Abstract
Abiotic stress tolerance in plants is pivotal to increase yield stability, but its genetic basis is still poorly understood. To gain insight into the genetic architecture of frost tolerance, this work evaluated a large mapping population of 1739 wheat (Triticum aestivum L.) lines and hybrids adapted to Central Europe in field trials in Germany and fingerprinted the lines with a 9000 single-nucleotide polymorphism array. Additive effects prevailed over dominance effects. A two-dimensional genome scan revealed the presence of epistatic effects. Genome-wide association mapping in combination with a robust cross-validation strategy identified one frost tolerance locus with a major effect located on chromosome 5B. This locus was not in linkage disequilibrium with the known frost loci Fr-B1 and Fr-B2. The use of the detected diagnostic markers on chromosome 5B, however, does not allow prediction of frost tolerance with high accuracy. Application of genome-wide selection approaches that take into account also loci with small effect sizes considerably improved prediction of the genetic variation of frost tolerance in wheat. The developed prediction model is valuable for improving frost tolerance because this trait displays a wide variation in occurrence across years and is therefore a difficult target for conventional phenotypic selection.
Key words: Association mapping, cross-validation, frost tolerance, genomic selection, Triticum aestivum, wheat.
Introduction
Wheat (Triticum aestivum L.) is one of the most important crops and is grown on 200 million hectares of farmland worldwide (Ortiz et al., 2008). In the northern hemisphere, wheat is commonly planted in the autumn to escape the potential drought stress in the summer (Vágújfalvi et al., 2012). Besides this, autumn-planted winter wheat possesses a higher yield potential than spring wheat owing to its longer growing period.
Frost tolerance is essential for autumn-planted wheat to survive freezing temperatures during winter in temperate zones (Sandve et al., 2011). Therefore, the genetic architecture of frost tolerance has been investigated for almost a century (Skinner and Garland-Campbell, 2008; Galiba et al., 2009). One prominent frost tolerance locus is Fr-1 (Sutka and Snape, 1989; McIntosh et al., 1998), which maps close to Vrn-A1 and thus has been supposed to represent a pleiotropic effect of Vrn-A1 (Dhillon et al., 2010). An additional locus underlying frost tolerance, Fr-A2, has been identified on chromosome 5A ~30 cM away from Vrn-A1 (Vágújfalvi et al., 2000, 2003). Homoeologous loci were reported for chromosomes 5B (Toth et al., 2003) and 5D (Snape et al., 1997). In addition to the major Fr-1 and Fr-2 loci, there are multiple genes with small effects underlying frost tolerance. Therefore, while some progress can be achieved by selecting for beneficial Fr-1 and Fr-2 alleles, once those are fixed in a breeding programme, additional tools are required to discover and deploy additional genes to further improve this trait.
Genomic selection has been suggested to predict phenotypes for traits that are controlled by multiple genes with small effects. In this approach, a large number of markers distributed across the genome are used simultaneously to train a prediction model (Meuwissen et al., 2001). Genomic selection has shown great success in livestock improvement (Hayes, 2009) and more recently also in plant breeding (Jannink et al., 2010). The simultaneous use of a large number of markers also has led to a substantially enhanced understanding of complex traits in human genetics (Lee et al., 2008).
In this study, a large and diverse mapping population of 1604 wheat hybrids and their 135 parental inbred lines was evaluated for frost tolerance and fingerprinted with a 9K single-nucleotide polymorphism (SNP) array (Würschum et al., 2013). Genome-wide association mapping was applied to study the genetic architecture of frost tolerance in winter wheat, and one major additive effect quantitative trait locus (QTL) was revealed on chromosome 5B. The scan for dominance effects determined their contribution to genetic variation, which was, however, considerably smaller than that of the additive effects. Similarly, several interaction effects were identified among genes, each explaining only a small proportion of the genetic variation of frost tolerance. In contrast to the genome-wide association mapping, the application of two genomic selection approaches that take into account also loci exhibiting small size effects led to a considerable increase in the accuracy of frost tolerance prediction in wheat.
