Abstract
Hydroxyapatite (HA) biomaterials and allograft bone are common alternatives to autogenous grafts, however these materials lack the strong osteoinductive potential of autologous bone. Previous studies have established that polyglutamate domains, which bind selectively to HA, can be engineered onto bioactive peptides as a mechanism for coupling osteoinductive signals onto HA and allograft. In the current investigation, we adapted the polyglutamate approach to tailor delivery of a model collagen-derived peptide, DGEA, by manipulating the number of glutamates in the HA binding domain. Specifically, DGEA was modified with diglutamate (E2-DGEA), tetraglutamate (E4-DGEA) or heptaglutamate (E7-DGEA), and it was found that initial peptide binding to HA and allograft was significantly enhanced as the number of glutamates increased. We also determined that the rate of release of polyglutamate-DGEA from substrates over a 5-day interval increased proportionally as the number of glutamate residues was decreased. Additionally, we tuned the peptide release rate by creating mixtures of E2-DGEA, E4-DGEA and E7-DGEA, and observed that release kinetics of the mixtures were distinct from pure solutions of each respective peptide. These collective results suggest that variable length polyglutamate domains provide an effective mechanism for controlled delivery of osteoregenerative peptides on HA-containing bone graft materials.
Keywords: Hydroxyapatite, allograft bone, polyglutamate domain, bioactive peptide, tunable peptide delivery
INTRODUCTION
Hydroxyapatite (HA) is one of the most commonly used biomaterials for bone regenerative therapies. HA is utilized in a variety of clinical treatments; for example, HA serves as a bone filler or component in bone cements, and is also applied as a coating on metal implants and screws to improve osteointegration 1. Additionally, HA is incorporated into degradable bone regenerative matrices, including electrospun scaffolds 2–5 and hydrogels 6–8, to serve as a bone mimetic molecule. HA is a calcium phosphate crystal found within native bone, and although it is highly osteoconductive, this material lacks osteoinductivity. Thus, many studies have been aimed at modifying HA surfaces to present osteoregenerative factors. In most investigations passive adsorption of biomodifiers onto HA was employed 9–12, however this approach is inefficient and offers little control over the dose and kinetics of delivery. More complex methods for coupling biologics to HA include silanizing the HA surface for covalent attachment of molecules 13–15, or chemically linking biologics to HA-binding bisphosphonate groups 16. While these strategies achieve effective coupling, silanization may destroy the osteoconductive surface of HA, whereas the use of bisphosphonates introduces a non-needed pharmacologically active agent to the implant site.
As an alternative method to couple regenerative molecules to HA, we and others have utilized acidic amino acid peptide domains that are known to selectively bind HA 17–25. Specifically, our group has engineered bioactive peptides with a seven amino acid glutamate sequence (E7) in order to improve peptide coupling to both synthetic HA 18,25,26 and allograft bone (via the biologic HA within bone) 17. Peptides modified with an E7 domain were loaded onto the HA surface in significantly greater quantities, and were retained for extended intervals. For instance, about 90% of E7-modified peptides bound to allograft were retained at 5 days following initial loading, while 50% of the unmodified peptides were lost within an hour 17. In prior studies polyacidic amino acid domains were used primarily to enhance anchoring of cell adhesive peptides, such as RGD 19,20,22,25 or the collagen-derived peptide DGEA 17,18,20. Given that RGD and DGEA peptides serve as surrogates for adhesion proteins normally embedded in the extracellular matrix, tight tethering to the HA surface is advantageous. However, the bioactivity of some types of biomodifiers, including chemoattractants and certain growth factors, depends on release from the HA surface in order to generate soluble gradients or influence the behavior of cells more distal to the implant. As an example, controlled release, with appropriate temporal kinetics, is important for the angiogenesis-inducing activities of VEGF and PDGF 27,28.
In the current study we hypothesized that by modifying the length of the polyglutamate domain, we could control the release of peptides from HA. To test this concept, the DGEA peptide was synthesized with variable length polyglutamate domains including: diglutamate “E2”, tetraglutamate “E4”, and heptaglutamate “E7”, and the effects of glutamate number on peptide loading and release were determined. DGEA was used as a model peptide due to its simple structure; however the fundamental strategy described is applicable for delivery of a variety of osteoregenerative molecules from HA. Moreover, results show that domains with variable glutamate number can be similarly used to control peptide binding and release on commercially available allograft. The use of acidic amino acid domains to facilitate delivery of bioactive molecules offers advantages when compared with more complex functionalization strategies because this system provides a tunable delivery mechanism, doesn’t destroy the osteoconductive surface of HA, and the polyglutamate-modified peptides are straightforward and relatively inexpensive to produce.
MATERIALS AND METHODS
Materials
Peptides: The following custom peptides were obtained from American Peptide Co., Inc. (Sunnyvale, California): the collagen-derived sequence DGEA, tagged with FITC (DGEA-K-FITC, 907.9 g/mol); polyglutamate-modified DGEA peptides E2-DGEA-FITC (EEDGEA-K-FITC, 1166.2 g/mol), E4-DGEA-FITC (EEEE-DGEA-K-FITC, 1424.4 g/mol), and E7-DGEA-FITC (EEEEEEE-DGEA-K-FITC, 1811.7 g/mol). All peptides were reconstituted in ddH20 to 1 mg/mL, aliquoted and stored at −20°C. HA disks: HA powder (MP Biomedicals, Solon, Ohio) was pressed into disks using a 15.875 mm die under 3000psi. All disks were sintered at 1000°C in a Thermolyne 48000 series furnace and allowed to cool in the furnace at decreasing intervals before storage under sterile conditions. Allograft: Commercial-grade, mineralized cortical particulate bone graft (OraGRAFT®) was obtained from LifeNet Health (Virginia Beach, Virginia) and stored at room temperature according to manufacturer’s instructions.
Imaging of peptide loading and retention
1 or 10µM peptide concentrations of FITC tagged DGEA, E2-DGEA, E4-DGEA, and E7-DGEA were coated onto HA disks or 25 mg of particulate allograft bone for 2 hours. Following coating, supernatants were aspirated completely and surfaces briefly washed 3 times for 1 minute with agitation (replacing Tris-buffered saline (TBS) with each wash). Relative peptide loading was qualitatively evaluated by fluorescent imaging using a Leica MZ16F fluorescent microscope with 1.0x objective. To evaluate peptide retention, HA disks and allograft bone were subjected to extensive wash steps and reimaged at 3 and 5 days using fluorescent microscopy. Of note, for HA disks all 4 samples were imaged in the same field; however because the allograft samples would not fit within a single field, overlapping images were taken at the same exposure and stitched together.
