Abstract
The TOR (target of rapamycin) protein, a large phosphatidylinositol 3-kinase-like protein kinase (PIKK) that is conserved in eukaryotes and is a central regulator of growth and metabolism. The analysis of function of TOR in plant growth and development has been limited by the fact that plants are very poorly sensitive to rapamycin. As the kinase domain of TOR is highly conserved, this study analysed the dose-dependent effect of three sets of first- and second-generation ATP-competitive inhibitors (called asTORis for active-site TOR inhibitors) recently developed for the human TOR kinase on Arabidopsis thaliana growth. All six asTORis inhibited plant root growth in a dose-dependent manner, with 50% growth inhibitory doses (GI50) of <10 μM and <1 μM for the first- and second-generation inhibitors, respectively, similarly to the values in mammalian cells. A genetic approach further demonstrated that only asTORis inhibited root growth in an AtTOR gene-dosage-dependent manner. AsTORis decreased the length of: (i) the meristematic zone (MZ); (ii) the division zone in the MZ; (iii) epidermal cells in the elongation zone; and (iv) root hair cells. Whereas meristematic cells committed to early differentiation, the pattern of cell differentiation was not affected per se. AsTORis-induced root hair growth phenotype was shown to be specific by using other growth inhibitors blocking the cell cycle or translation. AsTORis dose-dependent inhibition of growth and root hairs was also observed in diverse groups of flowering plants, indicating that asTORis can be used to study the TOR pathway in other angiosperms, including crop plants.
Key words: Arabidopsis, ATP-competitive mTOR kinase inhibitors, cell size, differentiation, Lotus, meristem, millet, Nicotiana, rice, root growth, root hairs, target of rapamycin.
Introduction
How plant and plant-cell growth is regulated, and particularly how regulatory systems inherited from the eukaryotic and prokaryotic ancestors of plants are integrated, are still open questions (Robaglia et al., 2012). Target of rapamycin (TOR) is a protein kinase that controls growth and metabolism in response to environmental cues in eukaryotes (Loewith and Hall, 2011; Laplante and Sabatini, 2012). TOR belongs to the family of phosphatidylinositol 3-kinase-like protein kinases (PIKKs) that includes the checkpoint kinases ATM (ataxia telangiectasia mutated) and ATR (ATM- and RAD3-related protein) (Templeton and Moorhead, 2005). The TOR pathway is the focus of large research efforts in yeast and animals due to its role in health, disease, cancer, and aging.
In both yeast and mammals, TOR, which is a very large protein, is present in at least two different multi-protein complexes, TORC1 and TORC2, that regulate different outputs required for cell and organism growth (Laplante and Sabatini, 2012). When active, yeast and mammalian TORC1 positively regulate protein synthesis, cell-cycle progression, and energy metabolism, while inhibiting stress responses, such as autophagy. The best-characterized targets of mammalian TORC1 are S6 kinase 1 (S6K1), which phosphorylates the ribosomal protein S6, and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1), which regulates the initiation of cap-dependent translation (Laplante and Sabatini, 2012). The main known function of TORC2 in yeast and mammals is to regulate spatial control of cell growth via the actin cytoskeleton. TORC1 and TORC2 regulate these important cellular processes in response to the environmental conditions perceived by the cell. In yeast, TORC1 is activated by nutrients through interactions with the vacuole, while starvation for carbon, nitrogen, phosphate, or amino acids mimics TORC1 inhibition (Loewith and Hall, 2011). Various stresses, including high salt, oxidative stress, and high temperature, are also sensed by TORC1 and TORC2. In addition, systemic signalling in multicellular organisms is integrated by the mTOR pathway (Laplante and Sabatini, 2012). Indeed, TORC1 and TORC2 are activated by insulin-like growth factors through the phosphatidylinositol 3- kinase (PI3K) pathway. Furthermore, TORC1 is inhibited by adenosine monophosphate-activated protein kinase (AMPK), the activity of which depends on the cell energy status (Laplante and Sabatini, 2012).
Complete genome sequence analysis indicates that some but not all of the proteins involved in the mammalian TOR pathway are found in all six major eukaryotic groups (Serfontein et al., 2010). These conserved proteins, PI3K, Phosphatase and TENsin homologue (PTEN), TOR, regulatory-associated protein of mTOR (RAPTOR), AMPK and S6K, may form the ancient eukaryotic TOR signalling pathway that has integrated particular inputs and outputs during the divergent evolution of each group. The finding that AtTOR loss-of-function mutants are embryo lethal and that AtTOR is expressed in meristems first indicated that the TOR pathway is essential for plant growth (Menand et al., 2002). Transgenic lines carrying induced RNA silencing of AtTOR experience impaired post-embryonic growth, a decrease in the ratio of polysomes to monosomes, lipid changes, and altered sensing of abiotic stresses (Deprost et al., 2007; Caldana et al., 2012; Xiong and Sheen, 2012). Even though these data validate the important role of AtTOR in growth regulation, such inducible AtTOR RNA-silencing lines are not easy to handle as they do not permit quantitative and kinetically controlled modulation of growth and/or AtTOR levels.
In yeasts and animals, rapamycin, which is produced by Streptomyces hygroscopicus, has been used extensively to dissect the TOR pathway due to its ability to specifically inhibit TOR activity through the formation of a ternary complex with TOR and the peptidyl-prolyl cis-trans isomerase FKBP12 (FK506 and rapamycin-binding protein of 12kDa) (Loewith and Hall, 2011). The use of rapamycin in plants is limited as different authors have reported that rapamycin does not affect Arabidopsis thaliana wild-type (WT) organ growth, even at concentrations up to the tens of micromolar range in solid medium (Sormani et al., 2007; Leiber et al., 2010; Ren et al., 2012). As A. thaliana FKBPs do not carry the amino acids critical for the interaction with rapamycin in animals and yeast, different groups have overexpressed yeast or mammalian FKBP12 proteins to create plants sensitive to rapamycin (Sormani et al., 2007; Leiber et al., 2010; Ren et al., 2012; Xiong and Sheen, 2012). These transgenic lines showed a partial inhibition of organ growth in response to rapamycin, but, in the sole study where plants were treated with a range of rapamycin concentrations, no clear-cut dose-dependent effect of rapamycin on growth was reported (Ren et al., 2012). Indeed, a plateau value of growth inhibition of about 50% was observed along the concentration range from 0.5 to 22 μM, which was associated with a phenotype of shortened root hairs (Ren et al., 2012). A similar phenotype was also reported for A. thaliana seedlings germinated in liquid medium with 10 μM rapamycin (Xiong and Sheen, 2012). However, these phenotypes observed under different growth conditions are hard to compare, preventing easy conclusions.
In addition, rapamycin only partially inhibits TORC1 and does not inhibit TORC2 in mammals (Feldman and Shokat, 2011; Laplante and Sabatini, 2012). Furthermore, unexpected molecular phenotypes unrelated to the AtTOR pathway might be generated by heterologous expression of FKBP12s due to its peptidyl-prolyl cis-trans isomerase activity (Gerard et al., 2011). Progress on studies of the TOR pathway depends strongly on the availability of specific and workable tools to manipulate its activity.
New mammalian TOR-specific inhibitors have been developed recently by several groups with the purpose of inhibiting the TOR pathway more efficiently than rapamycin for cancer therapy (Zhang et al., 2011). All these new TOR inhibitors are ATP competitive, as they target the ATP-binding pocket of the kinase domain and are called active-site TOR inhibitors (asTORis) (Dowling et al., 2010). They have been selected for their specificity by in vitro kinase assays with a wide range of protein kinases (Garcia- Martinez et al., 2009; Thoreen et al., 2009; Chresta et al., 2010; Garcia- Martinez et al., 2009; Q. Liu et al., 2010; Yu et al., 2009, 2010; Liu et al., 2011). In this study, we showed that asTORis clearly inhibit whole-plant growth in a concentration- and AtTOR gene-dosage-dependent manner. The phenotype of root inhibition is reported, i.e. reduction in organ growth, as well as early differentiation of meristematic cells leading to meristem size reduction and shortening of epidermal cells and root hairs without changes in the pattern of differentiation. We also showed that asTORis are potent and robust inhibitors in diverse angiosperms, including crops.
