Abstract
Metabolomics use in toxicology is rapidly increasing, particularly owing to advances in mass spectroscopy, which is widely used in the life sciences for phenotyping disease states. Toxicology has the advantage of having the disease agent, the toxicant, available for experimental induction of metabolomics changes monitored over time and dose. This review summarizes the different technologies employed and gives examples of their use in various areas of toxicology. A prominent use of metabolomics is the identification of signatures of toxicity – patterns of metabolite changes predictive of a hazard manifestation. Increasingly, such signatures indicative of a certain hazard manifestation are identified, suggesting that certain modes of action result in specific derangements of the metabolism. This might enable the deduction of underlying pathways of toxicity, which, in their entirety, form the Human Toxome, a key concept for implementing the vision of Toxicity Testing for the 21st century. This review summarizes the current state of metabolomics technologies and principles, their uses in toxicology and gives a thorough overview on metabolomics bioinformatics, pathway identification and quality assurance. In addition, this review lays out the prospects for further metabolomics application also in a regulatory context.
Keywords: metabolomics, pathways of toxicity, toxicity testing for the 21st century, analytical chemistry, bioinformatics, quality assurance
Introduction
Is there such thing as toxicometabolomics or is there only metabolomics use in toxicology? Is there something unique about this use that justifies coining a new word? The authors believe that mass spectroscopy-based metabolomics represents an enabling technology to drive toxicological progress within the current discussion of toxicology for the 21st century. The study of metabolomics, as applied to toxicology, has applications that are unique, e.g. the identification of pathways of toxicity (PoT), signatures of toxicity (SoT) for regulatory evidence and applications feeding into systems toxicology approaches, which are to some extent distinct from other areas in the life sciences. PoT, i.e. the molecularly defined cellular processes which connect a chemico-biological interaction of a toxicant with its adverse outcome, are a key concept for implementing the vision of Toxicity Testing for the 21st century (Tox-21c), the proposed change to a mechanistic toxicology by the NRC, 2007.
Metabolomics has been defined as ‘the quantitative measurement of time-related multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification’ (Nicholson et al., 1999). ‘Multiparametric’, however, may not fully represent the ‘omic’ notion of aiming for the entirety of aspects. While the hundreds to thousands of identifiable and currently detectable structures are already impressive enough, and it certainly serves us well at the moment, only a small fraction of all metabolites can be assessed in a single measurement. The ‘omics’ approach aims for larger coverage and a nearly complete detection of small molecules in a biological sample.
Metabolomics, as an emerging field of ‘omics’ research, primarily concerns comparative analysis of the endogenous metabolites present in any biological system or any specific physiological state. It aims to characterize and identify the metabolites – the end products of cellular metabolism. As an interdisciplinary field of science, metabolomics combines analytical chemistry, bioinformatics, statistics and biochemistry. When applied to toxicology studies, metabolomics also includes aspects of patho-biochemistry, systems biology and molecular diagnostics (Griffiths et al., 2010). In the past few years, metabolomics approaches have been widely used in toxicology studies, mostly because they have the ability to provide helpful information for better understanding the mechanisms of toxicity. Furthermore, metabolomics has the potential to identify biomarkers of toxic effects and build models for toxicity prediction (Craig et al., 2006; Heijne et al., 2005a; Ruepp et al., 2002; Schnackenberg et al., 2006, 2009). From an analytical perspective, the goal of metabolomics in toxicology studies is to ‘achieve a comprehensive measurement of the metabolome and how it changes in response to stressors, with biological payoff being an illumination of the relationship between the perturbations and affected biochemical pathways’ (Robertson, 2005).
The most common technologies currently employed for metabolomics are nuclear magnetic resonance (NMR) and mass spectrometry (MS), both of which allow for the phenotyping of a response. Pioneering work of the Imperial College group has favored NMR technologies in metabolomics in toxicology, but for the purpose of identifying unknown PoT via untargeted metabolomics, MS-based metabolomics appears the more suitable technology as it allows better deduction of a substance represented by a feature in the recorded spectrum, higher sensitivity and precision compared with NMR, and the ability to measure nearly the whole metabolome. These advances of MS have only been available for a few years. As the identification of metabolites for PoT deduction is especially critical for the implementation of Tox-21c, our review will predominantly address MS-based metabolomics, although many aspects can easily be translated to NMR.
Toxicology faces many challenges to meet the societal desire for safety science and its regulatory use (Hartung, 2009, 2011). Demanding legislation in Europe for chemicals and cosmetics (Hartung, 2008, 2010; Hartung and Rovida, 2009) prompts a need for change. This is in strong contrast to the lack of major methodological changes: toxicology is probably the only life science for which most experimental designs date back 40 to 60 years. Certainly, details have been refined and additional techniques have been added, but the vast majority of resources still go into treating rats and monitoring them for pathology. We have estimated that this amounts to animal testing costs of $3 billion worldwide per year (Bottini and Hartung, 2010). In spite of this expense, the return of investment is not satisfactory, with only 43% predictivity of rodent toxicology for human side effects in drug development (Olson et al., 2000) and the need to apply precautionary safety factors of 100–1000 on the results obtained. The testing capabilities to date, owing to costs and labor, do not allow for the assessment of the high number of substances, e.g. medicines, pesticides and consumer products, aiming to enter the market. A backlog of close to 100,000 substances in consumer products that have not been systematically tested cannot be addressed in the foreseeable future, although REACH in Europe and ongoing TSCA reauthorization in the US aim to reduce this burden (Hartung, 2010). This does not solve the problems of novel products (biological, functional food, cell therapies and nanoparticles), new hazards of concern (endocrine disruption, childhood asthma, atherosclerosis, obesity, diabetes etc.), mixture effects, or inter-individual differences (personalized toxicology). For all these, toxicology has little to offer and it is questionable whether an expansion of current approaches will solve the problem.
Many of these concerns culminated in the hallmark publication of the National Research Council (NRC, 2007), which proposed moving the science of toxicology towards a mechanistic understanding. This was also fully embraced in the development of a roadmap for a novel approach to systemic toxicity testing (Basketter et al., 2012). The basic concept is that drugs or toxicants cause perturbations of the concentrations and fluxes of endogenous metabolites involved in key biochemical pathways and the response of cells to toxicants or other stressors generally results in an adjustment of their intra- and/ or extracellular environment in order to maintain constancy of their internal environment (homeostasis). The principal question is: when does a normal adaptive response become abnormal (toxic)? There are two different views on what constitutes a toxicity pathway: one assumption is that drug- or toxicant-induced modulations are related to known metabolic pathways, which are deranged, and if the capacity for self-correction is exceeded, they turn into hazard manifestations. The opposing view is that the chemico–biological interaction leads to a cascade of events, which disrupts homeostasis. We tried to explain that these are simply different time-points in the chain of events, i.e. initially, pathways of toxicity derange whereas pathways of defense (PoD) counteract (Hartung et al., 2012). Then a new homeostasis under stress is established, which can result in damage if it is not stable or its defense capacities are exhausted by continued stress. Metabolomics is typically carried out measuring the latter, i.e. applying sub-toxic or borderline toxic doses/concentrations (or at least measuring before a manifestation of hazard), but at times at which the biological system has stabilized and the signatures of change can stably be measured. It will have to be shown which is the more telling condition and which can then be used for hazard prediction.
Principles and Types of Metabolomics
Metabolomics requires highly analytical instruments because it is dealing with thousands of small molecules with a diversity of chemical and physical properties. Many analytical technologies have been developed and applied for metabolomics in the past decade (Hollywood et al., 2006; Kim et al., 2011; Scalbert et al., 2009), but there are only two technologies which meet the high requirements for these studies: nuclear magnetic resonance (NMR) and mass spectrometry (MS).
NMR-based metabolomics
NMR technology has played an important role in metabolism studies for more than 30 years (Wishart, 2008). The NMR-based metabolomics approach in toxicology can be traced back to the early 1980s (Hanzlik et al., 1980; Robertson, 2005), and it remains one of the most extensively utilized analytical approaches in toxicometabolomics (Barba et al., 2008; Beger et al., 2010; Fukuhara et al., 2011; Liu et al., 2011; Neerathilingam et al., 2010 ). Compared with other analytical tools, NMR has several unique advantages, such as its simple quantitative readout and sampling capabilities. The technology, in particular, is non-destructive, requiring no sample alteration and only a small amount of material. Because NMR is non-destructive, it can even be used non-invasively, as it has been applied to detect molecules in vivo (Griffin and Kauppinen, 2007; Serkova et al., 2008). Furthermore, the technology requires no separation, provides structural information, and often has higher reproducibility than MS-based technology. With developments of new technologies (e.g. flow probe and automated shimming), more than 300 samples can be handled per day (Griffin, 2003). The major limitations of NMR for comprehensive metabolite profiling is its relatively low sensitivity and its unsuitability to detect low-concentration metabolites. The recent introduction of some new developments, such as cryoprobes, high field magnets (up to 900MHz) and low-volume microprobes, however, has pushed the limits of detection for modern NMR into the 10−5–10−6M range (Wishart, 2008). Sensitivity seems to be less of a concern today, but the time required to acquire spectra with adequate resolution is considerably longer compared to methods like MS. The possibility of automating metabolomics workflows is a further strength of both of these technologies. Considering all the above, NMR will continue to be a very important analytical tool for metabolomics study.
MS-based metabolomics
Over the past decade there has been a rapid growth of MS-based metabolomics publications (Robertson, 2005). Because of its high sensitivity, selectivity, and wide dynamic range, MS has emerged as a powerful tool in metabolomics studies. The wide applications of this technique for toxicological metabolomics have been reviewed (Beger et al., 2010; Kaddurah-Daouk et al., 2008; Patterson et al., 2010; Robertson, 2005; Roux et al., 2011). As biological samples can be introduced to the ion-source of MS either via direct-injection (DI) or after chromatographic/electrophoretic separation, two major methods, hyphenated method and direct injection method, have been used in MS-based metabolomics. Direct-injection or flow-injection analysis, especially when used on high-resolving mass spectrometers, provides an effective way to detect and quantify large numbers of metabolites, and was extensively utilized for metabolome analysis. An example of this technique is described in a study of Escherichia coli metabolism utilizing a DI-MS (Q-Tof mass spectrometry with a resolution of 8000–12 000 (FWHM) over the m/z 100–1000 range) that profiled small molecules in the impressive number of > 1400 biological extracts per day and successfully detected > 1500 distinct ions at a given mode from polar E. coli extractions (Fuhrer et al., 2011). The major drawback of the direct-injection technique is the presence of ion-suppression, which resulted in the failure to detect some metabolite ions with low ionization efficiencies. To minimize ion suppression, a hyphenated technique was developed to physically separate the complex analytes prior to exposure to an ion source by chromatography or electrophoresis so that individual or less complex metabolites can be eluted from the column/ capillary with different retention times and then analyzed with MS.