Materials and methods
Plant material and field experiments
This study was based on 1604 elite wheat hybrids and their 135 parental inbred lines adapted to Central Europe (Supplementary Table S1, available at JXB online). For hybrid seed production, the 135 parental inbred lines were classified into a male group consisting of tall and open-pollinating genotypes and a female group of semi-dwarf genotypes showing a short delay in flowering time compared to the male lines. The hybrids were derived by crossing 120 female and 15 male lines (Supplementary Fig. S1) using chemical hybridization agents. The 1739 genotypes were evaluated for frost tolerance in field trials at three locations in Germany (Seligenstadt: 50° 2′ N 8° 58′ E, 278 m above sea level, silt clay loam soil texture; Böhnshausen: 51° 51′ N 10° 57′ E, 146 m above sea level, sandy loam texture; Adenstedt: 52° 0′ N 9° 56′ E, 71 m above sea level, loam soil texture) in the year 2012. The experimental designs were partially replicated alpha designs and 29% of the hybrids were tested in two replications (Williams et al., 2010). The hybrids and lines were split into three trials linked with 10 common checks (Asdecouer, Genius, Hystar, JBAsano, Julius, Tuerkis, Colonia, Kredo, Tobak, and Tabasco). Sowing was done in the first 2 weeks of October and sowing densities ranged from 230 to 290 grains m–2 and plot sizes ranged from 7.5 to 9.7 m2. The plots were treated with fertilizers, fungicides, and herbicides according to standard agronomic practices for intensive wheat production. Frost tolerance was visually scored on a scale from 1 (no damage) to 9 (no plant survived).
Genotypic data
Genotyping was done with the 9K SNP array based on the Illumina Infinium assay (for details see Würschum et al., 2013). Quality checks for the SNP markers were performed to exclude those showing: (1) rate of missing values above 5%; (2) rate of heterozygosity in the parental inbred lines above 5%; or (3) minor allele frequency smaller than 0.05. In total, 1280 SNP markers were retained and missing genotypes were imputed following the approach suggested by Crossa et al. (2010; Supplementary Table S2). This work followed the suggestion of Bernardo (1993) and estimated the coancestry coefficients θ ij between inbreds i and j on the basis of marker data as θ ij = 1 + (S ij – 1)/(1 – T), where S ij is the proportion of marker loci with shared variants between inbreds i and j and T is the average probability that a variant from one parent of inbred i and a variant from one parent of inbred j are alike in state, given that they are not identical by descent. T was set as minimum of (1 – S ij) values.
Phenotypic data analyses
The phenotypic data of each environment were first analysed separately based on the statistical model
where y ijklm was the phenotypic performance for the ijth genotype in the mth incomplete block of the lth replication in the kth trial, µ was an intercept term, g ij was the genetic effect of the ijth genotype, t k was the effect of the kth trial, r lk was the effect of the lth replication in the kth trial, b mlk was the effect of the mth incomplete block in the lth replication of the kth trial, and e ijklm was the residual. Except b mlk, all effects were treated as fixed. Adjusted entry means were used in a second step to estimate the genetic variance components of hybrids and parental lines as well as the variance of genotype × location interactions. This work followed the suggestion of Möhring and Piepho (2009) and weighted each observation with one divided by their squared standard error. The variance of hybrids was further split into variance due to general and specific combining ability effects (Hallauer and Miranda, 1988). Significance of variance component estimates was tested by model comparison with likelihood ratio tests in which halved P-values were used as approximation (Stram and Lee, 1994). In addition, this work assumed fixed genetic effects and obtained the best linear unbiased estimates of the 1739 genotypes. The phenotypic data analyses were performed using the software ASReml 3.0 (Gilmour et al., 2009).
Genome-wide mapping
The additive and dominance design matrices for the hybrids and their parental lines were specified according to the F∞ metric of Falconer and Mackay (1996). Based on the adjusted entry means of the single locations, association mapping scans were performed for additive and dominance effects correcting for population stratification by fitting a polygenic effect for each individual as random, where the covariance for the polygenic effect is the kinship matrix estimated from the marker data (Yu et al., 2006). The kinship matrix for the parental lines was modelled using twice the estimated coancestry coefficients θ ij between inbreds i and j on the basis of marker data (Reif et al., 2011). The general combining ability effects reflect the additive effects of the hybrids (Hallauer and Miranda, 1988). Therefore, the kinship matrix for the hybrids modelled the covariance among general combining ability effects (Reif et al., 2013). This approach was contrasted with a Null model not correcting for population stratification and two models fitting an average heterotic effect (contrast of lines versus hybrids) considering and disregarding the kinship matrix. The significance of genome-wide association mapping scans for the main effects was estimated based on a false discovery rate (Benjamini and Hochberg, 1995) of 0.1. In addition, this work extended the tests for main effect and performed a two-dimensional genome scan for additive × additive, additive × dominance, dominance × additive, and dominance × dominance digenic epistatic effects. The extended biometrical model included the detected main-effect quantitative trait loci (QTL) as co-factors as well as the main and interaction effects of the marker pair under consideration (Würschum et al., 2011). The Bonferroni–Holm procedure (Holm, 1979) was applied to correct for multiple testing. The association mapping analyses were performed using the software ASReml 3.0 (Gilmour et al., 2009).