Material and methods
Plant material
A. thaliana WT plants used were from Columbia (Col-0) or Wassilewskija (WS) ecotypes. The ecotype used was Col-0, unless specified otherwise. The TOR/tor-1 (WS), TOR/tor-3 (Col-0), rhd6-2 (Lansberg erecta) and CYCB1;1:GUS (Col0) lines have been described previously (Colon-Carmona et al., 1999; Menand et al., 2002, 2007; Ren et al., 2011). Lotus japonicus cv. Gifu seeds were a gift from C. Vriet and T.L. Wang (John Innes Centre, Norwich, UK). Nicotiana benthamiana seeds were from the Tobacco Institute, SEITA, Bergerac, France). Panicum miliaceum (‘millet brun’) seeds were purchased from Moulin Meckert-Diemer (Krautwiller, France). Oryza sativa (cv. Nipponbare) seeds were from S. Jouannic (IRD, Montpellier, France).
In vitro plant growth
All products were purchased from Sigma unless stated otherwise. Seeds of all species were germinated and grown on a solid medium containing 5mM KNO3, 2.5mM KH2PO4, 2mM Mg(SO4)2, and 2mM Ca(NO3)2 as described by Estelle and Somerville(1987) with the microelements of Santoni et al. (1994) designed for A. thaliana. The medium contained 1% glucose with the pH adjusted to 5.5 and was solidified by 4g l–1 of PhytagelTM (see details in Supplementary Fig. S2 at JXB online) before autoclaving at 115 °C for 20min. The ammonium iron (III) citrate was added after autoclaving from a 2% stock solution that had been filter sterilized (0.22 μm). Plates were carefully poured and protected from desiccation under the flow bench. Transfer plates containing filter-sterilized DMSO at a final concentration of 0.1% with or without drug were stored in the dark for up to 1 week in plastic bags.
In all cases, 0.7% Tween 20 was added to the seed sterilization solution. A. thaliana and N. benthamiana seeds were surface sterilized for 10min in a solution containing 90% ethanol, 0.8 % sodium dichloroisocyanurate dihydrate (SDCD; 02 Javel-pastille, Richet, France) and then washed twice in absolute ethanol. L. japonicus seeds were surface sterilized in 0.8 % SDCD for 10min and washed twice in water. P. miliaceum seeds were surface sterilized in water containing 0.1% calcium hypochloride for 15min and washed six times with autoclaved water. O. sativa seeds were surface sterilized in a solution of 20% sodium hypochlorite and washed six times with water. After sowing on plates, A. thaliana seeds were incubated for 2 d at 4 °C in the dark before germination. After surface sterilization, O. sativa seeds were incubated in sterile water overnight at room temperature in the dark before plating. L. japonicus, N. benthamiana, and P. miliaceum seeds were sown and transferred to the growth chamber directly after surface sterilization. Seeds were germinated on solid medium for 2 d (P. miliaceum and O. sativa), 3 d (A. thaliana), 4 d (L. japonicus), or 5 d (N. benthamiana) before transfer to drug-containing medium, i.e. when the root length was <1cm. For O. sativa and P. miliaceum, plantlets were transferred to square Petri dishes (120×120×17mm) containing 50ml of solidified medium, whereas 90mm diameter round Petri dishes containing 25ml of medium were used for the other species. The Petri dishes were placed vertically to allow the roots to grow on the surface of the medium. Photoperiod of 16h at 23 °C (80 μmol photons m–2 s–1) was followed by 8h of dark at 18 °C.
Drug treatment
The suppliers and characteristics of the drugs are listed in Table 1. All drugs were stored and dissolved in DMSO according to the manufacturer’s instructions. Aliquots of stock solutions were stored at –20 °C at a concentration of 10 to 60mM depending on the asTORi. Plates containing drugs were generally prepared a couple of days before use. Plants were carefully transferred with jeweller’s forceps onto new plates containing drugs or DMSO. Details are given in Supplementary Fig. S2.
Table 1.
Characteristics of the drugs used in this study
| Drug name | Source | Target | Type of inhibition | GI50 in animal cells (μM)a | Selectivity | Tested in plants | Reference(s) | No. kinases tested |
|---|---|---|---|---|---|---|---|---|
| WYE-354 | Chemdea | mTOR | ATP competitive | 0.3–1 | No | Yu et al. (2009) | 260 | |
| WYE-132 | Chemdea | mTOR | ATP competitive | 0.01 | No | Yu et al. (2010) | ||
| KU-0063794 | Tocris Biosciences | mTOR | ATP competitive | 2.5–10 | At least 1000-fold specificity over other PIKKs and PI3-Ks | No | Garcia-Martinez et al. (2009); Chresta et al. (2010); Syed et al. (2013) | 76 |
| AZD-8055 | Chemdea | mTOR | ATP competitive | 0.03–0.1 | At least 1000-fold specificity over other PIKKs and PI3-Ks | No | Chresta et al. (2010) | 70 |
| Torin1 | Gift of Dr N. Gray and Tocris Biosciences | mTOR | ATP competitive | <0.25 | >300-fold selectivity over PI3K and PIKK | Nob | Thoreen et al. (2009); Q. Liu et al. (2010) | 442 |
| Torin2 | Gift of Dr N. Gray and Tocris Biosciences | mTOR | ATP competitive | 0.01–0.2 | 800-fold selectivity over PI3K | No | Liu et al. (2011; 2012b) | 440 |
| QL-IX-55 | Gift of Dr N. Gray | S. cerevisiae TORC1 | ATP competitive | 0.16 | No | Liu et al. (2012b) | 440 | |
| KU-55933 | Tocris Biosciences | ATM | ATP competitive | 10 | No TORC1 and C2 inhibition | Noc | Golding et al. (2009); Li and Yang (2010) | 229 |
| KU-60019 | Tocris Biosciences | ATM | ATP competitive | 0.1–0.3 | High below 1μM | No | Golding et al. (2009) | 229 |
| PI-103 | Tocris Biosciences | PI 3-K /mTORC1 | ATP competitive | 0.1–0.5 | High below 10 μM | No | Fan et al. (2006); Raynaud et al. (2007) | |
| LY294002 | Tocris Biosciences | Broad-spectrum PI3-K inhibitors; mTORC1 | ATP competitive | 4–15; 20–40 | Serum dependent | Yes | Gharbi et al. (2007); Lianguzova et al. (2007); Raynaud et al. (2007); Moon et al. (2009); Finlay and Griffin (2012) | |
| Cycloheximide | Sigma | Protein synthesis | Fixation on the E-site of ribosome | 1 | ND | Yes | X. Liu et al. (2010; Schneider-Poetsch et al. (2010) | |
| Roscovitin | Sigma | Pan CDK and pyridoxal kinase | ATP competitive | 10–50 | Tenfold higher than for ERK8 | Yes | Meijer et al. (1997); Planchais et al. (1997); Bach et al. (2005); Bain et al. (2007) | |
| Rapamycin | LC labs | TOR (only TORC1) | Allosteric through the formation of a complex with TOR and FKBP12 | 0.005–20 | Yes | Sormani et al. (2007); Ren et al. (2012); Xiong and Sheen (2012) Syed et al. (2013) |
ND, not determined.
aConcentration strongly depends on mammalian cell type.
bIn plants, the drug was only used in biochemical measurements and growth inhibition was not reported (Schepetilnikov et al., 2011; Xiong et al., 2013).
cThe drug was only used to increase γH2AX foci in plant nuclei (Amiard et al., 2010).
Root growth measurements
The position of the tip of each root was labelled every day on the back of the Petri dish. Six days after transfer to the new medium, all Petri dishes were scanned (EPSON Perfection 1250, EPSON TWAIN5 software) and root growth was measured with ImageJ software. Dose–response curves were expressed relative to the control growing on DMSO. The time course of the effect of inhibitors was expressed as the ratio of each daily length of treated roots compared with the corresponding length of the control growing on DMSO.
Microscopy
Pictures of the aerial parts of plants and the differentiated zone containing root hairs were taken directly from the Petri dish with a Zeiss stereomicroscope (Stereo Discovery V12). For meristem observations, roots were mounted in chloral hydrate solution (100g of chloral hydrate, 30ml of water, and 5ml of glycerine) and observed with a Zeiss microscope Axo imager M2 with a differential interference contrast illumination system. Cell surface area, cell length, and root hair measurements were done with ImageJ software and expressed relative to the control growing on DMSO, unless otherwise specified. The size of the meristematic zone (MZ), corresponding to the distance between the quiescent centre and the first elongated cortical cell, was measured as described previously (Perilli and Sabatini, 2010).