Peptide loading on HA disks and allograft bone
For all experiments, equimolar solutions of peptide were confirmed to have the same fluorescence by measuring values on a Biorad VersaFluor fluorometer. 0.1 µM solutions of each peptide (FITC-tagged DGEA, E2-DGEA, E4-DGEA, and E7-DGEA) were prepared, and the starting fluorescence of these solutions was determined by measuring fluorescence of 300 µL of coating solution in Biorad VersaFluor microcuvettes. Each solution was coated onto allograft bone (25mg) or HA disks. Depletion of solution fluorescence, measured by a decrease in fluorometry values, was evaluated at 30 minutes, 2 hours, or 6 hours during incubation with the coating solution. The values for residual fluorescence at these times points (unbound peptide) were subtracted from initial fluorescence values in order to obtain a quantitative measure of peptide binding to the substrate. Values were plotted as the percent of peptide bound (as compared with the starting solution).
Peptide release from HA disks and allograft bone
FITC-tagged peptides were coated onto HA or allograft for 2 hours at room temperature, and then values were calculated for the amount of peptide bound, as described above. The coating solutions were then aspirated completely and samples were subsequently washed briefly and placed in 1mL of either fresh TBS or DMEM media containing 10% fetal bovine serum. Samples were incubated over varying time intervals with continuous agitation. At 30 minutes, 2 hrs, 1 day and 5 days, the TBS and media solutions were monitored for the appearance of solution fluorescence, representing the release of FITC-tagged peptides from the substrates. The amount of peptide released, measured by fluorometry, was divided by the total amount initially bound to calculate a percentage of peptide released. These values were summed and plotted as the cumulative percent (%) of bound peptide released.
Loading and release of peptide mixtures from synthetic HA and allograft bone
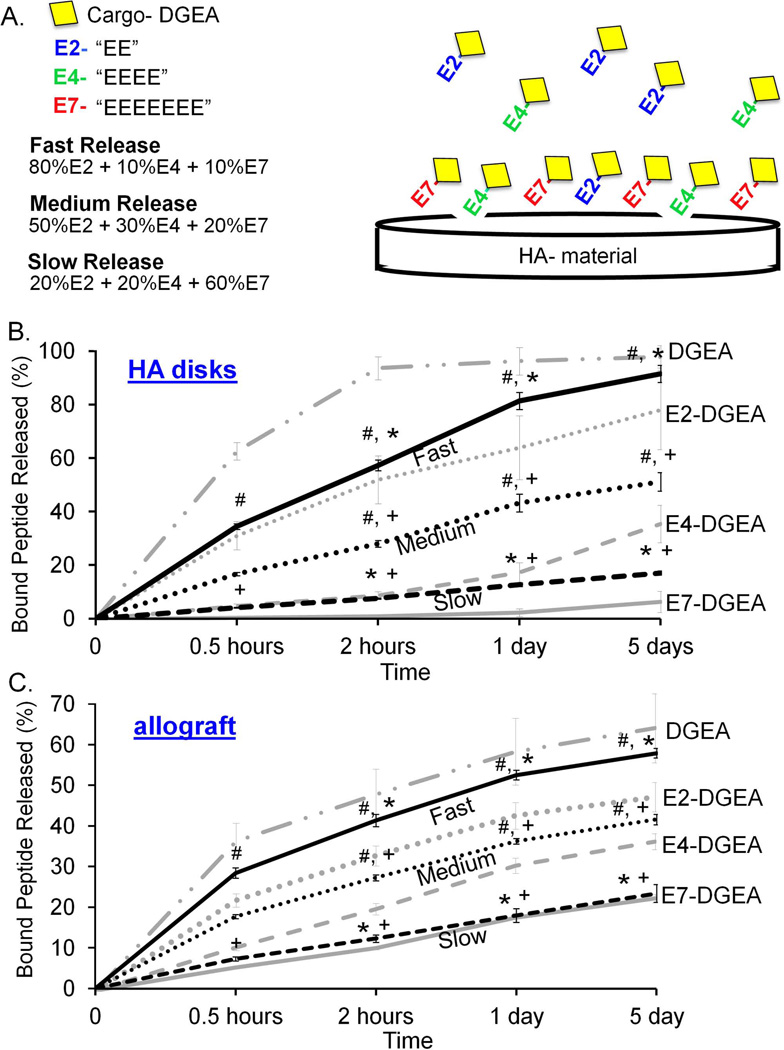
Peptide mixtures were prepared to the following specifications: (1) fast release [80% E2-DGEA, 10% E4-DGEA, and 10% E7-DGEA]; (2) medium release [50% E2-DGEA, 25% E4-DGEA, and 25% E7-DGEA]; and (3) slow release [10% E2-DGEA, 10% E4-DGEA, and 80% E7-DGEA]. The final total peptide concentration for each mixture was 0.1 µM. 1mL of each solution mixture was used to coat HA disks or 25mg of allograft bone. After measuring initial peptide binding, the coating solution was completely aspirated and replaced with fresh TBS. Samples were subjected to agitation and peptide release into the TBS solution was measured by fluorometry at 30 minutes, 2 hours, 1 day, and 5 days (as previously described). Cumulative release was graphed as a percent of peptide initially loaded, and in FIG 6, release profiles of 100% DGEA, E2-DGEA, E4-DGEA, and E7-DGEA are shown (in light gray) alongside the mixtures to demonstrate the distinct release characteristics of peptide mixtures.
Figure 6.
Release kinetics of peptide mixtures from synthetic HA and allograft bone. (A) Peptide mixtures were generated to facilitate “fast” release= 80% E2-DGEA + 10% E4-DGEA +10% E7-DGEA, “medium” release= 50% E2-DGEA + 30% E4-DGEA + 20% E7-DGEA, and “slow” release= 20% E2-DGEA + 20% E4-DGEA + 60% E7-DGEA. These mixtures were coated onto synthetic HA disks (B) or allograft bone (C) for 2 hours, and the amount of peptide binding was quantified. Coating solutions were then aspirated and replaced with fresh TBS buffer. Release of bound peptide into TBS was monitored by measuring solution fluorescence. Data are plotted as the percent of bound peptide released. To show that the peptide mixtures exhibited distinct release kinetics as compared with pure solutions, the release profiles for 100% DGEA, E2-DGEA, E4-DGEA, and E7-DGEA are included on the graphs (light gray coloring). Significant difference (p<0.05) between fast, medium and slow samples was denoted as follows: slow=#, medium=*, and fast=+.
Statistics
At least 3 independent experiments were performed for the quantitative peptide binding and retention assays, with each experiment performed in triplicate. All of the imaging studies were performed three independent times, and at least 2 distinct samples were evaluated per experiment. Data sets were assessed using one-way analysis of variance ANOVA. A confidence level of at least 95% (p < 0.05) was considered significant.