Based on the adjusted entry means of the 1739 genotypes, two approaches for genomic selection were applied considering additive and dominance effects: ridge regression best linear unbiased prediction (RR-BLUP; Whittaker et al., 2000) and BayesCπ (Dekkers et al., 2009; Habier et al., 2011). Details of the implementation of the models have been described in Zhao et al. (2013). All statistical procedures for the genomic selection approaches were executed using R (R Development Core Team, 2010).
The accuracy of the prediction of frost tolerance by association mapping and the two genomic selection approaches were evaluated using cross-validations. Since population structure in factorial crosses strongly influences prediction accuracy (Technow et al., 2012), a cross-validation strategy was used where training and validation sets were not related by common parental lines. This work sampled 100 times 610 hybrids and their 10 male and 80 female parental lines as the training set and estimated the additive and dominance effects. Hybrids based on the remaining parental lines formed the validation set in which predictions derived from the training set were tested for their accuracy. Prediction accuracy was estimated as Pearson’s correlation coefficient between the observed and the predicted hybrid performance.
Results
The 1739 entries were evaluated for frost tolerance occurring in the field under natural settings. Frost stress was severe at all three locations in the first half of February 2012, with minimum temperature down to –18 °C (Supplementary Fig. S2) concomitant with an absence of snow coverage and soil-borne diseases. The high correlations observed among the genotypic values estimated in the three locations (Supplementary Table S3) clearly suggested that a combined analysis across locations would not be biased by location-specific frost responses. The high frost stress revealed a large genetic variation for frost tolerance in the mapping population with high heritabilities (close to 90%; Table 1). The exceptional differentiation obtained under the natural setting is also underlined by the large difference observed in frost tolerance between the check varieties Julius and Tabasco (Supplementary Fig. S3). Julius is known to be frost tolerant and Tabasco frost susceptible.
Table 1.
First- and second-degree statistics for the 1604 hybrids and their 135 parental inbred lines evaluated for frost tolerance in field trials at three locations
| Source of variation | Frost tolerance |
|---|---|
| Lines | 5.51 (1.40–8.94) |
| Hybrids | 4.43 (1.28–8.58) |
| σ2 Lines | 3.14*** |
| σ2 Hybrids | 1.66*** |
| σ2 Additive-Female | 1.51*** |
| σ2 Additive-Male | 0.26*** |
| σ2 Dominance | 0.08*** |
| σ2 Lines × Location | 0.73*** |
| σ2 Hybrids × Location | 0.38*** |
| σ2 Additive-Male × Location | 0.36*** |
| σ2 Additive-Female × Location | 0.07*** |
| σ2 Dominance × Location | 0.04*** |
| σ2 e | 0.52 |
| Heritability Lines | 0.89 |
| Heritability Hybrids | 0.87 |
Values are mean (range) or variances. Frost tolerance was scored from 1 (no damage) to 9 (no plant survived).
*** Significantly different from zero at P ≤ 0.001.
This study observed a negligible correlation of 0.02 between frost tolerance and heading time. This indicated that the observed genotypic variation for frost tolerance was not likely to have been caused by differences in the transition from the vegetative to the generative growth phase. Nevertheless, as the transition from vegetative to generative growth was not measured directly, possible effects of the transition on frost tolerance cannot be ruled out completely.
Based on the genome-wide SNP marker data, there was no major population structure, but the presence of a family structure among the 135 parental lines (Supplementary Fig. S4). This can be explained by a steady interchange of lines among different wheat breeding programmes in their breeding history (Longin et al., 2012). The quantile–quantile plots for the four association mapping models revealed that, owing to the presence of family structures (Supplementary Fig. S4), population stratification had to be considered through the kinship matrix for the lines and hybrids for a single-locus regression model (Supplementary Fig. S5). Fitting an average heterotic effect, however, was not required. Mapping QTL in a combined population of hybrids and lines led to a substantially higher proportion of genotypic variance explained by the QTL (Supplementary Fig. S5). This held true even if the proportion of genotypic variation explained by SNPs was estimated separately for the parental lines and the hybrids. Therefore, the genome-wide mapping approaches were based on the combined population of hybrids and lines.
