Abstract
Human NK cells express cell surface class I MHC receptors (KIR) in a probabilistic manner. Previous studies have shown that a distal promoter acts in conjunction with a proximal bidirectional promoter to control the selective activation of KIR genes. We report here the presence of an intron 2 promoter in several KIR genes that produces a spliced antisense transcript. This lncRNA transcript contains antisense sequence complementary to KIR-coding exons 1 and 2 as well as the proximal promoter region of the KIR genes. The antisense promoter contains MZF-1 binding sites, a transcription factor found in hematopoietic progenitors and myeloid precursors. The KIR antisense lncRNA was only detected in progenitor cells or pluripotent cell lines, suggesting a function that is specific for stem cells. Overexpression of MZF-1 in developing NK cells led to decreased KIR expression, consistent with a role for the KIR antisense lncRNA in silencing KIR gene expression early in development.
Keywords: human, NK cells, KIR, antisense, transcription, lncRNA
Introduction
Natural Killer (NK) cells are an important component of the innate immune system wherein resides the ability to eliminate virally infected and tumor cells without prior sensitization1. The cytolytic activity of NK cells is modulated by the presence or absence of class I MHC molecules on target cells2. The interplay between activating and inhibitory receptors present on NK cells and their respective ligands on potential targets determines if cells are killed or spared.
NK cells use cell surface receptors for class I MHC to evaluate the status of target cells. The major type of class I receptor expressed by human NK cells is the killer cell immunoglobulin-like receptor (KIR) family3, 4. In contrast, the functionally analogous murine inhibitory receptors are members of the C-type lectin family (Ly49 receptor family)5. The expression of KIR and Ly49 on NK cells is variegated, with distinct subsets of NK cells displaying different combinations of receptors generated by a stochastic process6, 7. The ability of adjacent KIR genes to be differentially expressed is controlled by DNA methylation of the proximal promoter region8, 9. The KIR loci are fully methylated in NK progenitors, however, once activated, the proximal KIR promoter remains in a demethylated state and the gene is continuously transcribed. KIR expression is associated with fully differentiated, functional NK cells, and KIR have been shown to play a major role in the growth, differentiation, and function of NK cells10.
Previous studies from our group have demonstrated that the variegated expression of both Ly49 and KIR genes is determined by a stochastic switch operating in the promoter region11, 12, 13. In the murine Ly49 genes, a probabilistic bidirectional promoter active in immature NK cells is present upstream of the proximal promoter responsible for Ly49 expression in mature NK cells. The choice of forward transcription from this upstream element (Pro1) leads to activation of the proximal promoter. The expression of a Ly49 gene from the proximal promoter is dependent on distal transcription, since Pro1 deletion abrogates Ly49 transcription14. In contrast, the human KIR genes possess a proximal promoter with bidirectional transcriptional activity, whereas an upstream distal promoter is unidirectional. Similar to the Ly49 genes, the distal KIR promoter is active in committed NK progenitors and distal transcription is associated with activation of the proximal promoter15, 16. The location of the bidirectional promoter downstream of the distal promoter leads to the generation of opposing transcripts if antisense transcription is initiated from the proximal promoter17. The presence of dsRNA leads to the production of a 28 base antisense RNA with the properties of a Piwi RNA18. The Piwi class of small RNAs has been associated with gene silencing in germ cells, and recent studies have demonstrated the presence of these RNAs in somatic cell types as well19. Forced expression of proximal promoter antisense transcripts in developing NK cells leads to reduced KIR expression, and the 28 base element is essential for this suppression18.
The data presented in the current study reveals the presence of an additional antisense transcript in the 2DL1/S1 and 3DL1/S1 genes. The transcript is generated from a promoter in the second intron, and represents a spliced, polyadenylated RNA that appears to be non-coding. Overlap of this transcript with the proximal antisense transcript leads to the production of the previously characterized 28 base piRNA from this long noncoding RNA (lncRNA), and enforced expression of the distal antisense also leads to suppressed KIR expression. Our characterization of the promoter and transcript indicates activity only in pluripotent cells, suggesting a functional role for the antisense transcript in the initial silencing of the KIR loci.
