (Li et al., 2012). Briefly, frozen fine powder of roots (c.100mg) and leaves (c.50mg) was used for total RNA isolation. Total RNA was isolated and purified with a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, USA) and trace genomic DNA was digested by DNase I (E1091, Omega Bio-Tek) attached to the RNA extraction kit. The lack of trace genomic DNA in total RNA was confirmed by a control PCR using total RNA as templates. Aliquots of 1 μg total RNA were used for first-strand cDNA synthesis using a PrimeScript RT reagent kit (DRR037S, Takara, Dalian, China) in a 20 μl reaction. In this reaction system, random primers and oligo dTs were added according to the manufacturer’s instructions. Real-time PCR was performed in a 20 μl reaction using 10 μl 2× SYBR Green Premix Ex Taq II (DRR081A, Takara), 2.5 μl cDNA, and 0.2 μl of 20mM primers (Supplementary Table S1) in an IQ5 Real Time System (Bio-Rad, Hercules, CA, USA). To ensure the primer specificity, PCR products were sequenced and aligned with homologues from P. trichocarpa and other model plants (Supplementary Fig. S2). Actin2/7 was used as a reference gene (Brunner et al., 2004). PCR was performed in triplicate together with a dilution series of the reference gene. The efficiencies of all PCR reactions were between 95 and 105% (Supplementary Table S1).
Statistical analysis
Net flux data were calculated by Mageflux version 1.0 attached to the SIET system (Xu et al., 2006). Statistical tests were performed with Statgraphics (STN, St Louis, MO, USA). Data were tested for normality before further analysis. The effects of species and N treatment on variables were analysed by two-way ANOVAs. Differences between means were considered significant when the P-value of the ANOVA F-test was less than 0.05. The Cq values obtained from qPCR were normalized using the program proposed by Pfaffl et al. (2002) and the fold-changes of transcript levels were calculated in the program of REST (Pfaffl et al., 2002) as described by Li et al. (2012). For principal component analysis (PCA), data were standardized and computed by the command prcomp() in R (http://www.r-project.org/). The cluster analysis of gene expression was computed by command heatmap.2() with the package ‘gplots’ in R.
Results
Root morphology and photosynthesis
Root morphology and photosynthesis are sensitive to changes in resource (e.g. N) availability. Thus, these characteristics were analysed in both poplar species (Table 1). Pp and Pg had distinct root morphological characteristics under either the control N (1000 μM NH4NO3) or low N (100 and 10 μM NH4NO3) supply levels. Generally, Pg exhibited lower root biomass, total fine root length, and total fine root surface area, but higher root volume than Pp. Photosynthesis showed no differences between Pp and Pg under the control N level. Intrinsic water use efficiency was lower in Pg than that in Pp under the given N levels. As a stored photosynthate, foliar starch concentration was higher in Pg than those in Pp under the three N supply levels. Root starch concentration was similar in Pp and Pg.
Table 1.
Root morphological, photosynthetic characteristics, and starch of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 µM NH 4 NO 3
| Species | N treatment (μM) | Root biomass (g DW) | Total fine root length (m) | Total fine root surface area (cm2) | Total root volume (cm3) | A (mmol CO2 m–2 s–1) | WUEi (mmol CO2 mol–1 H2O) | PNUEi (mol CO2 (mg N)–1 s–1) | Foliar starch (mg (g DW)–1) | Root starch (mg (g DW)–1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Pp | 10 | 4.4±0.3bc | 21.9±2.0a | 123.3±14.4cd | 4.9±0.0a | 6.8±0.7a | 134.1±26.8d | 1.8±0.1a | 11.8±0.1b | 12.3±0.8ab |
| 100 | 4.5±0.3c | 39.9±2.2b | 158.2±18.5d | 4.6±0.8a | 9.8±0.3b | 120.7±3.2d | 2.4±0.2b | 11.2±1.2b | 13.6±0.2b | |
| 1000 | 3.5±0.3abc | 19.6±2.1a | 87.8±1.8bc | 6.2±0.5a | 17.1±0.8cd | 73.7±0.2c | 4.0±0.4c | 8.3±0.1a | 10.8±1.1a | |
| Pg | 10 | 2.5±0.4a | 22.0±8.5a | 71.2±11.8ab | 5.7±0.7a | 7.7±0.4a | 40.9±0.2a | 2.5±0.6b | 14.0±0.1c | 13.3±0.2b |
| 100 | 3.4±0.6a | 11.5±1.8a | 46.3±8.4a | 9.2±0.5b | 10.2±0.2b | 45.2±1.8ab | 2.8±0.5bc | 10.9±0.5b | 12.4±0.4ab | |
| 1000 | 2.5±0.3ab | 12.8±2.2a | 48.9±7.5a | 9.1±0.4b | 14.6±0.7c | 52.3±4.0b | 3.3±0.2c | 11.4±0.1b | 12.0±0.9ab | |
| P-values | Species | *** | ** | **** | **** | ns | **** | ns | ** | ns |
| N | Ns | ns | * | ** | *** | ns | ** | *** | ns | |
| Species × N | Ns | ** | * | * | * | * | ns | * | ns |
Data indicate mean ± SE (n = 6). Different letters in the same column indicate significant difference (P < 0.05). P-values of the ANOVAs of species, N treatment, and their interaction are indicated: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. Pp, Populus popularis; Pg, Populus alba × Populus glandulosa; A, leaf net photosynthetic rate; PNUEi, instantaneous photosynthetic N use efficiency; WUEi, intrinsic water use efficiency.
Low N availability affected root morphology and photosynthesis in both poplar species (Table 1). Total fine root surface area increased in Pp under both low N levels, but remained unaltered in Pg. Total root volume was unchanged in Pp but decreased in Pg under the lowest N supply level compared to that under the control N supply. Photosynthesis decreased stronger in Pp than those in Pg under N-limiting conditions. PNUEi was lower in both poplar species under limiting N supply levels in comparison with that under the control N condition. As a stored photosynthate, foliar starch concentration was higher in both species under low N levels than those under the control N level. N supply levels had no effects on root starch concentration.
These data show that limiting N levels induces distinct responses of root morphology and photosynthesis in Pp and Pg, which is probably associated with interspecific differences in N metabolism.
Net fluxes of NH4 +, NO3 –, and H+, activities of PM H+-ATPases, and accumulation of NH4 +, NO3 –, and NO2 –
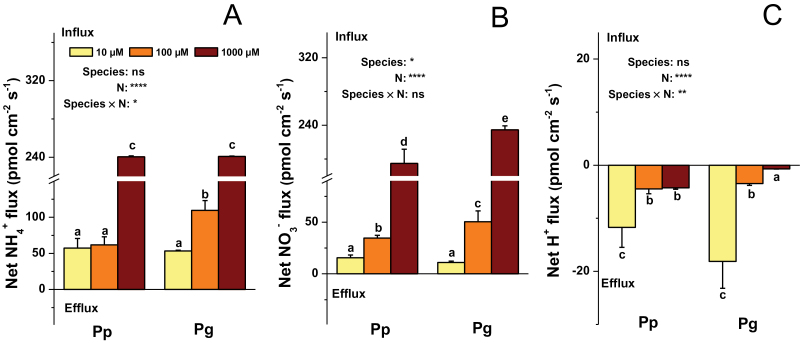
N uptake is the first crucial step, which may lead to distinct differences in N metabolism of Pp and Pg. Net fluxes of NH4 + and NO3 – measured along the root tips of Pg displayed large variation at different positions, and maximal net influxes of NH4 + and NO3 – occurred at the position of 15mm from the root apex (Supplementary Fig. S1B). Based on these findings in Pg and previous observations in Pp (Luo et al., 2013), net fluxes of NH4 +, NO3 –, and H+ were measured with greater detail at the position of 15mm from the root apex of both species (Fig. 2).
Fig. 2.
Net fluxes of NH4 + (A), NO3 – (B), and H+ (C) in 10min at 15mm from the root apex of fine roots of P. popularis (Pp) and P. alba × P. glandulosa (Pg). Data indicate mean ± SE (n = 6). The measuring solution (pH 5.5) contained 0.1mM KCl and 0.1mM CaCl2 as well as 10, 100, or 1000 μM NH4NO3. Bars labelled with different letters indicate significant difference between the treatments. P-values of the ANOVAs of species, N treatment, and their interaction are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant (this figure is available in colour at JXB online).