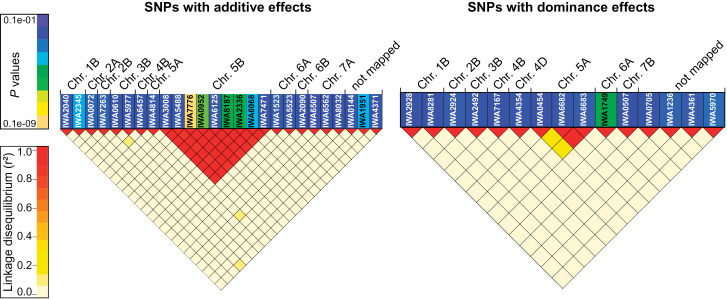
In the genome-wide association mapping scan, there were 26 and 15 SNPs which contributed significantly to the additive and dominance genetic variation for frost tolerance, respectively (Fig. 1). About 75% of these 41 SNPs exhibited significant (P < 0.01) interaction effects with the environments. A full two-dimensional scan for epistatic effects was performed, which revealed a total of 76 significant digenic epistatic effects. The distribution of the P-values revealed that, among the four types of digenic epistatic effects, additive × additive interactions were the most prevailing (Supplementary Fig. S6).
Fig. 1.
Heatplots of P-values of SNP markers contributing significantly to the additive and dominance genetic variation and linkage disequilibrium measured as squared Pearson’s correlation coefficients (r 2) among SNPs.
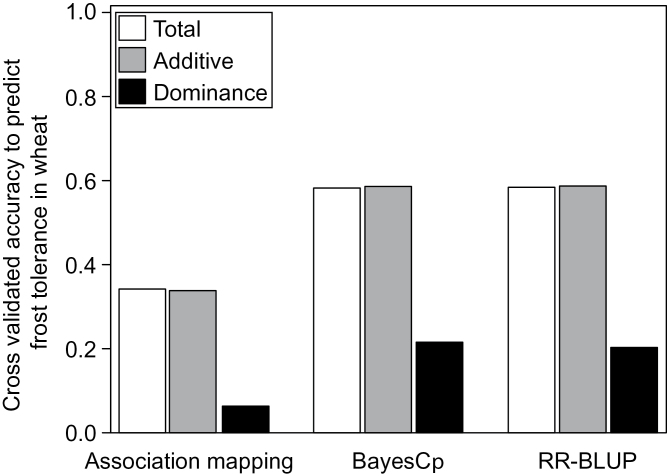
Using a robust cross-validation strategy in which training and validation sets were not linked through common parents, the phenotypic performance of unrelated hybrids could be predicted with an accuracy of 0.34 based on the SNPs detected in the association mapping scans (Fig. 2). By standardizing this with the square root of the broad-sense heritability (Dekkers, 2007), the accuracy increased up to 0.38. Employing only five linked SNPs that were significantly associated with frost tolerance in more than 90% of the cross-validation runs (Supplementary Table S4) was sufficient to explain 98% of the prediction accuracy of the total set of significant SNPs. These five SNPs all exhibit only significant additive effects, are located in the same genomic region on chromosome 5B, and have r 2 values amongst each other above 0.88.
Fig. 2.
Cross-validated accuracy to predict frost tolerance in wheat based on association mapping and the two genomic selection methods BayesCπ and RR-BLUP.
The location of these five SNPs suggests that this locus is proximal to Fr-B1/Vrn-B1, which is located roughly in the middle of the long arm (~70 cM from the centromere; Dubcovsky et al., 1998). By using previously developed PCR markers for this locus (Fu et al., 2005), this work confirmed that they are not linked to Fr-B1/Vrn-B1. Pearce et al. (2013) reported recently that a substantial proportion of the phenotypic variation of frost tolerance among spring wheats was associated with deletions involving CBF (C-repeat Binding Factor) genes at the Fr-B2 locus, also located on the long arm of chromosome 5B. The current work used the developed PCR markers for CBF-B9 and CBF-B14 (Pearce et al., 2013), which were segregating in the 135 parental wheat lines, and observed the absence of linkage disequilibrium between the alleles of the SNPs detected in the scan on chromosome 5B and the polymorphisms at the Fr-B2 locus with r 2 values smaller than 0.02.
The genomic selection approaches led to an accuracy of predicting the phenotypic performance of unrelated hybrids of 0.58, a substantial increase (nearly 60%) compared to the association mapping approach. BayesCπ and RR-BLUP showed negligible differences in the accuracy to predict frost tolerance despite the low posterior inclusion of markers in BayesCπ (20% additive and 6% dominance). There was an accuracy of prediction of frost tolerance with dominance effects based on RR-BLUP and BayesCπ of around 0.20. Nevertheless, a combined prediction considering additive and dominance effects was not superior to predicting frost tolerance exclusively by additive effects.
Discussion
A crucial factor in determining the quality of molecular data for genome-wide mapping studies is the extent of linkage disequilibrium. Theoretical findings (Hayes et al., 2009) suggest that a SNP marker density resulting in mean r 2 values among adjacent loci of above 0.2 is useful for genome-wide approaches to predict phenotypic performance. The current study’s companion paper (Würschum et al., 2013) estimated that the marker density underlying this study provided a magnitude of linkage disequilibrium measured as mean squared Pearson’s correlation coefficient (r 2) between adjacent SNPs of 0.32, which suggests that genome-wide mapping is promising with this experimental set up.