Results
Detection of a distal antisense KIR transcript
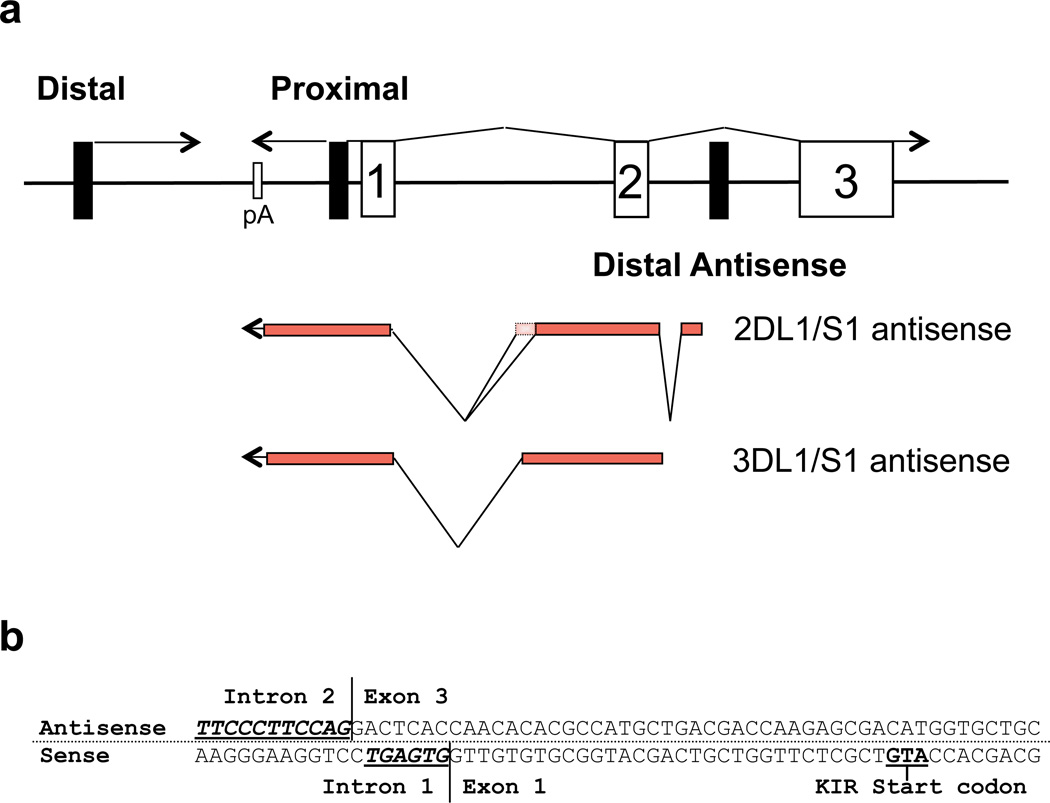
Our previous reports demonstrated that the human KIR genes all contain a proximal promoter that is bidirectional in nature12. Experiments designed to determine the 5’ start site for the proximal KIR2DL1 antisense transcript were conducted with RNA from the HEK293 cell line as a non-NK control. However, when HEK293 RNA was used, a transcript was identified that originated within intron 2 of the KIR2DL1/S1 gene. To determine if the antisense was also found in the 3D class of KIR, primers specific for the KIR3DL1 gene were also used to isolate antisense transcripts from the KIR3DL1/S1 genes. This novel antisense transcript is referred to as the KIR distal antisense in order to distinguish it from the proximal promoter-derived antisense transcripts that we have previously described12. The transcriptional start site for the KIR2DL1 distal antisense is located within the second intron, 181 nucleotides downstream of the second KIR-coding exon (Figure 1a). The KIR3DL1 distal antisense transcript starts 81 nucleotides downstream of exon 2. Two distinct alternatively spliced distal antisense transcripts of 710 and 781 nucleotides, each consisting of three exons, were cloned for the KIR2DL1 gene (GenBank accession numbers GQ422372 and GQ422373), whereas only one 825 nucleotide transcript consisting of two exons was cloned for the KIR3DL1 gene (GenBank accession number GQ422374). The distal antisense transcript has a complete overlap with exons 1 and 2 of the KIR coding transcript as well as the proximal antisense transcript (Figure 1a). Interestingly, the splice acceptor for the final antisense exon is only 7 bp downstream of the exon 1 splice donor of the sense KIR transcript, suggesting that the splice site definition signals or splicing enhancers are shared (Figure 1b). In addition, although the distal antisense transcripts have their respective start sites in intron 2, the polyadenylation signals are shared with the proximal antisense transcript12.
Figure 1.
Identification of KIR distal antisense transcripts. (a) A schematic of the organization of the 5’ region of the KIR genes is shown. Black rectangles represent promoter elements, and numbered rectangles represent exons. Lines represent KIR transcripts with their orientation indicated by arrows. The exon structure of the KIR2DL1/S1 and KIR3DL1/S1 distal antisense transcripts is indicated below. The additional exon sequence found in the KIR2DL1 alternative transcript (GenBank: GQ422373) is indicated by the dotted rectangle. (b) The nucleotide sequence of the region containing the KIR2DL1 distal antisense intron 2/exon 3 splice junction and the exon 1/intron 1 splice junction of the KIR2DL1 coding transcript is shown. The antisense strand is shown on top with the complementary coding sense strand shown on the bottom. Consensus splice acceptor (antisense) and splice donor (sense) sequences are underlined in bold.
KIR distal antisense transcripts are not present in differentiated cells
The presence of spliced distal antisense transcript was determined by RT-PCR with forward primers from regions not contained in sense or proximal antisense KIR2DL1 or KIR3DL1 transcripts (see Materials and methods). The non-spliced, proximal antisense transcripts were detected using the 3’-RACE technique previously described12. This technique produced positive signals in the cell types containing distal antisense due to the complete overlap of the distal antisense with the proximal transcript. However, the antisense transcripts were confirmed to originate only from the distal promoter by 5’-RACE analysis. Table 1 shows the results of a survey of various human tissues and cell lines for the presence of KIR2DL1 and KIR3DL1 distal antisense (first column, both genes detected when +ve) or antisense derived from the proximal promoter (second column, KIR gene of origin is indicated). Spliced KIR2DL1 and KIR3DL1 distal antisense transcripts were detected in RNA from several cell lines, including U937 (promonocytic leukemia), HEK293T (human embryonic kidney), Ntera-2 (human embryonic carcinoma), and H9 (human embryonic stem cell). The only freshly isolated tissue found to contain distal antisense was cord blood, possibly due to the presence of a small population of pluripotent cells. RNA from peripheral blood NK cells, monocytes, CD34-positive cord blood, or H9 ES cells differentiated into CD34+ cells did not express the distal antisense. Furthermore, cell lines representing differentiated lymphoid cell types such as YT (NK), Daudi (B) and Jurkat (T) did not contain distal antisense RNA. Interestingly, the NK3.3 cell line contained KIR2DL2 proximal antisense, whereas the Daudi and Jurkat lines contained an unusual proximal antisense transcript of the KIR3DP1 pseudogene that originated in intron 1 as determined by 5’-RACE analysis. The cell lines where the distal antisense transcript was detected represent early developmental stages. The distal antisense transcript was not detected in mature NK-like, B and T cell lines indicating that it may be present only in early progenitor cell types.