The most extensively used hyphenated MS approaches are GC (gas chromatography) and LC-MS (liquid chromatography-mass spectrometry). Gas chromatography combined with electron ionization (EI) mass spectrometry is the most developed technology for metabolite profiling. During electro-ionization, analyte molecules are ionized and fragmented in the gas phase (10−1 to 10−4 Pa) by interaction with 70 eV electrons generated from a hot filament and accelerated through a 70V electric field (Beran and Kevan, 1969). As the fragmentation is reproducible, these fragment ions form the ‘chemical fingerprints’ (‘features’) of the analyte. Therefore, many commercially available libraries with thousands of standard EI spectra can be used for rapid identification of analytes. Using this platform, more than 300 chemically diverse metabolites have been detected in a single run, including amino acids, organic acids, sugars and fatty acids (Jonsson et al., 2005). One prerequisite for GC-MS analysis is the need of volatile analytes that are stable at high temperature during the whole analysis. Generally, some non-volatile metabolites (e.g. sugars, amino acids) can be converted into volatile compounds by derivatization. Although chemical derivatization improves the coverage of metabolites, an additional source of variability in metabolomics may be introduced by such processing.
Liquid chromatography coupled to soft ionization mass spectrometry has also emerged as a powerful tool in metabolomics. Unlike GC-MS, which uses a high energy for ionization, LC-MS utilizes a so-called ‘soft ionization’ technique that preserves the intact molecular information, which is critical for identification. One can easily obtain the molecular weight of the intact molecule based on the protonated/deprotonated ions ([M + H]+, [M-H]-). Electrospray ionization (ESI) is the most widely used soft ionization technique in LC-MS. Owing to the development of high resolution mass analyzers over the past decade [e.g. time-of-flight (TOF), orbitrap and Fourier transform – ion cyclotron resonance (FT-ICR), the mass-to-charge ratio (m/z) of ions can be measured with accuracy within ppm range, which greatly decreases the number of elemental compositions of the ions and facilitates the identification based on the exact masses (Robertson, 2005). Compared with other platforms, LC-MS exhibits better sensitivity, selectivity, and a broader dynamic range. LC-MS, therefore, has been increasingly applied in metabolomics research over the past years, particularly in toxicology studies (Chen et al., 2007; Crockford et al., 2006; Singh, 2006; Wang et al., 2009; West et al., 2010). The major bottleneck for LC-MS based metabolomics studies is the availability of the searchable mass spectra libraries for identification of unknown compounds. Currently, there are few libraries (e.g. human metabolome database, METLIN and MassBank) that are publicly available for LC-MS-based metabolomics. The development of new databases/libraries based on mass accuracy, retention time, and MS/MS spectra will play a crucial role in global metabolite profiling.
The most recent development in the hyphenated MS technique is capillary electrophoresis-mass spectrometry (CE-MS). Compared with GC-MS and LC-MS, CE-MS has several unique advantages, such as small injection volume requirements, short analysis time and high separation power. In particular, since the metabolites are separated by charge and size, CE-MS can be used to analyze cations, anions, and even neutral molecules in a single run, which makes it a very promising technique for global metabolomics (Shulaev, 2006). CE-MS has been applied in both targeted and untargeted metabolomics research (Monton and Soga, 2007; Ramautar et al., 2009, 2011; Soga et al., 2003).
In addition to the most widely used techniques described above, several new techniques have been developed over the past few years. Desorption electrospray ionization (DESI) and direct analysis in real time (DART) are two techniques that ionize analytes under open atmospheric conditions and require almost no sample separation and minimal preparation. The applications of these two relatively new techniques in metabolomics have been reviewed (Gowda et al., 2008; Han et al., 2009; Werner et al., 2008). Another new technology, imaging mass spectrometry (IMS), has recently been used (Amstalden van Hove et al., 2010; Franck et al., 2009; Li et al., 2008; Sugiura and Setou, 2010). Using IMS, one can determine the distribution of hundreds of small molecules at complex surfaces (tissue, for example) in a single analysis without destroying the cellular and molecular integrity. Currently, there are two commonly used IMS techniques, secondary ion mass spectrometric (SIMS) imaging and matrix-assisted laser desorption ionization (MALDI) IMS. Both of these have been successfully applied in direct tissue imaging for assessing localization of metabolites, which may provide more accurate structure information for biomarker discovery. Although these new techniques have not been used as extensively as GC-MS and LC-MS, the apparent advantage in not needing chromatography makes these techniques so attractive that they could be useful analytical tools for high-throughput metabolomics.
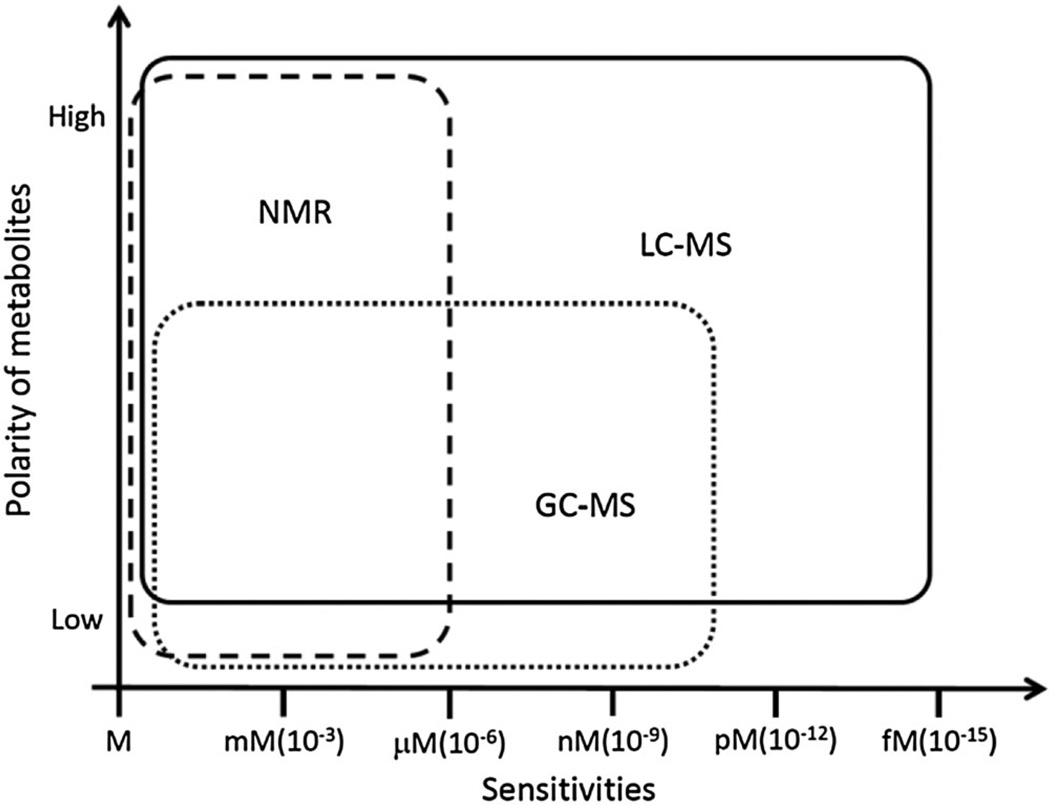
In summary, there is no single analytical tool, which could be suited to precisely identify and quantify thousands of small molecules of interest. The selection of the most suitable analytical tool is generally a compromise between sensitivity and selectivity. As illustrated in Fig. 1, NMR provides good selectivity and rapid analysis time, but has relatively low sensitivity, which limits this technology to the analysis of only concentrated metabolites. GC-MS is more sensitive than NMR, but suffers from lower selectivity. Additional chemical derivatization steps are needed for the analysis of some non-volatile/polar metabolites, which usually cost more time and introduce more variance. LC-MS offers both high sensitivity and selectivity. By choosing two complementary columns (reverse phase column and HILIC column), LC-MS can be used to analyze non-polar and polar metabolites. On the other hand, this technique requires relatively longer analysis time compared to NMR technology. Therefore, a combination of different analytical technologies is needed in order to obtain a better coverage of the metabolites with a broad range of polarities and molecular weights.
Figure 1.
Relative sensitivity and selectivity of different analytical tools for metabolomics.
Approaches for metabolomics
Generally, there are two major approaches used in metabolomics studies: targeted and untargeted (global). A targeted approach is used to determine the relative abundances and concentrations of specific sets of metabolites. In order to conduct this type of analysis, information about the exact structure of the metabolite must be available in advance (Cui et al., 2008; Kopka et al., 2005; Smith et al., 2005; Wishart et al., 2007). The targeted approach represents a quantitative analysis as it involves the comparison of the analytes to the authentic compounds. This method is well established in metabolomics and it also has many applications in toxicology (Begriche et al., 2011; Li et al., 2011; Lin et al., 2011a; Southam et al., 2011). One major disadvantage of the targeted approach is that one must know the metabolites of interest a priori so that the given compounds could be used as standards for absolute quantitation. Therefore, this approach is dependent on the availability of a purified form of the known metabolite and is not applicable for the identification of new metabolites.
In contrast, untargeted metabolomics is the comprehensive study of all metabolites in a biological sample, which is sometimes called ‘global metabolome analysis’ (Griffiths et al., 2010). This approach aims to find differing metabolites based on their relative quantitation and annotation of as many chromatographic/spectroscopic peaks (‘features’) as possible (Robertson, 2005). For example, in MS-based untargeted analysis, the initial raw data acquired with MS may produce hundreds even thousands of mass signals. This global metabolome analysis usually requires efficient data reduction using chemometric methods for identification of statistically significant metabolites. The untargeted approach is mainly used in mechanistic studies, hypothesis generation, biomarker discovery and diagnostics (Chen et al., 2011a; Issaq et al., 2011; Lucio et al., 2010; Wang et al., 2012; Woo et al., 2009). The disadvantage of untargeted metabolomics is that this approach is a relative quantification, not an absolute one. Furthermore, some of the significant features/peaks are not identifiable.
