Abstract
A rice cDNA, OsDEP1, encoding a highly cysteine (Cys)-rich G protein γ subunit, was initially identified as it conferred cadmium (Cd) tolerance on yeast cells. Of the 426 aa constituting OsDEP1, 120 are Cys residues (28.2%), of which 88 are clustered in the C-terminal half region (aa 170–426). To evaluate the independent effects of these two regions, two truncated versions of the OsDEP1-expressing plasmids pOsDEP1(1–169) and pOsDEP1(170–426) were used to examine their effects on yeast Cd tolerance. Although OsDEP1(170–426) conferred a similar level of Cd tolerance as the intact OsDEP1, OsDEP1(1–169) provided no such tolerance, indicating that the tolerance effect is localized to the aa 170–426 C-terminal peptide region. The Cd responses of transgenic Arabidopsis plants constitutively expressing OsDEP1, OsDEP1(1–169) or OsDEP1(170–426), were similar to the observations in yeast cells, with OsDEP1 and OsDEP1(170–426) transgenic plants displaying Cd tolerance but OsDEP1(1–169) plants showing no such tolerance. In addition, a positive correlation between the transcript levels of OsDEP1 or OsDEP1(170–426) in the transgenics and the Cd content of these plants upon Cd application was observed. As several Arabidopsis loss-of-function heterotrimeric G protein β and γ subunit gene mutants did not show differences in their Cd sensitivity compared with wild-type plants, we propose that the Cys-rich region of OsDEP1 may function directly as a trap for Cd ions.
Key words: cadmium tolerance, cysteine-rich protein, G protein γ subunit, heterotrimeric G protein signalling, Oryza sativa, OsDEP1.
Introduction
Cadmium (Cd) is one of the transition metals that is non-essential for almost all living organisms. It is also a noxious compound that inactivates and denatures structural and functional proteins of organisms by binding to free sulfhydryl groups, thereby inhibiting their growth and development. Another aspect of Cd toxicity is derived from its chemical similarity to metal co-factors or coordinated metals, such as Zn, Fe, and Ca, of enzymes, signalling intermediates, and transcription factors, especially the zinc-finger type (DalCorso et al., 2008; Verbruggen et al., 2009). Due to its high toxicity, Cd pollution poses serious problems both to our natural environment and to human health (Järup, 2003). To address such levels of Cd contamination, phytoremediation is being promoted as a promising approach, especially as it is more environmentally friendly and imposes a smaller financial burden than alternative physico-chemical approaches. Towards establishing easy-to-apply phytoremediation strategies, it is essential to identify important factors in the plant genome that can be exploited for such approaches.
To cope with Cd toxicity effects, plants are known to be equipped with the potential to chelate and extrude Cd, to sequester Cd into vacuoles, and to dissipate reactive oxygen species triggered by Cd. For the chelation of heavy metals, including Cd, various cysteine (Cys)-rich proteins are employed by plants. Small Cys-rich peptides, called metallothioneins (MTs), are the major chelators of Cd (Ecker et al., 1986; Freisinger, 2008). Recently, the class A heat-shock transcription factor HsfA4a was identified as a wheat clone that confers strong Cd tolerance in yeast and rice by upregulating the MT gene (Shim et al., 2010). Several other Cys-rich proteins, of various sizes and cellular localizations, have also been found to provide Cd tolerance (Willuhn et al., 1994; Song et al., 2004; Kuramata et al., 2009; Matsuda et al., 2009).
For Cd extrusion, AtPDR8, an ATP-binding cassette (ABC)-type transporter, has been found to be involved in Cd efflux at the plasma membrane (Kim et al., 2007). In addition, the wheat TM20 gene, which encodes a hydrophobic protein with 20 transmembrane domains, has been shown to stimulate Cd efflux when overexpressed in yeast cells, suggesting that TM20 functions to pump out Cd by an unknown mechanism (Kim et al., 2008). Furthermore, yeast cells and Arabidopsis plants constitutively expressing the Digitaria ciliaris CDT1 gene have been found to contain less than half the Cd levels of their respective controls, indicative of a reduced accumulation of Cd in these organisms, most probably through Cd extrusion (Kuramata et al., 2009). Finally, it is worth noting that tobacco plants grown in Cd-containing medium were found to secrete Cd-containing amorphous materials through the tips of their long trichomes, demonstrating the role that trichomes play in Cd extrusion (Choi et al., 2001).
For Cd sequestration to vacuoles, the Saccharomyces cerevisiae YCF1 (yeast cadmium factor 1), a vacuolar-localized ABC-type transporter, is known to function in the sequestration of both glutathione (GSH) conjugates and (GSH)2–Cd complexes into the vacuole; indeed, deletion of this gene (YCF1) renders the host yeast cells Cd hypersensitive (Li et al., 1996). Similarly in plants, two ABC-type vacuolar transporters that mediate arsenic and Cd tolerance have recently been identified in Arabidopsis (Mendoza-Cózatl et al., 2010; Song et al., 2010). P1B-ATPase subfamily members in both Arabidopsis (AtHMA3) and rice (OsHMA3) have been shown to be localized to the vacuolar membrane (tonoplast) and to be essential for the sequestration of Cd into vacuoles (Morel et al., 2009; Ueno et al., 2010; Miyadate et al., 2011). Tonoplast membrane-localized Cd2+/proton antiporters, such as AtCAX2 and AtCAX4, have also been shown to transport Cd without modification into vacuoles (Korenkov et al., 2007).
Finally, Cd is known to induce oxidative damage, such as through lipid peroxidation, which can lead to changes in membrane functionality and protein carbonylation (Romero-Puertas et al., 2002, 2004). Antioxidants and antioxidant-synthesizing enzymes have been implicated in the enhanced tolerance of plants to Cd toxicity (DalCorso et al., 2008; Verbruggen et al., 2009).
