Abstract
Water deficit is a serious environmental factor limiting the growth and productivity of plants worldwide. Improvement of drought tolerance and efficient water use are significant strategies to overcome this dilemma. In this study, a drought-responsive transcription factor, NUCLEAR FACTOR Y subunit B 7 (PdNF-YB7), induced by osmotic stress (PEG6000) and abscisic acid, was isolated from fast-growing poplar clone NE-19 [Populus nigra × (Populus deltoides × Populus nigra)]. Ectopic overexpression of PdNF-YB7 (oxPdB7) in Arabidopsis enhanced drought tolerance and whole-plant and instantaneous leaf water-use efficiency (WUE, the ratio of biomass produced to water consumed). Overexpressing lines had an increase in germination rate and root length and decrease in water loss and displayed higher photosynthetic rate, instantaneous leaf WUE, and leaf water potential to exhibit enhanced drought tolerance under water scarcity. Additionally, overexpression of PdNF-YB7 in Arabidopsis improved whole-plant WUE by increasing carbon assimilation and reducing transpiration with water abundance. These drought-tolerant, higher WUE transgenic Arabidopsis had earlier seedling establishment and higher biomass than controls under normal and drought conditions. In contrast, Arabidopsis mutant nf-yb3 was more sensitive to drought stress with lower WUE. However, complementation analysis indicated that complementary lines (nf-yb3/PdB7) had almost the same drought response and WUE as wild-type Col-0. Taken together, these results suggest that PdNF-YB7 positively confers drought tolerance and improves WUE in Arabidopsis; thus it could potentially be used in breeding drought-tolerant plants with increased production even under water deficiency.
Key words: Arabidopsis, drought tolerance, NF-YB, poplar, transcription factor, water-use efficiency.
Introduction
Poplar is an important tree species of great economic and ecological importance worldwide. It is also one of the fastest growing trees and its high productivity requires a high consumption of water (Ridge et al., 1986; Tschaplinski and Blake, 1989; Zsuffa et al., 1996; Bradshaw et al., 2000; Monclus et al., 2005). Environmental abiotic stress, such as drought, salinity, and cold, could have harmful effects on the development and growth of poplars (Tschaplinski et al., 1994; Renaut et al., 2005; Escalante-Pérez et al., 2009). North China is a mostly dry or semi-dry area. Water resources are key factors in plant yields (Sun et al., 2006). Water-use efficiency (WUE), measured as the biomass produced per unit transpiration, describes the relationship between water use and plant productivity. The basic physiological definition of leaf WUE is equal to the ratio of photosynthesis to transpiration, also referred to as transpiration efficiency (Karaba et al., 2007). High WUE could increase the total production of plants under variable soil water content in the soil as an adaptation to water scarcity.
Water-deficit-inducible genes can be classified into two major groups: the first group encodes function proteins, such as aquaporins, LEA proteins, and chaperones; the second group includes genes encoding regulatory proteins, such as transcription factors, protein kinases, hormones, and other signal molecules (Shinozaki et al., 2003; Valliyodan and Nguyen, 2006). Transcription factors play critical roles in controlling intrinsic developmental processes and responses to external stimuli by influencing the expression of downstream targets and have been confirmed to improve drought resistance in transgenic plants (Shinozaki et al., 2003). The transcription factors involved in drought-responsive pathways are distributed mainly in the AP2/ERF, bZIP, NAC, MYB, C2H2 zinc finger, and WRKY families (Abe et al., 1997; Kizis et al., 2001; Sakamoto et al., 2004; Lu et al., 2007; Xiang et al., 2008; Wang et al., 2009). AtMYB61, a member of the R2R3-MYB family of transcription factors in Arabidopsis thaliana, closes stomata to limit water loss while directing the establishment of water-conducting xylem vessels with larger vessel diameter and root system proliferation to better seek, acquire, and transport water (Romano et al., 2012). Overexpression of TaWRKY2 and TaWRKY19 wheat WRKY transcription factors could regulate drought stress tolerance in Arabidopsis by activating downstream target genes related to drought resistance, such as DREB, RD29, and COR6.6 (Niu et al., 2012). In addition, another transcription factor family, GTL1, could control stomatal density by transrepression of SDD1 and negatively regulate WUE and drought tolerance in Arabidopsis (Yoo et al., 2010).
Recently, the NUCLEAR FACTOR Y family (NF-Y) binding specifically to the CCAAT box, was identified in drought-responsive pathways and also named for the CCAAT-box binding factor (CBF) (Nelson et al., 2007; Stephenson et al., 2007; Li et al., 2008; Thirumurugan et al., 2008). NF-Y is a ubiquitous nuclear transcription factor that consists of three subunits: NF-YA (CBF-B, HAP2); NF-YB (CBF-A, HAP3); and NF-YC (CBF-C, HAP5). In yeast, the subunits are called HEME ACTIVATED PROTEIN (HAP) 2, 3, and 5, respectively, and are encoded by single genes (Mantovani, 1999). The CCAAT box is a cis-acting element widely found in the eukaryotic promoter region, which is bound specifically with proteins encoded by the NF-Y family and regulates a series of related gene expressions (Testa et al., 2005). Recent reports elucidate some functions of the NF-Y family. Several NF-YB and NF-YC members in Arabidopsis, tobacco, and wheat play roles in light regulation and flowering time (Kumimoto et al., 2008, 2010; Stephenson et al., 2010, 2011; Hackenberg et al., 2012). AtNF-YB6 (L1L) and AtNF-YB9 (LEC1) are involved in embryo development in seeds (Kwong et al., 2003; Yamamoto et al., 2009). In recent years, NF-Y family members in plants have been found to function in drought stress. Li et al. (2008) found that the plants overexpressing AtNF-YA5 display reduced stomatal aperture and leaf water loss and significantly promote drought resistance and AtNF-YA5 is regulated transcriptionally by abscisic acid (ABA) and posttranscriptionally by miR169. The overexpression of AtNF-YB1 confers improved performance in Arabidopsis with higher water potential and photosynthesis rates under drought treatments. Transgenic maize plants with ZmNF-YB2, the homologue of AtNF-YB1, show drought tolerance based on increased chlorophyll content, stomatal conductance, and photosynthesis rates and decreased leaf temperature under drought conditions and exhibit a grain yield advantage (Nelson et al., 2007). However, little has been reported about the relationship between NF-Y transcription factors and WUE.
This study identified a poplar drought-responsive NF-Y family member from the fast-growing black cottonwood with high WUE, which conferred drought tolerance and improved plant WUE under water deficit.
Materials and methods
Plant materials and growth conditions
The poplar genotype NE-19 [Populus nigra × (Populus deltoides × Populus nigra)] was used in this study. NE-19 cuttings with 15-cm-long stems were planted in April 2010, in the nursery of Beijing Forestry University, Beijing, China (40° 0′ 7.05′′ N 116° 15′ 1.60′′ E) for further gene analysis.
Arabidopsis Col-0 was selected as the wild-type control. Arabidopsis mutant nf-yb3 (stock name SALK_074951) was ordered from the Arabidopsis Biological Resource Center and the homozygous mutant for T-DNA insertion within AtNF-YB3 (AT4G14540) was verified by PCR. Arabidopsis seeds were sterilized by a 60-s 70% ethanol treatment followed by 1% NaClO within 10min and four washes in distilled water. Seeds were sown on half-strength Murashige and Skoog (MS) plates with 3% sucrose and 0.6% agar and stratified for 2 d at 4 °C before being transferred to the culture room at 22 °C under a 16/8 light/dark cycle. After germination, 10-d-old Arabidopsis seedlings were transplanted and grown at a density of four plants per 7×7 × 6.5cm pot containing a mixture of soil and vermiculite (2:1) at 22 °C under a 16/8 light/dark cycle (150 μmol m−2 s−1 and 70% relative humidity.