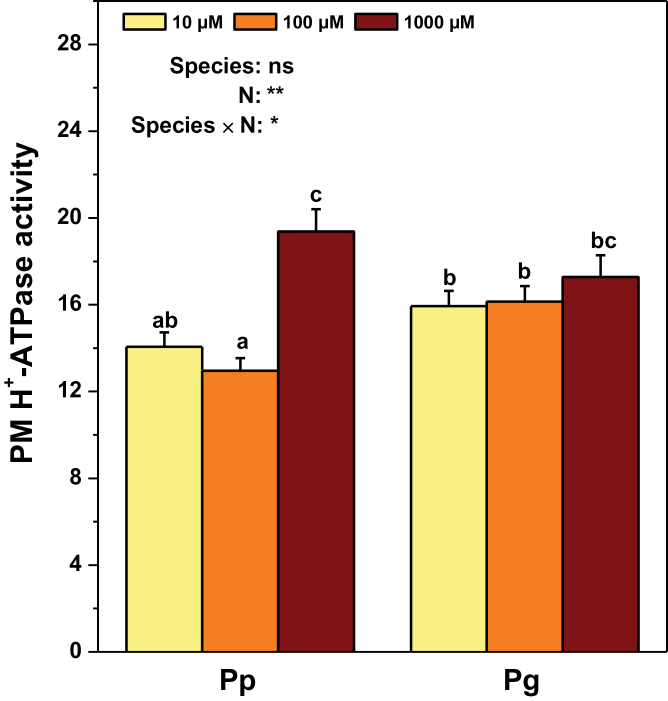
Under control N, net NH4 + influx was similar in roots of Pp and Pg (Fig. 2A), but net NO3 – influx was higher in roots of Pg than those of Pp (Fig. 2B). Under 100 μM NH4NO3, Pg exhibited higher net influxes of NH4 + or NO3 – than Pp (Fig. 2A, B). Since net influxes of NH4 + and NO3 – are associated with H+ fluxes (Luo et al., 2013), net H+ fluxes were also determined in Pp and Pg under the three N supply levels (Fig. 2C). The net H+ efflux was lower in Pg than in Pp under the control N level. Net H+ flux at the root surface is coupled with activities of PM H+-ATPases (Luo et al., 2013). Thus, PM H+-ATPase activities were analysed in isolated plasma membranes of fine roots. The PM H+-ATPase activity was similar in Pp and Pg under the control N condition (Fig. 3).
Fig. 3.
PM H+-ATPase activity (mmol Pi h–1 (mg protein)–1) in roots of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 μM NH4NO3. Bars indicate mean ± SE (n = 6). Different letters on the bars indicate significant difference. P-values of the ANOVAs of species, N treatment, and their interaction are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant (this figure is available in colour at JXB online).
Low N supply levels always resulted in significant decreases in net NO3 – influxes in Pp and Pg compared to the control N condition (Fig. 2B). Both low N levels led to increased net H+ efflux in Pg, but only the lowest N level elevated net H+ efflux in Pp in comparison with the control N supply (Fig. 2C). In Pp, the PM H+-ATPase activities decreased under low N supply compared to the control N condition, whereas no such effects were found in Pg (Fig. 3).
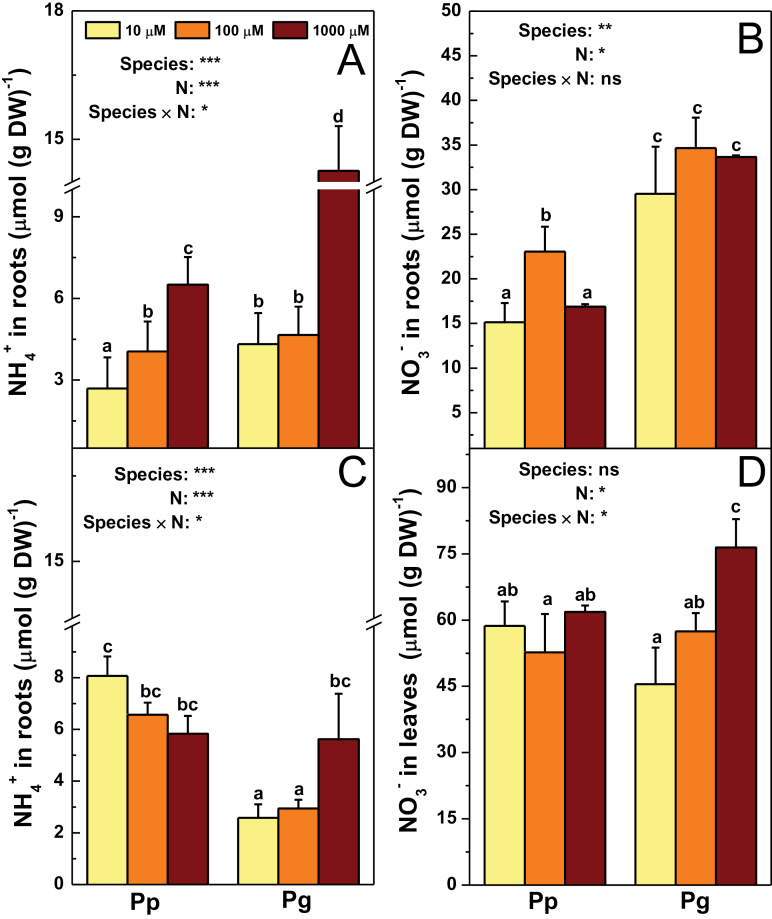
Different uptake rates of NH4 + or NO3 – at the root surface of Pg and Pp exposed to the three N levels may result in differences in NH4 +, NO3 –, and NO2 – content in plants. Therefore, these compounds were further analysed (Fig. 4 and Supplementary Fig. S3). NH4 + content was higher in roots of Pg than those of Pp (Fig. 4A). NO3 – content was higher in roots of Pg than those of Pp under the three N levels (Fig. 4B). Root NO2 – content was similar in both species (Supplementary Fig. S3). Foliar NH4 + content was similar in Pp and Pg under the control N supply, but lower in Pg than in Pp under limiting N conditions (Fig. 4C). Foliar NO3 – content was higher in Pg than that in Pp under the control N level (Fig. 4D).
Fig. 4.
NH4 + and NO3 – content in roots (A and B) and leaves (C and D) of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 μM NH4NO3. Bars indicate mean ± SE (n = 6). Different letters on the bars indicate significant difference. P-values of the ANOVAs of species, N treatment, and their interaction are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. DW, dryweight (this figure is available in colour at JXB online).
Low N supply affected NH4 +, NO3 –, and NO2 – content in poplars. In comparison with the control N level, NH4 + accumulation decreased in roots of both species in response to low N supply (Fig. 4A). NO3 – content increased in Pp roots under 100 μM NH4NO3 compared to that under the control N supply (Fig. 4B). The N treatment had no impacts on root NO2 – content (Supplementary Fig. S3). Foliar NH4 + content decreased in Pg in response to low N availability, but remained unaltered in Pp under the three N levels (Fig. 4C). Foliar NO3 – content decreased in Pg in response to low N supply (Fig. 4D).
Activities of enzymes involved in N assimilation, total N, δ15N, and mineral nutrients
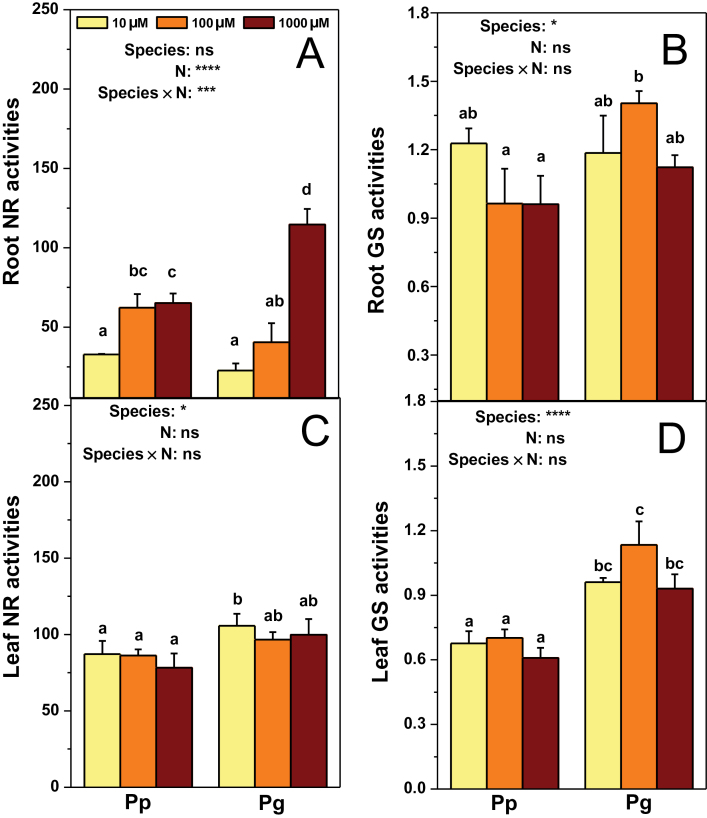
After the uptake of NH4 + and NO3 –, enzymes play important roles in N assimilation. Therefore, this study determined the activities of enzymes (NR, NiR, GS, GOGAT, and GDH) involved in N assimilation in Pp and Pg (Figs. 5 and Supplementary Fig. S4). Root NR activity was higher in Pg than that in Pp under the control N level (Fig. 5A). Root NiR activities were similar in Pp and Pg under the control N supply, but lower in Pg than in Pp under low N supply conditions (Supplementary Fig. S4). Root GS activity was higher in Pg than that in Pp under 100 μM NH4NO3 (Fig. 5B). Activities of GOGAT or GDH in roots were unaffected by species (Supplementary Fig. S4). In leaves, analysed enzyme activities of Pg were higher than those of Pp (Figs. 5C, D and Supplementary Fig. S4).