The genome-wide association mapping scan in combination with the robust cross-validation approach suggests that one gene with a major effect is present roughly in the middle of the long arm of chromosome 5B (Fig. 1). Interestingly, among the SNPs in that region, there were large differences in P-values, ranging from 4.4E–4 for IWA6125 to 8.5E–14 for IWA7776. This is surprising because this study observed high r 2 (above 0.9) between adjacent SNPs. Obviously the mapping resolution can still profit from the 10% of variation not explained by adjacent SNPs. The alleles of the SNPs detected in the scan on chromosome 5B were not in linkage disequilibrium with known polymorphisms at the Fr-B1 and Fr-B2 loci. Consequently, these findings suggested that a new frost stress locus had been detected on chromosome 5B.
Previous studies suggested that Vrn-A1 contributes substantially to the genetic variation of frost tolerance when differences are tested between winter and spring alleles (Sutka and Snape, 1989). However, significant effects associated to Vrn-A1 have been also detected among winter wheat varieties (Chen et al., 2009). One out of the 1280 SNPs (IWA0454) was tightly linked with two previously described SNPs in the Vrn-A1 gene (Chen et al., 2009; Díaz et al., 2012). Adding the marker IWA0454 in the cross-validation study resulted in an increase of 10% in accuracy compared to predicting frost tolerance based on the five SNPs described above. This underlines the possibility that VRN-A1 is involved in the determination of frost tolerance of elite wheat varieties, although with lower impact as expected from previous studies (Dhillon et al., 2010). The Fr-2 locus (Vágújfalvi et al., 2000) explained up to 40% of the phenotypic variation of frost tolerance in bi-parental QTL mapping studies based on contrasting parents (e.g. Båga et al., 2007). However, no major effect loci were observed on chromosome 5A. This can be explained either by fixation of the Fr-A2 locus in the lines adapted to Central Europe, a strong dependency of estimated marker effects on the genetic background (Miedaner et al., 2011), or by the lack of SNPs in linkage disequilibrium with the causal alleles of the Fr-A2 locus.
The association mapping scan yielded stable diagnostic markers only for one genomic region with a restricted potential to predict frost tolerance of wheat genotypes. This can be explained by a low number of large-effect QTLs underlying frost tolerance and the low power of association mapping strategies to tackle QTL with small to medium effect sizes. To overcome this limitation, two genomic selection approaches were used. While in BayesCπ only a fraction of 1 – π SNPs is assumed to contribute to frost tolerance, RR-BLUP assumes that all markers contribute to the trait. The accuracy in predicting frost tolerance was nearly 60% higher with BayesCπ (Fig. 2) as compared with association mapping. Frost tolerance was influenced by a subset of approximately 265 SNPs with additive effects and 74 SNPs with dominance effects. However, it is known from simulation studies (Habier et al., 2011) that these numbers tend to be overestimated. Assuming an infinitesimal model and applying RR-BLUP yielded similar accuracies to predict frost tolerance. This is surprising since BayesCπ is expected to tackle traits with a mixture of large-, medium-, and small-effect QTLs more appropriately than RR-BLUP (Daetwyler et al., 2010). A closer examination of the marker effects estimated for RR-BLUP, however, revealed that the large-effect QTL on chromosome 5B, for example, is captured through a number of linked SNPs with a sum of absolute values of the additive effects (0.55) comparable to BayesCπ (0.89).