Bioinformatics and Pathway identification
Omics techniques tend to generate variable-heavy data sets and clever bioinformatics and statistics are the keys to their success. It is no longer appropriate and possible to manually analyze these data. This section will therefore focus on data formats, data processing, statistics, available free software packages, and pathway analysis.
Along with several proprietary data formats that are platform-unique, several open file formats (e.g. netCDF, mzXML and mzDATA) have been developed which are more or less supported by the different platforms. The Seattle Proteome Center and Wikipedia host web pages with extensive overviews and descriptions of proprietary data formats, possible converters, and open file formats (Seattle Proteome Centre, 2012; Wikipedia, 2012).
Numerous statistical methods that are particularly appropriate for mining and interpreting metabolomics data and are especially suitable for the so-called ‘large P small N problem’ (more variables/metabolites than observations/samples) are available. Most of the methods are multivariate statistics methods for extracting information from large tables of data, but several machine learning approaches have become more and more popular. In general, the pattern recognition methods used in metabolomics can be divided into two categories: ‘unsupervised’ (clustering) and ‘supervised’ learning (classification). Both approaches have a long history in biometry. Whereas unsupervised learning is used to find structures in a collection of unlabeled (multivariate) observations, supervised learning approaches are used to find a mathematical function based on a training set that predicts the reference classifications (labels) of the observations in the training set and/or distinct test sets.
The typical unsupervised statistical techniques used in metabolomics are principal component analysis (PCA) and several clustering approaches. Typical supervised statistical techniques are partially least squares projections to latent structures (PLS) and machine learning approaches. Let X be a data matrix of N rows (observations like analytical samples, biological individuals, etc.) and P columns (variables may be of spectral or chromatographic origin, or other sources). In addition, for supervised learning a class label (classifier such as ‘control’ and ‘treated’) for each row may be available.
Preprocessing: normalization, transformation, missing values and filtering
Usually, the data matrix is first centered to the mean and scaled to the same variances, typically by a Z-transformation or a combination of mean centering and Pareto scaling (Eriksson et al., 1999). This is necessary as several multivariate methods such as PCA or PLS are maximum variance projection methods and non-normalized variables with large variance have stronger weights and are more likely to be addressed in the modeling than low variance variables. By scaling all variables to the same scale, no variable is allowed to dominate over the others. For the Z-scaling, the centering can be done by calculating the average value and standard deviation of each variable (column), subtracting the mean from the data, and dividing by the standard deviation. The Pareto scaling divides by the square root of the standard deviation instead of the standard deviation itself. Some people prefer the Pareto scaling because the Z-scaling can increase the noise of features of low signal variation. A more detailed overview of centering and scaling is given in van den Berg et al. (2006).
In addition, a log transformation of the data can help reduce heteroscedasticity, as the log transformation reduces large values more than small values. In addition, it renders a skewed distribution more symmetric. Presence of heteroscedasticity can invalidate inferential statistics that assume that variances of the error term do not vary with the effects being modeled (e.g. linear models such as linear regression, anova).
The last important preprocessing step is deciding how to deal with missing values, e.g. metabolites that could not be detected in a subgroup of samples. This is common in metabolomics and important because few types of statistics can cope with missing values and, in most cases, it is unclear if the reason for the missing value is a concentration below the detection limit or other experimental causes. Simple methods for replacing missing values are the replacement by the mean or median of the metabolite concentrations over all samples, which are in the same group (e.g. control group) or the imputation from the nearest neighbors. More advanced techniques include Probabilistic PCA, Bayesian PCA, or Singular Value Decomposition (Stacklies et al., 2007; Steinfath et al., 2008).
As with other omics technologies, preprocessing should include a (non-specific) filtering to improve the data quality, exclude non-relevant metabolites, and improve the power of the following statistics by reducing the multiple testing problem. An overview of possible filtering techniques and practical examples are given in Table 1.
Table 1.
Examples of preprocess filtering techniques including examples and rationale.
| Filter by | Examples | Rationale |
|---|---|---|
| Abundance | Features with low intensities are excluded (e.g. 5000 counts) | Improve data quality |
| Frequency | Features needs to be present in > 60% of the samples | Improve data quality |
| Feature variability | Features with high variability within group will be excluded (e.g. CV >30%) | Improve data quality |
| Fold change | Features with fold change of e.g. ≤ 1.2 between groups will be excluded | Samples with low variation across groups are not relevant for the further analysis |
| Annotation in pathway libraries | Features that are not annotated will be excluded | Pathway enrichment analysis are only possible if features are annotated |
Initial statistical analysis: unsupervised techniques
Initial analyses start usually with PCA (Fig. 2D) on the entire dataset to gain an overview of class separation, trends and outliers. The basic idea of PCA is dimension reduction, which means to present multivariate data tables as a low dimensional-plane, usually consisting out of two to three dimensions. PCA finds lines, planes, and hyperplanes in the P-dimensional space that approximate the data as well as possible.
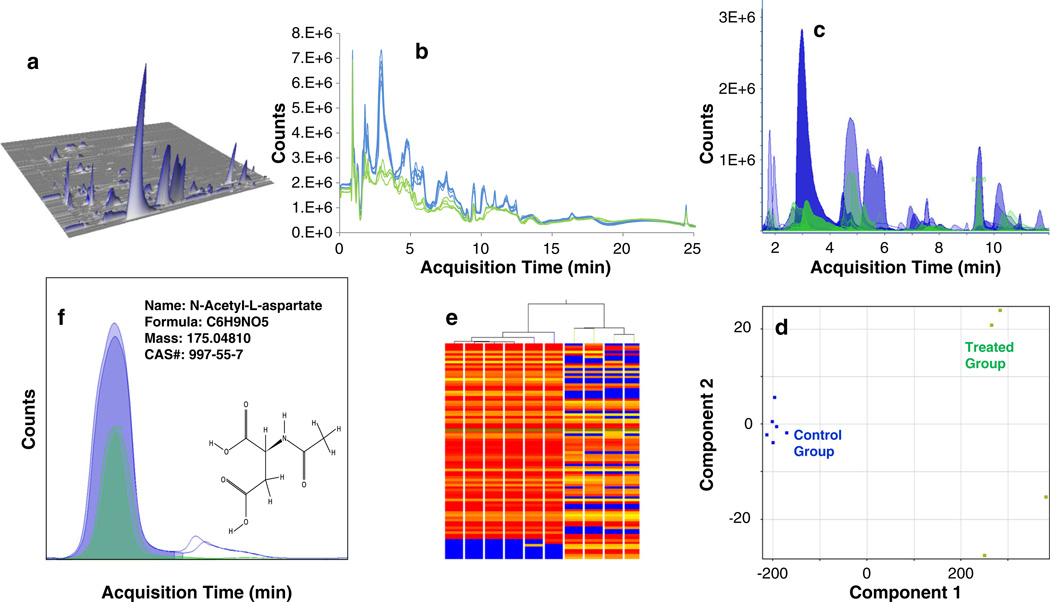
Figure 2.
Illustration of the large data reduction involved in a typical toxicometabolomics study. Metabolomics data are usually represented in their raw form as (A) chromatograms, i.e. 3D matrix of masses, retention times and intensities. Depending on the experiment study design and the strategy adopted (untargeted approach in this case), (B) biological replicates representing different conditions are acquired and using specialized software, (C) mass signals or “features” are extracted and aligned across all samples. Statistical analysis tools highlight variability and/or similarity between and within the different groups, e.g. (D) Principle components analysis (PCA) and (E) Hierarchical clustering The result may be displayed as a list of differentially abundant metabolites with corresponding p-value, fold change, etc. and (F) putative identity of the relevant metabolites established before confirmation using standard compounds and inclusion in a pathway enrichment analysis.
By using PCA, a data table X is modeled as X=T × P’ + E. The term T × P’ models the structure and E is the residual matrix that contains the noise. The principal component scores are the columns of the score matrix T. The meaning of the scores is given by the loadings of the first, second, …, components building up the loading matrix P. Two PC define a model plane, a window in the P-dimensional variable space. It is possible to visualize the P-dimensional variable space by projecting all observations onto this two-dimensional subspace. The co-ordinate values of observations on this plane are called scores and the plot is therefore called a score-plot. It can be used to show similarities and dissimilarities between the observations and to detect outliers. For an overview, the scores of the first two dimensions (principle components) are usually inspected. In addition, moderate outliers can be detected by inspecting the model residuals (Robertson and Lindon, 2005). A more detailed overview on PCA is given by Jackson (2003).
A second important statistical toolset for finding structures in unlabeled observations is cluster analysis (for a more detailed overview see Kaufman and Rousseeuw, 2005). The question behind cluster analysis is how to organize objects into groups whose members are in some way similar. There are two basic clustering strategies – non-hierarchical clustering, sometimes called partitioning methods, and hierarchical clustering. Hierarchical clustering can be further divided into agglomerative and divisive clustering. In agglomerative clustering, each object starts in its own cluster with one single element, and in each step of an iterative procedure the two closest clusters are merged until all objects are in one cluster. The result is a tree that can be shown as a dendrogram (Fig. 2E). To get a desired number of clusters, the tree is simply cut at the desired height. In contrast, divisive hierarchical clustering starts with all objects in a single cluster, and at each step of the iterative process the most heterogeneous cluster is divided into two separate clusters until all objects are in their own cluster.
Non-hierarchical clustering – e.g. k-means – partitions data on one level only by a series of iterations. Normally, single object exchanges or moves are proposed and the resulting change in some clustering criteria is then computed and the change is accepted if it improves the criteria. The process is repeated until a number of iterations are made. For example, the k-means method partitions the samples into k-groups, so that the sum of squared distances from the objects to the assigned cluster is minimized.
