Abstract
The bacterial endophyte Herbaspirillum frisingense GSF30T is a colonizer of several grasses grown in temperate climates, including the highly nitrogen-efficient perennial energy grass Miscanthus. Inoculation of Miscanthus sinensis seedlings with H. frisingense promoted root and shoot growth but had only a minor impact on nutrient concentrations. The bacterium affected the root architecture and increased fine-root structures. Although H. frisingense has the genetic requirements to fix nitrogen, only minor changes in nitrogen concentrations were observed. Herbaspirillum agglomerates were identified primarily in the root apoplast but also in the shoots. The short-term (3h) and long-term (3 weeks) transcriptomic responses of the plant to bacterial inoculation revealed that H. frisingense induced rapid changes in plant hormone signalling, most prominent in jasmonate signalling. Ethylene signalling pathways were also affected and persisted after 3 weeks in the root. Growth stimulation of the root by the ethylene precursor 1-aminocyclopropane 1-carboxylic acid was dose dependent and was affected by H. frisingense inoculation. Minor changes in the proteome were identified after 3 weeks. This study suggests that H. frisingense improves plant growth by modulating plant hormone signalling pathways and provides a framework to understand the beneficial effects of diazotrophic plant-growth-promoting bacteria, such as H. frisingense, on the biomass grass Miscanthus.
Key words: Biomass, diazotroph, endophyte, ethylene, Miscanthus, plant-growth-promoting bacteria.
Introduction
Many gramineous species maintain a close association with endophytic bacteria that are beneficial for plant growth and health (Reinhold-Hurek and Hurek, 1998). Diazotrophic, non-legume plant-growth-promoting bacteria are able to support plant growth at low nitrogen conditions by a combination of nitrogen fixation, increasing the availability of soil nutrients, promoting root growth by hormonal signalling, and controlling disease symptoms (Spaepen et al., 2009). In Brazilian sugarcane, diazotrophic bacteria substantially contribute between 25% and 60% to the nitrogen acquisition of plants, but the exact value is often vague (Boddey et al., 2001; Sevilla et al., 2001; Taule et al., 2012; Urquiaga et al., 2012). Evidence for substantial non-legume nitrogen fixation in fields of temperate climates is poor and variable (Bhattacharjee et al., 2008), and a better molecular understanding of the complex plant–bacterial interaction may help to increase nitrogen fixation rates.
The C4-fibre plant Miscanthus is a highly nutrient-efficient biomass plant and is one of the favourites for sustainable biomass production (Karp and Shield, 2008). It has been associated with the endophytic bacterium Herbaspirillum frisingense in the temperate climate of southern Germany. Herbaspirillum frisingense was isolated using nitrogen-free semi-solid medium (Kirchhof et al., 2001). Genome sequencing of H. frisingense showed that the bacterium has all the genomic requirements to fix nitrogen and lacks several factors that may contribute pathogenic characteristics that are found in other Herbaspirillum strains (Straub et al., 2013a ). Many Herbaspirillum isolates lack nitrogen-fixation genes, including isolates closely related to H. frisingense (Straub et al., 2013a ). The ability to fix nitrogen of some diazotrophic Herbaspirillum strains has been proven in association with wild rice but not with cultivated rice (Elbeltagy et al., 2001). Many Herbaspirillum spp. isolates have been obtained from tropical and subtropical conditions, with the nitrogen-fixing Herbaspirillum seropedicae investigated in most detail (Pedrosa et al., 2011). However, many H. seropedicae isolates are also potential pathogens on various hosts, which reduces their potential versatility as biofertilizers. By contrast, the genome structure (with the lack of many systems potentially involved in pathogenicity), the colonizing characteristics, and the production of plant-growth-promoting factors suggest that H. frisingense might be used as a potential biofertilizer for several C4 grasses (Monteiro et al., 2012b ).
The microaerobic diazotroph H. frisingense invades the intercellular spaces of Miscanthus and barley roots without apparent damage to the host (Rothballer et al., 2008). The potential nitrogen-fixing activity of this bacterial species was demonstrated using a acetylene reduction assay, PCR detection of nif genes (Rothballer et al., 2008), and genome sequencing (Straub et al., 2013a ). Herbaspirillum spp. strains differ in their capacity to synthesize the plant hormone auxin and other metabolites that regulate plant growth, such as N-acylhomoserine lactones (Schuhegger et al., 2006; von Rad et al., 2008). The ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) can be used as a nitrogen source, suggesting that a biosynthetic pathway for its production or catabolism exists (Glick et al., 2007; Rothballer et al., 2008). Genome sequencing of Herbaspirillum strains have shown that ACC degradation may be a common strategy of members of the genus Herbaspirillum to affect plant growth (Straub et al., 2013a ).
The molecular mechanisms of how plant-associated and/or growth-promoting Herbaspirillum bacteria suppress the plant immune system and specifically invade the host are still unclear (Pedrosa et al., 2011; Monteiro et al., 2012a ). The large variety of plant-growth-promoting bacteria and the high diversity in their genomic composition suggests that both common and strain-specific unique strategies for this interaction exist. However, the mechanisms of these potentially beneficial associations are still poorly understood.
Miscanthus belongs to the family Gramineae and its stem height reaches up to 4 m in one growth season. In particular, the variety Miscanthus×giganteus, a naturally occurring sterile hybrid of Miscanthus sinensis and Miscanthus sacchariflorus (Greef and Deuter, 1993) combines low nutrient, especially very low nitrogen, requirements with good agronomic properties and high biomass yields. In Germany, yields of 20–30 t ha–1 year–1 (Lewandowski and Kicherer, 1997) demonstrate the suitability of this grass as a renewable resource for energy production with an even CO2 balance. Many studies have confirmed that the use of a mineral fertilizer has little or no significant effect on Miscanthus×giganteus biomass production (Himken et al., 1997; Christian et al., 2008). According to Christian et al. (1997), only 19% of the total plant nitrogen was derived from the introduced fertilizer when nitrogen distribution and balance was examined after the application of an 15N-labelled chemical fertilizer (Christian et al., 1997). Model calculations on real biomass-yield data suggest a profound nitrogen input by nitrogen-fixing bacteria in the seasonal nitrogen balance (Davis et al., 2010). In addition to Herbaspirillum, alpha-, gamma- and deltaproteobacteria have been found associated with the leaves of Miscanthus (Straub et al., 2013b ).
In this study, the growth of young Miscanthus seedlings was found to benefit from inoculation with the betaproteobacterium H. frisingense. The growth-promoting potential of this bacterium was dependent on the nitrogen supply. H. frisingense affected the signalling of plant hormones, namely ethylene signalling, in root growth. Although nutrient concentrations in treated seedlings were not affected, the improved root growth resulted in improved overall nutrient acquisition and biomass production. These results identify plant-growth-promoting bacteria functions of this diazotrophic endophyte, which may be beneficial for sustainable biomass production with Miscanthus, especially for establishment from seeds and maintenance in marginal soils.
Material and methods
M. sinensis growth
Surface-sterilized M. sinensis seeds (10min in 70% ethanol, rinsed with sterile water, and dried) were germinated in quartz sand (0.3–0.8mm diameter), which was washed with HCl (rinsed with tap water, pH <1 adjusted with HCl, incubated overnight, rinsed with deionized water to pH >5) to wash out trace nutrients, biological contaminations, and dust. Prior to sowing, the sand was fertilized with modified Hoagland solution [1mM KH2PO4, 0.5mM MgSO4, 50 µM Na2EDTA, 50 µM FeSO4, 9 µM MnCl2, 0.765 µM ZnSO4, 0.32 µM CuSO4, 16nM (NH4)6Mo7O24, 46 µM H3BO3, 1mM CaCl2] containing 1mM ammonium nitrate. After 2 weeks, plants were transferred individually to pots (9cm diameter) with HCl-washed quartz sand and incubated under conditions of 14h light, 24 °C/10h dark, 19 °C. The pots were watered three times a week: twice with modified Hoagland solution without any nitrogen source and once a week with modified Hoagland solution containing 50 µM (low nitrogen) or 250 µM (higher nitrogen) ammonium nitrate. In some experiments, the nitrogen was supplied in a 10% 15N-enriched form. For ethylene growth tests, the plants were watered as above once a week with nutrient solution containing 50 µM ammonium nitrate, and twice a week with 0, 0.3, 1, 5, and 20 µM ACC (Sigma-Aldrich).