Poplar gene cloning, transformation, and expression analysis
Total RNA was extracted from the leaves of poplar NE-19 seedlings using the CTAB reagent method described by Chang et al. (1993). First-strand cDNA synthesis was performed using M-MLV Reverse Transcriptase and an oligo (dT) primer (Promega, Madison, WI, USA) according to the manufacturer’s instructions (Xing et al., 2011). The PdNF-YB7 cDNA sequence was amplified by PCR using the primers PdNFYB7f and PdNFYB7r (Supplementary Table S1, available at JXB online).
To obtain 35S:PdNF-YB7 and nf-yb3/PdB7 transgenic plants, the PdNF-YB7 cDNA was cloned into the pCAMBIA-1304 binary vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed into Arabidopsis Col-0 and mutant lines respectively by the floral dip method (Bechtold et al., 2003) using Agrobacterium tumefaciens GV3101. The transgenic lines were identified using half-strength MS plates containing 100mg l−1 hygromycin.
For promoter expression analysis, the PdNF-YB7pro:GUS construct, including a 2.3-kb fragment upstream from the initiation codon extracted from poplar NE-19 genomic DNA, was cloned into the pBI121 vector and transformed into Arabidopsis Col-0.
For subcellular localization of PdNF-YB7 in plant cells, GFP fusion proteins were observed using a confocal laser scanning microscope (DMI6000 CS; Leica, Wetzlar, Germany).
To analyse the expression levels of related genes, total RNA was extracted from transgenic, wild type, mutant, and complementation plants by the CTAB method. Real-time PCR analysis was performed using primers PdB7 and PdActin (Supplementary Table S1). Quantitative real-time PCR (qPCR) analysis followed the procedure described by Chen et al. (2009). SYBR Green was used to monitor the kinetics of PCR product formation in qPCR. The 18S rRNA transcript, as an internal control, was used to quantify the relative expression levels of genes in samples. The primer sequences are shown in Supplementary Table S2.
Histochemical staining analysis
To test the induction of GUS expression by osmotic stress, 10-d-old Arabidopsis seedlings were transferred from half-strength MS plates to half-strength MS liquid medium containing 25mM PEG6000 or 200mM mannitol for osmotic treatment. The controls were treated with half-strength MS liquid medium. GUS staining was performed by incubating the plants in GUS solution containing 100mM Na2HPO4 buffer, 1mM K3(Fe[CN]6), 1mM K2(Fe[CN]6), 10mM EDTA, 1% (v/v) Triton X-100 and 0.5mg ml–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid overnight at 37 °C in the dark, followed by clearing with 75% ethanol for another hour.
Physiological experiments
Three independent batches of seeds were used to confirm the germination rate. Twenty seeds for a line in one batch were used for germination comparison between oxPdB7s, Col-0, nf-yb3 and nf-yb3/PdB7 plants. Based on the diversity of germination time, the seeds would be separately sown on the plates to unify germination time. The Arabidopsis lines after germination were grown vertically for 8 d and the primary root length was measured. Rosette leaves were removed from 18-d-old seedlings grown in the soil and the leaf area was computed using Photoshop (Adobe Systems, San Jose, CA, USA). After transferring the plants to soil, plant height was measured every 3 d during the bolting period to calculate the average stem elongation rate.
Analysis of WUE
Instantaneous leaf WUE was defined as the ratio of the rate of CO2 assimilation (photosynthetic rate)/transpiration rate (Wong et al., 1978), which were measured using the Li-6400 Portable Photosynthesis System (Li-Cor, Lincoln, NE, USA). The young, fully expanded rosette leaves of 3-week-old plants were measured at an ambient CO2 concentration of 400 μmol mol−1, photosynthetic photon flux density of 600 μmol m−2 s−1, and a chamber temperature of 22 °C. Whole-plant WUE was measured by gravimetric analysis (Xing et al., 2011). Plants were cultured in pots of known weight filled with soil at saturated field capacity. During the experimental period, the pots were weighed daily (0.1g accuracy) and the difference in weight on subsequent days was corrected by adding water to maintain the saturated field capacity. Additionally, filled pots without plants were used to calculate water loss by evaporation. The water added during the experimental period from 22 to 42 d after germination was summed as the cumulative water transpired (CWT) minus evaporation. Representative plants from oxPdB7#1, #2, #3, Col-0, nf-yb3, and nf-yb3/PdB7 were sampled to measure the initial biomass (total biomass including rosette, inflorescence and root) at 22 d after germination (B22). The seedlings were oven-dried for 16h at 70 °C and weighed. The samples from 42 d after germination (B42) were measured as above. The whole-plant WUE was calculated as (B42 − B22)/(CWT between 22 and 42 d after germination).
Drought experiments
For stress treatment experiments, Arabidopsis seeds were sown on half-strength MS plates with 200mM mannitol and germination rates were recorded. The primary root length was compared on half-strength MS plates with 200mM mannitol where Arabidopsis seedlings were transplanted after germination on half-strength MS plates. The seedlings were watered for 15 d after transplanted into the soil, and then water was withheld. The pots were put on absorbent paper for a period of 10 d. After 10 d of water deficit, the pots were rewatered. Plant growth was determined 8 d after rewatering. The soil collected at the three stages (well-watered, drought, rewatered) were oven-dried to a constant weight for 16h at 90 °C and weighed for measurement of soil water content. The photosynthetic rates, instantaneous leaf WUE, and leaf water potential were measured at the three stages. The measurements of photosynthetic rates, instantaneous leaf WUE, and total biomass were described as previously. The leaf water potential was measured in situ nondestructively at the leaf surface of Arabidopsis seedlings using psychrometers (L-51A; WESCOR, Utah, USA) connected to the PSYPRO Water Potential System (WESCOR).
Water loss measurements
Rosette leaves of oxPdB7s, Col-0, nf-yb3, and nf-yb3/PdB7 plants, which were grown under normal conditions for 25 d after germination, were excised, weighed immediately (leaves weighing approximately 1g were harvested and used immediately for experiments), and incubated on a bench at room temperature and at 70% humidity and 150 μmol m−2 s−1. Losses in fresh weight were monitored at the times indicated (Ma et al., 2010). Water loss is expressed as the percentage of initial fresh weight.
Results
Identification and molecular characterization of differentially expressed genes
According to the microarray profile analysis of Populus euphratica response to drought stress (Yan et al., 2012a), several NF-YB family genes are induced by drought. Further qPCR analysis validated that PeNF-YB7 was especially upregulated in the leaves of drought-stressed poplars (Gray et al., 2009; Cao et al., 2011; Yan et al., 2012b). To study the role of the drought-related gene NF-YB in P. nigra × (P. deltoides × P. nigra), the current study characterized the poplar NUCLEAR FACTOR Y subunit B 7 (PdNF-YB7) (GenBank accession KC460319), the homologue of P. euphratica PeNF-YB7, for future research. The PdNF-YB7 cDNA is 672bp in length and encodes 223 amino acid residues with a predicted molecular mass of 24.644kDa and an isoelectric point of 7.56.