RESULTS
Binding of DGEA with variable-length polyglutamate domains to HA
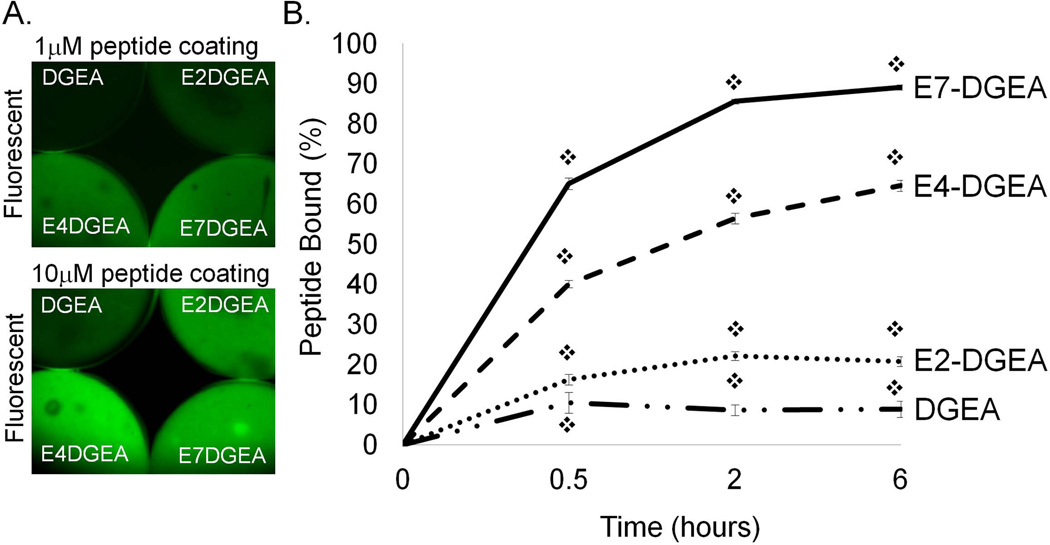
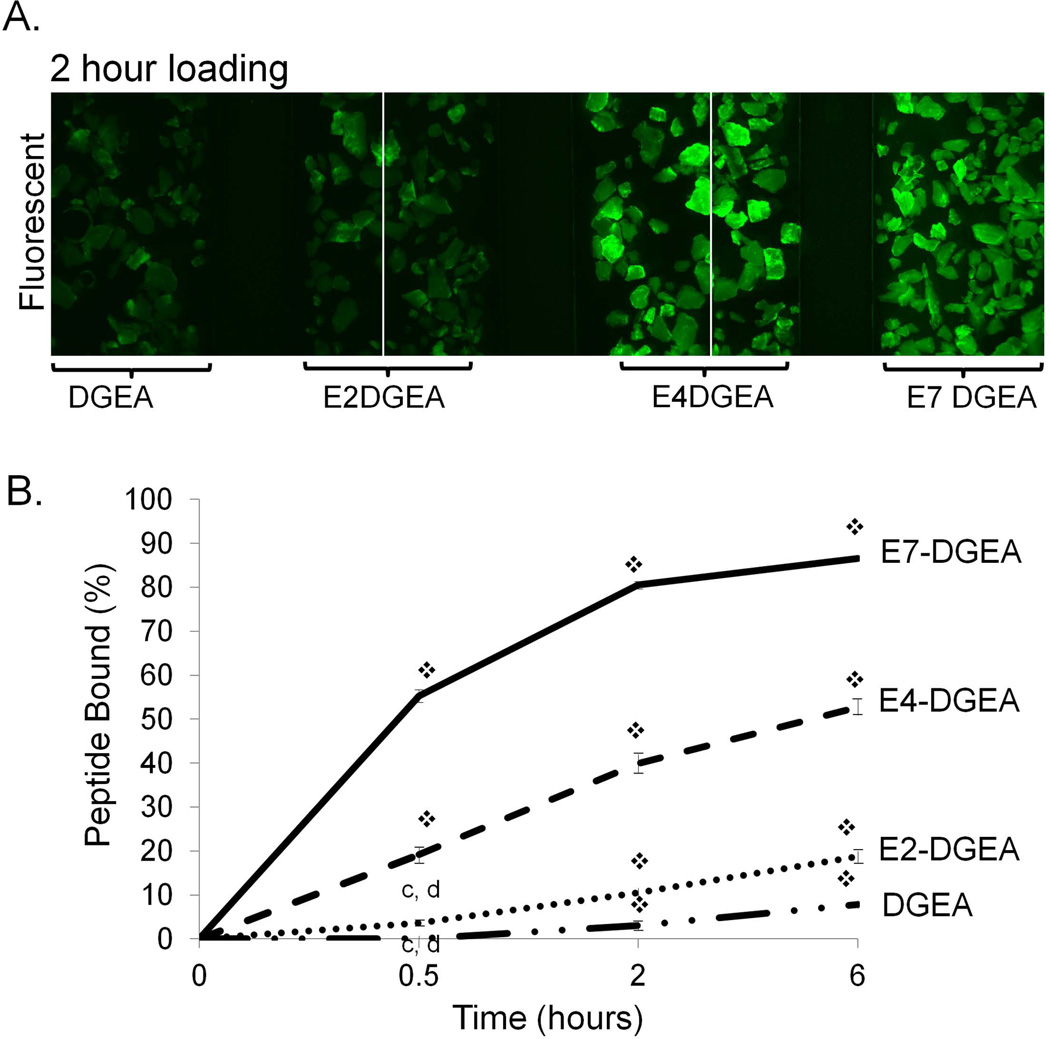
To evaluate the effect of polyglutamate domain length on peptide loading we prepared solutions of DGEA, E2-DGEA, E4-DGEA, and E7-DGEA (conjugated with FITC tags), and then coated equimolar concentrations of each peptide onto synthetic HA disks. Following 2 hours of coating, the coating solution was aspirated and disks were rinsed briefly and imaged (FIG 1A). We found that for two different peptide coating concentrations, peptide loading onto the HA surface appeared to increase as additional glutamates were incorporated. As a more quantitative measure of peptide loading, HA disks were coated with equimolar concentrations of peptide and the coating solution was monitored for depletion of fluorescence over time by fluorometry (with loss of solution fluorescence corresponding to peptide deposition on the HA surface). By subtracting the residual solution fluorescence (unbound peptide) from the fluorescence values obtained from the initial coating solution, we calculated the percent of peptide bound to HA disks (FIG 1B). At every time point there was a significant increase in the amount of peptide bound from solution as the number of glutamates was extended (FIG 1B).