Three-week-old plants were inoculated with H. frisingense GSF30T (Straub et al., 2013a ) The bacteria were grown for 2 d in liquid Luria–Bertani medium supplemented with 50mg l–1 of kanamycin and harvested by centrifugation for 5min at 4000rpm at 4 °C. The pellet was washed with water and resuspended in watering solution to a final OD600 of ~0.1). Each pot was watered with 40ml of solution containing bacteria at about 109 c.f.u. ml–1. For control plants, the bacterial solution was autoclaved. This inoculation step was repeated after a further 3 d. After 3 weeks, the plants were harvested. The sand was washed away with tap water, and the root was dried briefly with paper and weighed for fresh biomass determination. The significance was analysed using SAS System software Release 8.01, using one-way or two-way analysis of variance (ANOVA) with values of P as indicated.
Root morphology
Roots were excised from the sand culture, scanned, and analysed with a WinRHIZO system (Regent Instruments, Canada) for the number and length of roots of different diameter.
Element analysis
Roots and shoots were harvested separately, washed, dried for 2 d at 60 °C, and ground to a fine powder. The concentration of total nitrogen was determined using a EuroVector elemental analyser (HEKAtech, Wegberg, Germany). For the other elements, 10–18mg of tissue was dissolved in 65% HNO3 and heated in four increasing steps up to a maximal temperature of 220 °C at 160 bar in a microwave (UltraClave3; MLS, Germany). Elements were determined by inductively coupled plasma/optical emission spectrometry (PS1000; Leeman Labs, Lowell, MA, USA).
Sequencing
Plants were harvested at 1 p.m. and immediately frozen in liquid nitrogen and stored at –80 °C. The mRNA was isolated from 100mg of plant material with an RNeasy Plant Mini kit (Qiagen) and transcribed to cDNA, which was sequenced using Illumina technology at GATC Biotech AG (Germany). De novo assembly and mapping was also done at GATC Biotech AG.
Differentially expressed genes
Analysis of differentially expressed genes used a method similar to that described previously (Straub et al., 2013b ). Fold changes and relative read numbers were determined by DESeq version 1.8.3 (Anders and Huber, 2010) using Bioconductor. The procedure followed the instructions of the manufacturer (Analysing RNA-Seq data with the DESeq package; Working without any replicates: http://www.bioconductor.org/help/course-materials/2011/BioC2011/LabStuff/DESeq.pdf), and input were raw count tables. Contigs were filtered for meaningful expression by deleting contigs with the lowest DESeq base mean. The remaining 27.3% contigs represented more than 95% of reads.
Transcript annotation
Contigs were annotated with Mercator (http://mapman.gabipd.org/web/guest/mercator) and classified according to MapMan (http://mapman.gabipd.org/web/guest/mapman) functional plant categories (bins). Therefore, SwissProt/UniProt Plant Proteins (PPAP), TIGR5 rice proteins, clusters of orthologous eukaryotic genes database (KOG), conserved domain database (CDD) and Interpro scans were enabled, allowing multiple bin assignments. ‘Unassigned’ bins were considered with equal weight when assigning bin codes. Metabolic pathways were visualized with MapMan 3.5.1R2 and its built-in module PageMan, showing DESeq’s fold change values.
Real-time PCR
Plants were harvested at 1 p.m., immediately frozen in liquid nitrogen and stored at –80 °C. RNA was extracted from 100mg of fine-milled plant material with an innuPREP Plant RNA kit (Analytik Jena). Contaminating genomic DNA was removed with an RNase-Free DNase Set (Qiagen), according to the manufacturer’s instructions. High-quality RNA was chosen based on photometric measurements (NanoDrop 2000c; Thermo Scientific) and formaldehyde–agarose gel electrophoresis according to the RNeasy Mini Handbook, Appendix C (Qiagen) and was immediately transcribed to cDNA or frozen at –80 °C. SsoFast EvaGreen Supermix reaction cocktail (Bio-Rad) was used with a C1000TM thermal cycler chassis and a CFX384 Real-Time PCR Detection System (Bio-Rad) and evaluated with CFX Manager 2.1 software (Bio-Rad). Primers were selected with Primer3Plus (Untergasser et al., 2012). Primer sequences and expected product lengths of the selected reference genes (PP2AA2, Actin-1, and DnaJ; see Supplementary Materials and Methods at JXB online) and other genes can be found in Supplementary Table S1 in Supplementary at JXB online. Melting-curve analysis and agarose gel electrophoresis confirmed the specificity and quality of the PCR products.
Protein isolation and 2D separation of Miscanthus proteins
Total proteins were isolated from approximately 2.5g of frozen whole plants per biological replicate via acetone precipitation (Liu et al., 2006). Isoelectric focusing of proteins was performed with 1000 µg of protein extract using an IPGphor3 isoelectric focusing unit (GE Healthcare) and 24cm immobilized, non-linear pH 3–11 gradients (Imobiline Dry Strips; Amersham Biosciences). Rehydration was performed at 20V overnight. The voltage settings for the isoelectric focusing were 100V for 4h, 300V for 2.5h, 300–1000V gradient for 8h, 1000–8000V gradient for 8h, 8000V for 7h, and 8000–50V gradient to a final setting of 87 900 Vhs. Equilibration of the dry strips was performed as described previously (Sauer et al., 2006). Proteins in the equilibrated dry strips were then separated on the basis of their molecular weight by 12% SDS-PAGE using the Ettan DALTsix Electrophoresis System (GE Healthcare).
After electrophoresis, the proteins were stained with a modified colloidal Coomassie blue stain (Sauer et al., 2006) for 72h on an orbital shaker. Stained gels were imaged using a Typhoon Trio+ Imaging System (GE Healthcare). The resulting gel image files were exported to Progenesis SameSpots (Non-linear Dynamics, http://www.nonlinear.com). Proteins were accepted as differentially accumulated when they displayed a fold change of >1.8 and were significant in Student’s t-test at a significance level of P <0.05.
Microscopy
A green fluorescent protein (GFP)-tagged H. frisingense GSF30T strain was observed by confocal microscopy (Leica DMRE microscope equipped with a confocal head TCS SP; Leica, Wetzlar, Germany).
Results
Growth promotion and colonization of M. sinensis with H. frisingense
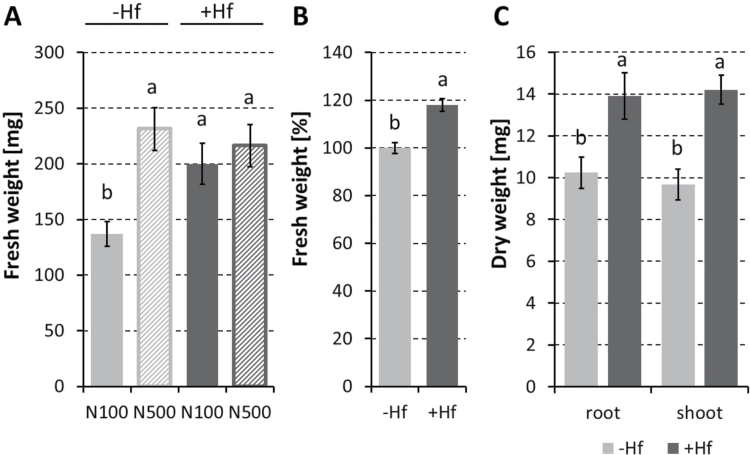
Seedlings of M. sinensis were grown in sand and watered with nutrient solutions that contained different concentrations of nitrogen. In low nitrogen conditions (100 µM total nitrogen watered once a week, N100), M. sinensis accumulated less biomass than in 500 µM nitrogen (N500), suggesting that the nitrogen supply limited the plant growth at low nitrogen (Fig. 1A). After 3 weeks of inoculation with H. frisingense GSF30T, the total fresh biomass of inoculated plants showed a significant increase to the same level as control plants fertilized with the higher N500 dosage. By contrast, the growth-promoting effect of H. frisingense GSF30T was absent when the plants received the higher nitrogen supply (Fig. 1A).
Fig. 1.