The rice DEP1 (DENSE AND ERECT PANICLE 1) locus was first identified by two independent research groups with quantitative trait loci analysis to control grain yield, grain numbers per panicle, and panicle morphology (Huang et al., 2009; Zhou et al., 2009). Deletion of the DEP1 gene during rice domestication was proposed to enhance meristematic activity and result in reduced inflorescence internode lengths that thereby increased grain numbers per panicle and, consequently, grain yields (Huang et al., 2009; Zhou et al., 2009; Taguchi-Shiobara et al., 2011). Recently, Arabidopsis AGG3, a DEP1 homologue, was identified as an Arabidopsis heterotrimeric GTP-binding protein (G protein) γ subunit (Chakravorty et al., 2011; Thung et al., 2012). Unlike the complex mammalian system, Arabidopsis has only one α (GPA1), one β (AGB1), and three γ (AGG1, AGG2, and AGG3) subunits as components of the heterotrimeric G protein system (Botella 2012; Thung et al., 2012). So far, AGG3 has been shown to be involved in both guard cell K+-channel regulation and morphological development (Chakravorty et al., 2011; Li et al., 2012a ,b).
Here, we identified OsDEP1 as a cDNA clone that confers Cd tolerance to yeast cells. The gene product, OsDEP1, is highly Cys-rich and is a component of the heterotrimeric G protein signalling pathway (Botella, 2012). Based on the results obtained, we discuss a functional role for this G protein subunit in the Cd stress response.
Materials and methods
Plant materials
Rice plants (Oryza sativa cv. Nipponbare) were grown hydroponically in 40% strength Hoagland’s solution #2 [2mM Ca(NO3)2.4H2O, 2mM KNO3, 0.8 μM MgSO4.7H2O, 0.0002% FeSO4.EDTA] or in soil in a greenhouse. Seeds of Arabidopsis thaliana ecotype Columbia (Col-0) and the loss-of-function heterotrimeric G protein gene mutants of Columbia background (gpa1-4, agb1-1, agb1-2, agg1-1C, agg3-1 and the triple mutant agg1-1C agg2-1 agg3-1), kindly provided by Professor J. Botella (Botella, 2012), were germinated and grown on vermiculite in a growth chamber at 22 °C under a 16h light/8h dark photocycle.
Yeast strains
The yeast strains used in this study were obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html), Frankfurt, Germany. The wild-type (WT) strain used was S. cerevisiae BY4742 with the relevant genotype (MATα; his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0). To identify cDNA clones conferring Cd tolerance on yeast cells, the Cd-sensitive BY4742 Δycf1 mutant (MATα; his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0; YDR135c::kanMX4) was employed. In addition, Δcup2 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, YGL166w::kanMX4) was used to test for Cu2+ tolerance. See Supplementary Methods at JXB online for details of the other metal-sensitive strains used.
Preparation of an O. sativa cDNA library
O. sativa cv. Nipponbare seedlings were grown in Murashige–Skoog medium (Murashige and Skoog, 1962) for 2 weeks. Total RNA was isolated from fresh rice seedlings by an SDS/phenol method (Shirzadegan et al., 1991), and mRNA was subsequently purified using an mRNA purification kit (QuickPrep mRNA Purification kit; GE Healthcare, Milwaukee, USA). The mRNA fraction was then converted to cDNA using a SMART cDNA library construction kit (BD Bioscience Clontech, Palo Alto, CA, USA). After SfiI enzyme digestion, the resulting cDNA was ligated into the SfiIA and SfiIB sites of a modified yeast expression vector, termed pGK1, of p112A1NE (Riesmeier et al., 1992), yielding a O. sativa cDNA library.
Isolation of Cd-tolerant clones from the rice cDNA library
To isolate Cd-tolerant clones from the rice cDNA library, we introduced the library into the yeast Δycf1 mutant cells using the lithium acetate method (Ito et al., 1983). Yeast colonies that grew on medium containing 20–60 μM CdCl2 were selected and their plasmids isolated, and these plasmids were then re-introduced into Δycf1 cells to reconfirm the Cd tolerance of the clones.
Construction of pOsDEP1 and its two derivatives
The fragment covering the ORF of OsDEP1 (Os09g0441900) was amplified using the primer pair Os09g0441900-F (5′-TTAGGCCATTACGGCCGTGAAGGCGGCGAGGGT-3′; the underlined sequence denotes the SfiIA restriction site in this study) and Os09g0441900-R (5′-TTAGGCCGAGGCGGCCTCAA CATAAGCAACCAC-3′, the underlined sequence denoting the SfiIB site in this study). The sequence-verified fragment was inserted into the SfiIA and SfiIB restriction sites of the yeast expression vector, pGK1, resulting in construct pOsDEP1. The OsDEP1 gene encodes a 426 aa residue protein that consists of two domains (Huang et al., 2009; Zhou et al., 2009; Taguchi-Shiobara et al., 2011): an N-terminal half (aa 1–169) and a Cys-rich C-terminal half (aa 170–426). Thus, two OsDEP1-derived fragments, covering the OsDEP1(1–169) and OsDEP1(170–426) regions, were amplified using the following primer pairs, respectively: Os09g0441900-F and OsDEP1(1–169)-R (5′-TTAGGCCGAGGCGGCCTCAGTTCGGTTTGCAG-3′), and OsDEP1(170–426)-F (5′-TTAGGCCATTACGGCCATGTGC TGTAAACCTAACTGCAG-3′) and Os09g0441900-R. The sequence-verified fragments were subcloned into the SfiIA and SfiIB sites of pGK1 vector, resulting in the respective constructs pOsDEP1(1–169) and pOsDEP1(170–426).
Cd and Cu tolerance assays in agar medium and in liquid culture
S. cerevisiae BY4742 Δycf1 or Δcup2 strain was transformed with either the pGK1 empty vector (EV), or with pOsDEP1, pOsDEP1(1–169), or pOsDEP1(170–426). The S. cerevisiae WT BY4742 strain transformed with EV was also used as a control. The turbidity of the SD-Ura (synthetic drop-out lacking uracil) liquid cultures, inoculated with the respective transformants, was adjusted to an optical density at 600nm (OD600) of 1.0, and tenfold serial dilutions then prepared aseptically. Subsequently, 5 μl of each of the dilution series was spotted onto SD-Ura agar medium with or without 50 μM CdCl2 or 300 μM CuCl2, and incubated at 30 °C for 3 d. In addition, growth of the yeast transformants described above in SD-Ura liquid medium supplemented with 40 μM CdCl2 was monitored at OD600.