All cluster methods have in common that the user has to specify, in advance, the distance (or similarity) algorithm between two measurements (e.g. city-block or Manhattan, Euclidian, Chebychev distances) and, for hierarchical techniques, the distance measured between each pair of objects (e.g. single, complete linkage). Before computing the distances, the data matrix needs to be centered and scaled as described above. For partitioning algorithms, the number of clusters typically needs to be specified as well. As it is well known that none of the clustering techniques are always optimal (or, in other words, how well the algorithm works depends on the data), several different clustering techniques should be applied in parallel and the results compared. Finally, the results are shown in a heatmap based on the computed between-object distances with dendrograms at the sides of the heatmap (Fig. 2E).
Initial statistical analysis: supervised techniques
Typical supervised statistical techniques applied in metabolomics are partial least squares projections to latent structures (PLS; Wold et al., 2001) and several supervised machine learning approaches (For a recent introductionary overview see Korman et al., 2012; Malley et al., 2011).
PLS has the ability to analyze data with many noisy, collinear, and even incomplete variables, modeling a relation between the measured data matrix X and Y in a linear multivariate model, whereas Y is normally an artificially designed matrix that describes the class label, doses, or time points. The data should be symmetrically distributed and have a constant error term. The data is often log-transformed, and z-transformed as described above.
Unfortunately, the power and robustness of traditional parametric statistical approaches – such as PCA, PLS, or linear models like t-tests, ANOVA, or other regression models – are dependent on assumptions about the (multivariate) distribution of the test data and, like any other parametric model, requires complete and correct specification of interaction and higher order terms. It is well known that violations of these assumptions or model specifications, such as non-normality, variance heterogeneity, non-specified variable interactions, or censored observations, can lead to inflated type I errors, loss of power and/or selection of wrong variables for the test statistics (see, for example, Kleensang et al., 2010). However, it is nearly impossible in complex metabolomics datasets to meet all these criteria and specify a parametric model including the complete and correct specification of interaction and higher order terms. Machine learning approaches are more tolerant of departures from a strict set of largely unverifiable assumptions (for an overview, see Malley et al., 2011) and have been applied successfully to various biomedical and other problems for variable selection and prediction, especially in cases of large numbers of predictors for a categorical response variable (Heidema et al., 2006). They are fully non-parametric, essentially make no distributional assumptions, and do not require a specified model as a starting point.
Moreover, the fact that they can be applied to a wide range of prediction problems, even if they are nonlinear and involve complex high-order interaction effects, makes them – within a very short period of time – important data analysis tools. These tools perform well in comparison to many other standard methods (Diaz-Uriarte and Alvarez de Andres, 2006; Heidema et al., 2006). A large variety of different machines are available which include random forests, k-nearest neighbors, support vector machines and neural networks, but will not be described here in more detail.
Free metabolomics analysis software packages
Several free software packages are available for the analysis of metabolic datasets. Some of them focus on the necessary preprocessing steps such as peak detection, alignments, and identification, and the implemented inferential statistical tools are more or less limited: AMDIS (Stein, 1999), MetAlign (Lommen, 2009), met-Idea (Broeckling et al., 2006) and mzMine 2 (Katajamaa et al., 2006; Pluskal et al., 2010).
Most of the recently developed software packages are implemented in the R system for statistical computing (R Development Core Team, 2006) as a combination of XCMS (Smith et al., 2005, 2006) for peak detection, peak grouping, and retention time correction and several other R add-on packages which are widely considered to be the most complete, up-to-date collection of statistical approaches. Several of these software packages are web-based to allow users to avoid learning the sometimes ‘beastly’ R programming language. Famous examples are MeltDB (Neuweger et al., 2008) from the Bielefeld University (Germany) and MetaboAnalyst (Xia et al., 2009) from the University of Alberta (Canada). Also, XCMS alone is available as a web-based application: XCMS online (Tautenhahn et al., 2012).
Current developments focus on pathway analysis and visualization tools – for example, MetaboAnalyst and MeltDB with its modules MetPA (Xia and Wishart, 2010a, 2010b; Xia et al., 2012) and MSEA (Persicke et al., 2012), and MPEA (Kankainen et al., 2011).
Pathway enrichment analysis
Through knowledge of biochemical pathways and gene regulatory networks, metablomics can be linked to pathway analysis. Already in 2002, Fiehn (2002) proposed that metabolomics results can be connected to the genotype through known biochemical pathways and gene regulatory networks. In comparison to other omics techniques, metabolomics is much closer to the phenotype than transcriptomics or proteomics, because these omics approaches only indicate the potential for pathophysiological or other changes. Several feedback mechanisms are simply not reflected in protein concentration or gene expression changes. Pathway analysis, however, can only be performed under the assumption that drug or toxin induced modulations are related to upfront known metabolic pathways. Several pathway information databases are available. Typical examples are The Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2006, 2008) and WikiPathways (Kelder et al., 2012). While enrichment analysis has become a very successful tool to link transcriptomics and proteomics results to pathway analysis (Mootha et al., 2003; Subramanian et al., 2005), Metabolite Set Enrichment Analysis (MSEA) are still in their infancy and only a few specialized software packages are available as listed before.
These approaches are in general based on two classes of statistics plus interactive visualization for the interpretation of the results, which are mainly based on the ideas and statistics from Gene Set Enrichment Analysis (GSEA). An overview of enrichment tools in the context of transcriptomics can be found in Huang et al. (2009).
The first class is singular enrichment analysis, where a set of ‘interesting’ metabolites is preselected (e.g. differentially expressed metabolites between control and sample selected by fold change or t-test p-value), and by comparing the amount/fraction of ‘interesting’ metabolites in a given pathway with the amount/fraction of ‘interesting’ metabolites that should occur by chance, a test of over- or underrepresentation in pathways can be calculated by Fisher’s exact test or hypergeometric distribution. However, applied to metabolomics it is difficult to decide about the ‘right’ metabolite reference background for the calculations. This is important because the estimation of the amount/fraction of ‘interesting’ metabolites occurring by chance directly impacts the enrichment p-value. The number of estimated metabolites in eukaryotic cells ranges from 4000 to 20 000 and up to 200 000 in the entire plant kingdom (Fernie, 2004, 2007). But how many of them can be detected depends on your experimental setting and platform.
The second approach takes into account all metabolites without selecting ‘interesting’ metabolites. The test statistic is calculated from the rank order based on fold change or t-test P-value of all metabolite members in the pathway, and p-values can be obtained by Monte-Carlo permutations of the test statistic.
One problem for both approaches compared with their use in transcriptomics is the much higher fraction of missing values for metabolomics, but it is largely unclear at the moment whether this only affects the power of the statistics or the robustness as well. In addition, as pointed out by Huang et al. (2009): ‘Due to the complexity of biological data-mining situations, in its current state, the analysis of large gene lists with the current enrichment tools is still more of an exploratory data-mining procedure rather than a pure statistical solution’. It is also unclear at the moment how, and to what extent, different omics sources can be combined systematically and statistically for pathway analysis.
Quality Assurance
Obtaining information-rich data sets from the emerging data streams, -omics, among others, is believed to produce important knowledge about disease and mechanisms of toxicity. This conceptual framework offers many opportunities for modern toxicology; many challenges, however, need to be addressed to ensure a sufficiently robust and informative outcome. Metabolomics information may be generated using diverse technological platforms, complex biological systems, and various analytical and computational methods (Castle et al., 2006), producing different data types and formats. It then becomes clear that measures and procedures for ensuring the quality of the data generated are necessary. We have to distinguish between quality assurance as best scientific practice for within-laboratory use and the regulatory use, which will typically require formal validation and acceptance of the method and a formal quality regime such as Good Laboratory Practice. Quality systems (QS) were developed originally for industrial production but they were increasingly introduced in many other sectors and organizations to establish a formal structure for establishing quality criteria. Examples of implementation in scientific and regulatory organizations are the US EPA Guidance for Developing Quality Systems for Environmental Programs (EPA, 2002) and the EPA Requirements for Quality Assurance Project Plans (EPA, 2001). Elements of a quality system include organizational structure, responsibilities, resources, methods, data management, processes and quality. We will focus on the quality elements. It is beyond the scope of this article to detail QS, and guidelines as well as an abundant review literature are available (such as the ISO 9000-series on quality management).
Transposing QS to research activities involving biological testing and analytical methods, method validation, and quality control would be the foundation of such a system. In general, method validation through laboratory studies ensures that the performance characteristics of the method meet the requirements for the intended application and provides an assurance of reliability during normal use. Method validation may be driven by regulatory requirements but it also provides good science, and allows quality control and assurance. While this is well accepted and widely applied in laboratory services and research and development (R&D), in academia many, if not most, researchers make little use of quality systems and method validation studies (Krull and Swartz, 1999). There are various reasons for this and they are detailed in (Mathur-De, 2000), where the author analyzes the scope and limitations of quality systems for research centers. It is not our intent to suggest that academic research should meet all regulatory guidelines, but we believe that in such interdisciplinary and rapidly evolving disciplines, the implementation of minimum and fit-for-purpose quality measures would be beneficial for monitoring and evaluating the quality of test methods and for assuring the accuracy and reliability of the test results.
In this part we will briefly describe the state-of-the-art in regulatory requirements for method validation and list some guidelines that may be of interest in metabolomics studies. We will then propose a general scheme for a quality system in a research project and advise some practical measures specific to metabolomics studies.
The principle of the validation of analytical methods is widespread in all the domains of activities where measurements are made. There are various regulatory documents and guidelines of normative character released by regulatory bodies [International Conference on Harmonization (ICH), the US Food and Drug Administration (FDA), the European Medicines Agency (EMEA), the International Organization for Standardization (ISO)] and other documents related to good practices [Good Laboratory Practices (GLP), Good Manufacturing Practices (GMP)]. For example, ICH has developed a text on the validation of analytical procedures (ICH, 1995). The FDA has proposed guidelines on analytical procedures and methods validation (FDA, 2000). A common ground for all these documents is the parameters for validation. They are defined by ICH (1996) and by other organizations and typically include: precision, accuracy, linearity, range, ruggedness, limit of detection, limit of quantitation, selectivity and specificity.
In addition, for toxicity testing, guidelines were primarily developed by three organizations: the Organisation for Economic Cooperation and Development (OECD), the European Centre for the Validation of Alternative Methods (ECVAM) (Hartung et al., 2004) and the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM). Criteria to be addressed in a validation exercise include (OECD, 2005): test definition (including purpose, need and scientific basis), relevance of the test method, repeatability and reproducibility, inter-laboratory transferability, predictive capacity and applicability domain. Although questions arise about whether the validation process, as it has been formalized over the last two decades, might meet the challenges of emerging methods and technologies especially in toxicity testing (Hartung, 2007, 2010; Leist et al., 2012), a pragmatic approach would be to adapt some of the principles and criteria listed above for ensuring some degree of quality in metabolomics studies.