Growth promotion of M. sinensis by H. frisingense GSF30T (Hf). The graphs show total biomass from 6-week-old plants. (A) Fresh weight [mg, ±standard error (SE)] of seedlings grown with (+Hf, dark grey bars) or without H. frisingense (–Hf, pale grey bars) under low (N100, 50 µM NH4NO3; 12 plants) or higher (N500, 250 µM NH4NO3; 13 plants) nitrogen levels. Different lower-case letters indicate a significant difference by ANOVA test (P < 0.05). (B) Fresh weight (% compared with –Hf, ±SE) following H. frisingense inoculation (n=11 independent experiments; total 223 plants +Hf and 259 plants –Hf) under N100 conditions. Different lower-case letters indicate a significant difference by ANOVA test (P < 0.001). (C) Dry weight (mg, ±SE) of 6-week-old seedlings, separated for roots and shoots, without or with H. frisingense inoculation under N100 conditions.
Although some variability in the plant-growth-promoting effect in individual experiments was apparent over a period of 4 years, growth promotion overall in more than 12 independent experiments revealed a significant increase of about 20% higher total fresh biomass (Fig. 1B, P < 0.001). In a typical successful growth-promoting experiment, the dry biomass of both roots and shoots increased by almost 40% (Fig. 1C, P < 0.01).
The successful colonization of roots and shoots of Miscanthus seedlings by H. frisingense GSF30T, or absence of the bacteria in seedlings grown from surface-sterilized seeds, was monitored using plant- and bacteria-specific primer pairs. The bacterial DNA was never detected from plants grown in the absence of the bacteria, but was always detected in those that showed beneficial growth effects following colonization by H. frisingense GSF30T (Supplementary Fig. S1 at JXB online).
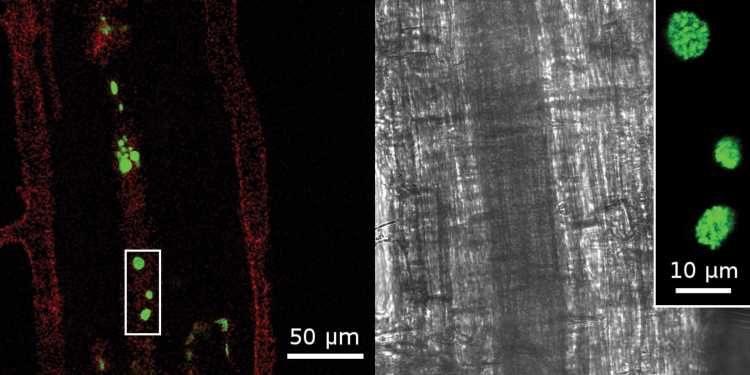
The localization of the growth-promoting bacteria in young seedlings was investigated using a GFP-labelled H. frisingense strain under confocal microscopy. The fluorescence of bacteria was detected in the plant apoplast, where aggregates of fluorescent spots were detected, both in roots (Fig. 2) and shoots (Supplementary Fig. S2 at JXB online). This spot-like aggregation differred from the colonization pattern of barley (Rothballer et al., 2008), where a stronger colonization and a more uniform apoplastic pattern has been observed.
Fig. 2.
Green fluorescent spots in M. sinensis roots inoculated with GFP-labelled H. frisingense. Bacterial aggregates were visible in the intercellular apoplastic space. Bright field image (right) and fluorescence image (left). The red colour indicates residual background fluorescence from the cell wall. The inset shows a magnification of the bacterial colony aggregates. (This figure is available in colour at JXB online.)
Effect on nutrient uptake and root morphology
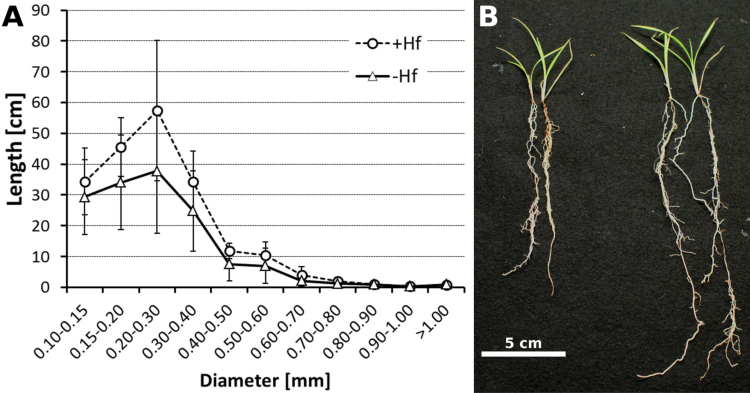
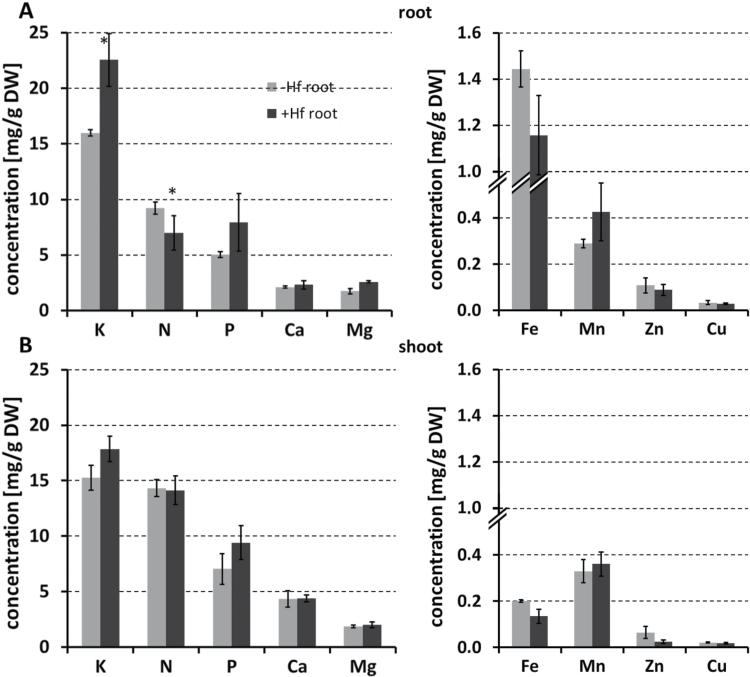
The beneficial effect of H. frisingense inoculation on root morphology was quantified from washed roots. These were scanned, and the total root length and number of laterals and fine roots were determined and analysed. Compared with controls that received autoclaved bacteria, inoculated seedlings showed a clear tendency towards an increase in fine-root structures, laterals, and in the total root length (Fig. 3), yielding a larger root system (Fig. 3B). Despite the larger surface area to absorb nutrients, the concentrations of macro- and micronutrients only showed marginal differences in potassium and nitrogen in roots (Fig. 4). While potassium was higher in roots in the presence of H. frisingense, a reduced nitrogen concentration was detected in roots upon inoculation. In the shoot, the nutrient concentrations were not affected by the diazotrophic H. frisingense. Due to the higher biomass in roots and shoots, however, the total content of all nutrients was increased, but this increase was only significant for K, P, Mg, and Mn in the shoot (Supplementary Fig. S3 at JXB online). When 15N-enriched nitrogen (10%) was applied for 3 weeks, the relative 15N/14N ratios in the tissue did not differ in non-inoculated and inoculated plants. Although a minor contribution of nitrogen via nitrogen fixation cannot rigorously be excluded by these experiments, a large contribution by nitrogen fixation appears unlikely, as a dilution effect of the 15N/14N ratio by potential atmospheric nitrogen fixation was not observed (Supplementary Fig. S4 at JXB online).
Fig. 3.
Root morphology and fine root structural changes of M. sinensis following inoculation with H. frisingense. (A) Root length of different diameter classes without inoculation (triangles) and with inoculation (circles). Quantitative root parameters were deduced using a WinRHIZO system. (B) Seedling appearance without inoculation (left) and with inoculation (right). Note the increased root system. (This figure is available in colour at JXB online.)
Fig. 4.
Differential element concentrations in roots (A) and shoots (B) without (grey bars) and with (black bars) H. frisingense inoculation. Macroelements (left graphs) and micronutrients (right graphs) are given in concentrations [mg/g dry weight (DW)]. Asterisks indicate significant differences at the P < 0.05 level (one-way ANOVA).