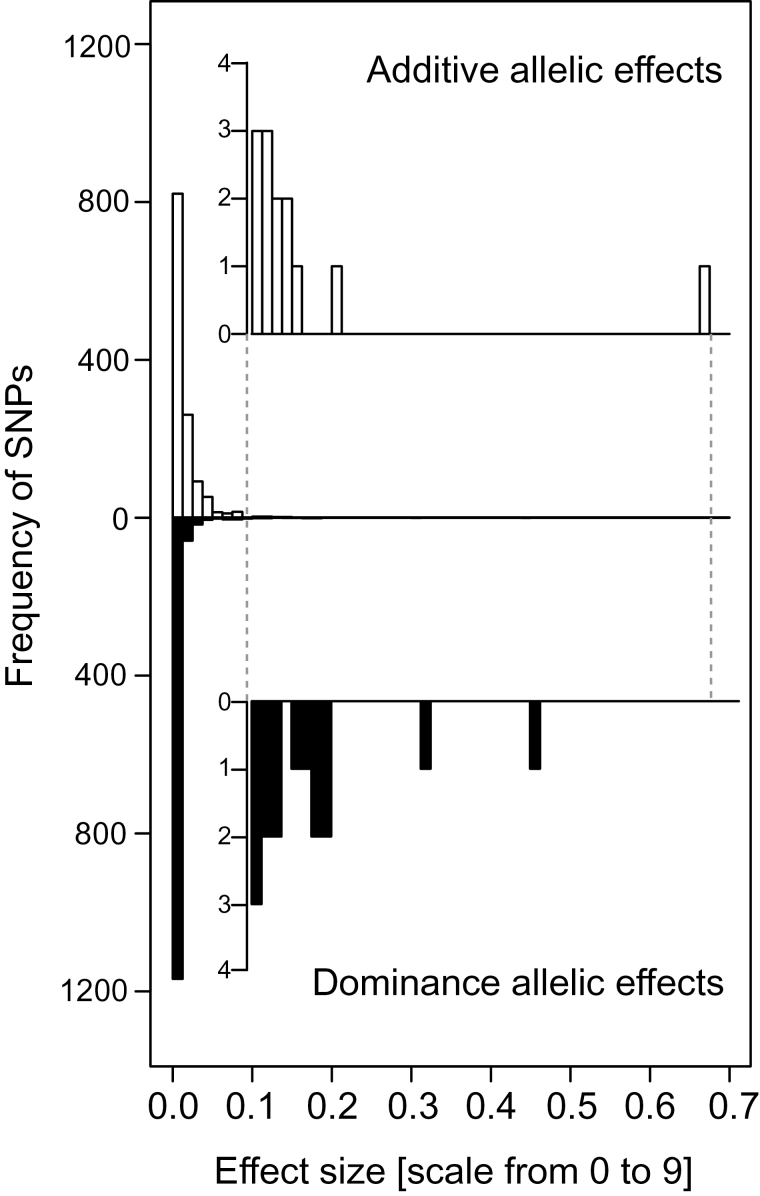
Across all mapping approaches, the accuracy of prediction of frost tolerance by dominance effects alone was smaller compared to the prediction exploiting exclusively additive effects (Fig. 2). This can be explained by the lower contribution of dominance effects to the genetic variation (Table 1; Figure 3), as expected for the autogamous crop wheat (Longin et al., 2012). In contrast to previous studies (Lee et al., 2008; Wittenburg et al., 2011), combining additive and dominance effects did not lead to higher accuracies of prediction of frost tolerance (Fig. 2). This seemed surprising as dominance captures a new source of variation, but can be explained by the large proportion of unexplained variation entering the prediction model when dominance effects are included (Zhao et al., 2013).
Fig. 3.
Distribution of marker effects estimated using BayesCπ for frost tolerance measured from a scale from 1 (no damage) to 9 (no plant survived).
The observed accuracy of prediction of frost tolerance (Fig. 2) suggests that there is still room for improvement. One possible approach to increase the ability to predict frost tolerance in wheat is to further enhance the marker density, for example by employing genotyping-by-sequencing approaches. This can be important to tackle the underlying causal polymorphisms with a higher precision compared to the use of a 9K SNP array. The assumption of an absence of epistasis is another potential reason for the still imperfect accuracy of prediction of frost tolerance (Zuk et al., 2012), as supported by previous studies on epistatic interactions among key players underlying frost tolerance (Galiba et al., 2009). To study this in more detail, the current work extended the main-effect association mapping approach towards a full two-dimensional scan for interaction-effects among SNPs. Among the four types of digenic epistatic effects, additive × additive effects were the most prevailing ones in terms of effect size (Supplementary Fig. S7) and skewness of P-values towards zero (Supplementary Fig. S6). This predominance of additive × additive effects is expected for selfing species such as wheat (Cockerham, 1984). The identified epistatic effects were, however, rather small, with a maximum contribution of a single effect to the phenotypic variation of 1.7% (Supplementary Fig. S7). Despite these small effect sizes, the large number of potential interaction effects can lead to a substantial contribution of epistasis to the phenotypic variation of frost tolerance. Consequently, the presence of epistatic effects is an explanation for this study’s inability to predict frost tolerance with a higher accuracy than observed.