β-Glucuronidase (GUS) staining
Plants were incubated at 37 °C in GUS staining solution (1mM 5-bromo-4-chloro-3-indolyl-glucuronide, 0.5mM potassium ferricyanide, 0.5mM potassium ferrocyanide, and 10mM sodium phosphate buffer, pH 7) for 2h for CYCB1;1:GUS, 4h for rhd6-2, and 16h for TOR/tor-1 lines.
Characterization of the progeny of TOR/tor-1 and TOR/tor-3 heterozygotes
The tor-1 and tor-3 mutants are embryo lethal, so the progeny of heterozygous plants produce only WT and heterozygous plants (Menand et al., 2002; Ren et al., 2011). As the tor-1 allele carries a translational fusion between the TOR N-terminal sequence and GUS, leading to GUS expression in meristems (Menand et al., 2002), we used GUS staining to distinguish TOR/tor-1 (GUS positive) and TOR/TOR (GUS negative) plants at the end of the growth measurements. The progeny of TOR/tor-3 plants were genotyped by PCR using combinations of the primers Tor-3R (5′-GGCAGTCAAACTATCAGCCTG-3′)/Tor-3L (5′-TGTCCCT GTAGATTGCTCCAC-3′) and Tor-3R/LBa1 (5′-TGGTTCACGT AGTGGGCCATCG-3′), as described previously (Ren et al., 2011).
Results
asTORis inhibit plant root growth in A. thaliana
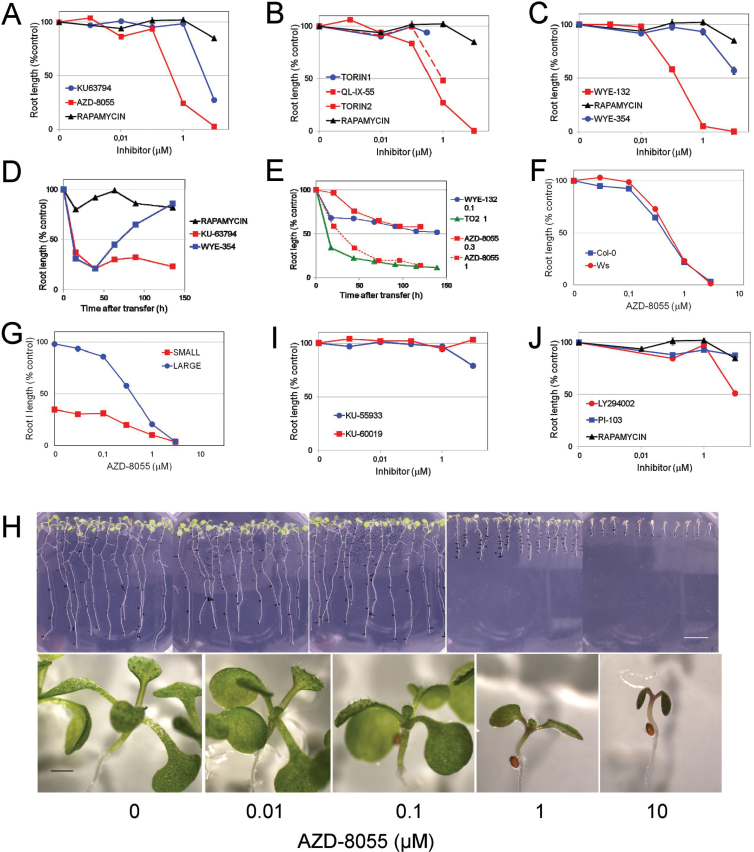
The amino acid residues of the TOR kinase domain that are important for ATP binding are largely conserved among A. thaliana and mammals (Supplementary Fig. S1 at JXB online). As new ATP-competitive inhibitors highly specific of mTOR have been developed (Zhang et al., 2011), we tested the dose-dependent effect of three sets of these asTORis on A. thaliana root growth. The main characteristics of the drugs used in our study are summarized in Table 1. Each set contained a first-generation inhibitor, namely KU63794, Torin1, and WYE-354 (Garcia-Martinez et al., 2009; Thoreen et al., 2009; Yu et al., 2009) and a second-generation counterpart that was developed to improve pharmacodynamics, namely AZD-8055, Torin2 and WYE-132, respectively (Chresta et al., 2010; Yu et al., 2010; Liu et al., 2011). We developed a sensitive and robust growth inhibition assay to test the effect of DMSO-dissolved drugs on primary root growth, as described in Supplementary Fig. S2. The dose–response curves for the effects of these drugs on primary root length showed that each asTORis inhibited root growth at concentrations ranging from 0.1 up to 10 μM. In contrast, rapamycin had almost no inhibitory effect on root growth over this concentration range (Fig. 1A–C). The second-generation molecules were about ten times more efficient at inhibiting growth than the first-generation inhibitors, in agreement with the results reported for mammalian cells (Table 1). Indeed, all second-generation drugs designed for mammals or for yeast (QL-IX-55; Liu et al., 2012b ) (Fig. 1B) clearly inhibited root growth at concentrations below 1 μM. Moreover, the time course of growth delay showed that all drugs except for WYE-354 triggered a rapid, constant, and reproducible level of growth reduction in a concentration-dependent manner as exemplified in Fig. 1D and E. The higher the concentration, the sooner the plateau value of inhibition was reached. These data indicated that the bioavailability and pharmacokinetics of the drugs tested enable their use in plant growth inhibition studies. This was not the case for rapamycin and WYE-354. These inhibitors easily precipitated in the medium at concentrations above 10 μM during the growing period and their growth inhibition capacity was lost after 1–2 d (Fig. 1D).
Fig. 1.
Inhibition of A. thaliana root growth by ATP-competitive inhibitors of TOR, ATM, and PI3Ks. Plants were grown for 3 d without drug or DMSO, transferred to drug-containing plates and grown for 6 d. (A–C) Dose-dependent effect of three sets of first-generation (blue) and second-generation (red) asTORis on primary root growth. Rapamycin (black) was tested at the same final concentration of 0.1% DMSO. (D–E) Time course of root growth delay of two first-generation asTORis (10 μM), compared with 10 μM rapamycin (D) and second-generation asTORis at the μM concentration indicated (E). (F, G) Dose–response curves of AZD-8055 on ecotypes WS and Col-0 (F) and on Col-0 plants germinated on medium with lower glucose (0.15% instead of 0.8%) to get a bimodal plant population size distribution (G). (H) Pictures of whole plants (upper panel) and aerial parts (lower panel) after growth on different AZD-8055 concentrations. Bars, 1mm (lower panel), 1cm (upper panel). (I) Dose-dependent effect of first-generation (blue) and second-generation (red) asATMis on root growth. (J) Dose-dependent effect of the PI3Ks inhibitors PI-103 (blue) and LY294002 (red) on root growth compared with rapamycin (black). Data are the mean of two experiments done with two or three plates containing ten seeds. The standard error was low (~2%) and is often hidden by the size of points.
AZD-8055, one of the potent second-generation asTORis, inhibited the growth of two ecotypes of A. thaliana (Fig. 1F) and of plant populations whose average size was ~1cm (large) or 0.5cm (small) before transfer to drug-containing medium (Fig. 1G). As the same 50% growth inhibitory dose (GI50) was observed, this suggested that asTORis impair growth independently of the plant size and of the ecotype. Concomitantly with roots shortening, the cotyledons and leaves stayed green and their development was severely delayed by AZD-8055 (Fig. 1H). These data suggested that the delayed growth at the whole-plant level might be due to a decreased meristem activity and possibly the reduced size of differentiated cells. In addition, growth was rescued when plants were transferred back to medium without drug, showing the reversibility of the effect of the drug.
AZD-8055 is at least a 1000-fold less potent towards the related PI3K and the PIKK kinases ATM and DNA-PK than TOR in mammalian cells (Chresta et al., 2010). Recently, ATP-competitive inhibitors of ATM (asATMis), another PIKK conserved between A. thaliana and Homo sapiens (Garcia et al., 2000), were developed for mammalian cells (Golding et al., 2009). The first- and second-generation asATMis KU-55933 and KU-60019, respectively, have similar if not identical ATM target specificity with little to no non-specific target effects and, notably, they do not inhibit mTOR (Golding et al., 2009). KU-55933 was reported to be active in plants through inhibiting γ-H2AX protein phosphorylation (Amiard et al., 2010) and AtATM mutants have no growth defect. We wished to check whether asATMis could affect plant growth. No growth inhibition was observed (Fig. 1I), indicating that asATMis action does not interfere with the asTORis action on growth inhibition. This suggested that the selectivity of the active-site inhibitors of ATM and mTOR is probably conserved towards the corresponding kinases in A. thaliana for a large range of concentrations.