Fig. 5.
Activities of nitrate reductase (NR, nmol NO3 – h–1 (mg protein)–1), and glutamine synthetase (GS, h–1 (mg protein)–1) in roots (A and B) and leaves (C and D) of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 μM NH4NO3. Bars indicate mean ± SE (n = 6). Different letters on the bars indicate significant difference. P-values of the ANOVAs of species, N treatment, and their interaction are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant (this figure is available in colour at JXB online).
Limiting N supply also influenced activities of enzymes implicated in N assimilation in both poplar species. Root NR activity reduced in both species in response to low N levels (Fig. 5A). Root NiR activity was stimulated in Pp by 10 μM NH4NO3 compared with that under the control N level, but remained unchanged in Pg under the three N levels (Supplementary Fig. S4). Activities of GOGAT or GDH in roots were unaffected by low N levels (Supplementary Fig. S4). In leaves, these enzyme activities in both species remained unaltered in response to low N availability except that foliar GDH activities decreased in Pg and stimulated in Pp exposed to 10 μM NH4NO3 (Figs. 5C, D and Supplementary Fig. S4).
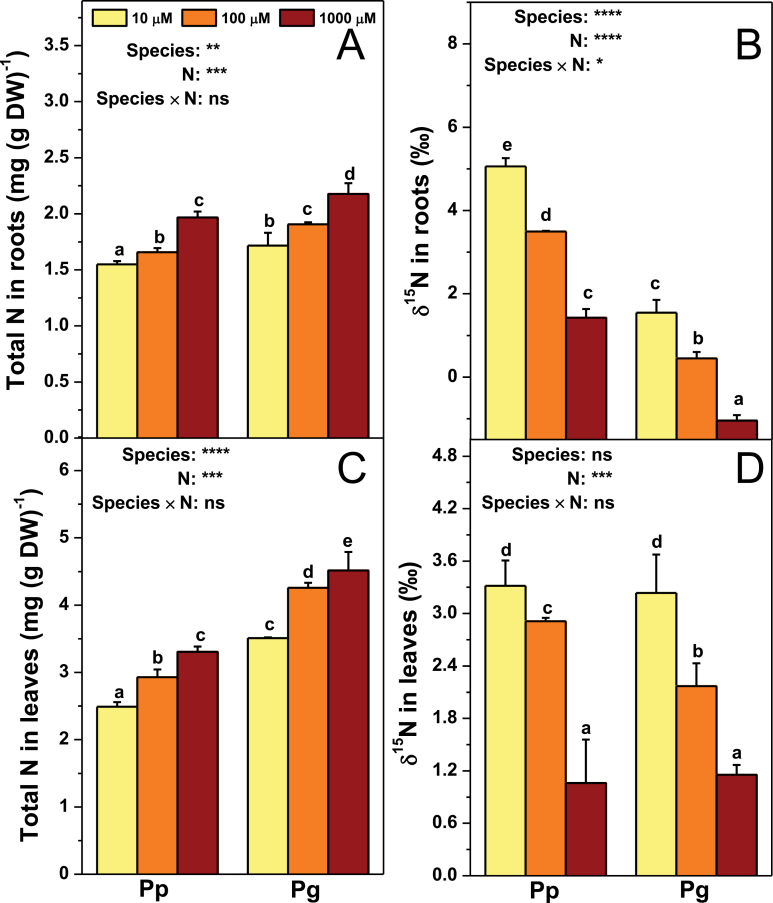
Irrespective of N forms, total N concentration in plants may mirror N availability. Moreover, fractionation of 15N may occur in different steps of N metabolism in plants, reflecting active status of N metabolism. Therefore, total N and δ15N were analysed in Pp and Pg (Fig. 6). Total N concentration in roots of Pg was higher than those of Pp under the same N level (Fig. 6A). In contrast, δ15N in roots of Pg was lower than that of Pp under the same N treatment (Fig. 6B). Foliar N concentration of Pg was also higher than those of Pp under the same N level (Fig. 6C). Foliar δ15N was unaffected by species (Fig. 6D).
Fig. 6.
Total N concentration and δ15N in roots (A and B) and leaves (C and D) of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 μM NH4NO3. Bars indicate mean ± SE (n = 6). Different letters on the bars indicate significant difference. P-values of the ANOVAs of species, N treatment, and their interaction are indicated. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. DW, dryweight (this figure is available in colour at JXB online).
Total N concentration in roots reduced with decreasing N supply levels in both species (Fig. 6A). On the contrary, δ15N in roots of both species increased in response to both low N levels (Fig. 6B). Foliar N concentration in both species was also lower under low N levels compared to those under the control N condition (Fig. 6C). In contrast, foliar δ15N in both species increased in response to low N availability (Fig. 6D).
As N availability can also affect uptake of other nutrients and carbon metabolism, mineral nutrients, soluble sugars, total C, δ13C, soluble protein, and phenolics were analysed in Pp and Pg (Supplementary Figs. S5 and S6, Supplementary Table S2). There were complex patterns in the responses of nutrient elements and carbon-bearing compounds to N supply levels.
PCA of morphological and physiological responses
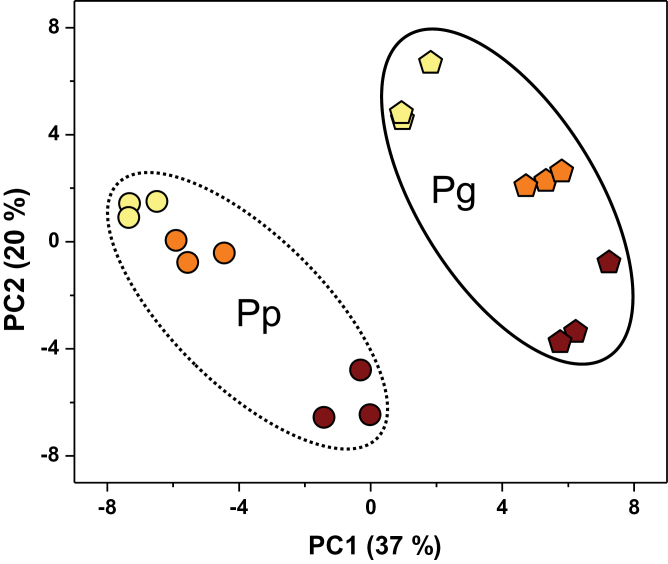
To unravel key parameters involved in the response patterns of both poplar species to N supply levels, a PCA was conducted using data of morphological and physiological parameters related to root morphology, photosynthesis, and N metabolism (Fig. 7, Supplementary Table S3). PC1 and PC2 accounted for 37 and 20% of the variation, respectively. PC1 clearly separated the variation of species effects, and PC2 uncovered the effects of N treatment levels. Foliar N concentration and root δ15N were key contributors to PC1, whereas A, net influxes of NH4 + and NO3 –, and foliar starch concentration was important factors to PC2. In the PCA plot, a greater distance between symbols associated with N treatment levels suggests a stronger responsiveness of morphological and physiological parameters to changes in N supply levels. Thus, the greater distance between symbols related to the control N level and 100 μM NH4NO3 in Pp compared to that in Pg indicates that Pp is more sensitive to decreasing N supply than Pg in the range of given N availability. These PCA results indicate that Pp and Pg exhibit distinct morphological and physiological responsiveness in acclimation to limiting N availability, which mainly results from differences of Pp and Pg in uptake of NH4 + or NO3 –, root 15N fractionation, foliar N and starch concentration, and A.
Fig. 7.