Transcriptome changes following inoculation
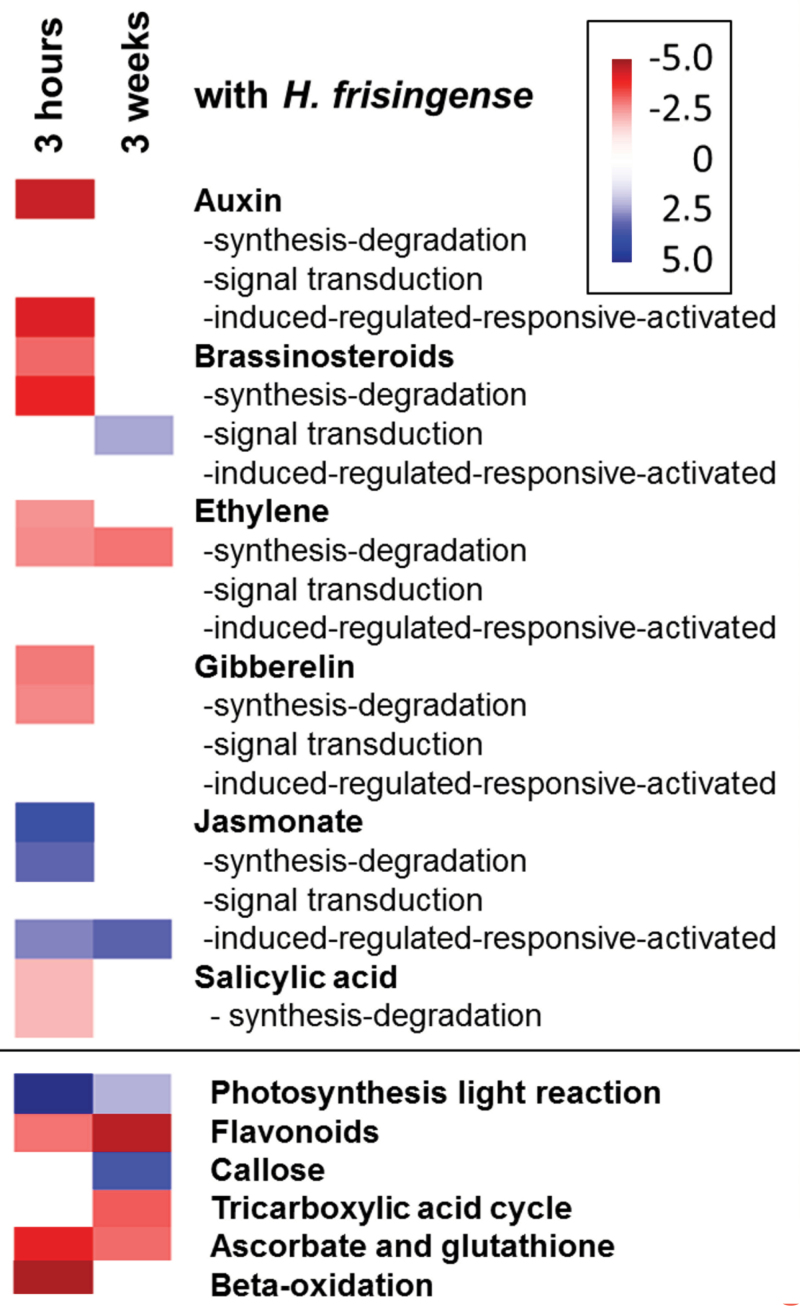
The early (3h) and long-term (3 weeks) changes in gene expression by H. frisingense inoculation were investigated with quantitative mRNA sequencing (RNA-Seq). The early response of M. sinensis to the inoculation with H. frisingense involved a consistent, prominent upregulation of genes involved in jasmonate signalling and biosynthesis, and in response genes (Fig. 5). Genes involved in the biosynthesis and signalling of other plant hormones were also affected in a concerted manner, but the individual gene expression changes were mostly only marginal. Despite this, absciscic acid, auxin, brassinosteroids, and gibberellin signalling appeared significantly lower at 3h after inoculation with H. frisingense (Fig. 5). However, the altered signalling apparently only persisted after 3 weeks for jasmonate and ethylene, whereas the expression changes appeared more variable for brassinosteroid signalling.
Fig. 5.
Differential expression of M. sinensis transcript categories related to hormones and key metabolic functions in response to H. frisingense inoculation. Red indicates lower expression of gene categories compared with non-inoculated plants, while blue indicates higher expression. Non-significant differences (z-score <1.96) are shown in white. Colouring is according to the z-scores of the bin-wise Wilcoxon test. A z-score of ±1.96 represents a P value of 0.05. The plot was generated using PageMan.
An overview of changes in various metabolic pathways 3h after inoculation showed a highly significant (P<0.00001) upregulation of photosynthesis-related genes and a downregulation of β-oxidation genes (Fig. 5 and Supplementary Fig. S5 at JXB online). Genes involved in terpene/flavonoid synthesis and redox metabolism involving ascorbate and glutathione were also collectively repressed, and these gene expression differences persisted after 3 weeks. By contrast, callose metabolism only appeared to be upregulated after 3 weeks, whereas genes involved in the tricarboxylic acid cycle were repressed after 3 weeks post-inoculation (P<0.003; Fig. 5 and Supplementary Fig. S6 at JXB online). Less prominent differences in gene expression categories indicated that several different metabolic traits were affected by inoculation, but, except for the jasmonate response, there were surprisingly few defence genes activated, which may explain why H.frisingense can effectively invade and colonize the plant (Supplementary Fig. S7 at JXB online).
Differential gene expression related to plant hormone signalling
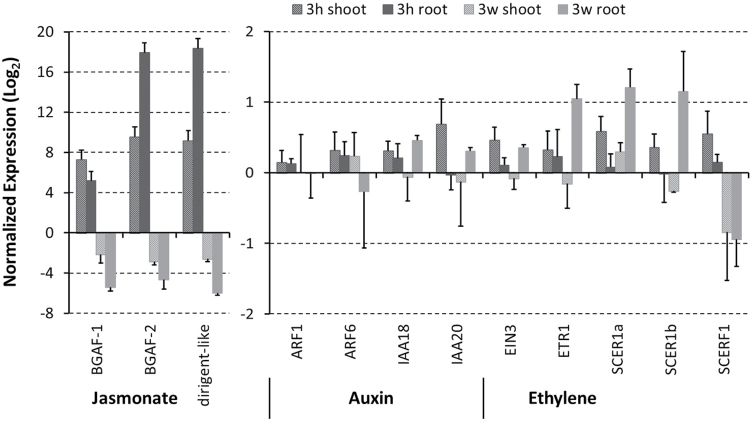
The potential involvement of jasmonate, auxin, and ethylene signalling in the rapid and long-term plant response to the bacterial inoculation was evaluated using quantitative real-time PCR (qRT-PCR) of selected marker genes. Marker genes responding to jasmonate in various plant species include two β-glucosidase aggregating factor genes (BGAF-1 and BGAF-2) and a dirigent-like protein gene. The expression of these genes was highly induced in the transcriptome dataset. When analysed by qRT-PCR, the expression of these genes was increased by several orders of magnitude after 3h of inoculation, in both roots and shoots. However, these genes were repressed after 3 weeks, especially in the roots (Fig. 6).
Fig. 6.
Normalized expression of M. sinensis transcripts related to hormones in response to H. frisingense (qRT-PCR, log2 scale). Note the different scale of jasmonate-regulated genes (BGAF-1, BGAF-2, and dirigent-like; left graph). Auxin-responsive genes were: auxin response factors ARF1 and ARF6, and the auxin-responsive IAA18 and IAA20 genes. Ethylene-related genes were ethylene insensitive 3 (EIN3), ethylene-resistant 1 (ETR1), the sugarcane ethylene receptor-like SCER1-like SCER1a and SCER1b, and the ethylene response factor SCERF1-like. A SCERF2-like gene (Cavalcante et al., 2007) was not identified in Miscanthus. 3h, 3h after inoculation; 3 w, 3 weeks after inoculation. Error bars: +SE of three (3h) or two (3 w) biological replicates. Qualitatively similar results were obtained from two further replicates.
By contrast, only very minor changes in the expression of several auxin-responsive genes, such as the moderately expressed auxin response factors ARF1, ARF6, IAA18, and IAA20 was observed at early and late stages, consistent with the minor expression changes in the transcriptome data set.