Table 1.
Detection of antisense transcripts
| Cell line/tissue | Distala | Proximalb |
|---|---|---|
| KIR−ve NK | − | + (all KIR) |
| KIR+ve NK | − | − |
| YT | − | − |
| NK92 | − | − |
| NK3.3 | − | + (2DL2) |
| Daudi (B) | − | + (3DP1) |
| Jurkat (T) | − | + (3DP1) |
| Monocytes | − | − |
| Ntera2 (EC) | + | − |
| H9 (ESC) | + | − |
| CD34+ve from H9 ESC | − | − |
| CD34+ve cord blood | − | − |
| Cord blood | + | − |
| HEK293 | + | − |
| U937 | + | − |
KIR2DL1 and KIR3DL1 transcripts detected with primers listed in Materials and Methods
KIR proximal antisense transcripts detected as previously described12. The identity of the KIR gene detected is listed in parentheses.
Characterization of the KIR intron 2 promoter elements
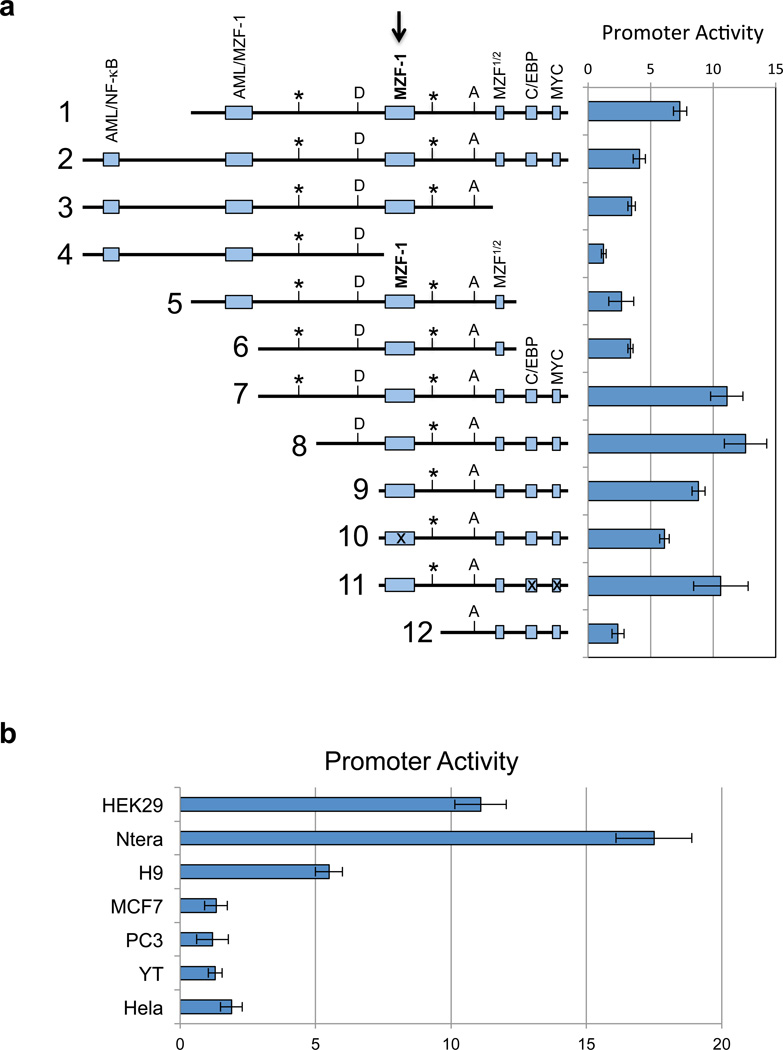
In order to gain insight into the observed preferential expression of KIR distal antisense in stem cell populations, the putative intron 2 promoter regions of the KIR2DL1 and KIR3DL1 genes were investigated. A large series of PCR-generated DNA segments in the vicinity of the observed transcriptional start sites was analyzed by cloning into the pGL3-basic luciferase reporter vector. Constructs were evaluated by transfection into the HEK293, Ntera2, and YT cell lines. None of the fragments tested showed any activity in the YT cell line, consistent with the lack of antisense transcripts in these cells (data not shown). Figure 2a show the results of transfection of KIR2DL1 promoter constructs into HEK293. The highest promoter activity was detected in fragments that contained a central binding site for the myeloid zinc finger 1 (MZF-1) transcription factor together with putative C/EBP and MYC sites located downstream (constructs 7–9). Addition of 5’ sequence containing additional putative combined AML-1/MZF-1 or NF-κB/AML-1 elements resulted in decreased activity (constructs 1–5). MZF-1 is a transcription factor belonging to the Krüppel family of zinc finger proteins, and it possesses two DNA binding domains20, 21. The first domain binds to the core sequence 5'-AGTGGGGA-3', and the second domain binds the core sequence 5'-CGGGNGAGGGGGAA-3'. MZF-1 is expressed by totipotent hematopoietic cells as well as myeloid progenitors, and therefore may contribute to the preferential activity of the distal antisense promoter in stem cells. Constructs lacking the central MZF-1 site (constructs 4 and 12) did not demonstrate significant promoter activity, indicating a key role for MZF-1. Core promoter activity was localized to a 135 bp region containing a putative binding site for the MZF-1 protein at the 5’ end, a MZF-1 half-site in the middle, and potential C/EBP and MYC binding sites at the 3’ end (construct 9). Mutation of the MZF-1 site produced a 30% decrease in promoter activity (construct 10). Although deletion of the 3’ region containing the C/EBP and MYC sites significantly decreased promoter activity, a core construct with mutations in the 3’ C/EBP and MYC site did not decrease promoter activity (construct 11), indicating that MZF-1 is likely the major transcription factor driving promoter activity. The specificity of the promoter for pluripotent cell types was examined by transfection of construct 7 into additional cell lines, Ntera2, H9 ES, MCF7, PC3, YT, and Hela (Figure 2b). As expected, promoter activity was only detected in cell lines that were shown to express KIR distal antisense transcripts by RT-PCR.