Principal component analysis (PCA) plot of the first two principal components in P. popularis (Pp) and P. alba × P. glandulosa (Pg). The analysis was conducted using data of physiological parameters of Pp and Pg exposed to 10 (yellow), 100 (orange), or 1000 (brown) μM NH4NO3, respectively (this figure is available in colour at JXB online).
Transcriptional regulation of genes involved in N metabolism
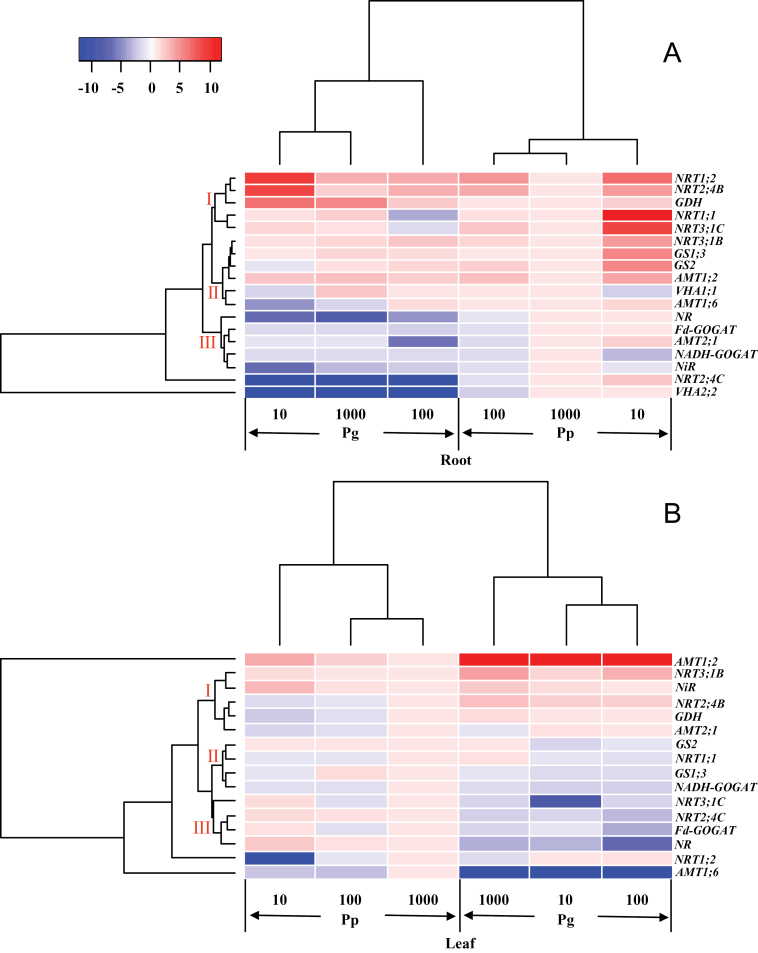
Since Pp and Pg demonstrated distinct patterns of morphological and physiological responses in acclimation to limiting N availability, interspecific differences may also be expected in the transcriptional regulation pattern of key genes implicated in N metabolism. Therefore, transcript levels of representative genes involved in N acquisition and assimilation were assessed in roots and leaves of both species (Fig. 8). The cluster analysis of transcript changes of N uptake- and assimilation-related genes clearly separated Pg and Pp based on their responsiveness to N supply levels (Fig. 8).
Fig. 8.
Cluster analysis of transcriptional fold-changes of key genes involved in N uptake and assimilation in roots (A) and leaves (B) of P. popularis (Pp) and P. alba × P. glandulosa (Pg) exposed to 10, 100, or 1000 μM NH4NO3. The colour scale indicates fold-changes of mRNAs. For each gene, the expression levels in roots or leaves of Pp exposed to 1000 μM NH4NO3 were defined as 1, and the corresponding fold-changes under 100 and 10 μM NH4NO3 were calculated (this figure is available in colour at JXB online).
In roots, NRT1;2, NRT2;4B, GDH, NRT1;1, and NRT3;1C formed a subcluster I (Fig. 8A). Under the control N level, the transcript abundance of genes in the subcluster I was higher in Pg than in Pp (Fig. 8A). The second subcluster consisted of NRT3;1B, GS1;3, GS2, AMT1;2, VHA1;1, and AMT1;6 and the transcript levels of these genes were similar or lower in Pg compared to those in Pp under the control N level (Fig. 8A). The subcluster III included NR, Fd-GOGAT, AMT2;1, NADH-GOGAT, and NiR, and the mRNA levels of these genes were lower in Pg than those in Pp under the control N level (Fig. 8A). The strongest differences existed for NRT2;4C and VHA2;2, which were strongly suppressed in Pg compared to those in Pp under the three N supply levels (Fig. 8A).
Limiting N supply affected transcript levels of genes involved in N metabolism in roots of both species (Fig. 8A). Generally, the transcript levels of genes from the subcluster I were increased in Pp in response to low N levels in comparison with those under the control N condition, but the transcript changes of these genes in Pg were diverse in response to low N availability (Fig. 8A). The transcriptional induction of genes from subcluster II except VHA 1;1 was detected in Pp in response to 10 μM NH4NO3 in comparison with those under the control N condition, but no such effects were found in Pg (Fig. 8A). The transcript levels of genes from subcluster III were suppressed in Pp in response to 100 μM NH4NO3 compared to those under the control N supply, but no such effects were observed in Pg (Fig. 8A). The strongest differences existed for NRT2;4C and VHA2;2 because NRT2;4C and VHA2;2 responded to N supply variation in Pp but not in Pg (Fig. 8A).
In leaves, NRT3;1B, NiR, NRT2;4B, GDH, and AMT2;1 formed a subcluster I (Fig. 8B). Under the control N level, the mRNA levels of genes in the subcluster I except AMT2;1 were higher in Pg than those in Pp (Fig. 8B). GS2, NRT1;1, GS1;3, and NADH-GOGAT constitute the second subcluster (Fig. 8B). The transcript levels of genes from this subcluster were similar or slightly lower in Pg than those in Pp (Fig. 8B). The third subcluster consisted of NRT3;1C, NRT2;4C, Fd-GOGAT, and NR (Fig. 8B). Under the control N level, genes from subcluster III showed lower transcript levels in Pg than those in Pp (Fig. 8B). The mRNA level of AMT1;2 was higher in Pg than that in Pp under the control N condition (Fig. 8B).
In leaves, transcript levels of genes related to N metabolism were also affected by N supply. NRT2;4B, GDH, and AMT2;1 from subcluster I had lower transcript levels in Pp in response to low N availability compared to those exposed to the control N supply, but the transcript levels of these genes were relatively stable in Pg under the three N levels (Fig. 8B). In the second subcluster, the mRNA levels of GS2 were stable in Pp in response to low N availability, but repressed in Pg under low N levels compared to that under the control N condition (Fig. 8B). In comparison with the control N supply, the transcript levels of genes from subcluster III were increased in Pp exposed to 10 μM NH4NO3, but suppressed or unaltered in Pg (Fig. 8B). The other three genes (i.e. NRT1;2, AMT1;6, and AMT1;2) displayed the strongest differences between Pp and Pg in response to N availability (Fig. 8B). NRT1;2 and AMT1;6 showed lower mRNA levels in Pp in response to low N availability, but no such effects were observed in Pg (Fig. 8B). The mRNA level of AMT1;2 was increased in Pp but not in Pg in response to low N availability (Fig. 8B).
To find out which genes are the most important ones in response to differences in species and/or N supply levels, a PCA was performed using data of fold-changes of transcripts in Pp and Pg under the three N levels (Supplementary Table S4). PC1 clearly separated species effects and PC2 the N treatment impacts. PC1 and PC2 accounted for 48 and 26% of the variation, respectively. Leaf NRT2;4C and AMT1;6, and root NRT2;4C were the most important contributors to PC1, whereas NADH-GOGAT, AMT1;2, and NRT1;2 in roots were essential factors in PC2.
Discussion
Differences between Pp and Pg in N metabolism under limiting N supply
The greater root biomass and larger fine root surface area of Pp compared with Pg suggest that root morphological features of Pp are more responsive to limiting N availability than those of Pg. More stimulation of root growth in Pp than in Pg can be critical for different acclimation patterns of both species to limiting N availability because N acquisition in these poplars depends on root characteristics. The greater root biomass and fine root surface area in Pp indicate that Pp may better exploit nutrient resources in rhizosphere in comparison with Pg. The higher growth rates of plants may need more N metabolites to support (Lawlor, 2002). Thus, growth can be a driving force for N metabolism of plants. Consequently, the higher root growth of Pp can lead to greater N demand, further triggering a stronger responsiveness to decreasing N availability. Despite lower total N concentration in Pp roots than in Pg roots, the greater root biomass of Pp resulted in higher N amount (14–38%) in Pp roots, suggesting that Pp can acquire more N to support higher root growth.