Finally, the gene expression of several ethylene-related genes was monitored. This included the highly abundant ethylene receptor ethylene insensitive 3 (EIN3) and several low-abundant ethylene-responsive genes. These genes included the ethylene-resistant 1 gene (ETR1), two sugarcane ethylene receptor 1-like genes (SCER1a and SCER1b), and the sugarcane ethylene response transcription factor 1-like (SCERF1), which had been analysed previously in a sugarcane–Herbaspirillum interaction (Cavalcante et al., 2007). While the expression of the ethylene receptor EIN3 was not affected by the inoculation, ETR1 and SCER1-like genes were upregulated in roots (Fig. 6). By contrast, SCERF1-like was repressed in roots and shoots after 3 weeks of growth with H. frisingense. These results suggested that the presence of the bacteria induced long-term differences in jasmonate and ethylene signalling, which were most prominent in the root. The relative changes in the qRT-PCR expression level of these marker genes correlated qualitatively, but not quantitatively, with the expression changes deduced from the transcriptomics data and varied slightly within biological replicates.
Growth promotion of M. sinensis with H. frisingense and ethylene
The identification of prominent differences in the ethylene signalling in Miscanthus roots by diazotrophic growth-promoting bacteria parallels results obtained with sugarcane (Nogueira et al., 2001; Cavalcante et al., 2007). Whether an external supply of ethylene or its precursor 1ACC altered root growth was therefore investigated. The root biomass of M. sinensis showed a dose-dependent ethylene sensitivity in pot experiments. The addition of (labile) ACC to the nutrient solution increased plant biomass only at an intermediate concentration, whereas seedlings inoculated with H. frisingense already had an increased biomass without the addition of ACC (Supplementary Fig. S8 at JXB online). The application of ACC to inoculated seedlings increased the biomass to a maximum when the concentration (5 µM) was higher than that in the absence of H. frisingense. This effect may partially be related to ACC deaminase of H. frisingense, which may decrease the amount of plant-available ethylene, which in turn affects root proliferation. This bell-shaped biomass distribution of total seedlings with respect to ACC supplementation was probably due to a primary effect on the root growth, which was visibly stimulated in the seedlings as a function of the ethylene availability (Supplementary Fig. S9 at JXB online). The differential sensitivity to external ethylene strengthens the findings of long-term differentially regulated ETR/ER and ERF genes in the roots (Fig. 6) and underlines the importance of ethylene in endophyte–plant interactions. However, these data also suggested that further plant-growth-promoting actions are conferred by H. frisingense, which are unrelated to ethylene.
Differential proteomics
Differentially abundant proteins (fold change >1.8, P<0.05) in M. sinensis seedlings grown without or with H. frisingense for 3 weeks were determined. Of the 14 protein spots identified, 12 were more abundant and two were less abundant after Herbaspirillum inoculation (Supplementary Fig. S10 at JXB online). The annotation of the differential spots is given in Table 1. Several differentially regulated proteins were related to primary metabolism, but none was related to plant hormone signalling. Three spots were annotated as fructose-bisphosphate aldolase: two of them were upregulated, whereas one was down-regulated. Two spots were close to each other, which could represent different phosphorylation states of the same protein, whereas the downregulated spot might indicate a lighter and smaller complex. Improved photosynthesis could be suggested by the higher abundance of the large subunit of ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) and serine–glycine hydroxymethyltransferase (Table 1). Aconitate hydratase was present in four differential spots, which differed only in their pI values (Table 1). These might therefore again represent different phosphorylation states. Two of the upregulated proteins were identified as β-tubulin and TCP-1 γ-chaperonin, which are both involved in cytoskeleton formation. Tubulin is a vital component of the cytoskeleton and TCP-1 γ-chaperonin folds as a complex with various proteins, including actin and tubulin. The differentially regulated cinnamyl alcohol dehydrogenase is involved in phenolpropanoid and lignin biosynthesis, whereas ascorbate peroxidases are involved in stress metabolism.
Table 1.
Differentially abundant proteins of M. sinensis grown 3 weeks with H. frisingense GSF30 TSpot number, significance level, fold changes, annotation, coverage, number of unique peptides, predicted mass, and the relative RNA expression differences (from RNA-Seq) of 14 differential spots are given. Localization of the individual spots in the 2D gels is shown in Supplementary Fig. S8
| Spot | ANOVA (P) | Fold | Protein annotation | Best hit | % Coverage | Unique peptides | kDa | Expression |
|---|---|---|---|---|---|---|---|---|
| 3156 | 0.004 | 4.9 | β-Tubulin | gi|293331107 | 41 | 24 | 55 | 0.0 |
| 961 | 0.005 | 1.9 | TCP-1 chaperonin, subunit gamma | gi|242036525 | 33 | 26 | 61 | 0.0 |
| 3096 | 0.002 | 2.9 | Cinnamyl alcohol dehydrogenase | gi|242049212 | 21 | 10 | 44 | –0.1 |
| 3068 | 0.000 | 4.6 | Rubisco large subunit | gi|48478779 | 31 | 18 | 53 | 0.1 |
| 1146 | 0.021 | 1.8 | Serine–glycine hydroxymethyltransferase | gi|242068375 | 37 | 21 | 52 | –0.3 |
| 3092 | 0.014 | –2.3 | Fructose-bisphosphate aldolase | gi|242059597 | 19 | 9 | 39 | –0.2 |
| 3102 | 0.007 | 2.4 | 39 | 20 | ||||
| 3139 | 0.030 | 2.1 | 57 | 23 | ||||
| 3074 | 0.033 | 3.3 | Aconitate hydratase | gi|242037013 | 26 | 30 | 107 | –0.1 |
| 3081 | 0.016 | 3.4 | 18 | 24 | ||||
| 3084 | 0.013 | 2.4 | 17 | 21 | ||||
| 3110 | 0.018 | 3.8 | 15 | 21 | ||||
| 2073 | 0.028 | 1.8 | l-Ascorbate peroxidase 2 | gi|226897533 | 23 | 5 | 27 | –0.2 |
| 2194 | 0.004 | –3.3 | l-Ascorbate peroxidase 1 | gi|242041317 | 28 | 9 |
Discussion
In this study, M. sinensis seedlings were found to profit from inoculation with the diazotrophic H. frisingense under conditions of a low nitrogen supply (Fig. 1). H. frisingense is genetically related to the sugarcane, rice, sorghum, and maize colonizer H. seropedicae, which, in contrast to H. frisingense, occasionally confers disease symptoms on some plant host genotypes (Monteiro et al., 2012b ). The absence of disease symptoms in Miscanthus inoculated with H. frisingense may be explained by the fact that H. frisingense lacks several genetic components that are involved in pathogenic functions (Straub et al., 2013a ). H. frisingense is also closely related to Herbaspirillum strains isolated from other habitats, such as strains that were isolated from the rhizosphere of Australian phragmites and from Japanese well water (Straub et al., 2013a ).
Among the non-legumes, the direct contribution of nitrogen fixation (25–60%) by diazotrophic endophytic communities to nitrogen nutrition is probably best verified in Brazilian sugarcane (Boddey et al., 2001; Taule et al., 2012; Urquiaga et al., 2012). The evidence for non-legume nitrogen fixation in temperate climates from field studies is less clear; for example, the diazotroph Azospirillum exerts its beneficial roles on growth, not via nitrogen fixation (Bashan et al., 1989). However, the extraordinary nitrogen efficiency in Miscanthus (Christian et al., 1997, 2008) suggests that, at least in this grass, diazotrophs contribute to this nitrogen efficiency. Our results suggest that, at least in very young seedlings, the potential contribution of nitrogen fixation to nitrogen nutrition is rather small, although it could not be rigorously excluded from the above experiments that some nitrogen fixation occurred. Significant nitrogen fixation may occur at later developmental stages, and it is interesting to note that alphaproteobacterial and rhizobacterial DNA sequences have been identified in field-grown adult Miscanthus leaves (Straub et al., 2013b ). While the nitrogen concentration was even reduced in inoculated roots, the potassium concentration was increased. The nutrient concentrations in the shoot were not affected by the inoculation (Fig. 4). Other plant-growth-promoting rhizobacteria, including the well-studied Azospirillum, increase nutrient concentrations under certain conditions (Spaepen et al., 2009). Whether H. frisingense can increase the availability of other soil nutrients, such as phosphorus, which was slightly but not significantly increased, should be analysed in future studies.