Generation of transgenic Arabidopsis plants
As the OsDEP1-coding region contained a single SacI site, we first removed this SacI site from the OsDEP1-coding region, without changing the amino acid sequence, by two-step PCR using OsDEP1 cDNA template, KOD-Plus DNA polymerase (Toyobo, Japan) and the following primer pairs. The first PCR was performed with two primer pairs, DEP-ox-F (5′- CGGTCTAGACAAGGAGATATAAC AATGGGGGAGGAGGCGGT-3′; the new XbaI site is underlined and the start codon is italic) and dep1-rr (5′- CGGGCTGCGCTCCTT CAAGGAAGT-3′, mutated site is underlined), and dep1-ff (5′-AAGGAGCGCAGCCCGTTTCTCGTT-3′; mutated site is underlined) and DFP1-ox-R (5′-CAGGAGCTCTCAACATAA GCAACCACT-3′; the new SacI site is underlined and the stop codon is italic). The second PCR was then performed on mixtures of the first PCR products using the primer pair DEP-ox-Fw and DEP-ox-R. In addition to the OsDEP1 (aa 1–426, SacI site mutated) fragment, the OsDEP1(1–169) and OsDEP1(170–426) fragments were also amplified with the following primer pairs; DEP1-ox-F and OsDEP1lackCys-rich-ox-R (5′-CAGGAG CTCTCAGTTCGGTTTGCAGCA-3′; SacI site is underlined), and OsDEP1Cys-rich-ox-F (5′-CGGTCTAGACAAGGAGATAT AACAATGTGCTGTAAACCTAA-3′; XbaI site is underlined) and DEP1-ox-R, respectively. The sequence-verified fragments encompassing the complete OsDEP1 ORF and OsDEP1(1–169) and OsDEP1(170–426) were digested with XbaI and SacI, and subcloned into the respective restriction sites of the binary vector, pBI121 (Clontech), yielding pBI121OsDEP1, pBI121OsDEP1(1–169), and pBI121OsDEP1(170–426), respectively. These plasmids were introduced by the freeze–thaw method into Agrobacterium tumefaciens GV3101 cells (Koncz and Schell, 1986), which were then used to transform A. thaliana ecotype Col-0 plants by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige–Skoog agar medium containing 50mg ml–1 of kanamycin (Km) and 50mg ml–1 of carbenicillin. T2 seeds obtained from self-fertilization of primary transformants were surface sterilized and grown on Km plates. Lines showing a 3:1 (resistant:sensitive) segregation ratio were selected and used to produce homozygous (KmR/KmR) T3 lines that were used for further study.
Reverse transcription-PCR (RT-PCR) analysis
Expression analysis of the transgene in transgenic Arabidopsis plants was performed by RT-PCR. Total RNA was extracted from whole seedlings, reverse transcribed, and then semi-quantitatively amplified using the following primer pairs: OsDEP1 forward, (5′-GTGAAGGCG GCGAGGGT-3′) and OsDEP1 reverse (5′-TCAACATAAG CAACCAC-3′); OsDEP1(1–169) forward, the same forward primer used for OsDEP1, and OsDEP1(1–169) reverse (5′-TCAGTTCG GTTTGCAG-3′); OsDEP1(170–426) forward (5′-ATGTGCTGTAA ACCTAACTGCAG-3′) and OsDEP1(170–426) reverse, the same reverse primer used for OsDEP1. As a control, the Arabidopsis tubulin gene was amplified using the following primers pair: tubulin forward (5′-CGTGGATCACAGCAATACAGAGCC-3′) and tubulin reverse (5′-CCTCCTGCACTTCCACTTCGTCTTC-3′).
Response of transgenic plants to CdCl2 and measurement of Cd contents
The seeds of control transgenics transformed with pBI121 (Clontech), and transgenics expressing full-length OsDEP1 or the OsDEP1(1–169) and OsDEP1(170–426) derivatives were surface sterilized, rinsed, and placed onto MRGL medium (Fujiwara et al., 1992; Kuramata et al., 2009) solidified with 1% (w/v) gellan gum either without CdCl2 (control) or with appropriate concentrations of CdCl2. After 2 weeks of incubation, seedling growth was analysed and root lengths were quantified using ImageJ software (National Institutes of Health, http://rsbweb.nih.gov/ij/). Five-d-old Arabidopsis seedlings grown on MRGL medium were transferred to deionized water containing 5 μM CdCl2 and incubated for a further 5 d. The seedlings (n=5) were washed thoroughly with sterilized water, blotted with a paper towel, and then dried at 65 °C for 1 d. Dried plant samples were digested with 60% nitric acid at 60 ºC, and the Cd contents were analysed by Zeeman atomic absorption spectrometry (AA240Z; Varian).
Statistical analysis
Data analysis was performed using the statistical tools (Student’s t-test) of Microsoft Excel software.
Results
Identification of OsDEP1 as a clone conferring Cd tolerance to yeast cells
Of the approximately 3.0×105 O. sativa cDNA clones analysed, six were identified as conferring Cd tolerance. Of these clones, three encoded class I MTs; the fourth clone was a homologue of Hypochaeris radicata HrCDT3 (GenBank accession no. AB454513) that has been implicated in Cd tolerance (published only in the NCBI database); the fifth clone encoded OsCDT1 (GenBank accession no. AK121052), which was also previously identified as a Cd tolerance-related clone (Kuramata et al., 2009; Matsuda et al., 2009); and the sixth clone was found to encode a protein that showed a high level of similarity to a keratin-associated protein (GenBank accession no. FJ039905). Interestingly, the gene corresponding to this latter clone was first identified as the causal gene of the rice panicle morphology mutant by two independent research groups (Huang et al., 2009; Zhou et al., 2009) and was thus termed OsDEP1 (O. sativa DENSE AND ERECT PANICLE 1). More importantly, a recent study has indicated that the gene product, OsDEP1, is an isoform of G protein γ subunits (Botella, 2012). As no studies on OsDEP1 in relation to heavy metal tolerance have been reported, we focused on this clone in this study.