This study provides insights into the genetic architecture of frost tolerance in wheat adapted to Central Europe. One major QTL on chromosome 5B segregating in European winter wheat was detected, which is of use for marker-assisted selection after thorough validation in independent experiments. In addition, for the first time it has been shown that using genomic selection approaches that assume a more complex genetic architecture represents a major step to improve understanding of the genetic architecture of frost tolerance. The developed calibration models are valuable to improve the frost tolerance of wheat, which is a trait that displays a wide variation in occurrence across years and is therefore a difficult target for conventional phenotypic selection.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Crossing scheme between the 120 female and 15 male parental wheat lines
Supplementary Fig. S2. Temperature profiles of the three locations
Supplementary Fig. S3. Distribution of the phenotypic values of 1739 genotypes for frost tolerance
Supplementary Fig. S4. Coefficients of parentage estimated for the 120 female and 15 male parental wheat lines from SNP marker data
Supplementary Fig. S5. Quantile–quantile plots for association mapping based on the combined population of hybrids and parental lines using four different biometrical approaches
Supplementary Fig. S6. Distribution of P-values for four different types of digenic epistatic effects
Supplementary Fig. S7. Box-whisker plots of the proportion of explained phenotypic variation for significant digenic epistatic effects
Supplementary Table S1. Description of the 135 parental lines used in this study
Supplementary Table S2. SNP data of the 135 parental lines
Supplementary Table S3. Phenotypic correlations of the adjusted entry means of 1604 hybrids and 135 lines of three environments
Supplementary Table S4. Summary of marker statistics for the three applied genome-wide mapping approaches
Acknowledgements
This research was conducted within the HYWHEAT project funded by BMBF (grant no. FKZ0315945D).
References
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN. 2007. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Functional and Integrative Genomics 7, 53–68 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57, 289–300 [Google Scholar]
- Bernardo R. 1993. Estimation of coefficient of coancestry using molecular markers in maize. Theoretical and Applied Genetics 85, 1055–1062 [DOI] [PubMed] [Google Scholar]
- Chen Y, Carver BF, Wang S, Zhang F, Yan L. 2009. Genetic loci associated with stem elongation and winter dormancy release in wheat. Theoretical and Applied Genetics 118, 881–889 [DOI] [PubMed] [Google Scholar]
- Cockerham CC. 1984. Additive by additive variance with inbreeding and linkage. Genetics 108, 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Campos G, Pérez P, et al. 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186, 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA. 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers JCM, Garrick DJ, Fernando RL. 2009. Use of high-density SNP genotyping for genetic improvement of livestock. Short course, Iowa State University, Ames, USA. [Google Scholar]
- Dekkers JCM. 2007. Prediction of response to marker-assisted and genomic selection using selection index theory. Journal of Animal Breeding and Genetics 124, 331–341 [DOI] [PubMed] [Google Scholar]
- Dhillon T, Pearce S, Stockinger E, Distelfeld A, Li C, Knox AK, Vashegyi I, Vágújfalvi A, Galiba G, Dubcovsky J. 2010. Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiology 153, 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. 2012. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7, e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. 1998. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics 97, 968–975 [Google Scholar]
- Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics , 4th edn Essex, UK: Longman [Google Scholar]
- Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Hayes P, Dubcovsky J. 2005. Large deletions in the first intron of the VRN-1 vernalization gene are associated with spring growth habit in barley and polyploid wheat. Molecular Genetics and Genomics 273, 54–65 [DOI] [PubMed] [Google Scholar]
- Galiba G, Vágújfalvi A, Li C, Solté A, Dubcovsky J. 2009. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Science 176, 12–19 [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. 2009. ASReml user guide release 3.0. Hemel Hempstead, UK: VSN International [Google Scholar]
- Habier D, Fernando RL, Kizilkaya K, Garrick DJ. 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer AR, Miranda JB. 1988. Quantitative genetics in maize breeding , 2nd edn Ames: Iowa State University Press [Google Scholar]
- Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. 2009. Invited review: genomic selection in dairy cattle: progress and challenges. Journal of Dairy Science 92, 433–443 [DOI] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6, 65–70 [Google Scholar]
- Jannink JL, Lorenz AJ, Iwata H. 2010. Genomic selection in plant breeding: from theory to practice. Briefings in Functional Genomics 9, 166–177 [DOI] [PubMed] [Google Scholar]