We also tested the dose-dependent effect on growth of two ATP-competitive inhibitors of mammalian PI3K, LY294002 and PI-103 (Table 1). LY294002, which was reported to be active also in plants (Lee et al., 2008), is a broad-spectrum inhibitor of PI3Ks that was not found to be exclusively selective for PI3K as it also inhibits other kinases including mTOR (Gharbi et al., 2007). PI-103 inhibits PI3K and mTORC1 (Fan et al., 2006; Raynaud et al., 2007). The effect of PI-103 could not be established as it slightly precipitated in the medium. LY294002 inhibited growth with a GI50 dose in the range of 10 μM (Fig. 1J), very close to that inhibiting growth of mammalian cells (Jiang et al., 2010).
Altogether, the ATP-competitive inhibitors of PI3K and PIKKs (ATM and TOR) designed for mammalian kinases that were workable in our conditions triggered plant growth inhibition (LY294002 and asTORis) or not (asATMis), with a specificity similar to that in mammalian cells. These results strongly suggest that the efficiency and specificity of kinase inhibition by ATP-competitive inhibitors is similar between plants and mammals.
AsTORis act in a TOR gene-dosage-dependent and specific manner in A. thaliana
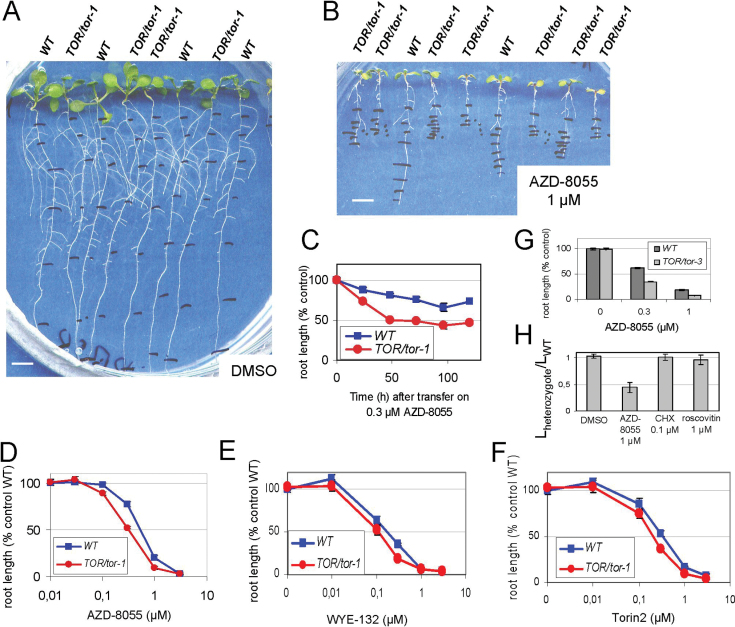
Studies in yeast and mammalian cells have demonstrated that lowering the dosage of a single gene encoding the target of a drug from two copies in a homozygous strain to one copy in a heterozygous strain results in increased drug sensitivity. This phenomenon of induced haplo-insufficiency has been used to identify the biological targets of drugs (Giaever et al., 1999; Lum et al., 2004; Kung and Shokat, 2005). To determine whether asTORis induce AtTOR haplo-insufficiency, we tested the dose-dependent effect of AZD-8055, WYE-132 and Torin2 on the growth of heterozygous AtTOR mutants. Loss-of-function homozygous mutants in the AtTOR gene are embryo lethal, but no phenotype has been described previously in heterozygous plants (Menand et al., 2002; Ren et al., 2011). The GUS reporter gene was fused in frame downstream of the coding sequence of the AtTOR gene, so the the tor-1 allele could be detected by following GUS expression in meristems (Menand et al., 2004). The progeny of TOR/tor-1 heterozygous plants that had been recorded individually for daily growth for 6 d were each subjected to GUS staining for genotyping. TOR/tor-1 heterozygous plants were not significantly different from WT when grown without the drug (Fig. 2A), whereas they were significantly smaller than WT plants when transferred to medium containing asTORis, as shown for AZD-8055 in Fig. 2B. Indeed, while the progression of the time course of inhibition was the same for WT and heterozygotes (Fig. 2C), the GI50 value was twofold lower in TOR/tor-1 plants than in WT (Fig. 2D–F). This suggested that all three second-generation asTORis could be used interchangeably. We performed a similar experiment using TOR/tor-3, another independent line that has a T-DNA insertion at the beginning of the TOR coding sequence (Ren et al., 2011). In a similar manner to TOR/tor1, TOR/tor-3 heterozygous plants, which were genotyped by PCR, grew like WT plants without drug and showed increased sensitivity compared with the WT when treated with AZD-8055 (Fig. 2G). Taken together, these data showed that the level of root growth inhibition by second-generation asTORis is dependent on the number of copies of the AtTOR gene.
Fig. 2.
AZD-8055, WYE-132, and Torin2 induce AtTOR haplo-insufficiency. Pictures and measurements were done 6 d after transfer of 3-d-old plants onto medium containing the indicated concentration of AZD-8055. (A, B) Whole plants from the progeny of TOR/tor1 heterozygotes grown vertically on DMSO (A) or 1 μM AZD-8055 (B). The genotypes of plants are indicated at the top of each plant after GUS expression was determined for each plant, whose growth was previously recorded. Bars, 500 μm. (C) Time course of root growth delay by 0.3 μM AZD-8055 of TOR/tor-1 and WT. (D–F) Dose-dependent effect of AZD-8055 (D), WYE-132 (E), and Torin2 (F) on the growth of TOR/tor1 and WT plants. (G) Differential sensitivity to AZD-8055 of root growth of TOR/tor-3 plants compared with WT. (H) Ratio of the root lengths of the TOR/tor1 on WT plants grown on DMSO or on the growth inhibitors AZD-8055, cycloheximide (CHX), or roscovitine at the indicated concentrations.
To show that the heterozygote hypersensitivity phenotype was exclusively dependent on asTORis, we tested the dose-dependent effect of other growth inhibitors that act on different intracellular targets. Cycloheximide, an inhibitor of protein synthesis, and roscovitine, a cyclin-dependent kinase inhibitor, were selected because they were previously tested in plants and mammalian cells (Wallace et al., 1970; Planchais et al., 1997). They both inhibited root growth in a concentration-dependent manner (Supplementary Fig. S3 at JXB online) and were stable over time (Supplementary Fig. S4 at JXB online). We observed no difference in sensitivity to cycloheximide and roscovitine between TOR/tor-1 heterozygous and WT plants (Fig. 2H). Therefore, these two compounds did not induce the haplo-insufficiency phenotype, revealing the selectivity of asTORis towards AtTOR.
Altogether, these data strongly suggest that asTORis-induced haplo-insufficiency is exclusively linked to the AtTOR gene and that asTORis inhibit root growth via inhibition of the AtTOR protein. As the AtTOR gene is unique and the GI50 dose of asTORis was twice as low for heterozygotes, it is tempting to suggest that the amount of TOR protein is twofold lower in heterozygotes. Therefore, whereas TOR/tor heterozygotes grow similarly to WT without asTORis, we assume that the reduced amount of TOR protein per se is not critical for growth.
AZD-8055 induces exit from the MZ and inhibits cell growth and root hair elongation
The A. thaliana primary root is spatially separated into three developmental zones; the MZ is where new cells are produced through active proliferation, and they increase in size progressively until they reach the elongation zone (EZ), where the cells elongate rapidly, and the differentiation zone (DZ), where root hairs develop by tip growth from epidermal cells (Dolan and Davies, 2004).