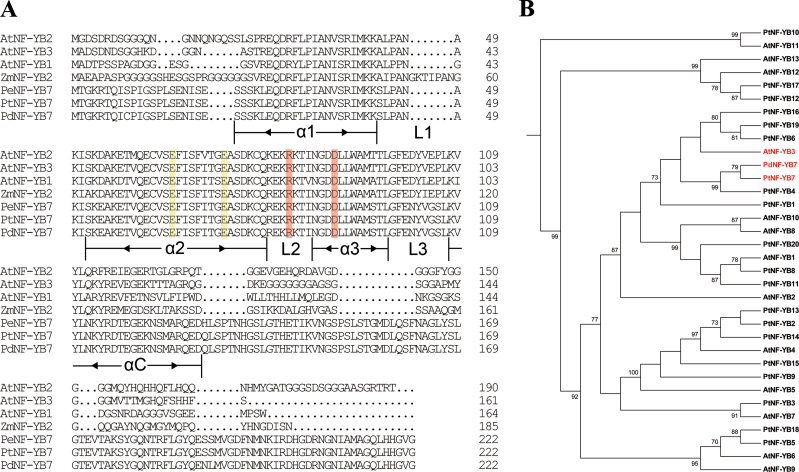
The protein structure alignment using InterPro (http://www.ebi.ac.uk/interpro/) showed that the PdNF-YB7 sequence domain includes a NF-YB transcription factor conserved site (IPR003956), NF-YB binding site (IPR003957), and NF-YB archaeal histone (IPR003958) (Supplementary Fig. S1). The results indicate that PdNF-YB7 is a member of the NF-YB transcription factor family. Multiple sequence alignment revealed that the PdNF-YB7 secondary structure has a basic helix−loop−helix motif composed of four helices and three loops as in NF-YB family conserved domains (Fig. 1A). This highly conserved domain plays a central role in the junction between the NF-Y transcription factor and DNA and interaction between NF-Y and proteins (Mantovani, 1999). Additionally, the Arg in loop 2 and Asp in helix 3, which function in combinations of NF-YB and NF-YC, are highly conserved in the PdNF-YB7 protein. The two Glu in helix 2 are important for NF-YA binding (Romier et al., 2003). The phylogenetic relationship between the poplar and Arabidopsis NF-YB family members was further analysed by amino acid sequence alignment (Dereeper et al., 2008, 2010). As shown in Fig. 1B, PdNF-YB7 did not cluster with known-function NF-YB proteins from Arabidopsis, such as drought-resistance protein, AtNF-YB1 (Nelson et al., 2007), and seed development-related proteins AtNF-YB9 and AtNF-YB6 (Kwong et al., 2003; Yamamoto et al., 2009). The results suggest that PdNF-YB7 differs from known-function NF-YB genes in Arabidopsis and has a specific function in poplars. The closest homologue to PdNF-YB7 was poplar protein PtNF-YB7 (NF-YB7 for Populus trichocarpa), which exhibits 99% identity. Moreover, this study found that, although not entirely orthologous to PdNF-YB7 (PtNF-YB6, PtNF-YB19, and PtNF-YB16 are even closer), AtNF-YB3 has the closest relationship to PdNF-YB7 among the Arabidopsis NF-YB family members using PdNF-YB7 as a phylogenetic control. For this reason, nf-yb3 was chosen for complementation experiments.
Fig. 1.
The NF-YB7 gene of P. nigra × (P. deltoides × P. nigra). (A) Multiple alignment of amino acid sequences of PdNF-YB7 and other plant NF-YBs. AtNF-YB1 (AT2G38880), AtNF-YB2 (AT5G47640), and AtNF-YB3 (AT4G14540) sequences were obtained from the TAIR database and ZmNF-YB2 (DQ333305), PeNF-YB7 (HQ161880), and PtNF-YB7 (POPTR_0007s06480) from NCBI and PopGenIE. The helix motifs are underlined in black (α1, α2, α3, and αC) and the loops are between two helixes (L1, L2, and L3). The Arg and Asp residues, highlighted in red, function in the combination of NF-YB and NF-YC. The two Glu residues, highlighted in yellow, function in the junction with NF-YA. (B) Phylogenetic relationships between poplar and Arabidopsis NF-YB family members. The Phylogeny.fr online web service was used for analysis of phylogenetic relationships. The Arabidopsis NF-YB family sequences were obtained from the TAIR database and the poplar NF-YB family sequences from the PopGenIE database.
Expression pattern of poplar PdNF-YB7
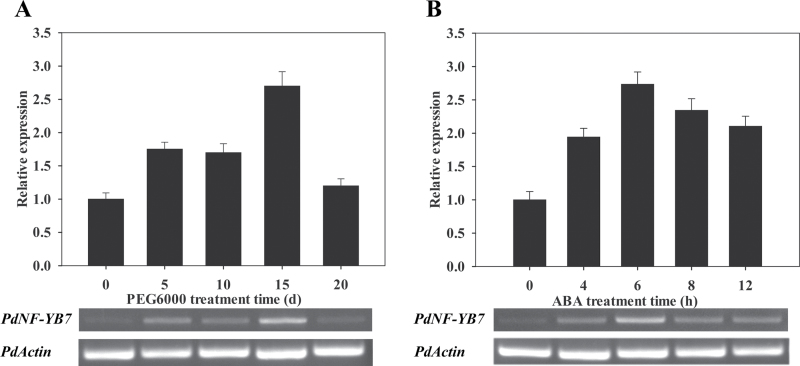
To investigate the involvement of PdNF-YB7 in poplar responses to osmotic stress, the expression level of PdNF-YB7 under 30% PEG6000 was tested by PCR (Yan et al., 2012b). The results indicate that the expression level of PdNF-YB7 rose gradually with increased stress intensity and peaked at 15 d of PEG treatment (Fig. 2A). ABA is an important secondary signalling molecule and its exogenous application can cause similar effects to osmotic stress and mediate some drought-responsive genes (Zhu, 2002). Thus, 200 μM ABA treatment was used to test the response of PdNF-YB7 in poplar (Chen et al., 2009). The expression of PdNF-YB7 increased ~2.7-fold by 6h, and then decreased slightly after 6h (Fig. 2B). The results indicate that ABA mainly functions in the early stage of osmotic stress. ABA-responsive stress signalling first modifies the constitutively expressed transcription factors, resulting in the expression of early response genes and then activates downstream stress tolerance effector genes. The early response genes typically encode transcription factors (Zhu, 2002).
Fig. 2.
Expression of PdNF-YB7 in response to osmotic stress and ABA in poplar. qPCR and PCR assay of accumulation of PdNF-YB7 transcripts in response to 30% PEG6000 (A) and 200 μM ABA (B). The expression levels were normalized to that of PdActin, and the level of PdNF-YB7 transcript in the control was sent at 1.0. Data are mean ± SE (n = 3 experiments).