The greater root growth in Pp, however, does not necessitate higher net influxes of N compared to those in Pg. Actually, Pp displayed lower net influxes of NH4 + and NO3 – compared to those of Pg under 100 μM NH4NO3, which is likely associated with PM H+-ATPase activities, activities of enzymes implicated in N assimilation, and functioning of AMTs and NRTs (Hawkins et al., 2008; Hawkins and Robbins, 2010; Alber et al., 2012; Luo et al., 2013). PM H+-ATPases play a central role in the uptake of NH4 + and NO3 – because H+ ions pumping to the apoplast through PM H+-ATPases create the proton motive force, driving the absorption of NH4 + and NO3 – in roots (Hawkins and Robbins, 2010; Luo et al., 2013). The lower activity of PM H+-ATPases is in line with the lower net influxes of NH4 + and NO3 – in Pp compared with Pg under 100 μM NH4NO3. Additionally, in most cases, the lower activities of enzymes involved in N assimilation and the lower total N concentration in roots and leaves of Pp versus Pg correspond well to the lower N uptake rates in Pp under 100 μM NH4NO3. In contrast to the lower total N concentration in Pp than in Pg, the higher δ15N in roots of Pp indicates that 15N is more rapidly enriched in the process of N metabolism in roots of Pp than of Pg. Since the processes of N metabolism in plants discriminate against the heavier N isotope leading to the depletion of 15N in plant dry mass compared with that in the soil (Tcherkez and Hodges, 2008; Falxa-Raymond et al., 2012; Gauthier et al., 2013), higher δ15N in roots of Pp indicates less fractionation of 15N occurs in Pp than in Pg. This is consistent with the lower net influxes and content of NH4 + and NO3 –, activities of NR and GS, and total N concentration in Pp compared with Pg. Based on morphological and physiological parameters related to N metabolism, the PCA results suggest that Pp is more sensitive to decreasing N supply than Pg under 100–1000 μM NH4NO3, which is mainly due to differences between Pp and Pg in the uptake of NH4 + or NO3 –, root 15N fractionation, foliar N and starch concentration, and A.
The distinct patterns of transcriptional regulation of genes implicated in N metabolism of Pp and Pg may be associated with the different morphological and physiological responses of both species to limiting N availability. This study group’s previous study suggests that, under N fertilization, differential expression of genes involved in N uptake (AMTs and NRTs) of Pp and Pg leads to accelerated N physiological processes in Pg than in Pp (Li et al., 2012). Under limiting N conditions, however, the current data show that transcriptional regulation of key genes involved in N metabolism of Pp is more responsive than that of Pg. These results indicate that Pp and Pg can differentially manage transcriptional regulation of key genes involved in N metabolism under low and high N availability. These results are consistent with previous studies. Arabidopsis plants manage N metabolism differently under deficient and sufficient N conditions (Lemaitre et al., 2008; Chardon et al., 2010; Ikram et al., 2012). These studies highlight that it is necessary to investigate the responses of plants not only to high N fertilization but also to low N availability.
Overall, Pp and Pg displayed different patterns of morphological, physiological, and transcriptional regulation in response to limiting N availability, which is mainly associated with the difference of both species in net influxes of NH4 + and NO3 –, root δ15N, foliar N and starch concentration, A, and the transcriptional regulation of genes (e.g. AMT1;2, NRT1:2, and NRT2;4C in roots and AMT1;6 and NRT2;4C in leaves) that are involved in N acquisition and assimilation.
The physiological and transcriptional regulation mechanisms of N metabolism of poplars in acclimation to low N availability
Root morphology responds highly plastic to N availability (Forde and Walch-Liu, 2009; Chapman et al., 2012). Increases in fine root surface area of poplars exposed to low N levels indicate that poplars stimulate growth of fine roots to forage for nutrients under limiting N availability. Consistently, Arabidopsis roots adopt an ‘active-foraging strategy’ by outgrowth of lateral roots under limiting N conditions, but a ‘dormant strategy’ by inhibited growth of lateral roots under sufficient N supply (Ruffel et al., 2011). Most plants can increase root growth, resulting in greater fine root surface area under short-term N deficiency, but exhibit stunted root growth under long-term limiting N availability due to lack of internal N (Kraiser et al., 2011; Shen et al., 2013). The current data indicate that poplar roots adopt an active-forage strategy to acquire N resources and other essential minerals under low N supply. Although poplar roots actively forage for nutrients under low N conditions, the acquired N appears insufficient for the biosynthesis of photosynthetic enzymes and metabolic precursors, leading to decreased A and PNUEi. In other higher plants, N deficiency also inhibits photosynthetic capacity and growth (Sardans and Penuelas, 2012).
Gradual decreases in net influxes of NH4 + and NO3 – at the root surface under decreased concentration of external NH4NO3 supply indicate that NH4 + and NO3 – concentration in the external solution play important roles in net influxes of these ions. This was also found for seedlings of Douglas fir (Pseudotsuga menziesii) and soybean (Glycine max) (Hawkins and Robbins, 2010). However, net influxes of NH4 +/NO3 – are greater at the root surface of white spruce (Picea glauca) exposed to 50 μM NH4NO3 than those of 1500 μM NH4NO3 (Alber et al., 2012). These results suggest that impacts of external NH4NO3 concentration on net fluxes of NH4 +/NO3 – are also related to plant species. Decreases in PM H+-ATPase activities and transcript levels of genes (VHA1;1 and VHA2;2) encoding PM H+-ATPases, root NH4 + and foliar NO3 – content, foliar GDH activity, total N concentration in roots and leaves, and soluble protein in roots and leaves of both poplar species are in agreement with lower net influxes of NH4 +/NO3 – under limiting N supply. Moreover, correlations between different enzymes involved in N metabolism, enzymes, gene expression levels, and transcript levels of different genes in both poplar species (Supplementary Fig. S7) indicate that poplar plants coordinate each step of N metabolism processes to acclimate to low N availability. These results indicate that the processes of N uptake and assimilation have slowed down in both poplar species in acclimation to low N availability. In plants, N and C metabolism is interconnected because the production of N metabolites such as amino acids needs C skeletons (Nunes-Nesi et al., 2010). The lower total C concentration in roots and foliar δ13C, the elevated starch concentration in leaves, and the correlations between δ13C and parameters of N metabolism in both poplar species under low N levels (Supplementary Figs. S5 and S7) suggest that C export and/or phloem transport is inhibited under N deficiency. Foliar starch accumulation is also observed in herbaceous plants in response to low N availability, which is ascribed to the reduced demand of C skeletons for N compounds such as amino acids and proteins under low N levels (Lemaitre et al., 2008; Ikram et al., 2012; Schluter et al., 2012). In the same line, earlier studies indicate that foliar starch accumulation is a consequence of photosynthesis exceeding the demands of respiration and growth under N deficiency conditions (Lawlor et al., 1987b; Lawlor, 2002).
Increases in δ15N in roots and leaves of poplars under low N levels are contrary to decreases in total N concentration, indicating that 15N is enriched in both poplar species under limiting N availability. N starvation also stimulates whole-plant δ15N in some genotypes of wild barley (Hordeum spontaneum) (Robinson et al., 2000). The higher δ15N values in both poplar species under low N conditions compared with that under the control N supply are probably associated with slowing down of N metabolism which leads to less depletion of 15N in poplars under low N availability. In other words, poplar plants are forced to utilize 15N to meet their N demands under limiting N conditions, leading to 15N enrichment in dry mass. Although the mechanisms underlying the variation in natural 15N are not completely known in plants under various environmental conditions (Cernusak et al., 2009; Tcherkez, 2011), 14N/15N fractionation can occur in the processes of N absorption, assimilation, recycling, and reallocation in plants and N release from plants (e.g. foliar NH3 volatiles and N-containing exudates of roots; Robinson et al., 2000; Ariz et al., 2011; Gauthier et al., 2013; Yousfi et al., 2013). Additionally, changes in environmental factors such as nutrients, can cause substantial alterations in δ15N in plants (Ariz et al., 2011; Gauthier et al., 2013; Yousfi et al., 2013). Furthermore, negative correlations between δ15N and transpiration rate were observed in wheat under salinity (Yousfi et al., 2013). Consistently, negative correlations occur between δ15N and transpiration rate, g s, A, or PNUEi in poplars (Supplementary Fig. S7). These results indicate that 15N enrichment in poplars exposed to low N levels is probably associated with (i) less active N metabolism in poplars under low N availability and/or (ii) elevated N release from roots and leaves. The latter possibility needs further studies.