Confirmation that OsDEP1 confers Cd tolerance
The fragment covering the full-length ORF of OsDEP1 was re-cloned into the pGK1 EV resulting in the recombinant plasmid pOsDEP1. Subsequently, pGK1 and pOsDEP1 were introduced into Cd-sensitive Δycf1 yeast cells, and the respective transformants then spotted onto SD-Ura agar plates with or without 50 μM CdCl2. The Δycf1 cells carrying pOsDEP1 grew well in Cd-containing medium, even better than the WT cells carrying EV (Fig. 1). OsDEP1 encoded a protein composed of 426 aa. Database searches revealed that OsDEP1 orthologues and paralogues were found in monocotyledonous plants, such as Triticum urartu (GenBank accession no. GQ324995; encoding a peptide of 283 aa), Hordeum vulgare (FJ039903; 295 aa) and Zea mays (NM_001158725; 408 aa), as well as in dicotyledonous plants, such as A. thaliana (AGG3, an isoform of γ subunits, NM_147870; 251 aa), Glycine max (BT095006; 209 aa), Ricinus communis (XM_002516219; 336 aa) and Vitis vinefera (CBI27799; 153 aa) (Supplementary Fig. S1 at JXB online). Amino acid sequence similarities between OsDEP1 and its dicotyledonous counterparts were mainly restricted to the N-terminal half (aa 40 to ~120–130) of OsDEP1.
Fig. 1.
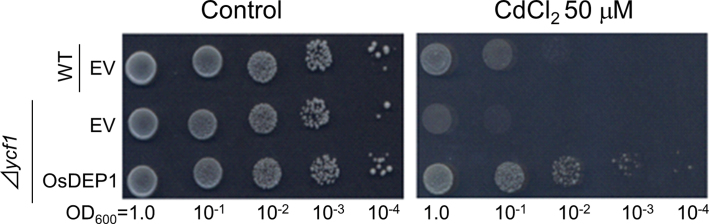
Identification of rice OsDEP1 as a clone conferring Cd tolerance to yeast cells. Cd tolerance of yeast cells expressing OsDEP1. Cells of S. cerevisiae strain BY4742 (Δycf1) carrying pGK1 (EV) or pOsDEP1, and its parental strain (WT, BY4742) carrying EV were grown in SD-Ura liquid medium for 16h. The OD600 of the cultures was adjusted to 1.0 from which tenfold dilution series (10–1, 10–2, 10–3, and 10–4) were prepared. Subsequently, 5 μl aliquots of each dilution were spotted onto SD-Ura control medium (left panel) or medium containing 50 μM CdCl2 (right panel), and the cells were allowed to grow for 3 d.
Impact of the C-terminal half of OsDEP1 on yeast Cd tolerance
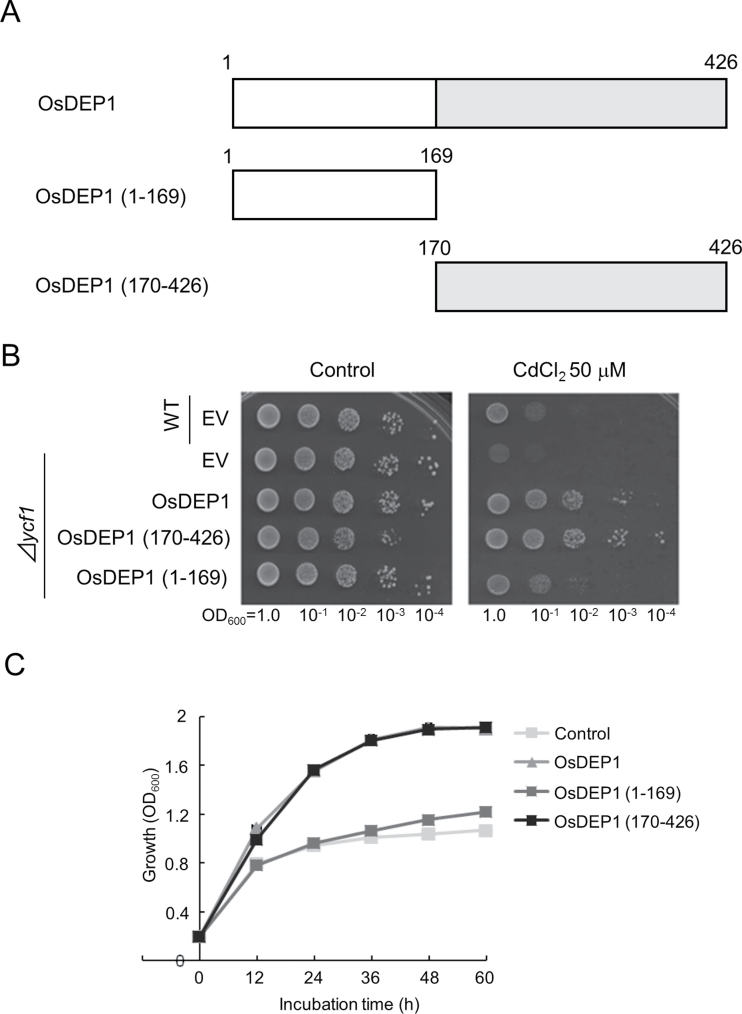
OsDEP1 is composed of two domains; an N-terminal half (aa 1–169) and a C-terminal half (aa 170–426). OsDEP1 is a highly Cys-rich protein, with 120 of the 426 aa (28.2%) being Cys residues, 88 of which are localized to the C-terminal region (Fig. 2A, Supplementary Fig. S2 at JXB online). Two truncated derivatives of OsDEP1, with fragments covering the OsDEP1(1–169) and OsDEP1(170–426) regions, were cloned into the pGK1 vector resulting in constructs pOsDEP1(1–169) and pOsDEP1(170–426), respectively. These plasmids were introduced into S. cerevisiae BY4742 (Δycf1) and assayed for Cd tolerance. The results demonstrated that OsDEP1(170–426) conferred the same level of Cd tolerance to yeast cells as the intact OsDEP1 clone, whereas OsDEP1(1–169) had only a slight effect on Cd tolerance (Fig. 2B). This result was further supported by turbidity growth assays, performed in SD-Ura liquid medium containing 40 μM CdCl2, in which host growth was monitored after the initial turbidity of the cultures was adjusted to OD600=0.2. After 24h of growth, hosts carrying intact OsDEP1 or OsDEP1(170–426) reached a cell density of OD600=1.6, whereas hosts carrying EV or OsDEP1(1–169) only attained an OD600=1.0 (Fig. 2C). These results clearly demonstrated that the OsDEP1(170–426) region is necessary and sufficient to confer Cd tolerance on host yeast cells.
Fig. 2.