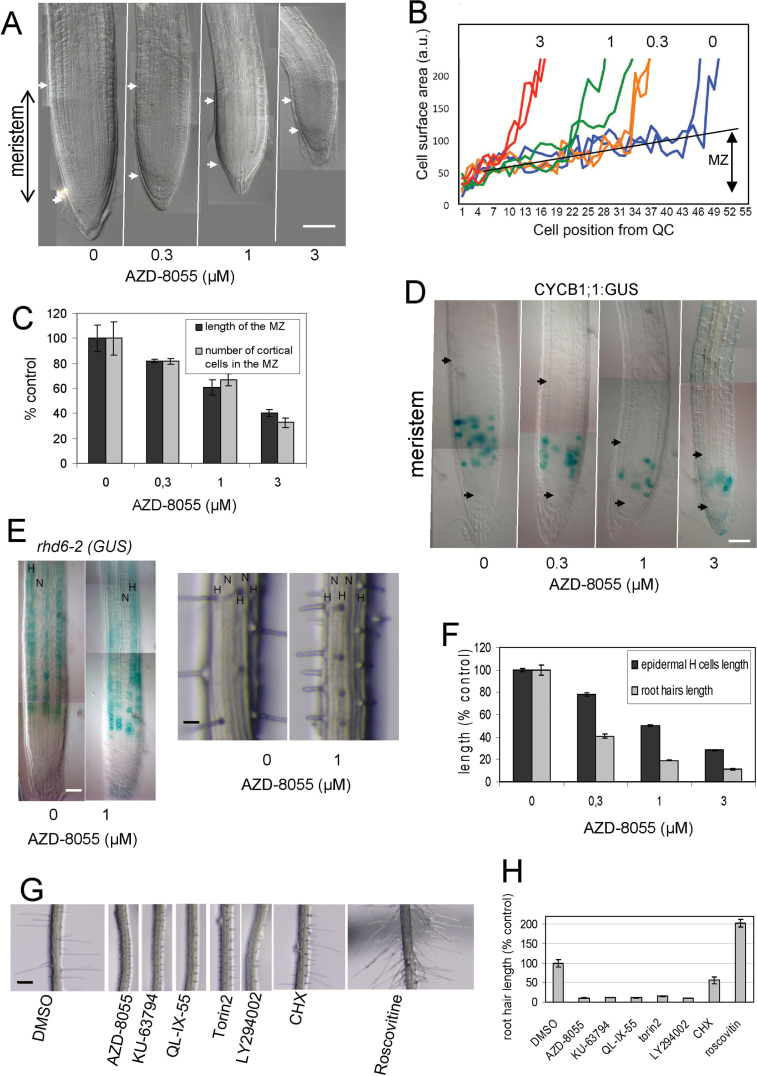
Two days after transfer of 3-d-old plants to AZD-8055, microscopic observations of root tips showed that AZD-8055 reduced the size of the MZ in a dose-dependent manner (Fig. 3A). Indeed, analysis of the surface area of cortical cells along the root, which reflects cell size, showed that the higher the AZD-8055 concentration, the smaller the size of cells leaving the MZ (Fig. 3B). In addition, the length of the MZ was linearly correlated with the number of cortical cells within the MZ (Fig. 3C). Therefore, the diminution of the MZ was due to a reduction in both the number of dividing cells and in cell growth in the MZ. CYCB1;1 is a marker of the G2/mitotic phase of the cell cycle and is expressed within the MZ in the zone of high mitotic activity (Colon-Carmona et al., 1999). CYCB1;1 accumulates when the cell cycle is blocked after stress revealing an increase in the number of cells whose division is blocked by G2 checkpoints (Ricaud et al., 2007). Such cell-cycle changes can be identified in plants expressing CYCB1;1:GUS by observing the density of blue cells. In CYCB1;1:GUS plants, we observed that increasing AZD-8055 reduced the zone of high mitotic activity (Fig. 3D). As the density of blue cells hardly changed within the zone of high mitotic activity, this suggested that G2 checkpoints that normally occur in a proliferating tissue are not impaired by asTORis. Altogether, our study indicated that AZD-8055 inhibits the cell proliferation capacity of the meristem by decreasing the number of cells in the MZ, mainly through promoting the acropetal differentiation of meristem cells.
Fig. 3.
AsTORis induce A. thaliana root phenotypes. Pictures and measurements were done 2 d after transfer of 3-d-old plants onto medium containing the indicated concentrations of AZD-8055. (A) Differential interference contrast microscopy picture of the MZ delimited by the arrows. (B) Cell surface area distribution of cortical cells relative to the quiescent centre (QC). Individual cells of two roots were measured for each indicated concentration of AZD-8055 (μM). (C) Length of the MZ and number of cortical cells of the MZ. (D) Expression of the marker CYCB1;1:GUS. (E) Expression of the rhd6-2 enhancer trap GUS gene (left panel) and pictures of epidermal cells in the differentiated area containing mature root hairs (right panel). Non-hair (N) and hair (H) cell files are indicated. (F) Dose response for AZD-8055 on the length of epidermal H cells and of root hairs. (G, H) Root hair phenotypes of plants grown with GI50 doses of the four indicated asTORis and LY294002 compared with cycloheximide (CHX) and roscovitine (G) and corresponding measurement of the inhibition relative to the DMSO control (H). Bars, 50 μm (A, D, E); 200 μm (G).
In A. thaliana roots, epidermal cells are arranged in alternative files of hair-bearing (H) and non-hair-bearing (N) cells. ROOT HAIR DEFECTIVE6 (RHD6) is a marker of H cells in the EZ before they produce hairs (Menand et al., 2007). The pattern of RHD6 expression in the EZ, and the alternation of H and N cell files in the differentiated root were also not affected by AZD-8055 (Fig. 3E). Cell size measurements in the DZ showed that AZD-8055 inhibited the length of H epidermal cells and of root hairs in a dose-dependent manner with a stronger effect on root hairs (Fig. 3 F). This indicated that AZD-8055 inhibits both the elongation of epidermal cells in the EZ and the root hair elongation that occurs in the DZ. Notably, the position of root hair emergence along epidermal cells was not affected (Supplementary Fig. S5 at JXB online). In summary, AZD-8055 induces MZ cells to differentiate and inhibits cell elongation and root hair growth but does not appear to affect the pattern of development per se.
Plants treated with four asTORis at concentrations close to their root GI50 showed a 90% shortening of root hairs (Fig. 3G, H). The effect of LY294002 was similar to that of asTORis whereas cycloheximide treatment resulted in a slight shortening of root hairs with an irregular distribution along the root. Conversely, roscovitine treatment resulted in very long root hairs. Therefore, we showed that several asTORis induced the same short root hair phenotype. This phenotype was distinct and separate from the root hair phenotypes caused by drugs that inhibit growth through blocking translation (cycloheximide) or the cell cycle (roscovitine). This evidence supports the notion that asTORis designed for mTOR specifically target AtTOR in A. thaliana. As LY294002, a broad-spectrum inhibitor of the kinase mPI3K that acts upstream of mTOR, induced the same root phenotype as asTORis, this suggests that the link between PI3K and TOR might be conserved in animals and plants.
AsTORis inhibit growth of diverse angiosperms
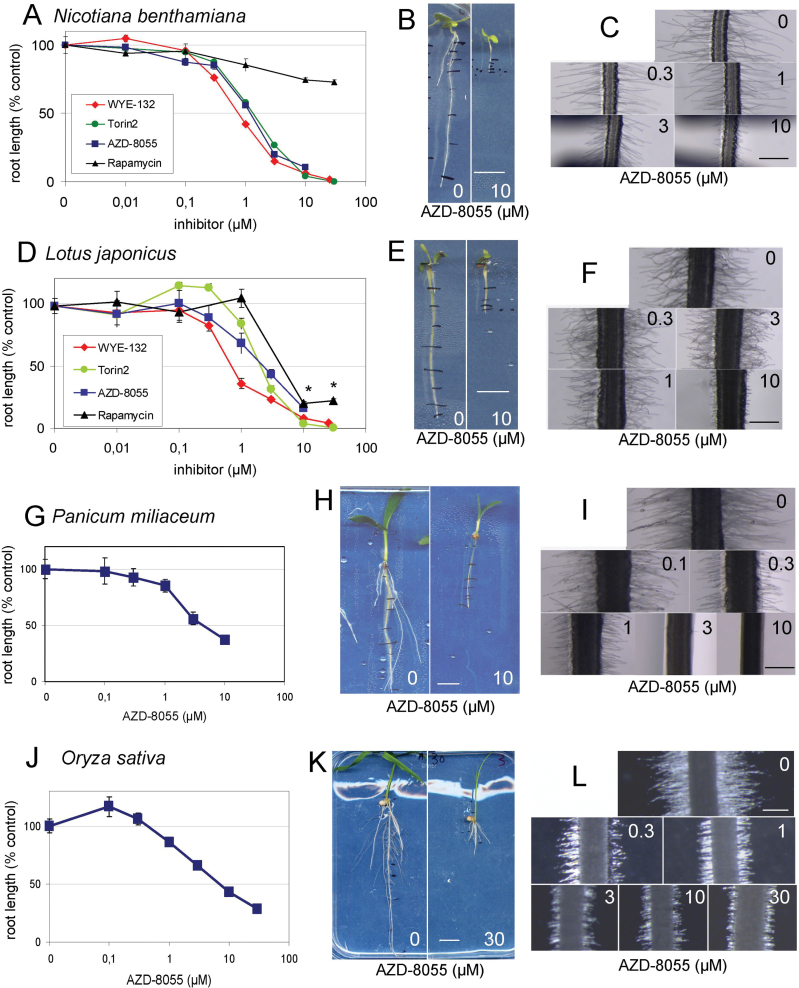
Angiosperms evolved approximately 150 million years ago and rapidly expanded to form the largest group of plants (Soltis et al., 2005). Angiosperms include monocots and eudicots. We selected species representative of diverse groups of angiosperms to test whether the sensitivity to asTORis was conserved. We selected two monocots, O. sativa (rice) and P. miliaceum (millet), and representatives of the two major clades of eudicots: the Rosids and Asterids. In addition to the Brassicale A. thaliana, L. japonicus, a second representative member of the Rosids, was chosen among the Fabales. We used the Solanales N. benthamiana as a representative of the Asterids.