Figure 2.
In vitro analysis of the KIR distal antisense promoter. (a) The left panel is a schematic representation of KIR2DL1 intron 2 fragments shown in an antisense orientation. Potential transcription factor binding sites are indicated by the labeled shaded rectangles. The central MZF-1 site is indicated by the bold arrow. Rectangles containing an “x” represent constructs wherein the respective site has been mutated. The position of the KIR2DL1 and KIR3DL1 distal antisense transcription start sites are indicated by vertical lines labeled with an asterisk. The MYC-binding site indicated at the 3’ end of the distal antisense promoter region overlaps with the exon 2/intron 2 junction on the sense strand. The boundaries of the first KIR2DL1 antisense intron are indicated by vertical lines labeled D (splice donor) and A (splice acceptor). The right panel shows the luciferase activity of pGL3 constructs containing the fragments depicted on the left. Constructs were transfected into HEK293 cells and relative luciferase activity was determined 48 hours post-transfection. Values represent the mean, and error bars indicate the standard deviation of at least 3 independent experiments. (b) Analysis of KIR2DL1 distal promoter activity (construct 7 in a) in various cell lines is shown. Values represent the mean and error bars indicate the standard deviation of at least 3 independent experiments.
EMSA analysis of the core MZF-1 site in YT and HEK293 cell extracts
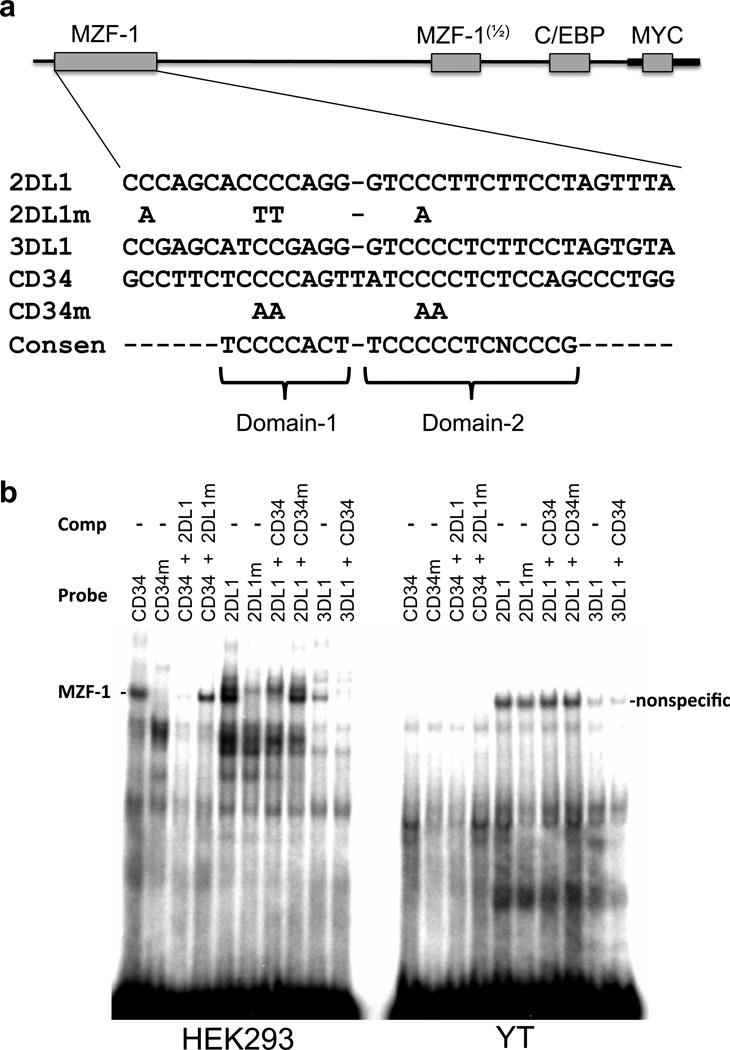
In order to investigate if the putative MZF-1 binding site could account for the tissue specificity of the distal antisense core promoter element, EMSA analysis of the putative MZF-1 site was performed with YT and HEK293 nuclear extracts. The previously characterized MZF-1 binding site from the CD34 promoter was used as a positive control22. Figure 3a shows the sequence of the oligonucleotide probes containing the predicted MZF-1 site in the KIR2DL1 and KIR3DL1 genes aligned with the functional MZF-1 site found in the human CD34 gene, as well as the consensus domain 1 and domain 2 sequences determined by oligonucleotide selection experiments21. Both of the MZF-1 binding domains contain central tracts of 4 cytidine residues, therefore mutated oligos contained disruptions of both domain 1 and domain 2 C-tracts. The KIR3DL1 element possesses several nucleotide differences that weaken the consensus domain-1 binding region, while KIR2DL1 has substitutions in the domain-2 consensus. Figure 3b shows that a distinct complex is formed in the HEK293 cell line with the CD34 and KIR probes, and this complex is absent from the YT cell nuclear extract. This complex was not generated when the mutated oligonucleotide probes were used. The higher intensity of the complex generated by the KIR2DL1 binding site may be due to the contiguous arrangement of the domain-1 and 2 consensus sequences, whereas there is an additional nucleotide separating these sites in the CD34 sequence. The reduced intensity of the complex formed by the KIR3DL1 probe is likely due to the significant disruption of the domain-1 binding consensus. Competition of the CD34 MZF-1 probe with unlabeled KIR2DL1 oligonucleotide resulted in the complete loss of the complex, whereas KIR2DL1 oligonucleotide with a mutated MZF-1 binding sequence had no affect. In addition, wild-type, but not mutated CD34 oligonucleotide inhibited KIR2DL1 and KIR3DL1 complex formation, demonstrating the