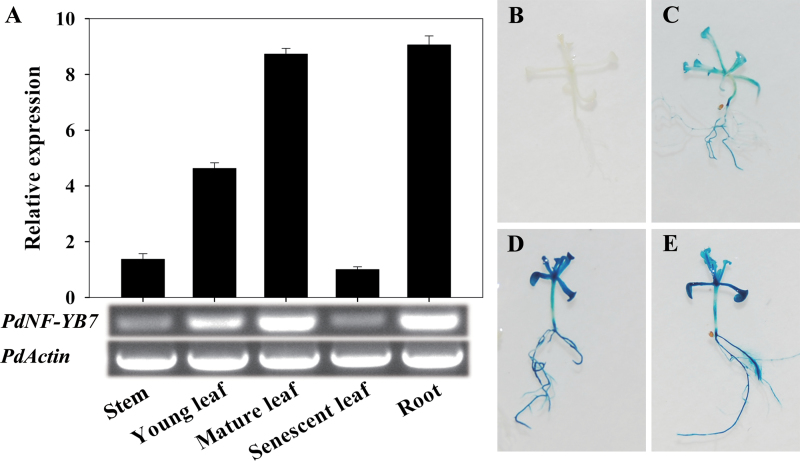
To study the tissue-specific presentation of expression of PdNF-YB7 in poplar, the expression of PdNF-YB7 was detected in roots, stems, young leaves, mature leaves, and senescent leaves of NE-19 under normal growth conditions. The results showed that PdNF-YB7 was expressed more highly in mature leaves, young leaves, and roots than in stems and senescent leaves (Fig. 3A).
Fig. 3.
Expression patterns of PdNF-YB7. (A) Tissue pattern of PdNF-YB7 in poplar. Total RNA was isolated from various tissues of seedlings grown in culture. The expression levels were normalized to that of PdActin. Data are mean ± SE (n = 3 experiments). (B–E) PdNF-YB7pro:GUS expression patterns in Arabidopsis: wild-type control Col-0 (B); control transgenic seedling (C); transgenic seedling treated with 25mM PEG6000 for 3h (D); transgenic seedling treated with 200mM mannitol for 3h (E) (this figure is available in colour at JXB online).
To assess the expression pattern of PdNF-YB7, this study cloned the promoter of PdNF-YB7 from poplar genomic DNA. A predicted analysis of the PdNF-YB7 promoter using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) revealed a series of water-related and abiotic stresses responsive elements, including ABRELATERD1, ACGTATERD1, CBFHV, DPBFCOREDCDC3, MYB, and MYC (Supplementary Table S3). The results suggest that PdNF-YB7 plays roles in the response to environmental stresses and in plant growth and development.
To analyse the regulatory activity of PdNF-YB7 during the stress response, this study constructed the PdNF-YB7pro:GUS expression vector and transformed Col-0 wild-type Arabidopsis. Then 25mM PEG6000 and 200mM mannitol were used to determine the osmotic stress response in transgenic plants. Histochemical staining analysis revealed that GUS was expressed in transgenic seedlings and that expression was enhanced in response to drought and osmotic treatment and observed throughout the entire plant (Fig. 3B–E).
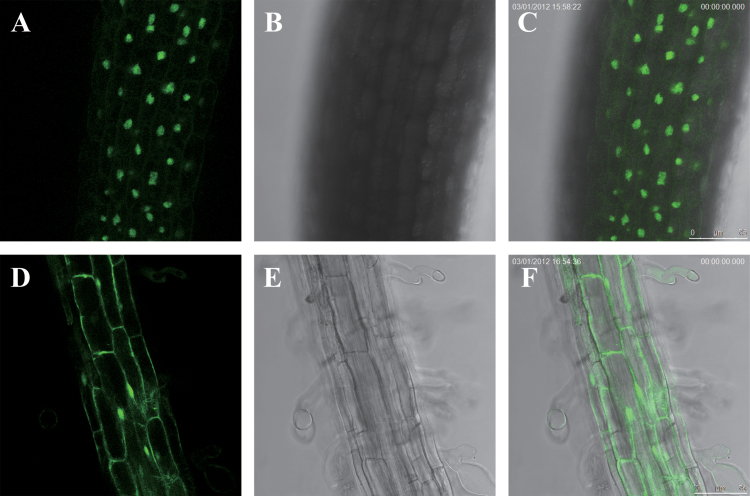
To test the subcellular localization of PdNF-YB7 in plant cells, the 35S:PdNF-YB7-GFP fusion was constructed and transformed into Arabidopsis Col-0. Expression of the PdNF-YB7-GFP fusion in Arabidopsis predominantly accumulated in the nucleus (Fig. 4), consistent with its function as a transcriptional regulator.
Fig. 4.
The subcellular localization of 35S:PdNF-YB7-GFP and control 35S:GFP expression in Arabidopsis cells. Cells were detected under a fluorescent or light field by a fluorescence microscope. (A–C) 35S:PdNF-YB7-GFP seedling in fluorescent light (A), bright light (B), and overlap image of fluorescent and bright light (C). (D–F) Control seedling in fluorescent light (D), bright light (E), and overlap image of fluorescent and bright light (F). Bars, 75 μm (this figure is available in colour at JXB online).
Phenotype of overexpressing lines under well-watered conditions
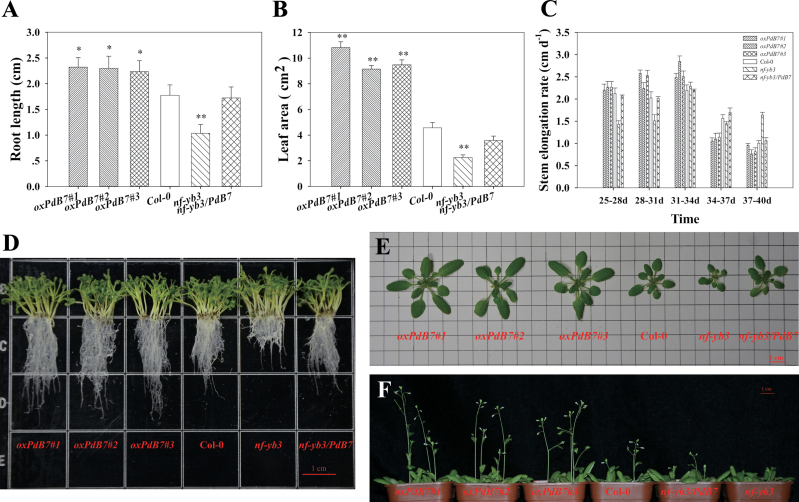
To evaluate the performance of overexpressing PdNF-YB7 (oxPdB7) lines grown in well-watered conditions, the growth phenotype of overexpressing Arabidopsis was observed during different developmental stages. The germination time for oxPdB7 was 1 or 2 d earlier than the wild-type control Col-0 and mutant nf-yb3. Eight days after germination, the primary root length of oxPdB7 also was longer than Col-0 (1.3-fold) and nf-yb3 (2.1-fold) (Fig. 5A, D). In 18-d-old seedlings grown in soil, transgenic plants had larger leaves than Col-0 and the mutant (Fig. 5E). The leaf area of oxPdB7 was about 1.00–1.37-fold larger than Col-0 and 3.07–3.82-fold larger than the mutant (Fig. 5B). The oxPdB7 lines bolted 22–23 d after germination, whereas the Col-0 and mutant plants bolted at approximately 23–24 d and 24–25 d, respectively. Additionally, the transgenic plants showed higher average stem elongation rate at the earlier shooting stage (25–34 d) compared to Col-0 and mutant (Fig. 5C, F). At 34 d, the inflorescence length of OxPdB7 varied from 28.2cm to 32.6cm, that of Col-0 varied from 21.3cm to 24.9cm and that of the mutant varied from 15.5cm to 18.3cm. However, at the late stage, Col-0 and nf-yb3/PdB7 (34–37 d), and nf-yb3 (37–40 d) displayed higher average elongation rate, respectively (Fig. 5C). The results indicate that the transgenic lines had faster stem elongation at the early shooting stage and earlier seeding establishment.
Fig. 5.