We observed that AZD-8055, WYE-132, and Torin2 inhibited the primary root growth of both N. benthamiana and L. japonicus in a dose-dependent manner (Fig. 4A, B, D, E). Interestingly, more drug was necessary to inhibit the growth of these plants compared with A. thaliana, but, as for A. thaliana, WYE-132 inhibited N. benthamiana and L. japonicus root growth at a lower GI50 than AZD-8055 and Torin2 (Figs 1A–C and 4A, D). This suggested that the relative efficiency of these drugs to inhibit TOR is conserved between these species. Indeed, the growth of the aerial part of all species tested, as illustrated with A. thaliana (Fig. 1H) and N. benthamiana (Supplementary Fig. S6 at JXB online), was delayed in a dose-dependent manner. Furthermore, experiments prolonged for up to 2 weeks indicated that asTORis robustly inhibited A. thaliana and N. benthamiana growth with time and in a dose-dependent manner (Supplementary Fig. S7 at JXB online). For P. milliaceum and O. sativa, which have more complex root architectures, including seminal roots, we recorded the length of the whole root system. Again, root growth inhibition by AZD-8055 was observed in a dose-dependent manner but with a higher GI50 than for the Rosids tested (Fig. 4G, H, J, K). A slight stimulation of root growth at low doses occurred for AZD-8055 with O. sativa (Fig. 4J) and Torin2 with L. japonicus (Fig. 4D). This is typical of hormesis, a universal but still poorly explained phenomenon of growth stimulation at low doses or under slight stress that has been described in plants (Calabrese and Blain, 2009). This indicated that more than one drug should be tested in pharmacological studies in plants. Noticeably, the short root hair phenotype that we observed in A. thaliana (Fig. 3F) was also observed in all species (Fig. 4C, F, I and L). Therefore, asTORis inhibited root growth in a dose-dependent manner in the five divergent angiosperms species that we tested in this study, indicating that they could probably be used as an inhibitor of TOR in most angiosperms. Although concentrations inhibiting growth in plants are generally higher than in mammalian cell cultures, a phenomenon probably related to the dynamics of influx/efflux of drugs in plant roots and/or to the root morphology, the conserved short root hair phenotype and the similar progression of the dose–response curves suggest that root growth inhibition by asTORis occurs through the similar mechanism of TOR inhibition in all species tested.
Fig. 4.
Dose–response curves for AZD-8055, WYE-132, Torin2, and rapamycin on root growth, and root hair and whole-plant phenotypes in divergent angiosperms: N. benthamiana (A–C), L. japonicus (D–F), P. miliaceum (G–I), and O. sativa (J–L). (A, D, G, J) Dose-dependent inhibition of root growth by AZD-8055, WYE-132, and Torin2 (A, D) or AZD-8055 (G, J). Asterisks indicate concentration of rapamycin resulting in precipitates observed in the plates and precluding clear conclusions. (B, E, H, K) Pictures of whole plants at selected concentrations of AZD-8055. (C, F, I, L) Pictures of roots in the differentiated area of plants grown on different concentrations of AZD-8055. Bars, 1cm (B, E, H, K), 500 μm (C, F, I, L). (This figure is available in colour at JXB online.)
Discussion
Inhibition of TOR in planta has been challenging since the first identification of TOR in A. thaliana 10 years ago (Menand et al., 2002; Dobrenel et al., 2011). Indeed, several distinct strategies have been developed involving transgenic plants because of the absence of a clear-cut dose-dependent effect of rapamycin. Here, we showed that several ATP-competitive inhibitors of TOR recently developed for animal cells efficiently inhibited A. thaliana growth. All but two (Torin1 and WYE-354) stably inhibited the growth of A. thaliana. We did not test high concentrations of Torin1, as it is poorly soluble in DMSO and the homogeneity of our experimental conditions would have been compromised. With regard to WYE-354, we observed a relaxation of inhibition and precipitates occurring during the growing period. This illustrated that the solubility of those compounds together with the stability and bioavailability (Golding et al., 2009; Liu et al., 2011) are key points that have to be analysed carefully in plant pharmacology studies.
Several groups have recently reported that asTORis, which inhibited mTOR but not PI3-K, provide a new pharmacological approach to selective mTOR inhibition (Guertin and Sabatini, 2009; Feldman and Shokat, 2011). Noticeably, we have shown here that the decrease in GI50 concentrations for growth inhibition by second-generation asTORis was 10 to 100 times slower compared with first-generation ones (Fig. 1A, C and Table 1). This is agreement with the results reported for mammalian cells. In addition, we showed that three second-generation asTORis, AZD-8055, WYE-132, and Torin2, induced haplo-insufficiency of the TOR/tor-1 heterozygote, i.e. the GI50 values for these asTORis were twice as low for the TOR/tor heterozygotes compared with the WT. AsTORis are highly selective to mammalian TOR compared with tens or hundreds of kinases tested including PI3Ks and other PIKKs, which are the closest structurally related kinases (Table 1 and references therein). Conversely, inhibitors of PI3Ks are barely selective to mTOR as exemplified by LY294002 (Gharbi et al., 2007; Zhang et al., 2011). Indeed, several asTORis including those used in our study have been developed from the lead compound PI-103, a dual inhibitor of PI3K/mTORC1 (Zhang et al., 2011). It is worth mentioning here that asATMis do not inhibit growth at concentrations that are active in plants (Amiard et al., 2010), showing that they are also specific in plants and probably do not compete with ATP in TOR. Due to conservation of the TOR protein, overall our data strongly support the suggestion that AtTOR is a target of asTORis in planta. Therefore, the haplo-insufficiency of AtTOR heterozygotes suggests that the amount of TOR protein inside heterozygote cells is twice as low as in the WT. With the stoichiometry observed, we hypothesize that half the amount of AtTOR protein is still sufficient to ensure heterozygote growth that is undistinguishable from the WT without asTORis. Altogether, our data suggest that animal and plant TOR proteins have a similar sensitivity to asTORis at the enzymatic level.
The range of concentrations of asTORis that inhibit growth is generally higher in plants than in mammalians cells (Table 1 and references therein) but much less than in yeast (Liu et al., 2012b ). The drug content in tissues is the result of the equilibrium between the drug concentration in the medium and in the tissue, and is a function of drug uptake, drug metabolism, drug efflux, and its ability to inhibit all complexes of the targeted protein. In our study, A. thaliana growth was inhibited by GI50 concentrations of 0.6 μM AZD-8055, 3 μM KU63794, and 10 μM WYE-354 (Fig. 1A–C). In yeast, growth inhibition by the same compounds was obtained with GI50 concentrations between 25 and 75 μM. Moreover, this was obtained by using a triple mutant, which lacks two ABC multidrug transporters and the sterol methyltransferase ERG6, showing the importance of efflux and/or metabolism of xenobiotics. Similarly, the ineffectiveness of Torin1 in yeast was probably due to a intracellular concentration that was too low, caused by export by efflux carriers (N.S. Gray, personal communication). Therefore, a number of Torin1 analogues were screened for inhibition at a dose of 10 μM, and QL-IX-55 was selected (Liu et al., 2012a ). The GI50 concentration for QL-IX-55 was nevertheless still higher than 10 μM with the WT and 163nM with the triple mutant, illustrating efflux or metabolism of xenobiotics. It is likely that plants have also efflux and metabolism mechanisms that would detoxify part of asTORis (Kreuz et al., 1996). This would explain the higher concentrations required to inhibit plant growth compared with mammalian cells and the different GI50 concentrations of asTORis for inhibition of angiosperm species. In addition, this would explain at least partially why rapamycin barely inhibits plant growth at 10 μM, even in FKBP-overexpressing lines, when inhibition occurs in the nanomolar range in yeast and mammalian cells (Sormani et al., 2007; Ren et al., 2012; Xiong and Sheen, 2012). In addition, the absence of a clear-cut dose-dependence effect of rapamycin on plant growth compared with asTORis probably reminds us that the sole inhibition of TORC1 but not all TOR complexes in mammals is a situation that might also occur in plants, underlining the potency of asTORis in both cases. Lastly, the dose–response curve of O. sativa/AZD-8055 and L. japonicus/Torin2 had an inverted U shape that is characteristic of hormesis (Calabrese and Blain, 2009). This indicates the importance of careful examination of the effect of several inhibitors on growth.