Figure 1.
Binding of peptides conjugated with variable length polyglutamate domains to synthetic HA. (A) FITC-conjugated DGEA, E2-DGEA, E4-DGEA, or E7-DGEA were coated onto HA disks at 1 or 10 µM and monitored for relative peptide binding by fluorescent microscopy. (B) FITC-conjugated peptides were coated at 0.1 µM onto synthetic HA disks for up to 6 hours. Time course for peptide binding was evaluated by measuring depletion of fluorescence from the coating solution over time. Residual solution fluorescence was subtracted from the starting fluorescence to calculate the percent of peptide bound.
Release of DGEA with variable-length polyglutamate domains from HA
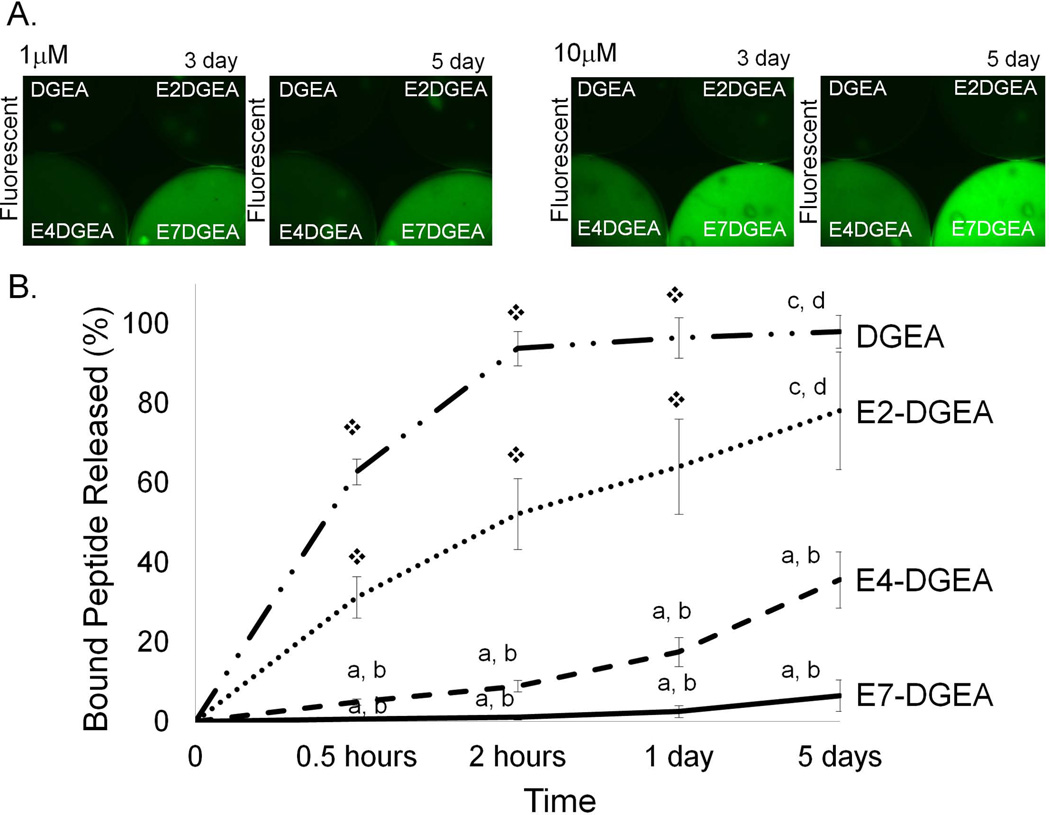
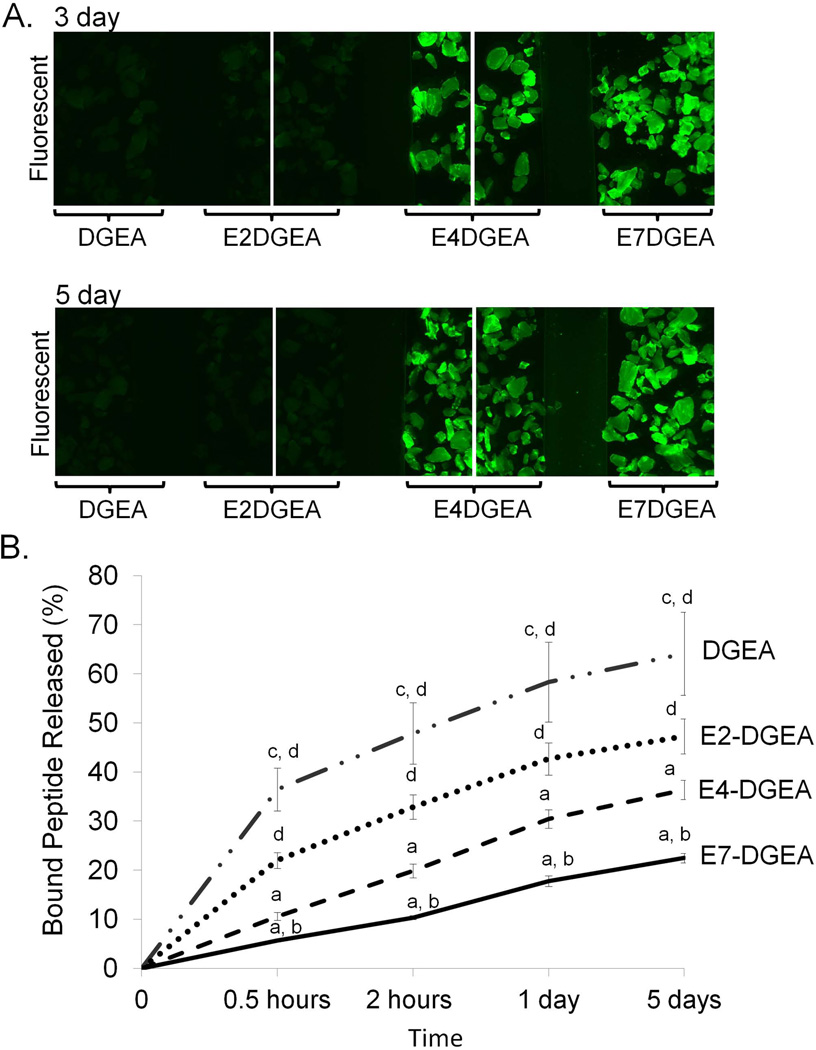
We hypothesized that shorter polyglutamate domains would result in faster release of peptide from the HA surface. HA disks were coated with DGEA, E2-DGEA, E4-DGEA, or E7-DGEA for 2 hours and then disks were washed briefly and placed in fresh TBS with agitation for up to 5 days. After 3 and 5 days of extensive wash steps, fluorescent imaging of the disks indicated that the number of glutamates in the binding domain correlated with the degree of peptide retention on HA (FIG 2A). To quantitatively measure release of polyglutamate-modified peptides, we coated HA disks with equimolar concentrations of DGEA, E2-DGEA, E4-DGEA, or E7-DGEA for 2 hours and calculated initial peptide loading. The coating solution was then aspirated and replaced with fresh TBS, and samples were incubated in TBS with agitation for up to 5 days. Fluorometry was used to monitor the appearance of fluorescence in solution, representing peptide release from the HA surface (note that all values are plotted as the percent of initial peptide loaded). We found a direct correspondence between the number of glutamate residues in the HA binding domains and the rate of peptide release (FIG 2B).