Diazotrophic bacterial grass colonizers, such as H. seropedicae and Gluconacetobacter diazotrophicus, promote root growth by manipulating plant hormone signalling. In this way, the root surface area available to absorb nutrients is increased and consequently allows the production of more total biomass. The transcriptome analysis identified that the jasmonate response and ethylene signalling were persistently altered by the presence of H. frisingense (Fig. 8). These plant hormones are well-known key players in abiotic and biotic stresses (Thatcher et al., 2005). The prominent upregulation of jasmonate-inducible defence proteins after 2 weeks of inoculation was reported for the closely related endophyte Azoarcus spp. upon invasion of Oryza sativa cultivars (Miche et al., 2006). In sugarcane, a partial overlap in the activation of resistance genes induced by methyl jasmonate and by colonization by H. seropedicae and Herbaspirillum rubrisubalbicans has been noted (Rocha et al., 2007), suggesting a conserved initial jasmonate signalling response in diverse endophyte–grass host interactions. This jasmonate response occurred before the establishment of an effective association (Rocha et al., 2007).
In the Miscanthus transcriptome and in qRT-PCR, three jasmonate-induced genes (BGAF-1, BGAF-2 and dirigent-like) were more highly expressed by orders of magnitude after 3h (Fig. 6). Three weeks after inoculation, the qRT-PCR data suggested that these jasmonate response genes were significantly repressed. BGAF is a Poaceae-specific protein that can bind β-glucosidase, which is involved in the jasmonate-dependent defence against pathogens in young maize plants. Herbivores release toxic aglycons from hydroxamic acid glucosides, which are one of the major defence compounds in members of the family Poaceae (Niemeyer, 1988). The predominant hydroxamic acid glucoside in maize is 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)-Glc, whose aglycone DIMBOA has a strong inhibitory effect on Agrobacterium tumefaciens (Sahi et al., 1990). BGAF aggregates and concentrates β-glucosidase, and protects BGAF and β-glucosidase from degradation, but has no effect on β-glucosidase activity (Esen and Blanchard, 2000). BGAF contains a jacalin-related lectin domain and a disease-response (dirigent protein) domain (Kittur et al., 2007).
The rapid, strong upregulation of these jasmonate-related genes after inoculation with H. frisingense suggested that a common early defence response in M. sinensis occurs. However, after establishment of the association, the respective genes were even suppressed by H. frisingense, which is well in accordance with its endophytic life style. Furthermore, little indication for further defence responses (except for changes in redox metabolism) were obtained from the transcriptome and proteome data, suggesting that H. frisingense is able to at least partially bypass or suppress the plant defence response. The weak colonization density (Fig. 2) may contribute to this minor defence reaction.
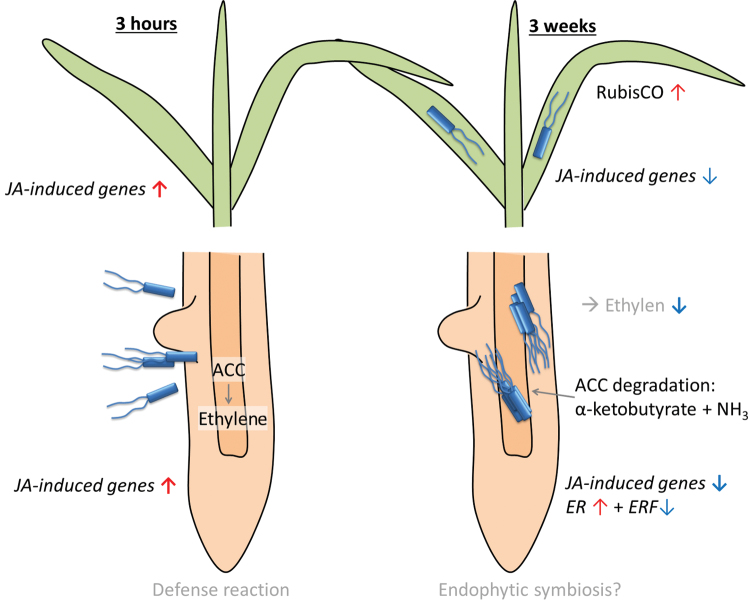
Ethylene is an essential signalling molecule for growth and developmental processes in plants (Wu et al., 2009). H. frisingense, as well as all other fully sequenced, plant-associated Herbaspirillum strains, produces the enzyme ACC deaminase (Rothballer et al., 2008). This enzyme degrades the ethylene precursor ACC and locally reduces plant-available ethylene. Divergent bacterial species are able to reduce the local ethylene levels via ACC deaminase activity and hence alter the root morphology (Contesto et al., 2008). Furthermore, ACC deaminase can protect plants from various stresses and in this way improve growth (Glick, 2004). In the established Miscanthus–Herbaspirillum interaction, ethylene receptors appeared specifically upregulated in roots, while ethylene response factors were repressed (Fig. 6). This is consistent with the assumption that H. frisingense can actively reduce ethylene levels in plant roots by ACC deaminase, which competes with the plant ACC oxidase for ACC (Glick, 2005). In rapeseed seedlings, the effects of ACC deaminase-containing bacteria on biomass gain is dependent on the nutritional status of the plants, and the presence of the bacteria often reduces the shoot nutrient content, which was not the case here (Belimov et al., 2002). Positive effects of plant root-growth-promoting bacteria on ethlyene signalling are well established in heavy metal stress (Safronova et al., 2006) and saline stress (Saravanakumar and Samiyappan, 2007), where the stress-induced root growth suppression is counteracted by ACC deaminase-containing bacteria. In sugarcane, ethylene-related genes are differentially affected by different diazotrophic bacterial strains (Nogueira et al., 2001; Cavalcante et al., 2007), in a similar way overall to here in Miscanthus. The lower availability of ethylene in Miscanthus roots is consistent with the upregulation of the ethylene receptor genes and repression of the response factors, but a SCER1 ethylene receptor is induced in sugarcane after inoculation with G. diazotrophicus or H. rubrisubalbicans, while an ethylene-responsive factor is only induced by the endophyte G. diazotrophicus (Cavalcante et al., 2007), which does not contain ACC deaminase to degrade the ethylene precursor (Straub et al., 2013a ). This persistent regulation of ethylene-responsive genes (including up- and downregulated genes) is similar in rice roots 7 d after inoculation with H. seropedicae (Brusamarello-Santos et al., 2012). However, besides this common regulation of jasmonate- and ethylene-responsive signalling (Fig. 7), only very little overall overlap in the endophyte-induced transcriptiome profile is seen with other grasses and Miscanthus. This may indicate endophyte strain-specific and plant species-specific responses during interaction with diverse bacterial species. Furthermore, there is also little overlap with grass responses to fungal endophytes, except for the differential regulation of genes involved in redox metabolism, although both endophytes colonize and feed from the apoplast (Tanaka et al., 2012).
Fig. 7.
Mechanistic scheme of the interaction of M. sinensis and H. frisingense. The beneficial effect of Herbaspirillum-induced root hormone signalling leads to higher root biomass, improved nutrient absorption, and, as a consequence, also increases the shoot biomass. Red arrows: up-regulated genes, blue arrow: down-regulated genes. JA, jasmonate; ER, ethylene receptor; ERF, ethylene response factor. (This figure is available in colour at JXB online.)
H. frisingense (as well as H. seropedicae) produces the plant growth-promoting factor indoleacetic acid (IAA) (Rothballer et al., 2008). IAA and ARF-like genes encode transcription factors that are expressed in a tissue-specific manner and respond to light and/or auxin (Cheong et al., 2002; Jain and Khurana, 2009; Brusamarello-Santos et al., 2012). A collective minor repression of IAA and ARF genes was observed in rice roots 7 d after inoculation with H. seropedicae, but there were genotype differences (cv. Cateto zebu and Nipponbare). Here, a differential expression of specific auxin-responsive genes was marginal and not persistent; the individual expression level differences were negligible (Fig. 6), suggesting only a minor change in the auxin signalling in the beneficial association. The cross-talk of auxin signalling and ethylene has also been studied with ACC deaminase-containing and -deficient bacteria in rapeseed. It was suggested that auxin, which activates ACC synthase in plants, must be balanced with ethylene to maintain optimal auxin signal transduction in the root (Stearns et al., 2012).