S6K is a conserved indicator of the TOR kinase activity that is phosphorylated at the T449 of AtS6K. It has been recognized by an antibody developed against the human S6K1 T(P)-389 (Xiong and Sheen, 2012). In another study, 15-d-old plants incubated for a couple of hours with 200nM Torin1 were associated with a decrease in AtS6K1 phosphorylation in extracts of the AT7 line that overexpress the cauliflower mosaic virus translation reinitiation factor TAV (Schepetilnikov et al., 2011). As the AT7 line was supposed to have a constitutive level at least eightfold higher that WT, the endogenous level of the phosphorylated AtS6K was barely detected in the WT. More recently, the phosphorylation of S6K1 T449 was shown to be inhibited when 100nM of Torin1 was added for 30min to transfected protoplasts transiently expressing AtS6K1–FLAG (Xiong et al., 2013). These concentrations are close to those used for mTORC1 and mTORC2 inhibition (Thoreen et al., 2009). If both examples illustrate the inhibition of AtTOR by Torin1, they also show that the endogenous levels of AtS6Ks are barely detectable with antibodies against human S6K in WT plants as already shown (Xiong and Sheen, 2012). The methodology of detection of endogenous plant S6Ks and their phosphorylated counterpart in WT plants has to be improved. For instance, it would be useful to develop antibodies against plant S6Ks and/or more sensitive detection methods such as ELISA (Ren et al., 2012).
The dose-dependent effect of asTORis on A. thaliana growth inhibition has allowed us to analyse in detail the progressive process of root growth delay and its consequences on organ development. As the asTORis dose increases, we observed the progressive diminution of the length of (i) the MZ; (ii) the division zone in the MZ; (iii) epidermal cells in the EZ; and (iv) root hair cells. Concomitantly, the density of cells that expressed the marker of the G2/mitotic phase, CYCB1;1–GUS, was unchanged. The effect of asTORis proceeded through a reduction in the number of MZ cells concomitant with their differentiation. The unchanged density of G2-marked cells when MZ size decreased indicated that the asTORis probably do not act on the G2 phase. It would be interesting to examine whether the other cell-cycle phases, and particularly the G1 and S phases, change. Indeed, during revision of this manuscript, a biochemical link between AtTOR inhibition and a marker of the S phase, transcription factor E2Fa, was reported in A. thaliana leaf protoplasts (Xiong et al., 2013). As our assay dealt with roots shifting from growth to inhibition of growth in the presence of asTORis and as the above-mentioned assay used protoplasts that are shifting from ‘quiescence’ of leaf cells to potential proliferation of protoplasts, it is difficult to speculate. It would certainly be interesting to follow the phosphorylation status of E2Fa in both situations in order to decipher the link between the cell cycle and TOR.
Along the root, no change in the patterning of cell lines such as hair-bearing and non- hair-bearing cells or of the patterning of root hair emergence was observed. This suggests that root growth inhibition by asTORis is a ‘mild’ process that does not trigger tissue dislocation, probably through a ‘mild’ effect on the cell cycle among other putative functions. Indeed, the size of the plants at the time of transfer did not change the dose–response curve. The cyclin-dependent kinase inhibitor roscovitine and the protein synthesis inhibitor cycloheximide did not phenocopy the asTORi-induced phenotype. This indicated that asTORis phenotypes would not be exclusively and/or directly due to inhibition of the cell cycle or translation. This suggests that the plant TOR inhibition phenotype results in coordinated and multifaceted changes.
Altogether, these phenotypes illustrate a slowdown in growth dynamics caused by TOR inhibition, as evidenced by a reduction in the number of divisions undergone by meristematic cells and reduced elongation as they approach the transition zone. Furthermore, the elongation of these prematurely differentiated cells was also decreased and subsequently the root hairs were also shortened. The development of the aerial part of all species studied was also slowed by asTORis, and the number and size of leaves decreased concomitantly with root shortening. However, no stress phenotypes could be observed. Therefore, the overall process of TOR inhibition is likely to operate by triggering a signal of growth delay or arrest that homogeneously affects all cells and does not interfere with the cell fate. The strength of this reversible slowdown signal is a function of the concentration of the asTORi and is of primary importance to further studies.
As AtTOR is expressed in the MZ (Menand et al., 2002), the use of asTORis opens new possibility of studying the connection between hormone signalling, cell fate, and the AtTOR pathway. Indeed, the asTORis-induced reduction of MZ resembles the effect of an increase in cytokinins and a decrease in gibberellic acid (Dello Ioio et al., 2007; Ubeda-Tomas et al., 2009). Assuming that the accumulation of hormones transporters and hormone synthesis are not disturbed, whereas proliferation slows down, the premature differentiation of asTORi-treated cells may be caused by an increase and shortening of the hormone gradient along the root (Muraro et al., 2013). This hypothesis is supported by the recent report that auxin and cytokinin signalling does not require AtTOR signalling and mitochondrial energy relay (Xiong et al., 2013). Lastly, as the PI3K inhibitor LY294002 generates a root phenotype close to asTORis, we suggest that, as in mammals (Laplante and Sabatini, 2012), PI3K may act upstream of AtTOR to integrate hormone signalling. However, more in-depth studies are required, as LY294002 was found not to be exclusively selective for the PI3Ks (Gharbi et al., 2007).
A reduction in the MZ and root hair length was also reported in FKBP12-overexpressing lines grown on 10 μM rapamycin (Ren et al., 2012), sharing part of the phenotypes with asTORis, although a rapamycin dose dependency was not reported. This argues in favour of the efficiency of asTORis in plants with the advantage of a clear-cut dose–response dependency of growth inhibition and tissue morphology in a swift and reproducible way. Therefore, asTORis allow inhibition of plant TOR without the production of chimaeric transgenic lines and/or RNA-silenced lines. Therefore, asTORis should help identify new targets of plant TOR kinase and establish the structure of the plant TOR pathway and its specific interconnection with cell energy status and hormone signalling. The finding that asTORis efficiently inhibit growth of different plant species shows that they could be used to decipher the degree of conservation of the TOR pathway among angiosperms.
Supplementary Material
Acknowledgements
This work was supported by the Agence Nationale de la Recherche grants ENVI-DEVO (ANR-08-JCJC-0054-01) and TRANSLATOR (ANR-BSV-010-01). We acknowledge Stefan Jouannic (IRD, Montpellier, France), Cécile Vriet (LGBP), Trevor L. Wang (John Innes Centre, Norwich, UK), and Muriel Reissolet (LGBP) for WT seeds, and Raju Datla (NRCC, Canada) for TOR/tor-3 seeds. We thank Nathanael S. Gray and Liu Qingsong for the generous gifts of the Torin1, Torin2, and QL-IX-55 inhibitors. We also acknowledge Coralie Horin for her help during the first experiments with asTORis. The microscopy was done in the IBDML imaging facility of Aix-Marseille University. The help of Elsa Crudeli and Cédric Matthews is acknowledged, as well as Alexandre Tremeau-Bravard, Ben Field, and Samantha J. England for critical review of the manuscript.