- Lee SH, van der Werf JHJ, Hayes BJ, Goddard ME, Visscher PM. 2008. Predicting unobserved phenotypes for complex traits from whole-genome SNP data. PLoS Genetics 4, e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longin CFH, Mühleisen J, Maurer HP, Zhang H, Gowda M, Reif JC. 2012. Hybrid breeding in autogamous cereals. Theoretical and Applied Genetics 125, 1087–1096 [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Hart GC, Demos KL, Gale MD, Rogers WW. 1998. Catalogue of gene symbols for wheat. In: Slinkard AE, ed, Proceedings of the 9th International Wheat Genetics Symposium. Saskatoon: University of Saskatchewan; pp. 123–127 [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedaner T, Wurschum T, Maurer HP, Korzun V, Ebmeyer E, Reif JC. 2011. Association mapping for Fusarium head blight resistance in European soft winter wheat. Molecular Breeding 28, 647–655 [Google Scholar]
- Möhring J, Piepho HP. 2009. Comparison of weighting in two-stage analysis of plant breeding trials. Crop Science 49, 1977–1988 [Google Scholar]
- Ortiz R, Sayre KD, Govaerts B, Gupta R, Subbarao GV, Ban T, Hodson D, Dixon JM, Ortiz-Monasterio JI, Reynolds M. 2008. Climate change: can wheat beat the heat? Agriculture, Ecosystem and Environment 126, 46–58 [Google Scholar]
- Pearce S, Zhu J, Boldizsár Á, Vágújfalvi A, Burke A, Garland-Campbell K, Galiba G, Dubcovsky J. 2013. Large deletions in the CBF gene cluster at the locus are associated with reduced frost tolerance in wheat. Theoretical and Applied Genetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Available at: www.R-project.org [Google Scholar]
- Reif JC, Maurer HP, Korzun V, Ebmeyer E, Miedaner T, Wurschum T. 2011. Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theoretical and Applied Genetics 123, 283–292 [DOI] [PubMed] [Google Scholar]
- Reif JC, Zhao Y, Würschum T, Gowda M, Hahn V. 2013. Genomic prediction of sunflower hybrid performance. Plant Breeding 132, 107–114 [Google Scholar]
- Sandve SR, Kosmala A, Rudi H, Fjellheim S, Rapacz M, Yamada T, Rognli OA. 2011. Molecular mechanisms underlying frost tolerance in perennial grasses adapted to cold climates. Plant Science 180, 69–77 [DOI] [PubMed] [Google Scholar]
- Skinner DZ, Garland-Campbell KA. 2008. Evidence of a major genetic factor conditioning freezing sensitivity in winter wheat. Plant Breeding 127, 228–234 [Google Scholar]
- Snape JW, Semikhodskii A, Fish L, Sarma RN, Quarrie SA, Galiba G, Sutka J. 1997. Mapping of frost tolerance loci in wheat and comparative mapping with other cereals. Acta Agronomica Hungarica 45, 268–270 [Google Scholar]
- Stram DO, Lee JW. 1994. Variance components testing in longitudinal mixed effects model. Biometrics 50, 1171–1177 [PubMed] [Google Scholar]
- Sutka J, Snape JW. 1989. Location of a gene for frost resistance on chromosome 5A of wheat. Euphytica 42, 41–44 [Google Scholar]
- Technow F, Riedelsheimer C, Schrag TA, Melchinger AE. 2012. Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theoretical and Applied Genetics 125, 1181–1194 [DOI] [PubMed] [Google Scholar]
- Toth B, Galiba G, Feher E, Sutka J, Snape JW. 2003. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theoretical and Applied Genetics 107, 509–514 [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. 2000. Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the Cor14b gene in frost-tolerant and frost-sensitive genotypes. Molecular and General Genetics 263, 194–200 [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. 2003. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Molecular Genetics and Genomics 269, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vágújfalvi A, Soltész A, Bálint AF, Vashegyi I, Tóth B, Kocsi B, Galiba G. 2012. Different approaches involving testing methods, gene mapping and transformation reveal new insights into cereal frost tolerance. Acta Agronomica Hungarica 60, 167–182 [Google Scholar]
- Whittaker JC, Thompson R, Denham MC. 2000. Marker assisted selection using ridge regression. Genetics Research 75, 249–252 [DOI] [PubMed] [Google Scholar]
- Williams ER, Piepho H-P, Whitaker D. 2010. Augmented p-rep designs. Biometrical Journal 53, 19–27 [DOI] [PubMed] [Google Scholar]
- Wittenburg D, Melzer N, Reinsch N. 2011. Including non-additive genetic effects in Bayesian methods for the prediction of genetic values based on genome-wide markers. BMC Genetics 12, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Langer S, Longin CFH, Korzun V, Akhunov E, Ebmeyer E, Schachschneider R, Kazman E, Schacht J, Reif JC. 2013. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theoretical and Applied Genetics 126, 1477–1486 [DOI] [PubMed] [Google Scholar]
- Würschum T, Maurer HP, Schulz B, Möhring J, Reif JC. 2011. Genome-wide association mapping reveals epistasis and genetic interaction networks in sugar beet. Theoretical and Applied Genetics 123, 109–118 [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zeng J, Fernando R, Reif JC. 2013. Genomic prediction of hybrid wheat performance. Crop Science 53, 802–810 [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences, USA 109, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.