Figure 2.
Release of peptides conjugated with variable length polyglutamate domains from HA. Peptides were loaded onto HA disks for 2 hours, and then samples were washed briefly and placed in fresh TBS for up to 5 days with agitation. (A) HA disks were evaluated by fluorescent microscopy at 3 and 5 days after loading to visualize the amount of peptide retained on disks. (B) Peptide release from disks was quantified by monitoring TBS solutions for the appearance of fluorescence using fluorometry. Data are plotted as the percent of peptide released relative to the amount initially bound. Significant difference (p<0.05) between samples was denoted as follows: relative to DGEA=a, E2-DGEA=b, E4-DGEA=c, E7-DGEA=d, and to all = ❖.
Release of DGEA with variable-length polyglutamate domains from HA in serum-containing media
Protein and lipid-rich extracellular fluids in the implant microenvironment can potentially influence peptide release from biomaterials. Thus, as a more stringent measure of polyglutamate-directed coupling, peptide release was examined in the presence of serum-containing media rather than TBS. Peptides were coated onto HA as before, and as previously observed, the amount of bound peptide correlated with the length of the polyglutamate domain (FIG 3A). Following peptide loading, samples were washed briefly, placed in DMEM containing 10% FBS and incubated for varying time intervals up to 5 days with agitation. At the end of the 5-day interval, disks were imaged, and it was clear that a greater amount of E7-DGEA peptide was present on the disks relative to all other peptides (FIG 3B). Quantitative fluorometric analyses revealed that the rate of peptide release corresponded with the length of the polyglutamate domain (FIG 3C), although greater release of E4 and E7-DGEA was observed in serum-containing media relative to release in TBS. Nonetheless, results in FIG 3 show that even in the presence of a solution containing proteins and lipids, the kinetics of peptide release can be controlled by varying the length of the polyglutamate domain. Furthermore, it is anticipated that if greater peptide retention were needed, this could be achieved by simply increasing the number of glutamate residues beyond seven.
Figure 3.
Release of peptides conjugated with variable length polyglutamate domains from HA in serum-containing media. (A) Peptides were coated onto HA disks as described previously, and the amount bound was calculated by measuring depletion of fluorescence from solution. (B) Following peptide coating, samples were washed briefly and placed in DMEM containing 10% fetal bovine serum for varying intervals up to 5 days with agitation. At the end of the 5-day interval, HA disks were imaged in order to visualize the amount of peptide remaining on the disks. (C) Peptide release from disks over the 5-day interval was quantified by monitoring the media solutions for the appearance of fluorescence using fluorometry. Data are plotted as the percent of peptide released relative to the amount initially bound. Significant difference (p<0.05) between samples was denoted as follows: relative to DGEA=a, E2-DGEA=b, E4-DGEA=c, E7-DGEA=d, and to all = ❖.
Binding of DGEA with variable-length polyglutamate domains to allograft bone
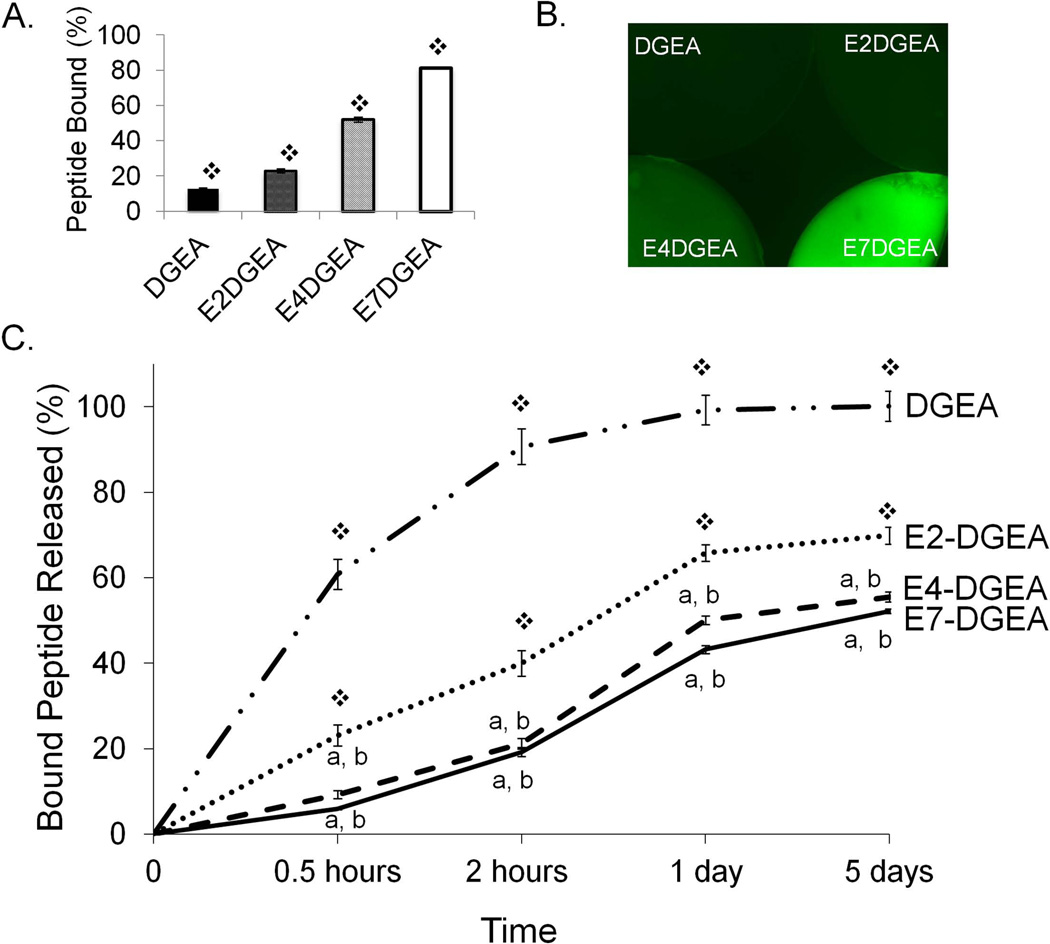
Given the importance of allograft materials in clinical treatments for bone repair, we evaluated whether the variable-length polyglutamate approach could be adapted to tailor peptide delivery on allograft via anchoring to the biologic HA within bone. To this end, we coated allograft bone with equimolar peptide solutions containing DGEA modified with E2, E4, or E7, and imaged for relative peptide loading. We found that, as with synthetic HA, greater peptide binding to allograft was observed as the number of glutamate residues was increased (FIG 4A). Additionally, we quantitatively evaluated differences in peptide loading over a period of 6 hours by monitoring depletion of fluorescence from peptide coating solutions (FIG 4B). We found that the percent of peptide loading increased with more glutamate residues present in the binding domain (E7>E4>E2).