In order to devise a quality system, let us first illustrate a scheme for the different stages of a scientific activity within a research project.
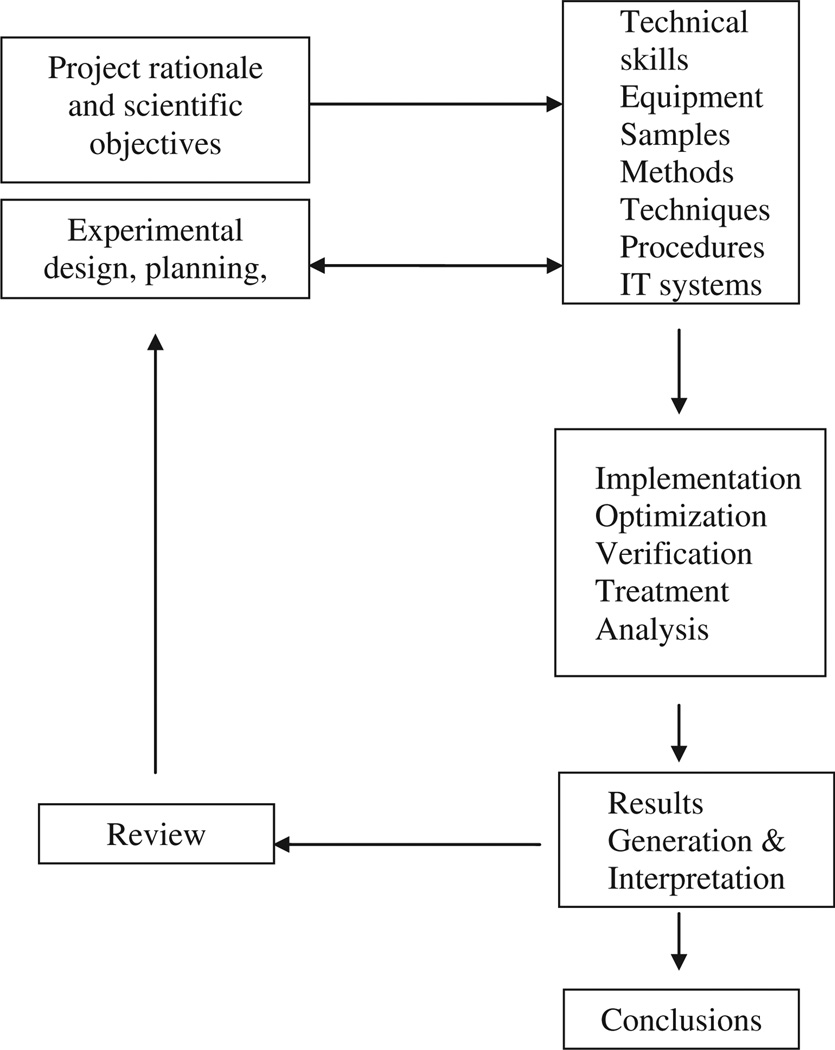
Figure 3 depicts the different elements of a quality controlled research project. It combines scientific and technical competences in addition to processing a task. It could also be divided into stages, starting with planning and management, then design and implementation, and finally analysis and review. The scientific rationale and objectives are the central part of the research activity and should be well-defined prior to any practical execution. A precise experimental design takes into account the resources, deadlines, and expected outcomes. It is also very dependent upon the available technical competences. We can note that this workflow is iterative, which illustrates the exploratory nature of scientific research. Indeed, once the results are generated, a process of review allows the modification, refinement, or optimization of the objectives and associated experimental design, and the drawing of conclusions in line with the projected objectives. Quality measures include a definition of a management organizational structure with identified roles and responsibilities. A well-documented project definition document notes the scientific rationale and objectives, all methods and standard operating procedures (SOP), methods and tools for results derivation, and any relevant information for an effective operation (such as the acceptance criteria and performance standards).
Figure 3.
General workflow of a research project adapted and modified from Mathur-De (2000).
Taking into account the structure of a research project and the principles of regulatory validation of analytical methods and of the validation of alternative methods, we can list some elements that lay the groundwork for quality control:
Definition of the scientific rationale, purpose, and objectives
Description of the scientific relevance
Applicability domain
Experimental design and planning
Technical description of test methods, including:
Optimization and specification of the test measurements
Development of standard operation procedures
Description of the methods and tools to derive and interpret results
Definition of the acceptance criteria
Implementation of quality assurance procedures
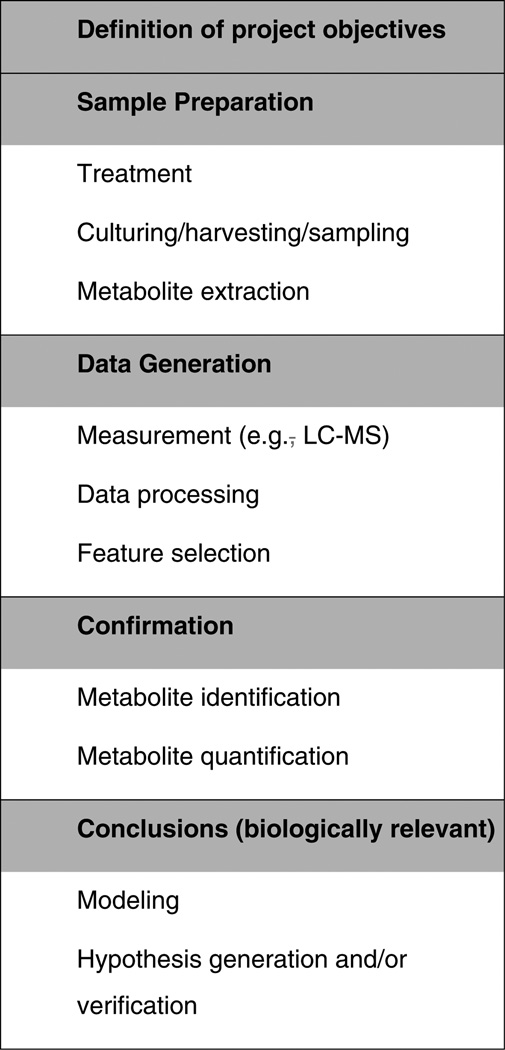
More specifically, for metabolomics studies, a typical workflow, as shown in Fig. 4, involves a series of tasks ranging from sample preparation, data acquisition, (pre-)processing, statistical analysis, metabolite identification and knowledge inference (such as identification and validation of pathways of toxicity).
Figure 4.
Typical metabolomics workflow.
The first step in the quality assurance process, as devised earlier, is to provide a clear description of the biological context, including the rationale, purpose and objectives, as well as the relevance of the metabolomics study (or test method). It also describes the method limitation and its applicability domain. The experimental design and planning will depend on the metabolomic approach adopted. Indeed, many approaches are currently used in metabolomics studies, ranging from fingerprinting to non-targeted profiling to targeted analysis (Robertson et al., 2011). In addition, clear and comprehensive standardized test method protocols are used together with standard operating procedures (SOP). This should include a description of the test system, sampling and treatment conditions, endpoint(s) assessed, measurements taken, specialized equipment or supplies that may be needed, measures of variability, the way in which the results are calculated and expressed, and the use of positive and negative controls and other performance checks. As numerous literature is available that gives valuable details of aspects related to the technical description of the test methods (OECD, 2005), we will focus on the implementation of quality procedures.
The metabolomics society has initiated an initiative to formulate a minimum of reporting standards that describe the experiments, called the Metabolomics Standards Initiative (MSI) (Castle et al., 2006). It follows the general ‘workflow’ model in metabolomics: from a description of the study design to sample workup, data acquisition, processing and export, and bound together by controlled vocabularies and relationships between the terms used. Five areas were identified that together describe metabolomics experiments: biological sample context, chemical analysis, data analysis, ontology and data exchange. A comparable initiative called ArMet (Jenkins et al., 2004) proposed a framework for the description of plant metabolomics experiments and their results with a formal data description for plant metabolomics that supports the results and the experimental metadata. We can note that these initiatives are in line with the proposed quality assurance process proposed here, especially with regard to definitions, specifications and reporting. Moreover, we suggest the drafting of a study definition document that includes all this relevant information to facilitate standardization and assist in data exchange.
Besides documentation and reporting, the quality measures should take into account the variability of the data generated. In general, variability reflects a combination of biological and analytical variability. Biological variability depends on factors inherent to the sample itself (genetics, environment, behaviorand health status), and effects introduced by the choice of the sampling procedure and its execution. Analytical variability results from issues in sample acquisition and storage, laboratory-related errors (including methodological errors, instrument imprecision, inconsistent or impure reagents and matrix effects), as well as errors related to the design of the experiment. These can result in bias or systematic errors and can also result in decreased reproducibility (Shurubor et al., 2005).
In this regard, many factors contribute to variability and influence the quality of the metabolomics data. The procedures used in sample collection, preparation and handling can affect the extracted metabolites. Optimized and consistent extraction protocols, as well as adequate sample storage, limits the variability in the metabolites extraction and analysis (Zhou et al., 2012). SOPs should be optimized for the specific metabolomic application and well documented. All the steps relevant to the sample preparation should be detailed enough to allow an unambiguous execution of the procedures. They should contain the important elements for the execution of the procedure, such as the equipment needed and a list of tasks in logical (chronological) order in accordance with the laboratory resources and practices. The SOPs contain the exact procedures of sampling, sample treatment, sample storage and metabolite extraction. Critical elements affecting the nature and quantities of the metabolites should be highlighted, such as the storage temperatures at different phases of the procedures, the exact quantities of sample and reagents used, etc. Any deviation from the SOP should be justified and documented.