OsDEP1 is composed of two domains; the C-terminal half is Cys rich and is sufficient to confer Cd tolerance to yeast cells. (A) Schematic representation of OsDEP1 and its two truncated derivatives. OsDEP1 consists of 426 aa, of which 120 are Cys residues, with 88 of these being localized to the OsDEP1(170–426) C-terminal half. (B) Cd tolerance of yeast cells expressing intact OsDEP1 or the truncated OsDEP1(1–169) and OsDEP1(170–426) derivatives. Experiments were performed as described in Fig. 1. (C) Growth curves of yeast strains in SD-Ura liquid medium supplemented with 40 μM CdCl2. The growth of the yeast cells was monitored at OD600. The data are means ±standard deviation (SD) from three independent experiments.
Metal specificity of OsDEP1 in yeast
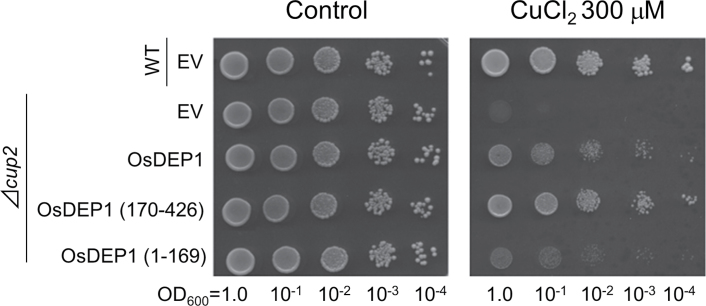
The specificity of OsDEP1-induced tolerance to specific metals in yeast was examined by an antibiotic assay method (see Supplementary Methods). The pOsDEP1 and control (EV) plasmids were introduced into the appropriate metal-sensitive yeast hosts, which were then added to molten top agarose media and immediately layered onto basal agar media. Subsequently, Cd, Cu, Co, Ni, Zn or Mn (all added as chloride salts) were infused into antibiotic assay discs and placed onto the solidified top agarose media. The diameters of the growth inhibition zones were measured after specific periods of incubation. In addition to Cd (Supplementary Fig. S3A at JXB online), yeast cells carrying pOsDEP1 also showed tolerance to Cu (Supplementary Fig. S3B) but not to the other four metals (Supplementary Fig. S3C–F). As the statistics did not support a significant difference in Cu response between cells (Δcup2) carrying EV or pOsDEP1, the Cu tolerance conferred by OsDEP1 expression in yeast was further confirmed by dilution spot tests. In the presence of 300 μM CuCl2, the Δcup2 yeast cells carrying pOsDEP1 grew far better than those carrying EV (Fig. 3). Furthermore, as with Cd tolerance, only the C-terminal OsDEP1(170–426) region was responsible for the observed Cu tolerance phenotype, whereas the N-terminal OsDEP1(1–169) region appeared to have only a marginal effect on Cu tolerance (Fig. 3).
Fig. 3.
Copper tolerance of yeast cells carrying pOsDEP1 and its two deletion derivatives. Experiments were performed as described in Fig. 1 except that the 50 μM CdCl2 was replaced with 300 μM CuCl2 (right panel).
Transgenic plants expressing OsDEP1 and OsDEP1(170–426) are Cd tolerant, but those overexpressing OsDEP1(1–169) are not
To determine the role of OsDEP1 in response to Cd in planta, we generated three series of Arabidopsis transgenic plants: those overexpressing full-length OsDEP1 and those overexpressing its two deletion derivatives, OsDEP1(1–169) and OsDEP1(170–426). Several independent homozygous lines expressing each of these constructs were obtained, and two lines each of the OsDEP1 and OsDEP1(170–426) transgenics were selected for further study. High expression levels of OsDEP1 in these transgenic lines were validated by RT-PCR analysis (Fig. 4A). In terms of Cd tolerance, lines #23 and #28 were tolerant to CdCl2 compared with control transgenic plants when tested on Cd assay plates (Fig. 4B, C). Similarly, lines Crich6 and Crich9 expressing OsDEP1(170–426) showed increased Cd tolerance (Fig. 4D, E). In contrast, none of the eight homozygous lines overexpressing OsDEP1(1–169), including ΔCrich1 and ΔCrich2, showed any such tolerance (Fig. 4F–H).
Fig. 4.
Generation of transgenic Arabidopsis lines expressing intact OsDEP1 and its truncated OsDEP1(1–169) and OsDEP1(170–426) derivatives, and their responses to CdCl2-induced stress. (A) RT-PCR analysis of OsDEP1 and OsDEP1(170–426) in each of two independent transgenic Arabidopsis lines. EV, control transgenic line; lines #23 and #28, transgenic lines carrying the intact OsDEP1 gene; lines Crich6 and Crich9, transgenic lines carrying the OsDEP1(170–426) Cys-rich domain. The Arabidopsis tubulin gene (GenBank accession no. NM_001203444, AtTUB) was used as a loading control. (B–E) Transgenic seeds were sown onto MGRL/1% gellan gum medium with or without 75 μM CdCl2 for 2 weeks, at which time pictures and root length measurements were taken. (B, C) Growth responses (B) and root lengths (C) of control transgenic (EV) and two independent OsDEP1 transgenic Arabidopsis lines, #23 and #28, in response to control 0 μM and 75 μM CdCl2. (D, E) Growth responses (D) and root lengths (E) of control transgenic (EV) and two independent transgenic Arabidopsis lines, Crich6 and Crich9, expressing the Cys-rich OsDEP1(170–426) region, to control (0 μM) and 75 μM CdCl2. Asterisks in (C) and (E) indicate that the difference from the control (EV) is statistically significant: *** P <0.001. (F) RT-PCR analysis of OsDEP1(1–169) in each of two independent transgenic Arabidopsis lines. EV, control transgenic line; lines ΔCrich1 and ΔCrich2, transgenic lines carrying the OsDEP1(1–169) domain. (G, H) Growth responses (G) and root lengths (H) of control transgenic (EV) and two independent transgenic Arabidopsis lines, ΔCrich1 and ΔCrich2, expressing the OsDEP1(1–169) region, to control (0 μM) and 75 μM CdCl2. (This figure is available in colour at JXB online.)