Although transcriptional regulation of genes involved in N metabolism plays a fundamental role in response to N deficiency or starvation in herbaceous plants (Hirai et al., 2004; Bi et al., 2007; Krouk et al., 2010; Patterson et al., 2010; Krapp et al., 2011; Ruffel et al., 2011; Engelsberger and Schulze, 2012; Kiba et al., 2012; Schluter et al., 2012), little is known on transcriptional regulation underlying N metabolism in trees under limiting N availability (Rennenberg et al., 2009, 2010). Transcriptional induction of several AMTs (e.g. AMT1;2) and NRTs (e.g. NRT1;2, NRT2;4B, and NRT3;1B) in poplar roots exposed to low N levels indicates that poplar roots increase mRNAs of key transporters for NH4 + and NO3 – as the result of acclimation to low N availability. Induced transcript abundance of AMT1;2 is also found in roots of P. tremula × tremuloides exposed to low N supply (Selle et al., 2005) and in P. tremula × alba under N starvation (Couturier et al., 2007). Similarly, transcription of OsAMT1;2 in roots of rice (Oryza sativa) is induced by NH4 + deficiency (Sperandio et al., 2011). NRT2;1 (i.e., NRT2;4C re-defined in this study) displays higher transcript levels in NO3 –-fed P. × canescens roots than in NH4 +-fed roots (Ehlting et al., 2007) and is induced upon application of NO3 – to N-deprived roots of peach (Prunus persica) seedlings (Nakamura et al., 2007). NRT3;1 (also called NAR2;1) is a key player in a two-component system including NRT2s for nitrate transport in Arabidopsis (Yong et al., 2010; Kotur et al., 2012) and rice (Yan et al., 2011). Correlation analysis between transcript levels of NRT3;1B or NRT3;1C and other NRTs (i.e., NRT1;1, NRT2;4B, and NRT2;4C) in Pp and Pg under normal and low N levels detected positive relationships (Supplementary Fig. S7). These correlations, combined with this study group’s previous findings where positive correlations also occurred under N-fertilizaiton conditions (Li et al., 2012), indicate that NRT3;1B and/or NRT3;1C may also act as partners of other NRTs for nitrate transport in poplars under various N levels. In contrast to induction of several AMTs and NRTs in poplar roots, reduced transcript levels of most AMTs and NRTs in leaves and genes (e.g. NR, NiR, GOGAT) involved in N assimilation in roots and leaves of poplars indicate that N assimilation (downstream processes after N uptake) is inhibited due to shortage of N-containing precursors under low N availability. These results suggest that overexpression of AMTs and NRTs in poplar roots and downregulation of most AMTs and NRTs in leaves and genes involved in N assimilation in roots and leaves of poplars play fundamental roles in acclimation to limiting N availability.
Taken together, increased fine root growth, slowed down N acquisition and assimilation, overexpressed transcripts of AMTs and NRTs in roots, and repressed transcript levels of AMTs and NRTs in leaves and key genes involved in N assimilation are primary mechanisms of both poplar species in acclimation to limiting N availability.
In summary, Pp exhibited greater root biomass and total fine root surface area, lower net influxes of NO3 – at the root surface, higher δ15N in roots, and more responsiveness of transcriptional regulation of 18 genes involved in N uptake and assimilation in roots and leaves than Pg under limiting N supply. These results indicate that N metabolism of Pp displays a stronger responsiveness to decreasing N availability than that of Pg. Under low N conditions, decreased net influxes of NH4 + and NO3 – at the root surface are consistent with lower root NH4 + and foliar NO3 – content, root NR activity, total N concentration in roots and leaves, and mRNA of most AMTs and NRTs in leaves and genes involved in N assimilation in roots and leaves. Moreover, low N supply levels increased fine root surface area, foliar starch accumulation, δ15N in roots and leaves, and transcript levels of several AMTs (e.g. AMT1;2) and NRTs (e.g. NRT1;2, NRT2;4B, and NRT3;1B) in roots of both poplar species. These data suggest that poplar species slow down processes of N acquisition and assimilation in acclimation to limiting N supply. These morphological, physiological, and molecular data suggest that poplar plants can differentially manage N metabolism under deficient and sufficient N conditions and that it is important to consider low N tolerance when selecting woody plants such as Populus spp. for energy plantations on nutrient-poor sites. Using technologies including genomics, transcriptomics (e.g. microarray, RNA sequencing). and metabolomics in future experiments, a deeper understanding of poplars in acclimation to low N availability may be obtained.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primers used for qRT-PCR.
Supplementary Table S2. Concentrations of mineral nutrients.
Supplementary Table S3. PCA of physiological parameters of both poplar species.
Supplementary Table S4. PCA of transcriptional changes of representative genes.
Supplementary Fig. S1. Net fluxes of NH4 + and NO3 – along the root tip.
Supplementary Fig. S2. Alignments of representative genes.
Supplementary Fig. S3. NO2 – content in roots and leaves.
Supplementary Fig. S4. Activities of NiR, GOGAT, and GDH in roots and leaves.
Supplementary Fig. S5. Soluble sugars, C concentration, and δ13C.
Supplementary Fig. S6. Soluble protein and phenolics.
Supplementary Fig. S7. Correlations of related parameters.
Acknowledgements
This work is supported by the State Key Basic Research Development Program (grant no. 2012CB416902), the National Natural Science Foundation of China (grant no. 31070539, 31100481, 31270647), the Fok Ying Tung Education Foundation (grant no. 121026), the Special Fund for Forest Science and Technology Research in the Public Interest (grant no. 201204210), and the Fundamental Research Funds for the Central Universities of China (grant no. YQ2013005). A.P. is grateful for financial support to the project BEST by the Bundesministerium für Forschung und Technology (BMBF). The authors are grateful to C. Kettner and G. Langer-Kettner for the nutrient element analysis and R. Langel at the Center for Stable Isotopes (KOSI) of the University of Göttingen for the isotope analysis.