Quality control procedures also include the estimation of the stability of the analytical procedure, the use of standards, error estimation of data reproducibility and criteria for data inclusion (and exclusion). Regular instrument calibration should be planned and performed. For MS, for example, resolution, sensitivity, retention time alignment, mass calibration and mass accuracy should be reported (Lindon et al., 2005a). Measures of quality are mainly contained in the experimental design, where the use of sufficient biological replicates helps minimize the random errors, estimate the overall method precision and allows a robust statistical analysis. Blank samples, which are analyte-free and prepared exactly as the test samples, give an idea of the overall levels of contamination and carryover. ‘QC samples’ may be used during a metabolomics experiment. Usually, they are prepared by mixing equal volumes of all samples present. This pooled sample provides a representative ‘mean’ of all analyzed metabolites (Gika et al., 2007). When injected at regular time intervals, it also provides a mean for estimating the stability of the analytical method. ‘QC samples’ may also be used for establishing acceptance criteria of the data. Another quality factor of the experimental design includes the randomization of the samples. This procedure ensures that no bias is introduced by preparing and analyzing replicate samples jointly. Nevertheless, care should be taken that randomization does not provide significant additional carry-over.
Besides analytical and biological variability, data quality is highly dependent on the data analysis, the algorithms applied and their parameters. Once the metabolite data are acquired, they are handled in order to prepare and reduce analytical instrument raw data (e.g. MS chromatograms) to data matrices for further analysis. The details of the steps involved were developed earlier in this paper and in another review paper (Sugimoto et al., 2012). Usually, a series of tasks are performed ranging from low-level processing (background correction, feature detection, normalization, alignment, etc.) to higher level processing consisting of various tools and methods for interpretation and visualization of the pre-processed data. This typically includes statistical analysis tools such as multivariate analyses to reveal, for example, differences between groups of samples. The most important sources of error in data analysis are the incorrect peak integration and alignment and the feature discrepancies in the biological replicates (Brodsky et al., 2010). Owing to the multiplicity of algorithms and approaches used and the variety of parameters applied, it is difficult to advise on a standard quality control procedure. However, for a quality controlled data processing workflow, all data manipulation steps should be documented and consistently applied when needed (e.g. in the same study). Adequate quality checks and manual curation are usually necessary in this type of processing. An example for quality control framework of data analysis is given in (Brodsky et al., 2010). Typically, the QC approach is based on discrepancies between replicate samples. After normalization, the overall quality of each replicate group is characterized by correlation coefficients between samples before removing sample outliers.
In summary, the value of information derived from metabolomics studies is directly related to the quality of data generated. Although many initiatives have been presented, such as for standardizing the reporting of metabolomics analyses or for ensuring quality in specific tasks of the metabolomics workflow, no general framework for quality assurance has been presented. By adapting some elements and the general principles from well established procedures, we provided here a general scheme for quality control implementation starting with a clear definition of the scientific basis of the study and the documentation and reporting of information on the methods and protocols used, whether they are biological, analytical or for data processing.
Practical Use of Toxicometabolomics
Metabolomics is increasingly used as a tool in toxicology. Compared with other omics approaches (genomics, transcriptomics and proteomics), it has several advantages. As mentioned earlier, it is closer to the phenotype and does not necessarily preprocess samples allowing further analyses. Metabolomics measures the final outcome after a toxic insult in a cascade of events, such as alterations of genes, transcripts, proteins and finally metabolites. Owing to the biological complexity, changes in one of them may or may not lead to changes in the others. Determining the final alteration (metabolites) enhances the possibility of understanding the actual toxicity and to associate the effects to adverse outcomes or phenotypic changes. Moreover, the time and costs for a single run are lower compared with other omics technologies. While chromatography adds some variability, mass detection is very accurate for enabling substance identification; the latter is further enhanced by MS/MS techniques. Besides, it is believed that the number of metabolites is fewer than the amounts of genes and proteins, making it easier to examine (van Ravenzwaay et al., 2007; Hartung et al., 2012). However, some can be difficult to identify owing to limitations of current databases.
Initially, metabolomics was mainly used for measurements of bio-fluids (e.g. blood, serum and urine), but it is also applicable to several other systems such as breast milk (Kalantzi et al., 2004), saliva (Bertram et al., 2009), sweat (Kutyshenko et al., 2011), spinal fluid (Crews et al., 2009), breath air (Kanoh et al., 2005), tissue (Coen et al., 2012) and cell cultures (Ellis et al., 2011; van Vliet et al., 2008; West et al., 2010). The use of metabolomics approaches has increased over the years for, in vivo, in vitro and in clinical applications. This chapter will introduce some applications of toxicometabolomics.
In vivo whole animal studies
Early 2000, Robertson and Bulera (2000) suggested the use of metabolomics as a new technique for rapid toxicity screening. Shortly after, the Consortium for Metabonomic Toxicology (COMET) was formed between five pharmaceutical companies and the Imperial College of Science, Technology and Medicine (IC), in London, UK (Lindon et al., 2005b). The consortium performed metabolomic studies to predict liver and kidney toxicity using urine and serum samples from rodents after 7 days of exposure to 147 compounds. The generated database still serves as a useful tool in the area of drug discovery and development within these companies (Bollard et al., 2010; Spagou et al., 2011). The chemical industry has also recognized metabolomics as a useful tool for toxicity testing. In 2012, Metanomics Health, a BASF Group company, launched their new MetaMap®Tox database (van Ravenzwaay et al., 2012). The database has been developed over several years, testing more than 500 chemicals (28-day rat studies) to generate metabolomic patterns for different toxicological targets (e.g. liver, kidney, thyroid, testes, blood, nervous system and endocrine systems). The metabolomic approach suggests toxicological mode of actions at an early state and can, therefore, speed up safety decisions and lower the cost through reduced animal studies. MetaMap®Tox is routinely used at BASF and is also offered as a customer service.
Blood, serum and urine are the most commonly-used samples for in vivo metabolomics studies, but the technique can be applied to other bio fluids or tissue samples. One example is a metabolomic study of liver samples from mice showing that polychlorinated biphenyls (PCBs) induce non-alcoholic fatty liver disease (Shi et al., 2012). The same association has been observed in epidemiological studies (Cave et al., 2010). In most studies however, tissue samples are only analyzed as a complement to bio-fluid measurements and give limited additional information (Coen et al., 2012; Shi et al., 2012).
Metabolomics is not only a useful tool for screening; it has increased the mechanistic knowledge within several areas of toxicology (Coen et al., 2007, 2012; van Ravenzwaay et al., 2007; Shima et al., 2011; Sieber et al., 2009; Coen et al., 2012) and the approach has been used to gain information about differences between genders, strains, age and species (Zhang et al., 2009a, 2009b; Qiao et al., 2011). In addition, toxico-metabolomics has shown to be a promising tool for safety assessment in newly identified challenges such as nano-materials (Bu et al., 2010; Tang et al., 2010) and dissimilar individual susceptibilities owing to diverse gut flora (Nicholson et al., 2012; Romick-Rosendale et al., 2009).
Although the probability is low that a single metabolite will be able to predict certain toxicity, metabolomic studies clearly show that a pattern of several changed metabolites can provide knowledge of target toxicity and give us a better mechanistic understanding. A strength of toxicometablomics studies lies in the context of systems biology (multi-omics) approaches, where metabolite profiles can help resolve questions that arise if one had only done a genomics-based study. One example of such integrated analysis software is the GeneSpring-Integrated Biology software platform (Agilent Technologies, Santa Clara, CA, USA) which allows the co-analysis of differential gene expression and metabolomics data. GeneSpring-IB is configurable for various multi-omics data analysis combinations. It imports raw data, performs a variety of statistical analyses, conducts pathway analyses, integration and visualization.
Clinical applications
Metabolomics is used extensively in clinics for diagnosis, prognosis and therapeutic evaluation of several diseases. The biggest impact of metabolomics in clinical applications is in the field of oncology (reviewed by Spratlin et al., 2009). Biofluids are more commonly used than tissue samples. Aside from the complexity in sample collection, tumors can be difficult to evaluate owing to their heterogeneity. Metabolomics analysis of samples from patients have identified promising biomarkers that can be used to detect several kinds of cancers such as bladder (Cao et al., 2012; Jobu et al., 2012), colorectal (Nishiumi et al., 2012), breast (Slupsky et al., 2010), ovarian (Garcia et al., 2011; Slupsky et al., 2010;), kidney (Ganti and Weiss, 2011; Lin et al., 2011b), liver (Chen et al., 2011b) and lung cancer (An et al., 2010). In addition, metabolomics has been useful in cancer research for therapeutic evaluations (Chung et al., 2003; Tenori et al., 2012) and in mechanistic understanding (Adinolfi et al., 2012; Denkert et al., 2012; Klawitter et al., 2011).
Furthermore, metabolomics has enhanced the research in other medical areas such as liver diseases (Kalhan et al., 2011; Wang et al., 2012), kidney diseases (Sato et al., 2011; Sun et al., 2012), respiratory diseases (Kanoh et al., 2005), diabetes (Ament et al., 2012; Brugnara et al., 2012; Lanza et al., 2010), obesity (Ament et al., 2012), cardiovascular diseases (Senn et al., 2012), multiple sclerosis (Smolinska et al., 2012), inflammatory bowel disease (Stephens et al., 2012) and neurodegenerative diseases (Johansen et al., 2009; Sato et al., 2011). Metabolomics has evidently shown to be a useful tool for examination, identification and for gaining information about numerous diseases. Toxicants contribute to many of these diseases, and their roles might be elucidated in the respective models and patient populations.
In vitro studies
In vitro approaches are increasingly recognized in toxicology. In 2007, the National Research Council published Toxicity Testing in the 21st Century: a Vision and a Strategy (NRC, 2007). The report proposed a paradigm shift in toxicology, where current animal tests should be replaced by in vitro cell systems combined with new, promising techniques such as metabolomics. In vitro and metabolomics are still not intensively applied but are increasingly used and have shown to be very promising in toxicology. One advantage of cell-based metabolomics is that both extracellular and intracellular metabolites can be measured. Metabolites detected in extracellular samples (cell culture media), can be associated to in vivo identified biomarkers from bio-fluids, while the intracellular samples (cell lysate) can provide information about the toxic mechanisms on the cellular level. The main studies are still performing measurements of cell culture media, but the assessment of intra-cellular metabolites is constantly increasing.