Few metabolic categories differed following inoculation, and this included stimulation of primary metabolism and repression of some secondary metabolism pathways (Fig. 5). Differences in the primary metabolism were suggested by the differential proteomes after 3 weeks (Table 1), which is consistent with the higher biomass following inoculation. The higher abundance of aldolase may increase the biomass by analogy to tobacco, where transgenes overexpressing plastid aldolase stimulated ribulose 1,5-bisphosphate regeneration and promoted CO2 fixation (Uematsu et al., 2012). Furthermore, the transcript analysis indicated a differential ascorbate and glutathionine redox metabolism, which was supported by the finding of the differential abundance of two ascorbate peroxidases. The higher amount of tubulin and a cytoskeleton chaperone may indicate differences in the cytoskeleton after inoculation, but no evidence for transcriptomic changes in cytoskeleton genes was observed. In Medicago, mycorrhizal symbiosis increases the β-tubulin gene MtTubb1 in infected root tissue, root nodules, the inner cortex, and the vasculature (Manthey et al., 2004). In alfalfa nodule formation, regulation of tubulin is due to the microtubular cytoskeleton changes during symbiosis (Timmers et al., 1998).
Phenylpropanoids and structural lignins are well known to be implicated in defence responses in plant–microbe interactions (Schenk et al., 2000; Cheong et al., 2002). In Arabidopsis, several members of the cinnamyl alcohol dehydrogenase gene family are induced in response to pathogenic Pseudomonas (Tronchet et al., 2010). This parallels the observation for the M. sinensis cinnamyl alcohol dehydrogenase after inoculation with H. frisingense. Finally, little similarity in the differential proteomes is apparent in other endophyte-grass responses, such as the sugarcane response to the plant-growth-promoting bacterium G. diazotrophicus after 1 d (Lery et al., 2011). This may partially be due to the fact that different sampling periods were used in the different studies, but as no general clues on plant responses in endophyte–plant interactions could be extracted from the transcriptomic responses, highly species-specific responses are suggested. Common effects include early jasmonate-related signalling in the entire plant, the local suppression of ethylene signalling to stimulate root growth, and changes in redox metabolism. Highly plant-specific interactions are also strongly supported by the fact that several studies identified genotype-specific responses within a single plant species.
Although not systematically analysed, we also noted unsuccessful inoculation attempts by Herbaspirillum. Low delivery rates are known to restrict the global potential of endophytes to colonize grasses in the field (Bhattacharjee et al., 2008). Combinations of diverse bacteria in the inoculum (Fischer et al., 2012) and/or mixtures of Herbaspirillum strains with other beneficial agents, such as humic acids (Canellas et al., 2012), may provide a solution to improve the colonization efficiency in the field. As H. frisingense has beneficial effects on Miscanthus seedling growth and establishment, whereas the plant defence upon inoculation is minimal, H. frisingense may gain a more widespread use as a cheap biofertilizer.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Exclusive detection of bacterial H. frisingense nitrogen-fixation regulator nifA in inoculated plants by PCR.
Supplementary Fig. S2. Localization of fluorescently labelled H. frisingense in the apoplast between leaf cells.
Supplementary Fig. S3. Increases in nutrient content (=dry biomass×nutrient concentration) of roots (A) and shoots (B) by H. frisingense inoculation.
Supplementary Fig. S4. Fertilization with 15N-enriched (10%) nutrient medium does not reveal nitrogen-fixation activities in H. frisingense-inoculated plants.
Supplementary Fig. S5. Transcriptome overview of M. sinensis grown for 3h with H. frisingense.
Supplementary Fig. S6. Transcriptome overview of M. sinensis grown for 3 weeks with H. frisingense.
Supplementary Fig. S7. Transcriptional categories affected by inoculation of M. sinensis with H. frisingense.
Supplementary Fig. S8. Influence of ethylene in growth promotion of M. sinensis by H. frisingense.
Supplementary Fig. S9. Effect of increasing ACC concentrations and H. frisingense on plant growth.
Supplementary Fig. S10. 2D gel electrophoresis of M. sinensis grown for 3 weeks without or with H. frisingense.
Supplementary Table S1. The primers used and predicted PCR fragment lengths.
Supplementary Materials and Methods. Details of the PCR conditions.
Acknowledgements
We thank BSc Hans-Uwe Balint for help with RT-PCR and Jens Katzmeier, Elke Dachtler, and Helene Ochott for nutrient analyses. The work was partially supported by the EU-grant ‘Biofector’.
Glossary
Abbreviations:
- ACC
1-aminocyclopropane-1-carboxylate
- IAA
indoleacetic acid
- qRT-PCR
using quantitative real-time PCR.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan Y, Singh M, Levanony H. 1989. Contribution of Azospirillum brasilense Cd to growth of tomato seedlings is not through nitrogen fixation. Canadian Journal of Botany 67, 2429–2434 [Google Scholar]
- Belimov AA, Safronova VI, Mimura T. 2002. Response of spring rape (Brassica napus var. oleifera L.) to inoculation with plant growth promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase depends on nutrient status of the plant. Canadian Journal of Microbiology 48, 189–199 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee RB, Singh A, Mukhopadhyay SN. 2008. Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Applied Microbiology and Biotechnology 80, 199–209 [DOI] [PubMed] [Google Scholar]
- Boddey RM, Polidoro JC, Resende AS, Alves BJR, Urquiaga S. 2001. Use of the 15N natural abundance technique for the quantification of the contribution of N2 fixation to sugar cane and other grasses. Australian Journal of Plant Physiology 28, 889–895 [Google Scholar]
- Brusamarello-Santos LCC, Pacheco F, Aljanabi SMM, Monteiro RA, Cruz LM, Baura VA, Pedrosa FO, Souza EM, Wassem R. 2012. Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae . Plant and Soil 356, 113–125 [Google Scholar]
- Canellas LP, Dobbss LB, Oliveira AL, Chagas JG, Aguiar NO, Rumjanek VM, Novotny EH, Olivares FL, Spaccini R, Piccolo A. 2012. Chemical properties of humic matter as related to induction of plant lateral roots. European Journal of Soil Science 63, 315–324 [Google Scholar]
- Cavalcante JJV, Vargas C, Nogueira EM, Vinagre F, Schwarcz K, Baldani JI, Ferreira PCG, Hemerly AS. 2007. Members of the ethylene signalling pathway are regulated in sugarcane during the association with nitrogen-fixing endophytic bacteria. Journal of Experimental Botany 58, 673–686 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. 2002. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis . Plant Physiology 129, 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian DG, Poulton PR, Riche AB, Yates NE. 1997. The recovery of 15N-labelled fertilizer applied to Miscanthus×giganteus . Biomass Bioenergy 12, 21–24 [Google Scholar]
- Christian DG, Riche AB, Yates NE. 2008. Growth, yield and mineral content of Miscanthus×giganteus grown as a biofuel for 14 successive harvests. Industrial Crops and Products 28, 320–327 [Google Scholar]
- Contesto C, Desbrosses G, Lefoulon C, Bena G, Borel F, Galland M, Gamet L, Varoquaux F, Touraine B. 2008. Effects of rhizobacterial ACC deaminase activity on Arabidopsis indicate that ethylene mediates local root responses to plant growth-promoting rhizobacteria. Plant Science 175, 178–189 [Google Scholar]
- Davis SC, Parton WJ, Dohleman FG, Smith CM, Del Grosso S, Kent AD, DeLucia EH. 2010. Comparative biogeochemical cycles of bioenergy crops reveal nitrogen-fixation and low greenhouse gas emissions in a Miscanthus×giganteus agro-ecosystem. Ecosystems 13, 144–156 [Google Scholar]
- Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T, Isawa T, Mitsui H, Minamisawa K. 2001. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Applied and Environmental Microbiology 67, 5285–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A, Blanchard DJ. 2000. A specific β-glucosidase-aggregating factor is responsible for the β-glucosidase null phenotype in maize. Plant Physiology 122, 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Pfitzner B, Schmid M, Simoes-Araújo S, Reis V, Martinez-Romero E, Baldani JI, Hartmann A. 2012. Molecular characterization of the diazotrophic bacterial community in non-inoculated and inoculated field grown sugar cane (Saccharum sp.). Plant and Soil 356, 83–99 [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng ZY, Duan J, McConkey B. 2007. Promotion of plant growth by bacterial ACC deaminase. Critical Reviews in Plant Sciences 26, 227–242 [Google Scholar]