References
- Alber A, Ehlting B, Ehlting J, Hawkins BJ, Rennenberg H. 2012. Net NH4 + and NO3 – flux, and expression of NH4 + and NO3 – transporters in roots of Picea glauc . Trees – Structure and Function 26, 1403–1411 [Google Scholar]
- Ariz I, Cruz C, Moran JF, Gonzalez-Moro MB, Garcia-Olaverri C, Gonzalez-Murua C, Martins-Loucao MA, Aparicio-Tejo PM. 2011. Depletion of the heaviest stable N isotope is associated with NH4 +/NH3 toxicity in NH4 +-fed plants. BMC Plant Biology 11, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banziger M, Betran FJ, Lafitte HR. 1997. Efficiency of high nitrogen environment for improving maize for low-nitrogen environment. Crop Science 37, 1103–1109 [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. 2007. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis . BMC Genomics 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau-Gauthier S, Pare D, Messier C, Belanger N. 2011. Juvenile growth of hybrid poplars on acidic boreal soil determined by environmental effects of soil preparation, vegetation control, and fertilization. Forest Ecology and Management 261, 620–629 [Google Scholar]
- Black BL, Fuchigami LH, Coleman GD. 2002. Partitioning of nitrate assimilation among leaves, stems and roots of poplar. Tree Physiology 22, 717–724 [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Brautigam A, Gagneul D, Weber AP. 2007. High-throughput colorimetric method for the parallel assay of glyoxylic acid and ammonium in a single extract. Analytical Biochemistry 362, 151–153 [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. 2004. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfapietra C, De Angelis P, Gielen B, et al. 2007. Increased nitrogen-use efficiency of a short-rotation poplar plantation in elevated CO2 concentration. Tree Physiology 27, 1153–1163 [DOI] [PubMed] [Google Scholar]
- Cao X, Jia JB, Li H, Li MC, Luo J, Liang ZS, Liu TX, Liu WG, Peng CH, Luo ZB. 2012. Photosynthesis, water use efficiency and stable carbon isotope composition are associated with anatomical properties of leaf and xylem in six poplar species. Plant Biology 14, 612–620 [DOI] [PubMed] [Google Scholar]
- Castro-Rodriguez V, Garcia-Gutierrez A, Canales J, Avila C, Kirby EG, Canovas FM. 2011. The glutamine synthetase gene family in Populus . BMC Plant Biology 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernusak LA, Winter K, Turner BL. 2009. Plant δ15N correlates with the transpiration efficiency of nitrogen acquisition in tropical trees. Plant Physiology 151, 1667–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman N, Miller AJ, Lindsey K, Whalley WR. 2012. Roots, water, and nutrient acquisition: let’s get physical. Trends in Plant Science 17, 701–710 [DOI] [PubMed] [Google Scholar]
- Chardon F, Barthelemy J, Daniel-Vedele F, Masclaux-Daubresse C. 2010. Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. Journal of Experimental Botany 61, 2293–2302 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Canovas FM, Man HM, Kirby EG, Mansfield SD. 2012. Enhanced expression of glutamine synthetase (GS1a) confers altered fibre and wood chemistry in field grown hybrid poplar (Populus tremula × alba) (717-1B4). Plant Biotechnology Journal 10, 883–889 [DOI] [PubMed] [Google Scholar]
- Cooke JEK, Weih M. 2005. Nitrogen storage and seasonal nitrogen cycling in Populus: bridging molecular physiology and ecophysiology. New Phytologist 167, 19–30 [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. 2007. The expanded family of ammonium transporters in the perennial poplar plant. New Phytologist 174, 137–150 [DOI] [PubMed] [Google Scholar]
- Dluzniewska P, Gessler A, Dietrich H, Schnitzler JP, Teuber M, Rennenberg H. 2007. Nitrogen uptake and metabolism in Populus × canescens as affected by salinity. New Phytologist 173, 279–293 [DOI] [PubMed] [Google Scholar]
- Ehlting B, Dluzniewska P, Dietrich H, et al. 2007. Interaction of nitrogen nutrition and salinity in grey poplar (Populus tremula × alba). Plant, Cell and Environment 30, 796–811 [DOI] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX. 2012. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. The Plant Journal 69, 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euring D, Lofke C, Teichmann T, Polle A. 2012. Nitrogen fertilization has differential effects on N allocation and lignin in two Populus species with contrasting ecology. Trees – Structure and Function 26, 1933–1942 [Google Scholar]
- Falxa-Raymond N, Patterson AE, Schuster WSF, Griffin KL. 2012. Oak loss increases foliar nitrogen, δ15N and growth rates of Betula lenta in a northern temperate deciduous forest. Tree Physiology 32, 1092–1101 [DOI] [PubMed] [Google Scholar]
- Finzi AC, Norby RJ, Calfapietra C, et al. 2007. Increases in nitrogen uptake rather than nitrogen-use efficiency support higher rates of temperate forest productivity under elevated CO2 . Proceedings of the National Academy of Sciences, USA 104, 14014–14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Walch-Liu P. 2009. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant, Cell and Environment 32, 682–693 [DOI] [PubMed] [Google Scholar]
- Gauthier PPG, Lamothe M, Mahe A, Molero G, Nogues S, Hodges M, Tcherkez G. 2013. Metabolic origin of δ15N values in nitrogenous compounds from Brassica napus L. leaves. Plant, Cell and Environment 36, 128–137 [DOI] [PubMed] [Google Scholar]
- Gobert A, Plassard C. 2007. Kinetics of NO3 – net fluxes in Pinus pinaster, Rhizopogon roseolus and their ectomycorrhizal association, as affected by the presence of NO3 – and NH4 + . Plant, Cell and Environment 30, 1309–1319 [DOI] [PubMed] [Google Scholar]
- Gogstad GO, Krutnes MB. 1982. Measurement of protein in cell suspensions using the Coomassie brilliant blue dye-binding assay. Analytical Biochemistry 126, 355–359 [DOI] [PubMed] [Google Scholar]
- Hawkins BJ, Boukcim H, Plassard C. 2008. A comparison of ammonium, nitrate and proton net fluxes along seedling roots of Douglas-fir and lodgepole pine grown and measured with different inorganic nitrogen sources. Plant, Cell and Environment 31, 278–287 [DOI] [PubMed] [Google Scholar]
- Hawkins BJ, Robbins S. 2010. pH affects ammonium, nitrate and proton fluxes in the apical region of conifer and soybean roots. Physiologia Plantarum 138, 238–247 [DOI] [PubMed] [Google Scholar]
- He J, Qin J, Long L, et al. 2011. Net cadmium flux and accumulation reveal tissue-specific oxidative stress and detoxification in Populus × canescens . Physiologia Plantarum 143, 50–63 [DOI] [PubMed] [Google Scholar]
- He JL, Ma CF, Ma YL, Li H, Kang JQ, Liu TX, Polle A, Peng CH, Luo ZB. 2013. Cadmium tolerance in six poplar species. Environmental Science and Pollution Research 20, 163–174 [DOI] [PubMed] [Google Scholar]
- Heinrichs H, Brumsack HJ, Loftfield N, Konig N. 1986. Verbessertes Druckaufschlussystem fur biologische und anorganische Materialien. Z Pflanzenernaehr Bodenkd 149, 350–353 [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. 2004. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 101, 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Le, Gouis J, Ney B, Gallais A. 2007. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany 58, 2369–2387 [DOI] [PubMed] [Google Scholar]
- Ikram S, Bedu M, Daniel-Vedele F, Chaillou S, Chardon F. 2012. Natural variation of Arabidopsis response to nitrogen availability. Journal of Experimental Botany 63, 91–105 [DOI] [PubMed] [Google Scholar]
- Jackson LE, Burger M, Cavagnaro TR. 2008. Roots nitrogen transformations, and ecosystem services. Annual Review of Plant Biology 59, 341–363 [DOI] [PubMed] [Google Scholar]
- Johnson DW. 2006. Progressive N limitation in forests: Review and implications for long-term responses to elevated CO2 . Ecology 87, 64–75 [DOI] [PubMed] [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, et al. 2012. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-straved plants. The Plant Cell 24, 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotur Z, Mackenzie N, Ramesh S, Tyerman SD, Kaiser BN, Glass ADM. 2012. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytologist 194, 724–731 [DOI] [PubMed] [Google Scholar]
- Koyama L, Kielland K. 2011. Plant physiological responses to hydrologically mediated changes in nitrogen supply on a boreal forest floodplain: a mechanism explaining the discrepancy in nitrogen demand and supply. Plant and Soil 342, 129–139 [Google Scholar]
- Kraiser T, Gras DE, Gutierrez AG, Gonzalez B, Gutierrez RA. 2011. A holistic view of nitrogen acquisition in plants. Journal of Experimental Botany 62, 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthome R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F. 2011. Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiology 157, 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. 2010. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biology 11, R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW. 2002. Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. Journal of Experimental Botany 53, 773–787 [PubMed] [Google Scholar]
- Lawlor DW, Boyle FA, Kendall AC, Keys AJ. 1987a. Nitrate nutrition and temperature effects on wheat: enzyme composition, nitrate and total amino acid content of leaves. Journal of Experimental Botany 38, 378–392 [Google Scholar]
- Lawlor DW, Boyle FA, Young AT, Keys AJ, Kendall AC. 1987b. Nitrate nutrition and temperature effects on wheat: photosynthesis and photorespiration of leaves. Journal of Experimental Botany 38, 393–408 [Google Scholar]
- Lawlor DW, Boyle FA, Keys AJ, Kendall AC, Young AT. 1988. Nitrate nutrition and temperature effects on wheat: a synthesis of plant growth and nitrogen uptake in relation to metabolic and physiological processes. Journal of Experimental Botany 39, 329–343 [Google Scholar]
- Lawlor DW, Kontturi M, Young AT. 1989. Photosynthesis by flag leaves of wheat in relation to protein, ribulose bisphosphate carboxylase activity and nitrogen supply. Journal of Experimental Botany 40, 43–52 [Google Scholar]