Figure 4.
Binding of peptides conjugated with variable length polyglutamate domains to allograft bone. DGEA, E2-DGEA, E4-DGEA and E7-DGEA peptides (conjugated with FITC) were coated onto 25 mg of allograft bone chips for up to 6 hours. (A) Following 2 hours of loading, allograft was imaged for relative peptide density using fluorescent microscopy. (B) Peptide coating solutions were monitored for depletion of fluorescence and percent of peptide bound to allograft was calculated. Significant difference (p<0.05) between samples was denoted as follows: DGEA=a, E2-DGEA=b, E4-DGEA=c, E7-DGEA=d, and to all = ❖.
Release of DGEA with variable-length polyglutamate domains from allograft bone
To evaluate the effect of the number of glutamate residues on the release of DGEA, allograft substrates were coated with equimolar concentrations of DGEA, E2-DGEA, E4-DGEA, and E7-DGEA, and then the coating solution was replaced with TBS and the samples incubated with agitation for up to 5 days. Fluorescent imaging revealed that E4 and E7-modified DGEA peptides were retained on allograft bone for at least 5 days (FIG 5A). Similarly, quantitative assays of solution fluorescence revealed that the degree of peptide release was directly correlated with the length of the polyglutamate domain (FIG 5B). Thus, variable length polyglutamate domains can be used to control peptide delivery from commercial allograft, as with synthetic HA.
Figure 5.
Release of peptides conjugated with variable length polyglutamate domains from allograft. Following a 2-hour coating with equimolar peptide concentrations, coating solutions were completely aspirated and samples were placed in fresh TBS with agitation for up to 5 days. (A) Fluorescent imaging was used to evaluate relative peptide retention densities at 3 and 5 days. (B) Peptide release was quantified by monitoring TBS solutions for the appearance of fluorescence using fluorometry. Data are plotted as the percent of bound peptide released. Significant difference (p<0.05) between samples was denoted as follows: DGEA=a, E2-DGEA=b, E4-DGEA=c, E7-DGEA=d, and to all = ❖.
Release characteristics of peptide mixtures with variable-length polyglutamate domains
Results shown in FIGs 1–5 confirmed that the length of the polyglutamate domain regulates both the binding and release of peptide from HA and allograft bone. We next hypothesized that we could utilize mixtures with varying lengths of polyglutamate domains to achieve finer tuning of release characteristics. To address this goal we generated 3 distinct mixtures of E2-, E4-, and E7-modified DGEA weighted for fast, medium, or slow release (FIG 6A). The percentages of each peptide were established based on binding characteristics; for example, fast release was highly weighted with E2-DGEA (80%) in combination with 10% E4-DGEA and 10% E7-DGEA. At the other end of the spectrum, the slow release mixture was composed of 20% E2-, 20% E4-, and 60% E7-DGEA, whereas the medium release solution was comprised of 50% E2-/30% E4-/20% E7-DGEA. These mixtures were coated onto synthetic HA (FIG 6B) or allograft bone (FIG 6C) for 2 hours. Coating solutions were then aspirated and replaced with fresh TBS, and peptide release into solution was subsequently measured by fluorometry as before. We found that over a 5 day-interval, peptide mixtures weighted for fast, medium, or slow release exhibited release kinetics that were distinct from solutions composed of 100% E2-, E4- or E7-DGEA (shown in graphs as light gray curves, FIG 6B&C). These results highlight the potential utility of combining peptides with variable length polyglutamate domains to tailor the amount and timing of peptide release from synthetic HA biomaterials and allograft bone.
DISCUSSION
Synthetic HA and allograft are widely used as alternatives for autografted bone, however these materials lack the strong osteoinductive capacity of autologous bone. Accordingly, reconstitution of HA and allograft with osteogenic proteins or peptides holds promise for improving clinical treatments utilizing these materials. The current dearth of efficient mechanisms to deliver osteoinductive signals from synthetic HA and allograft is the driving force behind the present study. HA offers limited functional groups for chemical coupling, therefore most studies have relied on passive adsorption of osteogenic molecules onto the HA surface 29. Because HA is highly adsorptive, passive adsorption of biologics has some efficacy, however this approach provides little control over the dosage or temporal kinetics of peptide/protein delivery, and achieving sustained delivery is a particular challenge. As an alternative method for delivery from HA-containing materials, we and others have investigated the use of HA-binding polyacidic amino acid domains to couple bioactive peptides onto HA and allograft 17–26.
This strategy was originally modeled on the method native bone binding proteins use to associate with bone mineral. Bone sialoprotein (BSP) 30, osteocalcin (OCN) 31, and statherin 32 utilize stretches of negatively charged amino acids, namely, aspartate and glutamate, to complex with the positively charged calcium within native HA. In one of the first studies to identify polyacidic amino acid sequences as HA-binding domains, Kuboki’s group adsorbed a bone-binding protein onto HA crystals. Subsequent digestion of the protein revealed that glutamate rich sequences remained bound to HA 33. Additional support for selective HA binding by polyglutamate was provided by studies of a modular peptide composed of a heptaglutamate domain (E7) attached to the cell adhesive motif, RGD 33. In this investigation, soluble E7 peptides, but not soluble RGD, could competitively inhibit binding of the E7-RGD peptide to the HA surface 33. In complementary studies, our group showed that soluble RGD, but not soluble E7, blocked the ability of HA functionalized with E7-RGD to support mesenchymal stem cell adhesion 25. These results indicate that, in modular peptides, E7 serves to anchor peptides to the HA surface, while the cell interaction domain is available for cell binding. Furthermore, Kuboki’s group evaluated the strength of E7-RGD binding to synthetic HA and found that the dissociation constant for E7-RGD was 13.5µM, 500x greater than full length BSP 33.