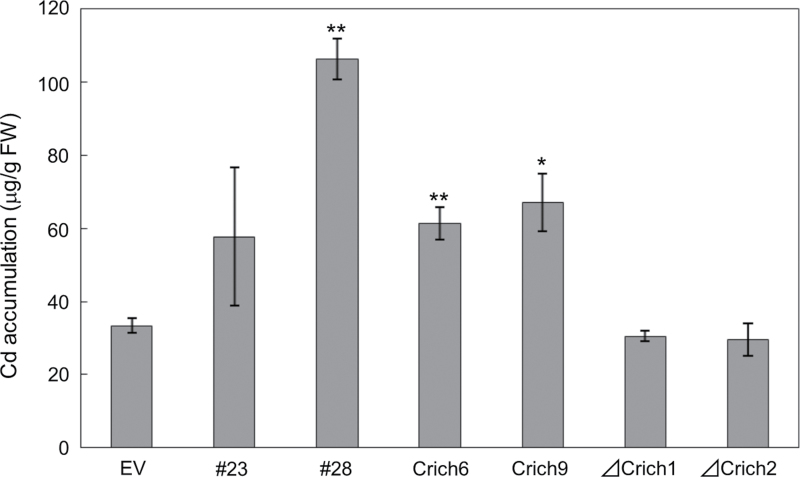
Transgenic plants expressing OsDEP1 and OsDEP1(170–426) accumulate more Cd
Seedlings of the above four Arabidopsis lines, #23, #28, Crich6 and Crich9, together with the pBI121 vector-transformed control transgenic line (EV), were treated with 5 μM CdCl2 for 5 d and their total tissue Cd levels then determined. All four lines, expressing either OsDEP1 or OsDEP1(170–426), accumulated more Cd compared with the control EV and OsDEP1(1–169) transgenics (Fig. 5). In particular, line #28, with the highest level of transgene expression, accumulated about threefold more Cd than control transgenics (Fig. 5).
Fig. 5.
Cd content in control and transgenic Arabidopsis seedlings carrying intact OsDEP1 or the truncated C-terminal OsDEP1(170–426) region. The control transgenics (pBI121, EV) and transgenics carrying either the intact OsDEP1 (lines #23 and #28) or the truncated OsDEP1(170–426) region (lines Crich6 and Crich9) were sown on MGRL/1.0% gellan gum medium and then transferred to 5 μM CdCl2 solution for 5 d before their total Cd contents were determined. The data are means ±SD from three independent experiments. Asterisks indicate that the difference from the control (EV) was statistically significant: *P <0.05; **P <0.01.
Cd responses of loss-of-function heterotrimeric G protein gene mutants
As described above, Arabidopsis has one α (GPA1), one β (AGB1), and three γ (AGG1, AGG2, and AGG3) subunits in its heterotrimeric G protein system (Thung et al., 2012; Botella, 2012). To address whether a heterotrimeric G protein signalling pathway is involved in the Cd response, we examined the Cd sensitivity of various loss-of-function heterotrimeric G protein gene mutants of Arabidopsis (Fig. 6A, Supplementary Fig. S4 at JXB online). Validation of the mutants was tested by RT-PCR analysis (Fig. 6B, C). Two allelic mutants of AGB1, agb1-1 and agb1-2, showed no differences in their Cd sensitivity compared with WT (Figs. 6D, G). Similarly, whereas the agg1-1C and agg3-1 single mutant plants showed hypersensitivity to Cd (Fig. 6E, F), the agg1-1C, agg2-1, agg3-1 triple mutant did not show any differences in its Cd sensitivity from WT plants (Fig. 6G). Based on the results of the β subunit mutant plant and triple mutant plant, we tentatively concluded that the βγ complex-mediated signalling pathway does not induce a cascade that leads to Cd tolerance. Interestingly, and in contrast, the α subunit mutant, gpa1-4, showed Cd hypersensitivity compared with WT lines (Fig. 6D).
Fig. 6.
Cd response of the Arabidopsis mutants of G protein subunit genes. (A) Schematic of the genomic organization of Arabidopsis G protein subunit genes. (B, C) Confirmation of the heteromeric G protein subunit gene mutants. Expression analysis of AtGPA1, AtAGB1, AtAGG1, AtAGG2, and AtAGG3 in WT (Col-0), gpa1-4, agb1-1, agb1-2, and agg1-1C (B), and agg3-1 and agg1-1C agg2-1 agg3-1 (C) (Arabidopsis seedlings was performed by RT-PCR using the primers listed in Supplementary Table S1 at JXB online. AtActin2 was used as a loading control. (D–G) Root lengths of control WT (parental line) and mutant lines in response to control (0 μM) and 75 μM CdCl2: WT (Col-0) (D), Gα (gpa1-4) and Gβ (agb1-2) mutants (D); WT (Col-0) and agg1-1C mutant (E); WT (Col-0) and agg3-1 mutant (F); WT (Col-0) and agb1-1 and the triple mutant agg1-1C agg2-1 agg3-1 (G). The data are means ±SD from three independent experiments. Asterisks indicate that the difference from the control (WT) was statistically significant: *** P <0.001. (This figure is available in colour at JXB online.)
Discussion
OsDEP1 is a useful genetic resource for phytoremediation of Cd and Cu pollution
By extensive screening of a rice cDNA library, aimed at identifying clones that could confer Cd tolerance to yeast cells, we obtained OsDEP1. The gene product, OsDEP1, consisted of 426 aa with a high Cys residue content (28.2%) clustered in the C-terminal half of the protein. In our studies, the Cys-rich C-terminal half (aa 170–426) of OsDEP1 was sufficient to provide exactly the same level of Cd tolerance to yeast as nascent OsDEP1, whereas the OsDEP1 N-terminal portion (aa 1–169), despite containing 32 Cys residues, could only confer limited Cd tolerance to host yeast cells (Fig. 2B, C). Similarly, in our transgenic Arabidopsis plants, expression of either the full-length OsDEP1 or its C-terminal Cys-rich region could provide enhanced tolerance to Cd toxicity (Fig. 4). In addition, OsDEP1 and its Cys-rich C-terminal half, but not its N-terminal half, could provide enhanced tolerance of yeast cells to Cu2+ but not to other heavy metals (Fig. 3, Supplementary Fig. S3). The transgenic plants expressing OsDEP1 or OsDEP1(170–426) were able to accumulate more Cd in their tissues than control plants. Indeed, there was a positive correlation between the levels of OsDEP1 or OsDEP1(170–426) expression and the amount of Cd accumulated (Fig. 5). Based on these results, we propose that OsDEP1, and even its C-terminal region alone, would be an extremely useful genetic resource for phytoremediation of Cd- and Cu-contaminated sites. Several rice cultivars are known to be Cd hyper-accumulators or Cd hypo-accumulators (Uraguchi et al., 2009; Ueno et al., 2011), and it will be interesting to examine whether there is any correlation between the levels and quality of OsDEP1 transcripts in these cultivars and their ability to accumulate Cd.