ability of this region to bind MZF-1. The lack of a detectable complex in YT cells is consistent with this binding site contributing to the observed promoter activity in HEK293 but not YT cells. The band detected with the KIR2DL1 probes in YT cells likely represents the slightly higher molecular weight band that is also present in HEK293 cells. However, this band is not affected by disruption of the MZF-1 consensus in the mutant oligonucleotides, and it is not competed by wild-type MZF-1 probe, and thus represents a non-MZF-1 binding protein.
Figure 3.
EMSA analysis of the putative MZF-1 binding sites of KIR2DL1 and KIR3DL1. (a) A schematic of the core promoter fragment is shown, with the shaded rectangles indicating potential transcription factor binding sites. The expanded sequence underneath shows the oligonucleotide probes from the predicted KIR2DL1 and KIR3DL1 MZF-1 binding sites compared with the known MZF-1 binding site in the human CD34 gene. The consensus nucleotides for MZF-1 binding are shown below (Consen). Brackets indicate the sequences bound by the first and second DNA-binding domains of MZF-1. Nucleotide substitutions present in mutated KIR2DL1 (2DL1m) and CD34 (CD34m) probes are shown below the respective sequences. (b) EMSA analysis of HEK293 (left panel) and YT (right panel) nuclear extracts using the oligonucleotide probes shown in (a). Unlabeled oligonucleotides used for cold competition are indicated (Comp), as are the labeled probes (Probe). The position of a HEK293 cell-specific MZF-1 complex (MZF-1 -) is indicated next to the left panel, and a non-specific complex formed in YT cells is indicated next to the right panel (nonspecific).
Enforced expression of MZF-1 decreases KIR expression in developing NK cells
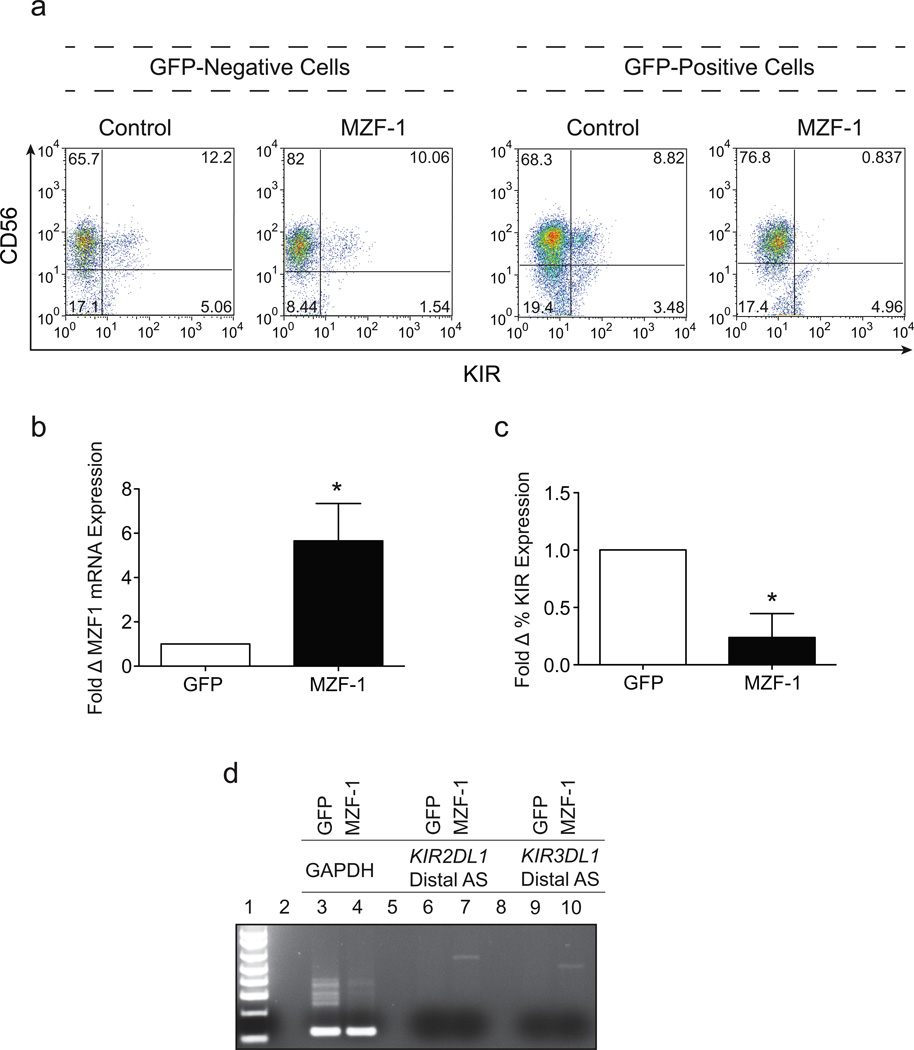
The complete overlap of the 3’ end of the distal antisense with the proximal antisense suggests that the distal transcript should also be capable of producing the 28 base piRNA, and mediate a similar KIR silencing activity. However, since distal antisense transcription is limited to very early progenitor cells, it might function to prevent premature opening of the KIR loci by distal forward transcript in stem cells. Previous studies have demonstrated that the KIR loci are heavily methylated at the NK progenitor stage, consistent with enforced silencing occurring in precursor populations23. Since in vitro analysis of the distal antisense promoter indicated a possible role for the MZF-1 transcription factor in progenitor cell-specific expression, we reasoned that enforced expression of MZF-1 would maintain distal antisense promoter activity in committed NK precursors and keep the KIR loci in their silent state. To address this hypothesis, we transduced primary CD34+ hematopoietic progenitor cells with a retroviral vector expressing MZF-1 and differentiated these cells into mature CD56+ NK cells in vitro. The expression of KIR on the surface of mature NK cells was analyzed by FACS after 21 days in culture. Over-expression of MZF-1 resulted in a 75% reduction in KIR expression compared with eGFP control cells or GFP negative cells present in the culture system. (Figure 4a). This effect was consistent and statistically significant in all donors tested (Figure 4c). Total RNA was harvested from eGFP control or MZF-1 over-expressing cells and used for RT-PCR with primer sets that specifically amplify either MZF-1 mRNA or distal antisense transcripts. As expected, MZF-1 mRNA levels were increased in cultures transduced with the MZF-1 expressing virus (Figure 4b). Distal antisense was detectable in MZF-1 containing cultures, but absent from control populations as previously observed (Figure 4d). The silencing effect of enforced MZF-1 expression is likely due to the continued presence of distal antisense transcripts in developing NK cells leading to the production of the 28 base piRNA and KIR gene silencing as previously shown for the proximal antisense transcript18.