- Lemaitre T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C. 2008. Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant and Cell Physiology 49, 1056–1065 [DOI] [PubMed] [Google Scholar]
- Li H, Li MC, Luo J, et al. 2012. N-fertilization has different effects on the growth, carbon and nitrogen physiology, and wood properties of slow- and fast-growing Populus species. Journal of Experimental Botany 63, 6173–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li BH, Kronzucker HJ, Shi WM. 2010. Root growth inhibition by NH4 + in Arabidopsis is mediated by the root tip and is linked to NH4 + efflux and GMPase activity. Plant, Cell and Environment 33, 1529–1542 [DOI] [PubMed] [Google Scholar]
- Lin CC, Kao CH. 1996. Disturbed ammonium assimilation is associated with growth inhibition of roots in rice seedlings caused by NaCl. Plant Growth Regulation 18, 233–238 [Google Scholar]
- Lukac M, Calfapietra C, Lagomarsino A, Loreto F. 2010. Global climate change and tree nutrition: effects of elevated CO2 and temperature. Tree Physiology 30, 1209–1220 [DOI] [PubMed] [Google Scholar]
- Luo J, Qin JJ, He FF, Li H, Liu TX, Polle A, Peng CH, Luo ZB. 2013. Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis . Planta 237, 919–931 [DOI] [PubMed] [Google Scholar]
- Luo ZB, Calfapietra C, Liberloo M, Scarascia-Mugnozza G, Polle A. 2006. Carbon partitioning to mobile and structural fractions in poplar wood under elevated CO2 (EUROFACE) and N fertilization. Global Change Biology 12, 272–283 [Google Scholar]
- Luo ZB, Calfapietra C, Scarascia-Mugnozza G, Liberloo M, Polle A. 2008. Carbon-based secondary metabolites and internal nitrogen pools in Populus nigra under Free Air CO2 Enrichment (FACE) and nitrogen fertilisation. Plant and Soil 304, 45–57 [Google Scholar]
- Luo ZB, Polle A. 2009. Wood composition and energy content in a poplar short rotation plantation on fertilized agricultural land in a future CO2 atmosphere. Global Change Biology 15, 38–47 [Google Scholar]
- Maizlich NA, Fritton DD, Kendall WA. 1980. Root morphology and early development of maize at varying levels of nitrogen. Agronomy Journal 72, 25–31 [Google Scholar]
- McAllister CH, Beatty PH, Good AG. 2012. Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnology Journal 10, 1011–1025 [DOI] [PubMed] [Google Scholar]
- Millard P, Grelet GA. 2010. Nitrogen storage and remobilization by trees: ecophysiological relevance in a changing world. Tree Physiology 30, 1083–1095 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umemiya Y, Masuda K, Inoue H, Fukumoto M. 2007. Molecular cloning and expression analysis of cDNAs encoding a putative Nrt2 nitrate transporter from peach. Tree Physiology 27, 503–510 [DOI] [PubMed] [Google Scholar]
- Novaes E, Osorio L, Drost DR, et al. 2009. Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytologist 182, 878–890 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. 2010. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Molecular Plant 3, 973–996 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Fukuoka H, Yano H, Ohkawa Y. 1999. Relationships between nitrite reductase activity and genotype-dependent callus growth in rice cell cultures. Plant Cell Reports 18, 576–581 [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. 2010. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant, Cell and Environment 33, 1486–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassard C, Guerin-Laguette A, Very AA, Casarin V, Thibaud JB. 2002. Local measurements of nitrate and potassium fluxes along roots of maritime pine. Effects of ectomycorrhizal symbiosis. Plant, Cell and Environment 25, 75–84 [Google Scholar]
- Plett D, Toubia J, Garnett T, Tester M, Kaiser BN, Baumann U. 2010. Dichotomy in the NRT gene families of dicots and grass species. PLoS One 5, e15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle A, Douglas C. 2010. The molecular physiology of poplars: paving the way for knowledge-based biomass production. Plant Biology 12, 239–241 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Dannenmann M, Gessler A, Kreuzwieser J, Simon J, Papen H. 2009. Nitrogen balance in forest soils: nutritional limitation of plants under climate change stresses. Plant Biology 11, 4–23 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Wildhagen H, Ehlting B. 2010. Nitrogen nutrition of poplar trees. Plant Biology 12, 275–291 [DOI] [PubMed] [Google Scholar]
- Robinson D, Handley LL, Scrimgeour CM, Gordon DC, Forster BP, Ellis RP. 2000. Using stable isotope natural abundances (δ15N and δ13C) to integrate the stress responses of wild barley (Hordeum spontaneum C. Koch.) genotypes. Journal of Experimental Botany 51, 41–50 [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences, USA 108, 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardans J, Penuelas J. 2012. The role of plants in the effects of global change on nutrient availability and stoichiometry in the plant–soil system. Plant Physiology 160, 1741–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter U, Mascher M, Colmsee C, Scholz U, Brautigam A, Fahnenstich H, Sonnewald U. 2012. Maize source leaf adaptation to nitrogen deficiency affects not only nitrogen and carbon metabolism but also control of phosphate homeostasis. Plant Physiology 160, 1384–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selle A, Willmann M, Grunze N, Gessler A, Weiss M, Nehls U. 2005. The high-affinity poplar ammonium importer PttAMT1.2 and its role in ectomycorrhizal symbiosis. New Phytologist 168, 697–706 [DOI] [PubMed] [Google Scholar]
- Shen JB, Li CJ, Mi GH, Li L, Yuan LX, Jiang RF, Zhang FS. 2013. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. Journal of Experimental Botany 64, 1181–1192 [DOI] [PubMed] [Google Scholar]
- Sorgona A, Lupini A, Mercati F, Di, Dio L, Sunseri F, Abenavoli MR. 2011. Nitrate uptake along the maize primary root: an integrated physiological and molecular approach. Plant, Cell and Environment 34, 1127–1140 [DOI] [PubMed] [Google Scholar]
- Sperandio MVL, Santos LA, Bucher CA, Fernandes MS, de Souza SR. 2011. Isoforms of plasma membrane H+-ATPase in rice root and shoot are differentially induced by starvation and resupply of NO3 – or NH4 + . Plant Science 180, 251–258 [DOI] [PubMed] [Google Scholar]
- Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE. 2011. Lignin content in natural Populus variants affects sugar release. Proceedings of the National Academy of Sciences, USA 108, 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain T, Goldstein JL. 1964. The quantitative analysis of phenolic compounds. In: Pridham JB, ed, Methods in polyphenol chemistry. Oxford: Pergamon Press; pp 131–145 [Google Scholar]
- Tcherkez G. 2011. Natural 15N/14N isotope composition in C3 leaves: are enzymatic isotope effects informative for predicting the 15N-abundance in key metabolites? Functional Plant Biology 38, 1–12 [DOI] [PubMed] [Google Scholar]
- Tcherkez G, Hodges M. 2008. How stable isotopes may help to elucidate primary nitrogen metabolism and its interaction with (photo)respiration in C3 leaves. Journal of Experimental Botany 59, 1685–1693 [DOI] [PubMed] [Google Scholar]
- Toledo Marchado A, Silvestre Fernandes M. 2001. Participatory maize breeding for low nitrogen tolerance. Euphytica 122, 567–573 [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 [DOI] [PubMed] [Google Scholar]
- Xu GH, Fan XR, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182 [DOI] [PubMed] [Google Scholar]
- Xu Y, Sun T, Yin LP. 2006. Application of non-invasive microsensing system to simultaneously measure both H+ and O2 fluxes around the pollen tube. Journal of Integrative Plant Biology 48, 823–831 [Google Scholar]
- Wang L, Zhou QX, Ding LL, Sun YB. 2008. Effect of cadmium toxicity on nitrogen metabolism in leaves of Solanum nigrum L. as a newly found cadmium hyperaccumulator. Journal of Hazardous Materials 154, 818–825 [DOI] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF. 2012. Uptake, allocation and signaling of nitrate. Trends in Plant Science 17, 458–467 [DOI] [PubMed] [Google Scholar]
- Weih M. 2004. Intensive short rotation forestry in boreal climates: present and future perspectives. Canadian Journal of Forest Research-Revue Canadienne De Recherche Forestiere 34, 1369–1378 [Google Scholar]
- Werner RA, Bruch BA, Brand WA. 1999. ConFlo III – an interface for high precision δ13C and δ15N analysis with an extended dynamic range. Rapid Communications in Mass Spectrometry 13, 1237–1241 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. 2009. Genotype and time of day shape the Populus drought response. The Plant Journal 60, 703–715 [DOI] [PubMed] [Google Scholar]
- Yan M, Fan XR, Feng HM, Miller AJ, Shen QR, Xu GH. 2011. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant, Cell and Environment 34, 1360–1372 [DOI] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. 1954. The estimation of carbohydrates in plant extracts by anthrone. The Biochemical Journal 57, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong ZH, Kotur Z, Glass ADM. 2010. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. The Plant Journal 63, 739–748 [DOI] [PubMed] [Google Scholar]
- Yousfi S, Serret MD, Araus JL. 2013. Comparative response of δ13C, δ18O and δ15N in durum wheat exposed to salinity at the vegetative and reproductive stages. Plant, Cell and Environment 36, 1214–1227 [DOI] [PubMed] [Google Scholar]
- Yousfi S, Serret MD, Marquez AJ, Voltas J, Araus JL. 2012. Combined use of δ13C, δ18O and δ15N tracks nitrogen metabolism and genotypic adaptation of durum wheat to salinity and water deficit. New Phytologist 194, 230–244 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Di T, Xu G, Chen X, Zeng H, Yan F, Shen Q. 2009. Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant, Cell and Environment 32, 1428–1440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.