In addition to RGD 19–22,25, polyacidic amino acid domains have been used to couple a variety of other cell instructive peptides to synthetic HA, including proteoglycan-binding peptides KRSR and FHRRIKA 26, collagen mimetic peptide DGEA 17,18, and a BMP-2 derived peptide 17,24. These studies have unequivocally shown that polyacidic amino acid domains increase peptide loading and retention on HA, and in turn, better peptide coupling consistently elicits an enhanced cell response. Also important, polyacidic amino acid domains are effective in anchoring bioactive peptides to diverse forms of HA including sintered HA disks, as well as composite HA-containing biomaterials including nano-HA/polycaprolactone (PCL) electrospun scaffolds 18 and HA-coated poly (lactide-co-glycolide) (PLG) films 24. Polyglutamate domains have also been used successfully to improve coupling of osteoinductive peptides on allograft bone 17, suggesting a potential mechanism to reconstitute allograft bone with regenerative molecules normally lost during processing steps required for allograft transplantation. Finally, recent work from our group has established the feasibility of modifying nanoparticles with polyglutamate domains 34. Specifically, cargo-loaded P22 nanocages containing polyglutamate substitutions were bound in greater quantities to synthetic HA and allograft bone than unmodified P22 nanocages, addressing the potential for utilizing polyacidic amino acid domains to endow drug-delivering nanocages with selectivity for HA 34.
The principal focus of prior research concerning polyacidic amino acid domains has been on achieving tight anchoring of peptides on the HA surface, which is of obvious importance for peptides that promote cell adhesion to material surfaces. However in the current study, the polyglutamate coupling strategy was adapted to enable tunable release of a model peptide into solution, in recognition of the fact that some types of biologics require gradient release for activity. To accomplish this goal, DGEA peptides were engineered with E2, E4, or E7 domains and it was found that the length of the polyglutamate sequence directly regulated the rate at which peptides were released. Longer polyglutamate domains, such as E7, exhibited slow release from HA and allograft bone, while short polyglutamate domains (E2) were released more rapidly. Additionally, intermediate release kinetics were achieved by mixing E2-, E4-, and E7-modified peptides at different ratios. Variable length polyglutamate domains may be useful for controlling the dose and timing of bioactive peptide or recombinant protein delivery, and may also hold promise for delivering two (or more) distinct peptides/proteins with differing optimal temporal kinetics. In sum, this study provides an advance by identifying a technically straightforward and cost-effective method for tailoring delivery of bone regenerative molecules from the surface of HA-containing biomaterials and allograft bone.
ACKNOWLEDGEMENTS
This research was supported by a grant from the Osseointegration Foundation (SLB). WMW was supported by the UAB Medical Scientist Training Program, PPB was supported by NIH T32 training grant, GM 008111-25 and BKC was supported by NIH/NIDCR predoctoral fellowship 1F31DE021613.
REFERENCES
- 1.Giannoudis PV, Dinopoulos H, Tsiridis E. Bone substitutes: an update. Injury. 2005;36(Suppl 3):S20–S27. doi: 10.1016/j.injury.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Phipps MC, Clem WC, Catledge SA, Xu Y, Hennessy KM, Thomas V, Jablonsky MJ, Chowdhury S, Stanishevsky AV, Vohra YK, et al. Mesenchymal stem cell responses to bone-mimetic electrospun matrices composed of polycaprolactone, collagen I and nanoparticulate hydroxyapatite. PLoS One. 2011;6(2):e16813. doi: 10.1371/journal.pone.0016813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phipps MC, Xu Y, Bellis SL. Delivery of platelet-derived growth factor as a chemotactic factor for mesenchymal stem cells by bone-mimetic electrospun scaffolds. PLoS One. 2012;7(7):e40831. doi: 10.1371/journal.pone.0040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xing ZC, Han SJ, Shin YS, Koo TH, Moon S, Jeong Y, Kang IK. Enhanced osteoblast responses to poly(Methyl Methacrylate)/Hydroxyapatite electrospun nanocomposites for bone tissue engineering. J Biomater Sci Polym Ed. 2012 doi: 10.1163/156856212X623526. ePub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Li G, Jiang J, Liu X, Luo L, Nan K. Electrospun fibrous scaffold of hydroxyapatite/poly (epsilon-caprolactone) for bone regeneration. J Mater Sci Mater Med. 2012;23(2):547–554. doi: 10.1007/s10856-011-4495-0. [DOI] [PubMed] [Google Scholar]
- 6.Bongio M, van den Beucken JJ, Nejadnik MR, Leeuwenburgh SC, Kinard LA, Kasper FK, Mikos AG, Jansen JA. Biomimetic modification of synthetic hydrogels by incorporation of adhesive peptides and calcium phosphate nanoparticles: in vitro evaluation of cell behavior. Eur Cell Mater. 2011;22:359–376. doi: 10.22203/ecm.v022a27. [DOI] [PubMed] [Google Scholar]
- 7.Lin G, Cosimbescu L, Karin NJ, Tarasevich BJ. Injectable and thermosensitive PLGA-g-PEG hydrogels containing hydroxyapatite: preparation, characterization and in vitro release behavior. Biomed Mater. 2012;7(2):024107. doi: 10.1088/1748-6041/7/2/024107. [DOI] [PubMed] [Google Scholar]
- 8.Song W, Markel DC, Jin X, Shi T, Ren W. Poly(vinyl alcohol)/collagen/hydroxyapatite hydrogel: properties and in vitro cellular response. J Biomed Mater Res A. 2012;100(11):3071–3079. doi: 10.1002/jbm.a.34240. [DOI] [PubMed] [Google Scholar]
- 9.Hennessy KM, Clem WC, Phipps MC, Sawyer AA, Shaikh FM, Bellis SL. The effect of RGD peptides on osseointegration of hydroxyapatite biomaterials. Biomaterials. 2008;29(21):3075–3083. doi: 10.1016/j.biomaterials.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennessy KM, Pollot BE, Clem WC, Phipps MC, Sawyer AA, Culpepper BK, Bellis SL. The effect of collagen I mimetic peptides on mesenchymal stem cell adhesion and differentiation, and on bone formation at hydroxyapatite surfaces. Biomaterials. 2009;30(10):1898–1909. doi: 10.1016/j.biomaterials.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono I, Gunji H, Kaneko F, Saito T, Kuboki Y. Efficacy of hydroxyapatite ceramic as a carrier for recombinant human bone morphogenetic protein. J Craniofac Surg. 1995;6(3):238–244. doi: 10.1097/00001665-199505000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Ono I, Gunji H, Suda K, Kaneko F, Murata M, Saito T, Kuboki Y. Bone induction of hydroxyapatite combined with bone morphogenetic protein and covered with periosteum. Plast Reconstr Surg. 1995;95(7):1265–1272. doi: 10.1097/00006534-199506000-00019. [DOI] [PubMed] [Google Scholar]