Figure 4.
Enforced expression of MZF-1 decreases KIR expression in developing NK cells. (a) FACS analysis of day 21 in vitro differentiated cord blood NK cells transduced with either a control GFP expressing lentiviral vector or a lentiviral vector expressing GFP-IRES-MZF-1. Cells gated on the GFP−ve subset are shown on the left, and cells gated on the GFP+ve subset are shown on the right. Cells were stained with a cocktail of APC-conjugated NCAM16.2 (CD56) and PE-conjugated DX9 (KIR3DL1), EB6 (KIR2DL1/S1) and GL183 (KIR2DL2/S2/L3). (b) The average fold increase in MZF-1 mRNA expression in cells over-expressing MZF-1 relative to control GFP cells calculated from 3 independent donors. Differences in MZF-1 mRNA expression were statistically significant (p=0.0189) as determined by two-tailed Student’s t test. (c) The average fold reduction of KIR expression in cells over-expressing MZF-1 relative to control GFP cells calculated from 3 independent donors. Differences in KIR expression were statistically significant (p=0.0274) as determined by two-tailed Student’s t test. (d) PCR analysis of GAPDH (Lanes 3 & 4), KIR2DL1 distal antisense transcript (Lanes 6 & 7) and KIR3DL1 distal antisense transcript (Lanes 9 & 10) in sorted GFP and MZF-1 over-expressing day 21 in vitro-derived NK cells. Data is representative of 3 donors tested.
Discussion
Global transcriptome analysis has revealed that the majority (75%) of the human genome is transcribed into RNA, of which only ~1% codes for protein24. There is an abundance of long noncoding RNA, and it has been shown to function in many aspects of gene regulation such as genomic imprinting, gene silencing, and gene activation25. The novel KIR antisense transcript identified in this study belongs to the gene body-associated class of lncRNA. A recent study of lncRNA expressed by human embryonic stem cells (hESCs) revealed that the majority of lncRNA (89%) are associated with the promoters, enhancers, and bodies of protein-coding genes26. Furthermore, 5% of the lncRNAs were antisense transcripts originating within protein-coding genes like the KIR distal transcript identified in this study. The expression of the KIR distal antisense lncRNA by hESC and other cell types with stem cell properties, but not mature differentiated cell types, suggests a role in the initial silencing of KIR loci. In support of this hypothesis, differentiation of hESC into CD34-expressing cells resulted in a loss of distal antisense transcription. It is of interest to note that the MZF-1 transcription factor-binding site implicated in the function of the KIR distal antisense promoter is also present in the CD34 promoter. This suggests that additional factors contribute to the preferential expression of distal antisense in stem cells. MZF-1 is a bi-functional transcriptional regulator capable of activating transcription in cells of hematopoietic origins, while repressing transcription in other cell types20.