Comparison of OsDEP1 and other Cys-rich proteins involved in Cd tolerance
Several other studies have previously identified Cys-rich proteins that can provide enhanced tolerance to Cd toxicity. DcCDT1 from D. ciliaris is a 55 aa peptide of which 15 residues (27%) are Cys. The protein is localized to the cytoplasmic membrane and appears to function in the chelation and possible extrusion of Cd, as transgenic DcCDT1 plants accumulate considerably less Cd than controls (Kuramata et al., 2009). The 25kDa CRP protein of earthworm, Enchytraeus buchholzi, contains 27% Cys residues that are present predominantly in a Cys–X–Cys and Cys–Cys arrangement. The gene encoding CRP is Cd inducible, suggestive of a defensive role in Cd-induced damage (Willuhn et al., 1994). Similarly, OsDEP1 contains 28% Cys residues and its Cys arrangement resembles that of CRP. As with DcCDT1, the OsDEP1 protein localizes to cytoplasmic membranes and/or nuclei (Huang et al., 2009; Zhou et al., 2009; Taguchi-Shiobara et al., 2011), but, in contrast to DcCDT1, transgenic Arabidopsis plants expressing OsDEP1 or OsDEP1(170–426) accumulated more Cd than controls. Considering that OsDEP1 is a Gγ subunit, it is likely that it is localized to the inside of cytoplasmic membranes, whereas DcCDT1 may be oriented to the outside of the cytoplasmic membrane. Such a possibility would explain the observed differences in Cd uptake between the DcCDT1- and OsDEP1-expressing transgenic plants. Further work is required to substantiate this hypothesis.
Does OsDEP1 activate the Cd tolerance system via heterotrimeric G protein signalling?
OsDEP1 was formerly identified through quantitative trait loci analysis as a gene that controls panicle erectness, grain number per panicle and consequently grain yield (Huang et al., 2009; Zhou et al., 2009; Taguchi-Shiobara et al., 2011). Although the functional OsDEP1 allele was found to result in drooping panicles, deletion mutations in the Cys-rich region caused semi-dwarfism, increased spikelet numbers, and erect panicles (Huang et al., 2009; Zhou et al., 2009; Taguchi-Shiobara et al., 2011). As reported recently, the Arabidopsis DEP1 homologue, AGG3, is a γ subunit of heterotrimeric G proteins, and regulates guard cell K+-channel activity and influences organ size and shape (Chakravorty et al., 2011; Li et al., 2012a,b). Furthermore, based on the understanding that OsDEP1 is an AGG3 homologue, and thus a G protein γ subunit, Botella (2012) discussed how the OsDEP1 mutation could lead to increased grain yields. The N-terminal 100 aa region of OsDEP1 is a γ-domain that interacts with the β subunit of heterotrimeric G protein complexes to transduce extracellular signals via cell-surface receptors to downstream effectors (Botella, 2012). The Gβγ subunits are associated with GDP-bound Gα subunits in an inactive state. Once the extracellular portion of the G protein-coupled receptor binds its ligand, G protein signalling is activated: the activated G protein-coupled receptor triggers dissociation of the trimeric complex to Gβγ and Gα subunits, with the latter becoming activated by GTP binding. The resulting Gα and Gβγ subunits then further transmit these signals to their own effectors (Temple and Jones, 2007). As OsDEP1(170–426), which lacks the γ-domain essential for Gβ subunit association, is still able to effectively enhance Cd tolerance, it may be that OsDEP1 does not exert its Cd tolerance effect via G protein signalling.
Given that OsDEP1 is a Gγ protein, there are two possibilities for how OsDEP1 confers Cd tolerance to host plants: one is that OsDEP1 and OsDEP1(170–426) trap and detoxify these Cd ions directly, while the other possibility is that OsDEP1 activates the heterotrimeric G protein signalling pathway. Although the Arabidopsis AGG1 (=Gγ1) and AGG3 (=Gγ3) mutant plants were hypersensitive to Cd (Fig. 6E, F), the two allelic Gβ mutant plants and the Gγ triple mutant plant did not show any differences in root growth in the presence of Cd (Fig. 6D, G), indicating that the Gβγ pathway does not affect Cd sensitivity. On the other hand, the GPA1 (=Gα) mutant plant showed Cd hypersensitivity compared with the WT plant (Fig. 6D). As GPA1 has been implicated in guard cell K+-channel regulation (Wang et al., 2001; Chakravorty et al., 2011), it may be possible that the Gα protein signalling pathway regulates certain ion channel(s) that recognize heavy metals such as Cd and Cu ions. Of course, further study is needed to determine whether the Gα pathway is really involved in the observed Cd response. If this is the case, then functional coupling of the heterotrimeric G protein pathway to Cd responses opens a new door for exploring signal cascades in plants upon exposure to Cd or other heavy metals.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Phylogenetic relationships between OsDEP1 and its orthologs and paralogs inother monocot and dicot plants.
Supplementary Fig. S2. Schematic representation of OsDEP1 and its amino acid sequence.
Supplementary Fig. S3. Response of yeast cells carrying pOsDEP1 to various heavy metals.
Supplementary Fig. S4. Growth responses of the loss-of-function mutants of heterotrimeric G protein(s).
Table S1. Primers used to analyse the expression of heterotrimeric G protein genes in Arabidopsis.
Supplementary Methods. (i) Yeast strain used. (ii) Metal specificity of OsDEP1 in yeast.
Acknowledgements
Dr José Ramón Botella (University of Queensland, Australia) is acknowledged for kindly providing the seeds of the Arabidopsis G protein subunit mutants. This work was supported by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) (#7609 to S.K. and #6816 to T.M.) and a grant from the Saito Gratitute Foundation to T.M. and S.K. S.K. and T.M. are recipients of the Research Fellowship for Young Scientists from JSPS.