Phenotypes of overexpressing PdNF-YB7 lines under well-watered conditions. (A) Difference in the primary root length at 8 d after germination. (B) Difference in plant leaf area of 18-d-old seedlings. (C) Plant average stem elongation rate in the time course of growth. (D) Morphological comparisons of the primary root length under half-strength MS for 8 d. (E) Morphology of rosettes of 18-d-old seedlings. (F) Morphology of 23-d-old seedlings grown under well-watered conditions. Data are mean ± SE (n = 30). Asterisks denote significant differences: *P ≤ 0.05; **P ≤ 0.01 (this figure is available in colour at JXB online).
Overexpressing PdNF-YB7 improves WUE in Arabidopsis
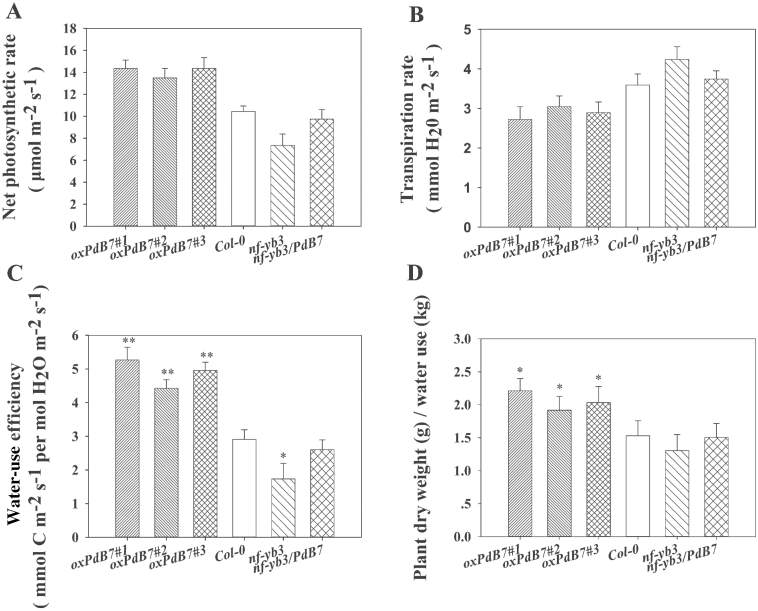
The oxPdB7 lines had higher net photosynthetic rates than Col-0 and the mutant under the same conditions and the mutant, with the slowest net photosynthetic rate, had a rate equivalent to Col-0 after complementation with PdNF-YB7 (Fig. 6A). For the transpiration rate, the oxPdB7 lines showed slower rates than Col-0 and the mutant (Fig. 6B). Based on the higher photosynthetic capability and lower transpiration level, the oxPdB7 lines had higher leaf WUE than Col-0 and the mutant (Fig. 6C). In addition, whole-plant WUE of oxPdB7 lines was higher than Col-0 and the nf-yb3 mutant (Fig. 6D).
Fig. 6.
Gas exchange analysis of overexpressing PdNF-YB7 lines shows that PdNF-YB7 improved WUE in Arabidopsis. (A–C) Net photosynthetic rate (A), transpiration rate (B), and instantaneous leaf WUE (C) of 3-week-old seedlings. (D) Plant dry weight variation under different water consumption (20 d). Data are mean ± SE (n = 15). Asterisks denote significant differences: *P ≤ 0.05; **P ≤ 0.01.
Overexpressing PdNF-YB7 increases drought tolerance under water deficit
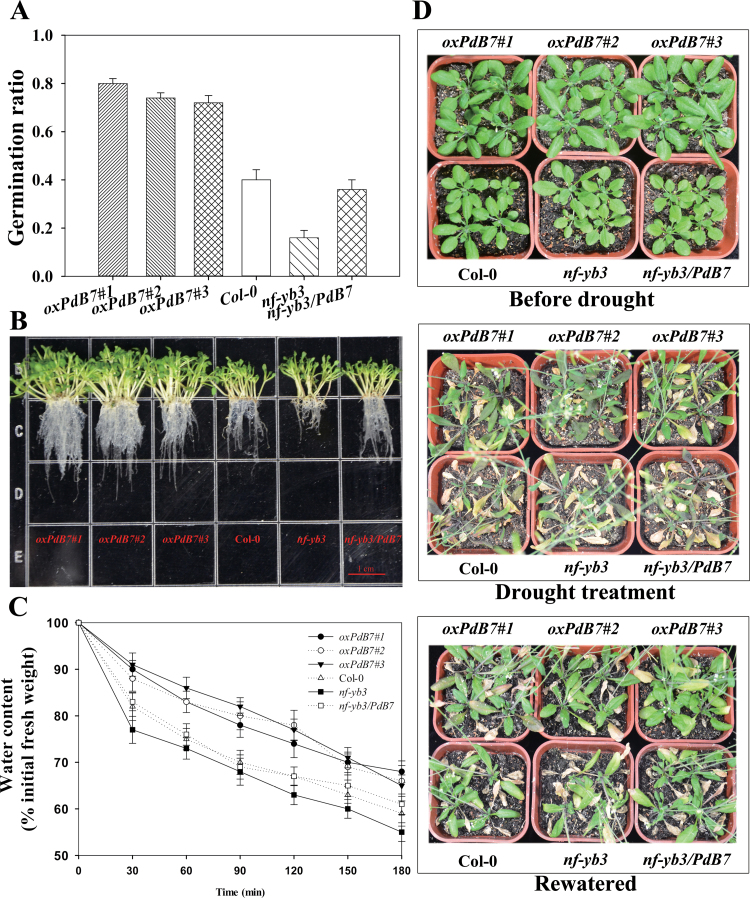
To decipher the mechanism by which water deficit affects plant development and growth, various experimental set ups were developed. The seeds of transgenic lines, Col-0, nf-yb3, and the complemented line nf-yb3/PdB7 were sown on half-strength MS culture with 200mM mannitol for osmotic stress. After 4 d, oxPdB7 lines had more vigorous germination (75.3%) than that of Col-0 (40.4%) and the mutant (16.3%) (Fig. 7A). However, by comparison, all seeds of transgenic lines, Col-0, nf-yb3, and nf-yb3/PdB7 had sprouted after 4 d under normal conditions (data not shown). Additionally, the primary root lengths of 8-d-old seedlings were also different. The oxPdB7 plants had considerably longer (1.4- and 2.4-fold, respectively) primary roots than that of Col-0 and nf-yb3 (Fig. 7B). Compared to the primary roots of Arabidopsis under well-watered conditions, primary root lengths of oxPdB7 lines decreased by 21.7% to 27.3% while that of Col-0 decreased by 35.3% and that of nf-yb3 reduced by 44.8% under drought conditions. Additionally, the detached leaves of transgenic plants lost water more slowly than Col-0 and the mutant (Fig. 7C).
Fig. 7.
Overexpression of PdNF-YB7 confers drought tolerance in Arabidopsis. (A) Difference in germination ratio among oxPdB7s, Col-0, nf-yb3, and nf-yb3/PdB7 plants grown on half-strength MS with 200mM mannitol. (B) Morphological differences in the primary root length of 8-d-old seedlings under 200mM mannitol. (C) Water loss from detached leaves; water loss is expressed as the percentage of initial fresh weight of detached leaves; data are means from five leaves for each of four independent experiments. (D) Morphological differences in drought experiments; the seedlings were grown in soil for 15 d under well-watered conditions; thereafter, water was withheld for 10 d; then plants were rewatered for 8 d. Data are mean ± SE (n = 50) (this figure is available in colour at JXB online).