The distal antisense transcript contains the 28 base element previously shown to mediate silencing initiated by the proximal antisense, predicting a similar silencing activity for the distal transcript. Enforced expression of MZF-1 during in vitro NK cell differentiation resulted in detectable levels of distal antisense and reduced levels of KIR expression consistent with a gene silencing activity of the distal transcript. Therefore, the distal antisense may be generated in progenitor cells to ensure that the locus is silenced, ensuring that KIR are not expressed in other cell lineages during development, whereas the proximal antisense controls the probabilistic expression of KIR in mature NK. The presumed transient role of the KIR lncRNA in hESC may not be a general phenomenon of gene body-associated antisense lncRNAs, since differentiation of hESC into endoderm did not result in a decrease in the number of gene body-associated antisense lncRNAs26. The KIR gene family may represent a special case where it is important that the KIR genes are completely silenced in NK progenitors so that they do not negatively impact the process of NK cell differentiation. It is also of interest to note that the distinct unspliced antisense transcript derived from a promoter in intron 1 of the KIR3DP1 pseudogene that was detected in Daudi and Jurkat cells (Table 1), was also found in NK cells of the intermediate CD56-bright phenotype (data not shown). CD56-bright NK cells represent the immediate precursor of mature CD56-dim NK cells that express KIR. This KIR3DP1 antisense lncRNA was highly expressed in CD56-bright NK, and undetectable in mature NK cells. Perhaps KIR3DP1 antisense transcripts are generated at this later point in development to ensure that the pseudogene remains in a silent state.
In summary, we have shown that KIR antisense lncRNAs derived from a promoter in intron 2 are only expressed at the earliest stages of cell differentiation, and represent a distinct class of lncRNA capable of silencing genes in stem cells.
Materials and methods
Cell lines
H9 hESCs were obtained from WiCell (Madison, WI). Ntera2 cells were kindly provided by Dr. Peter Andrews (University of Sheffield, UK). HEK293T, Hela, and Ntera2 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin (P/S), sodium pyruvate, and L-glutamine. MCF7 cells were cultured in DMEM as above with the addition of 0.01 mg/ml human recombinant insulin. YT, Daudi, Jurkat, PC3, and U937 cells were cultured in RPMI 1640 media containing 10% FBS, P/S, and L-glutamine.
NK Cell and CD34+ HPC Isolation
The use of all human tissue was approved by the Committee on the Use of Human Subjects in Research at the University of Minnesota, and informed consent was secured in accordance with the Declaration of Helsinki. Umbilical cord blood was obtained from full-term consenting mothers at Memorial Blood Bank (Minneapolis, MN, USA), Placental Blood Program of the New York Blood Center (New York, NY, USA), Saint Louis Cord Blood Bank (Saint Louis, MO, USA), or local obstetric units. Total mononuclear cells were separated from the peripheral blood of healthy donors by Histopaque® (Sigma-Aldrich, Saint Louis, MO, USA) gradient centrifugation. CD56+ NK cells were column-isolated by negative selection using magnetic beads (Miltenyi Biotech, Auburn, CA, USA). CD34+ HPCs were isolate from umbilical cord blood donors by Histopaque® (Sigma-Aldrich) gradient centrifugation followed by a double-column positive selection using anti-CD34 microbeads (Miltenyi).
CD34+ HPC viral transduction and in vitro NK cell differentiation
The Uracil-Specific Excision Reagent (USER) cloning protocol (New England Biolabs, Beverly, MA, USA) was used to create the MZF-1-eGFP pELNS vector. The following primers were used for PCR amplification of MZF-1 from CD34+ hematopoietic progenitor cells: Forward 5’-ggagacaucaccatgaggcctgcggtgctgg-3’ and reverse 5’-gggaaaguctactcggcgctgtggacgc. High-titer lentivirus was used to transduce freshly isolated cord blood CD34+ hematopoietic progenitor cells, which were subsequently sorted based on eGFP expression. Sorted cells were cultured for 21 days on the EL-08-1D2 stromal line27. The culture medium and supplemented cytokines for NK cell differentiation were as previously described16.
RT-PCR of antisense KIR transcripts
Total RNA was purified from 1×107 cells with the RNeasy kit (Qiagen, Valencia, CA, USA). The SuperScript First Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) was used to generate cDNA from 0.5µg of total cellular RNA. For PCR reactions, 35 cycles of 94°−20s, 60° −20s, 73°−20s was performed on 2µl of cDNA using Hot-Start Taq Mastermix (Denville, South Plainfield, NJ, USA). For KIR2DL1-derived transcripts, the forward primer was: 5'-AGAACTTGACTCTGCCAAGGGAATG-3’. For KIR3DL1 antisense, the forward primer was: 5'-ATCCCCCGACAGGACTTCCCTC-3’. A common reverse primer was used for both genes: 5’-TGAGTTGGTCATAGTGAAGGAC-3’. All PCR products were cloned into the pCR2.1-topo vector (Invitrogen) and sequenced in order to verify the identity of the transcript and the KIR gene responsible for the product.