- 13.Borcard F, Staedler D, Comas H, Juillerat FK, Sturzenegger PN, Heuberger R, Gonzenbach UT, Juillerat-Jeanneret L, Gerber-Lemaire S. Chemical functionalization of bioceramics to enhance endothelial cells adhesion for tissue engineering. J Med Chem. 2012;55(18):7988–7997. doi: 10.1021/jm301092r. [DOI] [PubMed] [Google Scholar]
- 14.Durrieu MC, Pallu S, Guillemot F, Bareille R, Amedee J, Baquey CH, Labrugere C, Dard M. Grafting RGD containing peptides onto hydroxyapatite to promote osteoblastic cells adhesion. J Mater Sci Mater Med. 2004;15(7):779–786. doi: 10.1023/b:jmsm.0000032818.09569.d9. [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Cheng K, Weng W, Yang C. Immobilization of RGD peptide on HA coating through a chemical bonding approach. J Mater Sci Mater Med. 2009;20(11):2349–2352. doi: 10.1007/s10856-009-3794-1. [DOI] [PubMed] [Google Scholar]
- 16.Uludag H, Kousinioris N, Gao T, Kantoci D. Bisphosphonate conjugation to proteins as a means to impart bone affinity. Biotechnol Prog. 2000;16(2):258–267. doi: 10.1021/bp990154m. [DOI] [PubMed] [Google Scholar]
- 17.Culpepper BK, Bonvallet PP, Reddy MS, Ponnazhagan S, Bellis SL. Polyglutamate directed coupling of bioactive peptides for the delivery of osteoinductive signals on allograft bone. Biomaterials. 2013;34(5):1506–1513. doi: 10.1016/j.biomaterials.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culpepper BK, Phipps MC, Bonvallet PP, Bellis SL. Enhancement of peptide coupling to hydroxyapatite and implant osseointegration through collagen mimetic peptide modified with a polyglutamate domain. Biomaterials. 2010;31(36):9586–9594. doi: 10.1016/j.biomaterials.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujisawa R, Mizuno M, Nodasaka Y, Kuboki Y. Attachment of osteoblastic cells to hydroxyapatite crystals by a synthetic peptide (Glu7-Pro-Arg-Gly-Asp-Thr) containing two functional sequences of bone sialoprotein. Matrix Biol. 1997;16(1):21–28. doi: 10.1016/s0945-053x(97)90113-x. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert M, Giachelli CM, Stayton PS. Biomimetic peptides that engage specific integrin-dependent signaling pathways and bind to calcium phosphate surfaces. J Biomed Mater Res A. 2003;67(1):69–77. doi: 10.1002/jbm.a.10053. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert M, Shaw WJ, Long JR, Nelson K, Drobny GP, Giachelli CM, Stayton PS. Chimeric peptides of statherin and osteopontin that bind hydroxyapatite and mediate cell adhesion. J Biol Chem. 2000;275(21):16213–16218. doi: 10.1074/jbc.M001773200. [DOI] [PubMed] [Google Scholar]
- 22.Itoh D, Yoneda S, Kuroda S, Kondo H, Umezawa A, Ohya K, Ohyama T, Kasugai S. Enhancement of osteogenesis on hydroxyapatite surface coated with synthetic peptide (EEEEEEEPRGDT) in vitro. J Biomed Mater Res. 2002;62(2):292–298. doi: 10.1002/jbm.10338. [DOI] [PubMed] [Google Scholar]
- 23.Lee JS, Lee JS, Murphy WL. Modular peptides promote human mesenchymal stem cell differentiation on biomaterial surfaces. Acta Biomater. 2010;6(1):21–28. doi: 10.1016/j.actbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy MB, Hartgerink JD, Goepferich A, Mikos AG. Synthesis and in vitro hydroxyapatite binding of peptides conjugated to calcium-binding moieties. Biomacromolecules. 2007;8(7):2237–2243. doi: 10.1021/bm070121s. [DOI] [PubMed] [Google Scholar]
- 25.Sawyer AA, Weeks DM, Kelpke SS, McCracken MS, Bellis SL. The effect of the addition of a polyglutamate motif to RGD on peptide tethering to hydroxyapatite and the promotion of mesenchymal stem cell adhesion. Biomaterials. 2005;26(34):7046–7056. doi: 10.1016/j.biomaterials.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Sawyer AA, Hennessy KM, Bellis SL. The effect of adsorbed serum proteins, RGD and proteoglycan-binding peptides on the adhesion of mesenchymal stem cells to hydroxyapatite. Biomaterials. 2007;28(3):383–392. doi: 10.1016/j.biomaterials.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Chen RR, Silva EA, Yuen WW, Mooney DJ. Spatio-temporal VEGF and PDGF delivery patterns blood vessel formation and maturation. Pharm Res. 2007;24(2):258–264. doi: 10.1007/s11095-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 28.Richardson TP, Peters MC, Ennett AB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19(11):1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 29.Bose S, Tarafder S. Calcium phosphate ceramic systems in growth factor and drug delivery for bone tissue engineering: a review. Acta Biomater. 2012;8(4):1401–1421. doi: 10.1016/j.actbio.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganss B, Kim RH, Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10(1):79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- 31.Hoang QQ, Sicheri F, Howard AJ, Yang DS. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature. 2003;425(6961):977–980. doi: 10.1038/nature02079. [DOI] [PubMed] [Google Scholar]
- 32.Stayton PS, Drobny GP, Shaw WJ, Long JR, Gilbert M. Molecular recognition at the protein-hydroxyapatite interface. Crit Rev Oral Biol Med. 2003;14(5):370–376. doi: 10.1177/154411130301400507. [DOI] [PubMed] [Google Scholar]
- 33.Fujisawa R, Wada Y, Nodasaka Y, Kuboki Y. Acidic amino acid-rich sequences as binding sites of osteonectin to hydroxyapatite crystals. Biochim Biophys Acta. 1996;1292(1):53–60. doi: 10.1016/0167-4838(95)00190-5. [DOI] [PubMed] [Google Scholar]
- 34.Culpepper BK, Morris DS, E PP, Bellis SL. Engineering nanocages with polyglutamate domains for coupling to hydroxyapatite biomaterials and allograft bone. Biomaterials. 2013;34(10):2455–2462. doi: 10.1016/j.biomaterials.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]