References
- Amiard S, Charbonnel C, Allain E, Depeiges A, White CI, Gallego ME. 2010. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis . Plant Cell 22, 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach S, Knockaert M, Reinhardt J, et al. 2005. Roscovitine targets, protein kinases and pyridoxal kinase. Journal of Biological Chemistry 280, 31208–31219 [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. 2007. The selectivity of protein kinase inhibitors: a further update. Biochemistry Journal 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese EJ, Blain RB. 2009. Hormesis and plant biology. Environmental Pollution 157, 42–48 [DOI] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. 2012. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana . The Plant Journal 73, 897–909 [DOI] [PubMed] [Google Scholar]
- Chresta CM, Davies BR, Hickson I, et al. 2010. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Research 70, 288–298 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. 1999. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. The Plant Journal 20, 503–508 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17, 678–682 [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. 2007. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports 8, 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Sormani R, Moreau M, Mozzo M, Montane MH, Menand B, Robaglia C, Meyer C. 2011. Regulation of plant growth and metabolism by the TOR kinase. Biochemical Society Transactions 39, 477–481 [DOI] [PubMed] [Google Scholar]
- Dolan L, Davies J. 2004. Cell expansion in roots. Current Opinion in Plant Biology 7, 33–39 [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Fonseca BD, Sonenberg N. 2010. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochimica et Biophysica Acta 1804, 433–439 [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Molecular Genetics & Genomics 206, 200–206 [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. 2006. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell 9, 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Shokat KM. 2011. New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor kinase domain inhibitors (TORKinibs). Current Topics in Microbiology and Immunology 347, 241–262 [DOI] [PubMed] [Google Scholar]
- Finlay MR, Griffin RJ. 2012. Modulation of DNA repair by pharmacological inhibitors of the PIKK protein kinase family. Bioorganic & Medicinal Chemistry Letters 22, 5352–5359 [DOI] [PubMed] [Google Scholar]
- Garcia V, Salanoubat M, Choisne N, Tissier A. 2000. An ATM homologue from Arabidopsis thaliana: complete genomic organisation and expression analysis. Nucleic Acids Research 28, 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. 2009. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochemistry Journal 421, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard M, Deleersnijder A, Demeulemeester J, Debyser Z, Baekelandt V. 2011. Unraveling the role of peptidyl-prolyl isomerases in neurodegeneration. Molecular Neurobiology 44, 13–27 [DOI] [PubMed] [Google Scholar]
- Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, Waterfield MD. 2007. Exploring the specificity of the PI3K family inhibitor LY294002. Biochemistry Journal 404, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. 1999. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nature Genetics 21, 278–283 [DOI] [PubMed] [Google Scholar]
- Golding SE, Rosenberg E, Valerie N, et al. 2009. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin AKT and ERK prosurvival signaling, and inhibits migration and invasion. Molecular Cancer Therapeutics 8, 2894–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. 2009. The pharmacology of mTOR inhibition. Science Signaling 2, pe24. [DOI] [PubMed] [Google Scholar]
- Jiang H, Fan D, Zhou G, Li X, Deng H. 2010. Phosphatidylinositol 3-kinase inhibitor (LY294002) induces apoptosis of human nasopharyngeal carcinoma in vitro and in vivo. Journal of Experimental & Clinical Cancer Research 29, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuz K, Tommasini R, Martinoia E. 1996. Old enzymes for a new job (herbicide detoxification in plants). Plant Physiology 111, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C, Shokat KM. 2005. Small-molecule kinase-inhibitor target assessment. ChemBioChem 6, 523–526 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y. 2008. Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiology 147, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. 2010. The TOR pathway modulates the structure of cell walls in Arabidopsis . Plant Cell 22, 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang DQ. 2010. The ATM inhibitor KU-55933supp resses cell proliferation and induces apoptosis by blocking Akt in cancer cells with overactivated Akt. Molecular Cancer Therapeutics 9, 113–125 [DOI] [PubMed] [Google Scholar]
- Lianguzova MS, Chuykin IA, Nordheim A, Pospelov VA. 2007. Phosphoinositide 3-kinase inhibitor LY294002 but not serum withdrawal suppresses proliferation of murine embryonic stem cells. Cell Biology International 31, 330–337 [DOI] [PubMed] [Google Scholar]
- Liu Q, Chang JW, Wang J, et al. 2010. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benz o[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. Journal of Medicinal Chemistry 53, 7146–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kirubakaran S, Hur W, et al. 2012a. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. Journal of Biological Chemistry 287, 9742–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ren T, Fresques T, Oppliger W, Niles BJ, Hur W, Sabatini DM, Hall MN, Powers T, Gray NS. 2012b. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae . ACS Chemical Biology 7, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, Sabatini DM, Gray NS. 2011. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. Journal of Medicinal Chemistry 54, 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang JM, Zhang SS, Liu XY, Liu DX. 2010. Induction of cell cycle arrest at G1 and S phases and cAMP-dependent differentiation in C6 glioma by low concentration of cycloheximide. BMC Cancer 10, 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, et al. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116, 121–137 [DOI] [PubMed] [Google Scholar]
- Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. European Journal of Biochemistry 243, 527–536 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA 99, 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Meyer C, Robaglia C. 2004. Plant growth and the TOR pathway. Current Topics in Microbiology and Immunology 279, 97–113 [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer D, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480 [DOI] [PubMed] [Google Scholar]
- Moon SW, Chung EJ, Jung SA, Lee JH. 2009. PDGF stimulation of Muller cell proliferation: contributions of c-JNK and the PI3K/Akt pathway. Biochemical and Biophysical Research Communications 388, 167–171 [DOI] [PubMed] [Google Scholar]
- Muraro D, Byrne H, King J, Bennett M. 2013. The role of auxin and cytokinin signalling in specifying the root architecture of Arabidopsis thaliana. Journal of Theoretical Biology 317, 71–86 [DOI] [PubMed] [Google Scholar]
- Perilli S, Sabatini S. 2010. Analysis of root meristem size development. Methods in Molecular Biology 655, 177–187 [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Trehin C, Perennes C, Bureau JM, Meijer L, Bergounioux C. 1997. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2cell suspension. The Plant Journal 12, 191–202 [DOI] [PubMed] [Google Scholar]
- Raynaud FI, Eccles S, Clarke PA, et al. 2007. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Research 67, 5840–5850 [DOI] [PubMed] [Google Scholar]
- Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. 2011. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis . Plant Physiology 155, 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, et al. 2012. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis . Plant Cell 24, 4850–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montane MH. 2007. ATM-mediated transcriptional and developmental responses to γ-rays in Arabidopsis . PLoS One 2, e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. 2012. Sensing nutrient and energy status by SnRK1 and TOR kinases. Current Opinion in Plant Biology 15, 301–307 [DOI] [PubMed] [Google Scholar]
- Santoni V, Bellini C, Caboche M. 1994. Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis mutants. Planta 192, 557–566 [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. 2011. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO Journal 30, 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature Chemical Biology 6, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfontein J, Nisbet RE, Howe CJ, de Vries PJ. 2010. Evolution of the TSC1/TSC2-TOR signaling pathway. Science Signaling 3, ra49. [DOI] [PubMed] [Google Scholar]
- Soltis D, Soltis P, Endress P, Chase M. 2005. Phylogeny and evolution of angiosperms. Sunderland: Sinauer Associates [Google Scholar]
- Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C. 2007. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biolog 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed F, Sanganee HJ, Bahl A, Bayat A. 2013. Potent dual inhibitors of TORC1 and TORC2 complexes (KU-0063794 and KU-0068650) demonstrate in vitro and ex vivo anti-keloid scar activity. Journal of Investigative Dermatology 133, 1340–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton GW, Moorhead GB. 2005. The phosphoinositide-3-OH-kinase-related kinases of Arabidopsis thaliana . EMBO Reports 6, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. Journal of Biological Chemistry 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomas S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. 2009. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Current Biology 19, 1194–1199 [DOI] [PubMed] [Google Scholar]
- Wallace A, Ashcroft RT, Leo MW, Wallace GA. 1970. Effect of cycloheximide, gamma irradiation, and phosphorus deficiency on root pressure exudation in tobacco. Plant Physiology 45, 300–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2012. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. Journal of Biological Chemistry 287, 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. 2013. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Shi C, Toral-Barza L, et al. 2010. Beyond rapalog therapy: preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Research 70, 621–631 [DOI] [PubMed] [Google Scholar]
- Yu K, Toral-Barza L, Shi C, Zhang WG, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, et al. 2009. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Research 69, 6232–6240 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Duan Y, Zheng XF. 2011. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discovery Today 16, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.