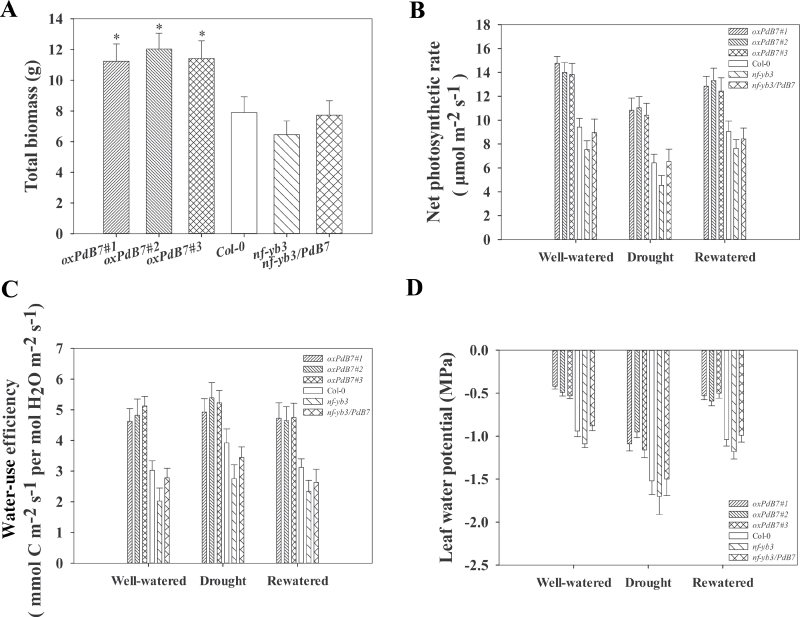
After the seedlings were transplanted to soil, water deficit was imposed for 10 d, followed by a rewatering period of 8 d (Fig. 7D). Analysis of soil water status for the genotypes explained that soil water contents of three phases in drought experiment were 53.20±1.19% (well-watered), 6.45±1.00% (drought), and 46.01±0.95% (rewatered), respectively. During water deprivation, Col-0 and the nf-yb3 mutant withered and showed more severe wilting than overexpressing plants, but the oxPdB7 lines exhibited continued development and growth resulting in more biomass (Fig. 8A). Photosynthesis analysis showed that transgenic lines maintained a significantly higher photosynthetic rate than Col-0 and the mutant under stress treatment (Fig. 8B), resulting in an increase in instantaneous leaf WUE (Fig. 8C). The leaf water potential of Arabidopsis seedlings showed significant differences among the transgenic, Col-0, nf-yb3, and nf-yb3/PdB7 seedlings under drought. The transgenic lines had higher leaf water potential compared to Col-0 and the mutant under drought stress (Fig. 8D). Thus, the expression of PdNF-YB7 was demonstrated to be sufficient to improve tolerance to water scarcity in Arabidopsis.
Fig. 8.
Physiological analysis of overexpressing PdNF-YB7 lines under drought experiments. (A) Plant total biomass variation for 20 d. (B–D) Net photosynthetic rate (B), instantaneous WUE of leaves under three different stages (C), and leaf water potential (D) under three different stages. Data are mean ± SE (n = 15). Asterisks denote significant differences: *P ≤ 0.05.
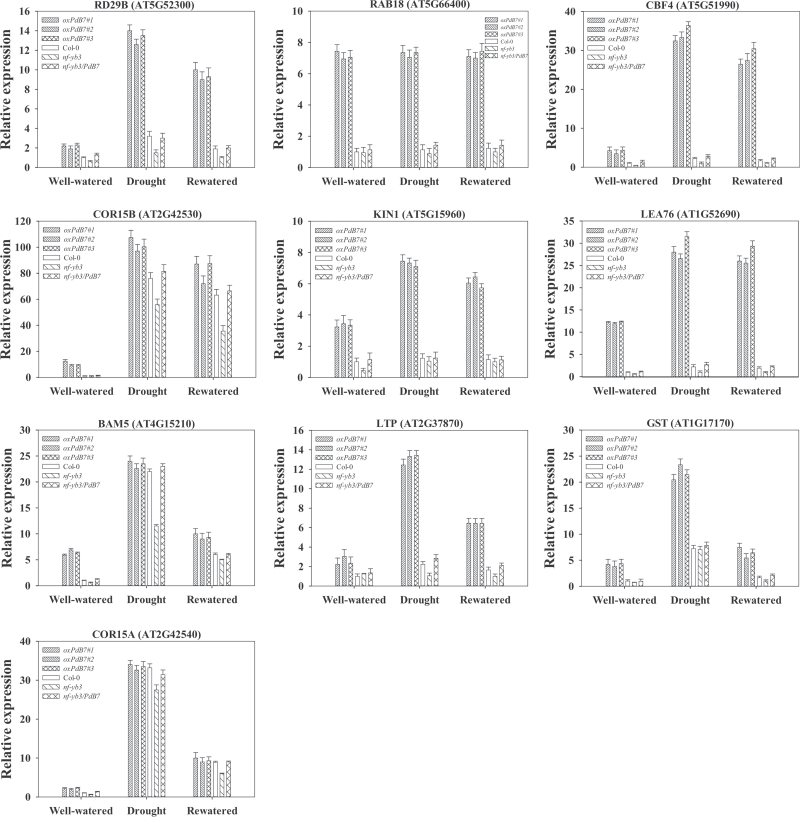
Expression analysis of stress-responsive genes regulated by the PdNF-YB7 transcription factor
To determine the improved drought tolerance by altered expression of PdNF-YB7, the expression levels of some drought-related genes in the leaves were analysed by qPCR in an independent experiment using the oxPdB7 lines, Col-0, mutant, and complementation under well-watered, drought, and rewatered conditions. The results showed that ABA pathway markers (RD29B, RAB18, and CBF4), CBF pathway markers (COR15B, KIN1, and LEA76)(Nelson et al., 2007), and several predicted candidate target genes of AtNF-YA5 (BAM5, LTP, GST, and COR15A) (Li et al., 2008) were differentially expressed in the oxPdB7 lines compared to the 35S:NF-YB1 and 35S:NF-YA5 lines (Fig. 9). CBF4, COR15B, LEA76, BAM5, and GST were more highly expressed in the oxPdB7 lines under well-watered conditions. Under water deficit, these stress-responsive genes were strongly induced in transgenic plants. However, the majority of these genes did not significantly change in nf-yb3, suggesting that for many of these genes, PdNF-YB7 was required for induction by dehydration.
Fig. 9.
Transcript level analysis of drought-related genes in oxPdB7s, Col-0, nf-yb3, and nf-yb3/PdB7 plants. Quantitative real-time PCR was used to analyse expression. The seedlings were sampled on day 14 after being transferred to soil under well-watered conditions on day 6, after drought treatment, and on day 5 after rewatering. 18S rRNA was used as the internal control. Data are mean ± SE (n = 3 experiments).
Discussion
Plants have evolved regulatory mechanisms to adapt to environmental water deficit. Transcription factors regulate expression of the stress-responsive genes by binding specifically to the motif of the promoters to modulate resistance to drought and lower productivity loss (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). NF-Y is an important transcription factor family controlling drought tolerance in plants (Nelson et al., 2007; Stephenson et al., 2007; Li et al., 2008). In a previous study, a drought-responsive NF-YB family member PeNF-YB7 was screened from P. euphratica with an Affymetrix Poplar GeneChip microarray and was further identified by qPCR (Yan et al., 2012a). To study drought tolerance of fast-growing poplar, PdNF-YB7, the homologue of PeNF-YB7, was identified from the high WUE poplar genotype NE-19 [P. nigra × (P. deltoides × P. nigra)] (Yin et al., 2007; Hao et al., 2011). In the current experiments, expression analysis showed that PdNF-YB7 was differentially expressed and concomitantly induced in response to water deficit in the NE-19 seedlings (Fig. 2A). Overexpression of PdNF-YB7 in Arabidopsis exhibited earlier seedling establishment, longer primary roots, larger leaf areas, and increased photosynthetic rate that conferred drought tolerance and improved WUE in transgenic plants.