5’ RACE (Rapid Amplification of cDNA ends)
5’ RACE was performed on 10 ug of total HEK293T RNA according to manufacturer’s protocol using FirstChoice® RLM-RACE Kit (Ambion Inc). cDNA synthesis was performed using primers: 5’-CTTTCACGTTAGCACAG-3’ for KIR2DL1 and 5’-AAGGGCCTGGCTGCCAAGACGCAC-3’ for KIR3DL1. The cDNA amplification was performed using a 2DL1 gene-specific outer primer: 5’-AAGGGCCTGGCTACCAAGACTCAC-3’ and inner primer: 5’-CATCCATCATGATCTTTCTTTCC-3’. The outer and inner primers used for KIR3DL1 amplification: are 5’-TGCCCTGGTTTGCCTGCAGATG-3’ and 5’-GGTCCATCATGATCTTTCTTTCTAG-3’. Linker primers were used as provided by the kit.
Generation of luciferase reporter constructs
Promoter fragments were generated by PCR using oligonucleotide primers and cloned into the pCR2.1- Topo vector (Invitrogen). Inserts were excised with SacI/XhoI enzymes and cloned into the corresponding sites in the pGL3-basic firefly luciferase reporter vector (Promega, Madison, WI, USA).
Cell transfection and luciferase assays
YT cells were transfected by electroporation using a BTX ECM 830 (Genetronics, San Diego, CA, USA). YT transfections were conducted using 5×106 cells in serum free RPMI medium with 10 µg of pGL3 constructs plus 50 ng of Renilla luciferase pRLSV40 vector and electroporated at 250V, with three pulses of 7ms at an interval of 100 ms. HEK293T and Ntera2 cells were transfected with 1 µg of pGL3 constructs plus 1 ng of Renilla luciferase pRLSV40 using FuGene 6 (Roche Molecular Biochemicals, Indianapolis, IN, USA) following the manufacturer’s protocol. Luciferase activity was assayed at 48 h (YT, HEK293 T and Ntera-2) using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized relative to the Renilla luciferase activity for each transfection.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from HEK293T and YT cells using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich). Protein concentration was measured with a Bio-Rad protein assay (Hercules, CA), and samples were stored at −70°C until use. DNA oligonucleotide probes corresponding to the predicted MZF-1 binding sequences of the KIR2DL1 and KIR3DL1 core antisense promoters and the previously characterized CD34 MZF-1 site were synthesized. Probes were labeled with [α-32P]deoxycytidine triphosphate (3000 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) by fill-in using the Klenow fragment of DNA polymerase I (Invitrogen). 32P-labeled double-stranded oligonucleotides were purified using mini Quick Spin Oligo Columns (Roche Diagnostics, Mannheim, Germany). DNA-protein binding reactions were performed in a 10-µL mixture containing 5 µg nuclear protein and 1 µg poly(dI-dC) (Sigma-Aldrich) in 4% glycerol, 1 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5). Nuclear extracts were incubated with 1 µL 32P-labeled oligonucleotide probe (10,000 cpm) at room temperature for 20 minutes and then loaded on a 5% polyacrylamide gel (37:5:1). Electrophoresis was performed in 0.5× TBE for 2 hours at 130 V, and the gel was visualized by autoradiography.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute (NCI), NIH, under contract HHSN261200800001E and NCI grant P01 111412 (JSM, SKA). This research was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Carrington M, Norman P. The KIR Gene Cluster. Bethesda, MD: National Library of Medicine (US), NCBI; 2001. [Google Scholar]
- 4.Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 5.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 6.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 7.Held W, Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur J Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 8.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 9.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 11.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, et al. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanamachi DM, Moniot DC, Cado D, Liu SD, Hsia JK, Raulet DH. Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J Immunol. 2004;172:1074–1082. doi: 10.4049/jimmunol.172.2.1074. [DOI] [PubMed] [Google Scholar]
- 15.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 16.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, et al. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cichocki F, Miller JS, Anderson SK. Killer immunoglobulin-like receptor transcriptional regulation: a fascinating dance of multiple promoters. J Innate Immun. 2011;3:242–248. doi: 10.1159/000323929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bamezai S, Rawat VPS, Buske C. Concise Review: The Piwi-piRNA axis: pivotal beyond transposon silencing. Stem Cells. 2012;30:2603–2611. doi: 10.1002/stem.1237. [DOI] [PubMed] [Google Scholar]
- 20.Hromas R, Davis B, Rauscher FJ, 3rd, Klemsz M, Tenen D, Hoffman S, et al. Hematopoietic transcriptional regulation by the myeloid zinc finger gene, MZF-1. Curr Top Microbiol Immunol. 1996;211:159–164. doi: 10.1007/978-3-642-85232-9_16. [DOI] [PubMed] [Google Scholar]
- 21.Morris JF, Hromas R, Rauscher FJ., 3rd Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol Cell Biol. 1994;14:1786–1795. doi: 10.1128/mcb.14.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris JF, Rauscher FJ, 3rd, Davis B, Klemsz M, Xu D, Tenen D, et al. The myeloid zinc finger gene, MZF-1, regulates the CD34 promoter in vitro. Blood. 1995;86:3640–3647. [PubMed] [Google Scholar]
- 23.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer immunoglobulin-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 24.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 26.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, et al. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]