Glossary
Abbreviations:
- ABC
ATP-binding cassette
- Cd
cadmium
- Cys
cysteine
- EV
empty vector
- Km
kanamycin
- MT
metallothionein
- ORF
open reading frame
- RT-PCR
reverse transcription-PCR
- SD
standard deviation
- WT
wild type.
References
- Botella JR. 2012. Can heterotrimeric G proteins help to feed the world? Trends in Plant Science 17, 563–568 [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR. 2011. An atypical heterotrimeric G-protein γ subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana . The Plant Journal 67, 840–851 [DOI] [PubMed] [Google Scholar]
- Choi YE, Harada E, Wada M, Tsuboi H, Morita Y, Kusano T, Sano H. 2001. Detoxification of Cd in tobacco plants: formation and active excretion of crystals containing Cd and calcium through trichomes. Planta 213, 45–50 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Farinati S, Maistri S, Furini A. 2008. How plants cope with cadmium: staking all on metabolism and gene expression. Journal of Integrative Plant Biology 50, 1268–1280 [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Butt TR, Sternberg EJ, Neeper MP, Debouck C, Gorman JA, Crooke ST. 1986. Yeast metallothionein function in metal ion detoxification. Journal of Biological Chemistry 261, 16895–16900 [PubMed] [Google Scholar]
- Freisinger E. 2008. Plant MTs—long neglected members of the metallothionein superfamily. Dalton Transactions 47, 6663–6675 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Yokota-Hirai M, Chino M, Komeda Y, Naito S. 1992. Effects of sulfur nutrient on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiology 99, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X. 2009. Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics 41, 494–497 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. Journal of Bacteriology 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järup L. 2003. Hazards of heavy metal contamination. British Medical Bulletin 68, 167–182 [DOI] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y. 2007. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. The Plant Journal 50, 207–218 [DOI] [PubMed] [Google Scholar]
- Kim YY, Kim DY, Shim D, Song WY, Lee J, Schroeder JI, Kim S, Moran N, Lee Y. 2008. Expression of the novel wheat gene TM20 confers enhanced cadmium tolerance to bakers’ yeast. Journal of Biological Chemistry 6, 15893–15902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular Genetics and Genomics 204, 383–396 [Google Scholar]
- Korenkov V, Hirschi K, Crutchfield JD, Wagner GJ. 2007. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 226, 1379–1387 [DOI] [PubMed] [Google Scholar]
- Kuramata M, Masuya S, Takahashi Y, Kitagawa E, Inoue C, Ishikawa S, Youssefian S, Kusano T. 2009. Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant and Cell Physiology 50, 106–117 [DOI] [PubMed] [Google Scholar]
- Li S, Liu W, Zhang X, Liu Y, Li N, Li Y. 2012a. Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control. Plant Signaling and Behavior 7, 1357–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, et al. 2012b. The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana . New Phytologist 194, 690–703 [DOI] [PubMed] [Google Scholar]
- Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA. 1996. The yeast Cd factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. Journal of Biological Chemistry 271, 6509–6517 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Kuramata M, Takahashi Y, Kitagawa E, Youssefian S, Kusano T. 2009. A novel plant cysteine-rich peptide family conferring cadmium tolerance to yeast and plants. Plant Signaling and Behavior 4, 419–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cózatl DG, Zhai Z, Jobe TO, et al. 2010. Tonoplast-localized Abc2 transporter mediates phytochelatin accumulation in vacuoles and confers cadmium tolerance. Journal of Biological Chemistry 285, 40416–40426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadate H, Adachi S, Hiraizumi A, et al. 2011. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytologist 189, 190–199 [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. 2009. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis . Plant Physiology 149, 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum 15, 473–497 [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. 1992. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO Journal 11, 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Palma JM, Gomez LA, Del Rio LA, Sandalio LM. 2002. Cadmium causes oxidative modification of proteins in plants. Plant, Cell and Environment 2, 677–686 [Google Scholar]
- Romero-Puertas MC, Rodriguez-Serrano M, Corpas FJ, Gomez M, Del Rio LA, Sandalio LM. 2004. Cadmium-induced subcellular accumulation of O2− and H2O2 in pea leaves. Plant, Cell & Environment 27, 1122–1134 [Google Scholar]
- Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y. 2010. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadegan M, Christie P, Seemann JR. 1991. An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Research 19, 6055–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Martinoia E, Lee J, Kim D, Kim DY, Vogt E, Shim D, Choi KS, Hwang I, Lee Y. 2004. A novel family of cys-rich membrane proteins mediates Cd resistance in Arabidopsis . Plant Physiology 135, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Park J, Mendoza-Córzatl DG, et al. 2010. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proceedings of the National Academy of Sciences, USA 107, 21187–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Kawagoe Y, Kato H, et al. 2011. A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breeding Science 61, 17–25 [Google Scholar]
- Temple BR, Jones AM. 2007. The plant heterotrimeric G-protein complex. Annual Review of Plant Biology 58, 249–266 [DOI] [PubMed] [Google Scholar]
- Thung L, Trusov Y, Chakravorty D, Botella JR. 2012. Gγ1+Gγ2+Gγ3=Gβ: the search for heterotrimeric G-protein γ subunits in Arabidopsis is over. Journal of Plant Physiology 169, 542–545 [DOI] [PubMed] [Google Scholar]
- Ueno D, Koyama E, Yamaji N, Ma JF. 2011. Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. Journal of Experimental Botany 62, 2265–2272 [DOI] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF. 2010. Gene limiting cadmium accumulation in rice. Proceedings of the National Academy of Sciences, USA 107, 16500–16505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S. 2009. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. Journal of Experimental Botany 60, 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. Mechanisms to cope with arsenic or cadmium excess in plants. Current Opinion in Plant Biology 12, 364–372 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM. 2001. G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292, 2070–2072 [DOI] [PubMed] [Google Scholar]
- Willuhn J, Schmitt-Wrede HP, Greven H, Wunderlich F. 1994. cDNA cloning of a Cd-inducible mRNA encoding a novel cysteine-rich, non-metallothionein 25-kDa protein in an enchytraeid earthworm. Journal of Biological Chemistry 269, 24688–24691 [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Li Z, Yi C, Liu J, Zhang H, Tang S, Gu M, Liang G. 2009. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.