Nuclear factor Y is one of the largest transcription factor gene families in plants. A number of NF-Y proteins have been identified as regulators of drought tolerance in different plant species. AtNF-YB1, ZmNF-YB2, and TaNF-YB2, were reported to confer drought resistance in Arabidopsis, maize, and wheat, respectively, and to increase crop productivity under drought field tests (Nelson et al., 2007; Stephenson et al., 2007). PdNF-YB7 was inferred to have the same effect on Arabidopsis because of the relatively close evolutionary relationship and genetic distance (Yan et al., 2012b). In addition, another NF-Y subunit family member, AtNF-YA5, plays a role in drought resistance in Arabidopsis (Li et al., 2008). Apart from these, other NF-YB family members, such as AtNF-YB9 (LEC1) and AtNF-YB6 (L1L), are essential factors controlling embryonic development and are phylogenetically and functionally distinct from other NF-YB family members, such as AtNF-YB1 and PtNF-YB7 (Kwong et al., 2003; Yamamoto et al., 2009; Yan et al., 2012b). Most of the above findings were reported for herbaceous plants such as Arabidopsis and maize. However, reports of NF-YBs in woody plants are still sparse. The PdNF-YB7 gene studied here is the first reported NF-YB gene in fast-growing black poplar.
According to the phylogenetic analysis, PdNF-YB7 shared high sequence similarity and clustered with a member of the poplar NF-YB family genes. The conserved domain analysis of the multiple sequence alignment showed that PdNF-YB7, PtNF-YB7, and AtNF-YB3 are very highly conserved. In Arabidopsis, AtNF-YB3 plays an important role in the promotion of flowering specifically under inductive long-day photoperiodic conditions. Consistent with this, the overexpression of PdNF-YB7 in Arabidopsis caused earlier seedling germination time and enhanced the development of both vegetative and reproductive organs (Fig. 5F). Notably, in these experiments, the transcript levels of PdNF-YB7 were upregulated by drought stress, resembling those of AtNF-YB1 in Arabidopsis (Nelson et al., 2007). Different from AtNF-YB1, transcript levels of PdNF-YB7 were also affected by ABA treatment. Promoter expression analysis of PdNF-YB7 provided further support for its role in stress tolerance; GUS gene expression was enhanced by drought and was observed throughout the entire plant after stress treatment. Element analysis of the promoter indicated that several ABA-responsive elements were included in PdNF-YB7 promoter region (Supplementary Table S3). This is the same result of AtNF-YA5 in Arabidopsis; two ABA-responsive element sequences could be found in the promoter region of AtNF-YA5, which is proved to be involved in drought resistance (Li et al., 2008). Additionally, increasing evidence is being found for NF-YB/bZIP interactions, and bZIP proteins are well known to be involved in ABA signaling (Liu and Howell, 2010). Interestingly, a tissue-specific expression analysis indicated another difference between PdNF-YB7 and AtNF-YB3. Previous research revealed that AtNF-YB3 is expressed more highly in flowers and young leaves, but is absent in roots (Siefers et al., 2009), while the current experiments suggested that PdNF-YB7 is highly expressed in root. This is consistent with AtNF-YA5, overexpression of which is proved to increase drought tolerance in Arabidopsis (Li et al., 2008).
To better understand the regulatory mechanisms of drought tolerance conferred by overexpressing PdNF-YB7, this study confirmed the expression patterns of genes that may potentially be regulated by NF-Ys according to previous research on AtNF-YB1 and AtNF-YA5. For the ABA pathway markers (CBF4, RD29B, and RAB19) or CBF pathway markers (COR15B, KIN1, and LEA76), none of them showed differences in expression between AtNF-YB1 overexpression plants and controls (Nelson et al., 2007). However, in the current study, RD29B and CBF4 showed significant and consistent differences expression in 35S:PdNF-YB7 plants, indicating that the ABA-dependent dehydration response was regulated by PdNF-YB7. Drought-inducible genes encoding functional proteins such as KIN1, LEA76, LTP, and GST were also highly expressed in transgenic plants, especially in response to water deficit, suggesting that PdNF-YB7 potentially increased the accumulation of protective proteins under drought conditions.
Transgenic Arabidopsis overexpressing poplar NF-YB7 showed significantly higher biomass under well-watered and drought conditions. Increased photosynthetic leaf area enhanced carbon assimilation, resulting in more biomass accumulation. Several studies have implicated NF-Y in controlling photosynthesis by regulating the chloroplast ATP synthase (Kusnetsov et al., 1999) and some nuclear-encoded photosynthesis genes such as RBCS and CAB (Miyoshi et al., 2003). Transgenic wheat with TaNF-YB3 has a significant enhancement in leaf chlorophyll content and photosynthesis rate (Stephenson et al., 2011).
Recent studies have shown that root growth is closely connected with drought tolerance (Pennisi, 2008). In the current study, overexpressing PdNF-YB7 in Arabidopsis increased primary root length that led to expansion of the root surface area. Ballif et al. (2011) also found that overexpressing AtNF-YB2 enhanced primary root elongation due to a faster cell division and/or elongation. Additionally, PdNF-YB7 overexpression transgenic plants also maintained higher leaf water potential than that of wild type under water-deficit conditions. Often during drought conditions, plants avoid low soil water potential by achieving a balance between water absorption and loss, for example, by decreasing the stomatal aperture while maintaining root growth (Verslues et al., 2006). Decrease of stomatal apertures resulted in decreased transpiration rate thus reduced water loss. Meanwhile, increased root length improved absorption of water and mineral solutes. Equality between uptake and loss of water and thereby maintenance of constant leaf water potential is assisted by stomatal changes, which appear to be in response to conditions in the root (Aston and Lawlor, 1979). Altogether, these physiological phenotypes conferred by PdNF-YB7 support that the gene could be potentially used in breeding drought-tolerant plants and promoting plant production under drought conditions. Further experiments are now needed to characterize the effects of PdNF-YB7 in poplar to elucidate its regulatory mechanisms in woody plants.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences used for cloning of PdNF-YB7 cDNA.
Supplementary Table S2. Primers used for PCR and qPCR.
Supplementary Table S3. Putative elements of the promoter of PdNF-YB7.
Supplementary Fig. S1. The structural alignment result of the PdNF-YB7 protein.
Acknowledgements
The research was supported by grants from the Ministry of Science and Technology of China (2011BAD38B01, 2009CB119101), the National Natural Science Foundation of China (31070597, 31270656, 30972339), and the Scientific Research and Graduate Training Joint Programs from BMEC (Regulation of Tree WUE and Stress Resistance Mechanism of Poplar).