- Glick BR. 2004. Bacterial ACC deaminase and the alleviation of plant stress. Advances in Applied Microbiology 56, 291–312 [DOI] [PubMed] [Google Scholar]
- Glick BR. 2005. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiology Letters 251, 1–7 [DOI] [PubMed] [Google Scholar]
- Greef JM, Deuter M. 1993. Syntaxonomy of Miscanthus×giganteus Greef-Et-Deu. Angewandte Botanik 67, 87–90 [Google Scholar]
- Himken M, Lammel J, Neukirchen D, Czypionka-Krause U, Olfs HW. 1997. Cultivation of Miscanthus under West European conditions: seasonal changes in dry matter production, nutrient uptake and remobilization. Plant and Soil 189, 117–126 [Google Scholar]
- Jain M, Khurana JP. 2009. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS Journal 276, 3148–3162 [DOI] [PubMed] [Google Scholar]
- Karp A, Shield I. 2008. Bioenergy from plants and the sustainable yield challenge. New Phytologist 179, 15–32 [DOI] [PubMed] [Google Scholar]
- Kirchhof G, Eckert B, Stoffels M, Baldani JI, Reis VM, Hartmann A. 2001. Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. International Journal of Systematic and Evolutionary Microbiology 51, 157–168 [DOI] [PubMed] [Google Scholar]
- Kittur FS, Lalgondar M, Yu HY, Bevan DR, Esen A. 2007. Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. Journal of Biological Chemistry 282, 7299–7311 [DOI] [PubMed] [Google Scholar]
- Lery LMS, Hemerly AS, Nogueira EM, von Kruger WMA, Bisch PM. 2011. Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium Gluconacetobacter diazotrophicus and sugarcane. Molecular Plant–Microbe Interactions 24, 562–576 [DOI] [PubMed] [Google Scholar]
- Lewandowski I, Kicherer A. 1997. Combustion quality of biomass: practical relevance and experiments to modify the biomass quality of Miscanthus×giganteus . European Journal of Agronomy 6, 163–177 [Google Scholar]
- Liu Y, Lamkemeyer T, Jakob A, Mi G, Zhang F, Nordheim A, Hochholdinger F. 2006. Comparative proteome analyses of maize (Zea mays L.) primary roots prior to lateral root initiation reveal differential protein expression in the lateral root initiation mutant rum1 . Proteomics 6, 4300–4308 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Puhler A, Perlick AM, Kuster H. 2004. Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Molecular Plant–Microbe Interactions 17, 1063–1077 [DOI] [PubMed] [Google Scholar]
- Miche L, Battistoni F, Gernmer S, Belghazi M, Reinhold-Hurek B. 2006. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Molecular Plant–Microbe Interactions 19, 502–511 [DOI] [PubMed] [Google Scholar]
- Monteiro RA, Balsanelli E, Tuleski T, et al. 2012a. Genomic comparison of the endophyte Herbaspirillum seropedicae SmR1 and the phytopathogen Herbaspirillum rubrisubalbicans M1 by suppressive subtractive hybridization and partial genome sequencing. FEMS Microbiology Ecology 80, 441–451 [DOI] [PubMed] [Google Scholar]
- Monteiro RA, Balsanelli E, Wassem R, et al. 2012b. Herbaspirillum–plant interactions: microscopical, histological and molecular aspects. Plant and Soil 356, 175–196 [Google Scholar]
- Niemeyer HM. 1988. Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defence chemicals in the Gramineae. Phytochemistry 27, 3349–3358 [Google Scholar]
- Nogueira EM, Vinagre F, Masuda HP, Vargas C, Pádua VLM, Silva FP, Santos RV, Baldani JI, Ferreira PCG, Hemerly AS. 2001. Expression of sugarcane genes induced by inoculation with Gluconacetobacter diazotrophicus and Herbaspirillum rubrisubalbicans . Genetics and Molecular Biology 24, 199–206 [Google Scholar]
- Pedrosa FO, Monteiro RA, Wassem R, et al. 2011. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genetics 7, e1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. 1998. Life in grasses: diazotrophic endophytes. Trends in Microbiology 6, 139–144 [DOI] [PubMed] [Google Scholar]
- Rocha FR, Papini-Terzi FS, Nishiyama MY, Jr, et al. 2007. Signal transduction-related responses to phytohormones and environmental challenges in sugarcane. BMC Genomics 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer M, Eckert B, Schmid M, Fekete A, Schloter M, Lehner A, Pollmann S, Hartmann A. 2008. Endophytic root colonization of gramineous plants by Herbaspirillum frisingense . FEMS Microbiology Ecology 66, 85–95 [DOI] [PubMed] [Google Scholar]
- Safronova VI, Stepanok VV, Engqvist GL, Alekseyev YV, Belimov AA. 2006. Root-associated bacteria containing 1-aminocyclopropane-1-carboxylate deaminase improve growth and nutrient uptake by pea genotypes cultivated in cadmium supplemented soil. Biology and Fertility of Soils 42, 267–272 [Google Scholar]
- Sahi SV, Chilton MD, Chilton WS. 1990. Corn metabolites affect growth and virulence of Agrobacterium tumefaciens . Proceedings of the National Academy of Sciences, USA 87, 3879–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanakumar D, Samiyappan R. 2007. ACC deaminase from Pseudomonas fluorescens mediated saline resistance in groundnut (Arachis hypogea) plants. Journal of Applied Microbiology 102, 1283–1292 [DOI] [PubMed] [Google Scholar]
- Sauer M, Jakob A, Nordheim A, Hochholdinger F. 2006. Proteomic analysis of shoot-borne root initiation in maize (Zea mays L.). Proteomics 6, 2530–2541 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. 2000. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proceedings of the National Academy of Sciences, USA 97, 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R, Ihring A, Gantner S, et al. 2006. Induction of systemic resistance in tomato by N-acyl-l-homoserine lactone-producing rhizosphere bacteria. Plant, Cell & Environment 29, 909–918 [DOI] [PubMed] [Google Scholar]
- Sevilla M, Burris RH, Gunapala N, Kennedy C. 2001. Comparison of benefit to sugarcane plant growth and N-15 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and Nif(–) mutant strains. Molecular Plant–Microbe Interactions 14, 358–366 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Okon Y. 2009. Plant growth-promoting actions of rhizobacteria. Plant Innate Immunity 51, 283–320 [Google Scholar]
- Stearns JC, Woody OZ, McConkey BJ, Glick BR. 2012. Effects of bacterial ACC deaminase on Brassica napus gene expression. Molecular Plant–Microbe Interactions 25, 668–676 [DOI] [PubMed] [Google Scholar]
- Straub D, Rothballer M, Hartmann A, Ludewig U. 2013a. The genome of the endophytic bacterium H. frisingense GSF30T identifies diverse strategies in the Herbaspirillum genus to interact with plants. Frontiers in Microbiology 4, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub D, Yang H, Liu Y, Ludewig U. 2013b. Transcriptomic and proteomic comparison of two Miscanthus genotypes: high biomass correlates with investment in primary carbon assimilation and decreased secondary metabolism. Plant and Soil(10.1007/s11104-013-1693-1). [Google Scholar]
- Tanaka A, Takemoto D, Chujo T, Scott B. 2012. Fungal endophytes of grasses. Current Opinion in Plant Biology 15, 462–468 [DOI] [PubMed] [Google Scholar]
- Taule C, Mareque C, Barlocco C, Hackembruch F, Reis VM, Sicardi M, Battistoni F. 2012. The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant and Soil 356, 35–49 [Google Scholar]
- Thatcher LF, Anderson JP, Singh KB. 2005. Plant defence responses: what have we learnt from Arabidopsis? Functional Plant Biology 32, 1–19 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, de Billy F, Truchet G. 1998. Nod factor internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa. Development 125, 339–349 [DOI] [PubMed] [Google Scholar]
- Tronchet M, Balague C, Kroj T, Jouanin L, Roby D. 2010. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis . Molecular Plant Pathology 11, 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu K, Suzuki N, Iwamae T, Inui M, Yukawa H. 2012. Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. Journal of Experimental Botany 63, 3001–3009 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Research 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquiaga S, Xavier RP, de Morais RF, et al. 2012. Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant and Soil 356, 5–21 [Google Scholar]
- von Rad U, Klein I, Dobrev PI, Kottova J, Zazimalova E, Fekete A, Hartmann A, Schmitt-Kopplin P, Durner J. 2008. Response of Arabidopsis thaliana to N-hexanoyl-dl-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229, 73–85 [DOI] [PubMed] [Google Scholar]
- Wu CH, Bernard SM, Andersen GL, Chen W. 2009. Developing microbe–plant interactions for applications in plant-growth promotion and disease control, production of useful compounds, remediation and carbon sequestration. Microbial Biotechnology 2, 428–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.