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. 1997. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. The Plant Cell 9, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston M, Lawlor D. 1979. The relationship between transpiration, root water uptake, and leaf water potential. Journal of Experimental Botany 30, 169–181 [Google Scholar]
- Ballif J, Endo S, Kotani M, MacAdam J, Wu Y. 2011. Over-expression of HAP3b enhances primary root elongation in Arabidopsis . Plant Physiology and Biochemistry 49, 579–583 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Jolivet S, Voisin R, Pelletier G. 2003. The endosperm and the embryo of Arabidopsis thaliana are independently transformed through infiltration by Agrobacterium tumefaciens . Transgenic Research 12, 509–517 [DOI] [PubMed] [Google Scholar]
- Bradshaw H, Ceulemans R, Davis J, Stettler R. 2000. Emerging model systems in plant biology: poplar (Populus) as a model forest tree. Journal of Plant Growth Regulation 19, 306–313 [Google Scholar]
- Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt BF., 3rd 2011. Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon . PloS One 6, e21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116 [Google Scholar]
- Chen J, Xia X, Yin W. 2009. Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica . Biochemical and Biophysical Research Communications 378, 483–487 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Pérez M, Lautner S, Nehls U, Selle A, Teuber M, Schnitzler JP, Teichmann T, Fayyaz P, Hartung W, Polle A. 2009. Salt stress affects xylem differentiation of grey poplar (Populus × canescens). Planta 229, 299–309 [DOI] [PubMed] [Google Scholar]
- Gray J, Bevan M, Brutnell T, et al. 2009. A recommendation for naming transcription factor proteins in the grasses. Plant Physiology 149, 4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Keetman U, Grimm B. 2012. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. International Journal of Molecular Sciences 13, 3458–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Zhao T, Xia X, Yin W. 2011. Genome-wide comparison of two poplar genotypes with different growth rates. Plant Molecular Biology 76, 575–591 [DOI] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, Krishnan A, Nataraja KN, Udayakumar M, Pereira A. 2007. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proceedings of the National Academy of Sciences, USA 104, 15270–15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Lumbreras V, Pages M. 2001. Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Letters 498, 187–189 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ. 2008. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis . Planta 228, 709–723 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF., 3rd 2010. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana . The Plant Journal 63, 379–391 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Landsberger M, Meurer J, Oelmüller R. 1999. The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. Journal of Biological Chemistry 274, 36009–36014 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. 2003. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. The Plant Cell 15, 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Lida K, Lu XY, Cui X, Jin H, Zhu JK. 2008. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell 20, 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. 2010. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis . The Plant Cell 22, 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC. 2007. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis . Plant Molecular Biology 63, 289–305 [DOI] [PubMed] [Google Scholar]
- Ma H-S, Liang D, Shuai P, Xia X-L, Yin W-L. 2010. The salt-and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana . Journal of Experimental Botany 61, 4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ito Y, Serizawa A, Kurata N. 2003. OsHAP3 genes regulate chloroplast biogenesis in rice. The Plant Journal 36, 532–540 [DOI] [PubMed] [Google Scholar]
- Monclus R, Dreyer E, Villar M, Delmotte FM, Delay D, Petit JM, Barbaroux C, Le Thiec D, Bréchet C, Brignolas F. 2005. Impact of drought on productivity and water use efficiency in 29 genotypes of Populus deltoids × Populus nigra . New Phytologist 169, 765–777 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, et al. 2007. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences, USA 104, 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu CF, Wei W, Zhou QY, et al. 2012. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell and Environment 35, 1156–1170 [DOI] [PubMed] [Google Scholar]
- Pennisi E. 2008. Plant genetics. Getting to the root of drought responses. Science 320, 173. [DOI] [PubMed] [Google Scholar]
- Renaut J, Hoffmann L, Hausman JF. 2005. Biochemical and physiological mechanisms related to cold acclimation and enhanced freezing tolerance in poplar plantlets. Physiologia Plantarum 125, 82–94 [Google Scholar]
- Ridge C, Hinckley T, Stettler R, Van Volkenburgh E. 1986. Leaf growth characteristics of fast-growing poplar hybrids Populus trichocarpa × P. deltoides . Tree Physiology 1, 209–216 [DOI] [PubMed] [Google Scholar]
- Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang KY, Li E, Douglas CJ, Western TL. 2012. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytologist 195 774–786 [DOI] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. 2003. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. Journal of Biological Chemistry 278, 1336–1345 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiology 136, 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. 2003. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology 6, 410–417 [DOI] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WEt, Tayrose G, Holt BF., 3rd 2009. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiology 149, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. 2007. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum . Plant Molecular Biology 65, 77–92 [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. 2010. TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Functional and Integrative Genomics 10, 265–276 [DOI] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue GP. 2011. TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum . Functional and Integrative Genomics 11, 327–340 [DOI] [PubMed] [Google Scholar]
- Sun HY, Liu CM, Zhang XY, Shen YJ, Zhang YQ. 2006. Effects of irrigation on water balance, yield and WUE of winter wheat in the North China Plain. Agricultural Water Management 85, 211–218 [Google Scholar]
- Testa A, Donati G, Yan P, Romani F, Huang TH, Vigano MA, Mantovani R. 2005. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. Journal of Biological Chemistry 280, 13606–13615 [DOI] [PubMed] [Google Scholar]
- Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. 2008. Identification, characterization and interaction of HAP family genes in rice. Molecular Genetics and Genomics 279, 279–289 [DOI] [PubMed] [Google Scholar]
- Tschaplinski T, Blake T. 1989. Water relations, photosynthetic capacity, and root/shoot partitioning of photosynthate as determinants of productivity in hybrid poplar. Canadian Journal of Botany 67, 1689–1697 [Google Scholar]
- Tschaplinski T, Tuskan G, Gunderson C. 1994. Water-stress tolerance of black and eastern cottonwood clones and four hybrid progeny. I. Growth, water relations, and gas exchange. Canadian Journal of Forest Research 24, 364–371 [Google Scholar]
- Valliyodan B, Nguyen HT. 2006. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Current Opinion in Plant Biology 9, 189–195 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. 2006. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. The Plant Journal 45, 523–539 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X. 2009. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230, 1155–1166 [DOI] [PubMed] [Google Scholar]
- Wong SC, Cowan IR, Farquhar GD. 1978. Leaf conductance in relation to assimilation in Eucalyptus pauciflora Sieb. ex Spreng: Influence of irradiance and partial pressure of carbon dioxide. Plant Physiology 62, 670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. 2008. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiology 148, 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HT, Guo P, Xia XL, Yin WL. 2011. PdERECTA, a leucine-rich repeat receptor-like kinase of poplar, confers enhanced water use efficiency in Arabidopsis . Planta 234, 229–241 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57, 781–803 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. 2009. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. The Plant Journal 58, 843–856 [DOI] [PubMed] [Google Scholar]
- Yan DH, Fenning T, Tang S, Xia X, Yin W. 2012a. Genome-wide transcriptional response of Populus euphratica to long-term drought stress. Plant Science 195, 24–35 [DOI] [PubMed] [Google Scholar]
- Yan DH, Xia X, Yin W. 2012b. NF-YB family genes identified in a poplar genome-wide analysis and expressed in Populus euphratica are responsive to drought stress. Plant Molecular Biology Reporter 31, 363–370 [Google Scholar]
- Yin WW, Wan XQ, Xia XL. 2007. Relations between stable carbon isotope discrimination and water use efficiency as well as growth for poplar. Scientia Silvae Sinicae 8, 004 [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV. 2010. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. The Plant Cell 22, 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsuffa L, Giordano E, Pryor L, Stettler R. 1996. Trends in poplar culture: some global and regional perspectives. In: Stettler RF, Bradshaw HD, Jr., Heilman PE, Hinckley TM, eds, Biology of Populus and its implications for management and conservation. Ottawa: NRC Research Press; pp. 515–539 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.