One promising approach is the use of human embryonic stem cells in combination with extracellular metabolomic analysis. The model has shown potential to predict several teratogenic compounds (Cezar et al., 2007; Kleinstreuer et al., 2011; West et al., 2010). The use of human cells is of high value, as it can provide information about human toxicity and will prevent the need for inter-species extrapolation.
Moreover, measurements of intracellular metabolites have shown promising results in the assessment of neurotoxicity (van Vliet et al., 2008), renal toxicity (Ellis et al., 2011), hepatotoxicity (Ruiz-Aracama et al., 2011), mitochondrial toxicity (Balcke et al., 2011), lung toxicity (Vulimiri et al., 2009) and the toxicity of cell-penetrating peptides (Kilk et al., 2009). One of the main challenges of using cell lysate is the extraction procedure. Several groups, including ours, have spent a lot of time exploring the best methods to achieve robust, reproducible and effective data (Dietmair et al., 2010; Meyer et al., 2010).
The latest concept in in vitro toxicology is the development of human-on-a-chip (Hartung and Zurlo, 2012). Multiple cell culture chambers with diverse cell types are connected with fluidic channels to mimic multi-organ interactions. The interaction of the organs is essential for their function and the chip is foreseen to mimic the human body and to better predict the toxicity of chemicals and drugs (Sung et al., 2010). The approach has also been applied in the ‘metabolomics-on-a-chip’ assay where liver and kidney cells have been successfully co-cultured and are able to predict organ specific toxicity (Shintu et al., 2012).
Pathways of toxicity
Combinations of omics techniques, for example, metabolomics and transcriptomics, are believed to be valuable in the identification of PoT or adverse outcome pathways (Hartung and McBride, 2011). The term PoT was coined to distinguish molecularly defined pathways from the toxicity pathways/adverse outcome pathways, which are typically reported in a narrative form. Metabolites that are changed can be associated with metabolic pathways using available databases such as KEGG and WikiPathways. Adding information from transcriptomics or proteomics will confirm and give a broader knowledge about the perturbed pathways (Parman et al., 2011). One of the first integrated transcriptomics and metabolomics studies showed that the combination was more sensitive in detecting chemically induced hepatotoxicity and enabled the generation of new hypotheses on the cellular mechanism (Heijne et al., 2005b). However, the use of metabolomics alone has already shown to reveal information on possible PoT in several areas such as, ecotoxicology (Ankley et al., 2009; Santos et al., 2010), obesity (Xie et al., 2012), developmental toxicity (West et al., 2010), hepatotoxicity (Shintu et al., 2012), cardio toxicity (Wang et al., 2012) and toxicity of cell-penetrating peptides (Kilk et al., 2009).
Metabolic flux analysis is another recently used tool in in vitro toxicometabolomics studies, though it is more advanced in plant physiology. Cells in flux studies are fed with carbon-labeled substrates that, owing to metabolic activity, are distributed throughout the metabolic pathways. Analysis of the labeled metabolites can provide a better insight into cellular physiology. Furthermore, it can provide information about interactions between cellular metabolism and detoxification pathways (Boslem et al., 2011; Iyer et al., 2010).
Metabolomics in combination with in vitro systems is a promising tool in the search for PoT to better predict and understand toxicity in humans. It can lay the foundation of a Systems Toxicology approach (Hartung et al., 2012).
Discussion
Metabolomics use in toxicology is rapidly increasing. As a technology capturing the phenotypic change on molecular level, it lends itself to characterization of the impact of an agent on the body’s physiology. Table 2 compares it to the other most prominent omics technologies currently in use in toxicology. It can be used on bio-fluids as well as tissues and their cultures. The small sample volume requirements are ideally suited for work with small rodents or tissue cultures. In contrast to transcriptomics, the costs of single measurements are much lower, allowing more replicates, concentrations or time-points to be assessed. The relatively small number of metabolites and the very well-established biochemical pathways connecting them allow for a superior background for interpretation. This review focuses on LC-MS and GC-MS as bioanalytical analysis techniques and not on NMR. However, many aspects of quality assurance and applications in toxicology can be directly translated.
Table 2.
Comparison of advantages, limitations and developmental needs for different omics technologies used in toxicology.
| Transcriptomics / genomics | Proteomics | Metabolomics | |
|---|---|---|---|
| Advantages |
|
|
|
| Limitations |
|
|
|
| Developmental needs |
|
|
|
Modified from van Vliet, 2011. Current standing and future prospects for the technologies proposed to transform toxicity testing in the 21st century. ALTEX 28: 17–44.
Metabolomics makes a true Systems Toxicology assessment possible – one that is not possible by traditional animal tests. However, we need to be aware that metabolic responses are in a flux with timing of metabolic responses probably also being characteristic for individual toxins and their concentrations. This is very different than the much longer turnover of mRNA and proteins. For modeling in Systems Toxicology approaches, this flux needs to be addressed, but the respective ‘fluxomics’ is only emerging.
The relatively high throughput of metabolomics enables us to ask more questions in parallel. For example more doses, replicates or time-points from close to the onset of initial biomolecular events (chemico-biological interaction) to the manifested toxicological effect, can be assessed. It also provides very direct mechanistic insights. Studies can focus on intracellular changes, but also on other bio-fluids, allowing less invasive measurements and incorporation into current guideline studies and human trials. Low dose effects that do not give results in animals on the level of (histo)pathology can be monitored. But as with other omics data, this might indicate that non-relevant effects of agents are being discerned. While it could be an extremely powerful tool to identify alerts of biological responses overlooked in the respective apical tests, it presents a significant danger of misclassification. It will have to be shown whether the advantages and disadvantages outweigh each other.
Regulatory acceptance for metabolomics data is largely unclear, but interest is high. New guidelines for submission and use of such data will be necessary, which will take some time. Taken together, metabolomics represents an enabling technology – not a game-changer on its own, but a core technology for implementing Tox-21c, the most promising revolution in regulatory toxicology in decades.
Acknowledgments
NIH transformative research grant ‘Mapping the Human Toxome by Systems Toxicology’ (RO1ES020750) and FDA grant ‘DNTox-21c Identification of pathways of developmental neurotoxicity for high throughput testing by metabolomics’ (U01FD004230).
References
- Adinolfi E, Raffaghello L, Giuliani AL, Cavazzini L, Capece M, Chiozzi P, Bianchi G, Kroemer G, Pistoia V, Di Virgilio F. Expression of P2X7 Receptor Increases In Vivo Tumor Growth. Cancer Res. 2012;72:2957–2969. doi: 10.1158/0008-5472.CAN-11-1947. [DOI] [PubMed] [Google Scholar]
- Ament Z, Masoodi M, Griffin JL. Applications of metabolomics for understanding the action of peroxisome proliferator-activated receptors (PPARs) in diabetes, obesity and cancer. Genome Med. 2012;4:32. doi: 10.1186/gm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstalden van Hove ER, Smith DF, Heeren RM. A concise review of mass spectrometry imaging. J. Chromatogr. A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- An Z, Chen Y, Zhang R, Song Y, Sun J, He J, Bai J, Dong L, Zhan Q, Abliz Z. Integrated ionization approach for RRLC-MS/MS-based metabonomics: finding potential biomarkers for lung cancer. J. Proteome Res. 2010;9:4071–4081. doi: 10.1021/pr100265g. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bencic DC, Breen MS, Collette TW, Conolly RB, Denslow ND, Edwards SW, Ekman DR, Garcia-Reyero N, Jensen KM, Lazorchak JM, Martinoviå D, Miller DH, Perkins EJ, Orlando EF, Villeneuve DL, Wang R-L, Watanabe KH. Endocrine disrupting chemicals in fish: developing exposure indicators and predictive models of effects based on mechanism of action. Aquat. Toxicol. 2009;92:168–178. doi: 10.1016/j.aquatox.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Balcke GU, Kolle SN, Kamp H, Bethan B, Looser R, Wagner S, Landsiedel R, van Ravenzwaay B. "Linking energy metabolism to dysfunctions in mitochondrial respiration--a metabolomics in vitro approach". Toxicol. Lett. 2011;203:200–209. doi: 10.1016/j.toxlet.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Barba I, Fernandez-Montesinos R, Garcia-Dorado D, Pozo D. Alzheimer’s disease beyond the genomic era: nuclear magnetic resonance (NMR) spectroscopy-based metabolomics. J. Cell. Mol. Med. 2008;12:1477–1485. doi: 10.1111/j.1582-4934.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basketter DA, Clewell H, Kimber I, Rossi A, Blaauboer B, Burrier R, Daneshian M, Eskes C, Goldberg A, Hasiwa N, Hoffmann S, Jaworska J, Knudsen TB, Landsiedel R, Leist M, Locke P, Maxwell G, McKim J, McVey EA, Ouédraogo G, Patlewicz G, Pelkonen O, Roggen E, Rovida C, Ruhdel I, Schwarz M, Schepky A, Schoeters G, Skinner N, Trentz K, Turner M, Vanparys P, Yager J, Zurlo J, Hartung T. A roadmap for the development of alternative (non-animal) methods for systemic toxicity testing. ALTEX. 2012;29:3–89. doi: 10.14573/altex.2012.1.003. [DOI] [PubMed] [Google Scholar]
- Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol. Appl. Pharmacol. 2010;243:154–166. doi: 10.1016/j.taap.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Begriche K, Massart J, Robin MA, Borgne-Sanchez A, Fromenty B. Drug-induced toxicity on mitochondria and lipid metabolism: mechanistic diversity and deleterious consequences for the liver. J. Hepatol. 2011;54:773–794. doi: 10.1016/j.jhep.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Beran JA, Kevan L. Molecular Electron Ionization Cross Sections at 70-Ev. J. Phys. Chem. 1969;73:3866–3876. [Google Scholar]
- Bertram HC, Eggers N, Eller N. Potential of human saliva for nuclear magnetic resonance-based metabolomics and for health-related biomarker identification. Anal. Chem. 2009;81:9188–9193. doi: 10.1021/ac9020598. [DOI] [PubMed] [Google Scholar]
- Bollard ME, Contel NR, Ebbels TMD, Smith L, Beckonert O, Cantor GH, Lehman-McKeeman L, Holmes EC, Lindon JC, Nicholson JK, Keun HC. NMR-based metabolic profiling identifies biomarkers of liver regeneration following partial hepatectomy in the rat. J. Proteome Res. 2010;9:59–69. doi: 10.1021/pr900200v. [DOI] [PubMed] [Google Scholar]
- Boslem E, MacIntosh G, Preston AM, Bartley C, Busch AK, Fuller M, Laybutt DR, Meikle PJ, Biden TJ. A lipidomic screen of palmitate-treated MIN6 beta-cells links sphingolipid metabolites with endoplasmic reticulum (ER) stress and impaired protein trafficking. Biochem. J. 2011;435:267–276. doi: 10.1042/BJ20101867. [DOI] [PubMed] [Google Scholar]
- Bottini AA, Hartung T. The economics of animal testing. ALTEX Special Issue. 2010;27:67–77. doi: 10.14573/altex.2009.1.3. [DOI] [PubMed] [Google Scholar]
- Brodsky L, Moussaieff A, Shahaf N, Aharoni A, Rogachev I. Evaluation of peak picking quality in LC-MS metabolomics data. Anal. Chem. 2010;82:9177–9187. doi: 10.1021/ac101216e. [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 2006;78:4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- Brugnara L, Vinaixa M, Murillo S, Samino S, Rodriguez MA, Beltran A, Lerin C, Davison G, Correig X, Novials A. Metabolomics approach for analyzing the effects of exercise in subjects with type 1 diabetes mellitus. PLoS One. 2012;7:e40600. doi: 10.1371/journal.pone.0040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Yan G, Deng P, Peng F, Lin H, Xu Y, Cao Z, Zhou T, Xue A, Wang Y, Cen X, Zhao Y-L. NMR-based metabonomic study of the sub-acute toxicity of titanium dioxide nanoparticles in rats after oral administration. Nanotechnology. 2010;21:125105. doi: 10.1088/0957-4484/21/12/125105. [DOI] [PubMed] [Google Scholar]
- Cao M, Zhao L, Chen H, Xue W, Lin D. NMR-based metabolomic analysis of human bladder cancer.”. Anal. Sci. 2012;28:451–456. doi: 10.2116/analsci.28.451. [DOI] [PubMed] [Google Scholar]
- Castle AL, Fiehn O, Kaddurah-Daouk R, Lindon JC. Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Brief. Bioinform. 2006;7:159–165. doi: 10.1093/bib/bbl008. [DOI] [PubMed] [Google Scholar]
- Cave M, Appana S, Patel M, Falkner KC, McClain CJ, Brock G. Polychlorinated biphenyls, lead, and mercury are associated with liver disease in American adults: NHANES 2003-2004. Environ. Health Perspect. 2010;118:1735–1742. doi: 10.1289/ehp.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezar GG, Quam JA, Smith AM, Rosa GJM, Piekarczyk MS, Brown JF, Gage FH, Muotri AR. Identification of small molecules from human embryonic stem cells using metabolomics. Stem Cells Dev. 2007;16:869–882. doi: 10.1089/scd.2007.0022. [DOI] [PubMed] [Google Scholar]
- Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/ mass spectrometry-based metabonomic method. Anal. Bioanal. Chem. 2011a;401:1899–1904. doi: 10.1007/s00216-011-5245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang X, Cao R, Lu X, Zhao S, Fekete A, Huang Q, Schmitt-Kopplin P, Wang Y, Xu Z, Wan X, Wu X, Zhao N, Xu C, Xu G. Serum 27-nor-5beta-cholestane-3,7,12,24,25 pentol glucuronide discovered by metabolomics as potential diagnostic biomarker for epithelium ovarian cancer. J. Proteome Res. 2011b;10:2625–2632. doi: 10.1021/pr200173q. [DOI] [PubMed] [Google Scholar]
- Chung Y-L, Troy H, Banerji U, Jackson LE, Walton MI, Stubbs M, Griffiths JR, Judson IR, Leach MO, Workman P, Ronen SM. Magnetic resonance spectroscopic pharmacodynamic markers of the heat shock protein 90 inhibitor 17-allylamino,17-demethoxygeldanamycin (17AAG) in human colon cancer models. J. Natl. Cancer Inst. 2003;95:1624–1633. doi: 10.1093/jnci/djg084. [DOI] [PubMed] [Google Scholar]
- Coen M, Goldfain-Blanc F, Rolland-Valognes G, Walther B, Robertson DG, Holmes E, Lindon JC, Nicholson JK. Pharmacometabonomic investigation of dynamic metabolic phenotypes associated with variability in response to galactosamine hepatotoxicity. J. Proteome Res. 2012;11:2427–2440. doi: 10.1021/pr201161f. [DOI] [PubMed] [Google Scholar]
- Coen M, Hong YS, Clayton TA, Rohde CM, Pearce JT, Reily MD, Robertson DG, Holmes E, Lindon JC, Nicholson JK. The mechanism of galactosamine toxicity revisited; a metabonomic study. J. Proteome Res. 2007;6:2711–2719. doi: 10.1021/pr070164f. [DOI] [PubMed] [Google Scholar]
- Craig A, Sidaway J, Holmes E, Orton T, Jackson D, Rowlinson R, Nickson J, Tonge R, Wilson I, Nicholson J. Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J. Proteome Res. 2006;5:1586–1601. doi: 10.1021/pr0503376. [DOI] [PubMed] [Google Scholar]
- Crews B, Wikoff WR, Patti GJ, Woo H-K, Kalisiak E, Heideker J, Siuzdak G. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal. Chem. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK. Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies. Anal. Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- Denkert C, Bucher E, Hilvo M, Salek R, Orešič M, Griffin J, Brockmöller S, Klauschen F, Loibl S, Barupal DK, Budczies J, Iljin K, Nekljudova V, Fiehn O. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4:37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmair S, Timmins NE, Gray P, Nielsen LK, Krömer JO. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Ellis JK, Athersuch TJ, Cavill R, Radford R, Slattery C, Jennings P, McMorrow T, Ryan MP, Ebbels TMD, Keun HC. Metabolic response to low-level toxicant exposure in a novel renal tubule epithelial cell system. Mol. Biosyst. 2011;7:247–257. doi: 10.1039/c0mb00146e. [DOI] [PubMed] [Google Scholar]
- EPA. EPA Requirements for Quality Assurance Project Plans. U.S. Environmental Protection Agency (USEPA; 2001. [Google Scholar]
- EPA. Guidance For Developing Quality Systems For Environmental Programs. U.S. Environmental Protection Agency (USEPA; 2002. [Google Scholar]
- Eriksson L, Johansson E, Kettapeh WS. Sweden: Umetrics, Umea; 1999. Introduction to Multi- and Megavariate Data Analysis Using Projection Methods (PCA & PLS) p. 425. [Google Scholar]
- FDA. Analytical Procedures and Methods Validation: Guidance for Industry. Rockville, MD: Food and Drug Administration; 2000. [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Fernie AR. The future of metabolic phytochemistry: Large numbers or metabolites, higher resolution, greater understanding. Phytochemistry. 2007;68:2861–2880. doi: 10.1016/j.phytochem.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:55–171. [PubMed] [Google Scholar]
- Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, Vinatier D, Wisztorski M, Day M, Fournier I, Salzet M. MALDI imaging mass spectrometry: state of the art technology in clinical proteomics. Mol. Cell Proteomics. 2009;8:2023–2033. doi: 10.1074/mcp.R800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- Fukuhara K, Ohno A, Ando Y, Yamoto T, Okuda H. A 1H NMR-based metabolomics approach for mechanistic insight into acetaminophen-induced hepatotoxicity. Drug Metab. Pharmacokinet. 2011;26:399–406. doi: 10.2133/dmpk.dmpk-11-rg-005. [DOI] [PubMed] [Google Scholar]
- Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol. Oncol. 2011;29:551–557. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E, Andrews C, Hua J, Kim HL, Sukumaran DK, Szyperski T, Odunsi K. Diagnosis of early stage ovarian cancer by 1H NMR metabonomics of serum explored by use of a microflow NMR probe. J. Proteome Res. 2011;10:1765–1771. doi: 10.1021/pr101050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gika HG, Theodoridis GA, Wingate JE, Wilson ID. Within-day reproducibility of an HPLC-MS-Based method for metabonomic analysis: Application to human urine. J. Proteome Res. 2007;6:3291–3303. doi: 10.1021/pr070183p. [DOI] [PubMed] [Google Scholar]
- Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics . Expert Rev. Mol. Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr. Opin. Chem. Biol. 2003;7:648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Kauppinen RA. A metabolomics perspective of human brain tumours. FEBS J. 2007;274:1132–1139. doi: 10.1111/j.1742-4658.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Targeted metabolomics for biomarker discovery. Angew. Chem. Int. Ed Engl. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- Han J, Datla R, Chan S, Borchers CH. Mass spectrometry-based technologies for high-throughput metabolomics. Bioanalysis. 2009;1:1665–1684. doi: 10.4155/bio.09.158. [DOI] [PubMed] [Google Scholar]
- Hanzlik RP, Bhatia P, Stitt R, Traiger GJ. Biotransformation and excretion of methylcyclopentadienyl manganese tricarbonyl in the rat. Drug Metab. Dispos. 1980;8:428–433. [PubMed] [Google Scholar]
- Hartung T. Toxicology for the twenty-first century. Nature. 2009;460:208–212. doi: 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- Hartung T, McBride M. Food for Thought … on mapping the human toxome. ALTEX. 2011;28:83–93. doi: 10.14573/altex.2011.2.083. [DOI] [PubMed] [Google Scholar]
- Hartung T. From alternative methods to a new toxicology. Eur. J. Pharmaceutics Biopharmaceutics. 2011;77(3):38–349. doi: 10.1016/j.ejpb.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Hartung T, van Vliet E, Jaworska J, Bonilla L, Skinner N, Thomas R. Food for thought… systems toxicology. ALTEX. 2012;29:119–128. doi: 10.14573/altex.2012.2.119. [DOI] [PubMed] [Google Scholar]
- Hartung T, Zurlo J. Food for thought … alternative approaches for medical countermeasures to biological and chemical terrorism and warfare. ALTEX. 2012;29:251–260. doi: 10.14573/altex.2012.3.251. [DOI] [PubMed] [Google Scholar]
- Hartung T. Food for thought… on alternative methods for chemical safety testing. ALTEX. 2010;27:3–14. doi: 10.14573/altex.2010.1.3. [DOI] [PubMed] [Google Scholar]
- Hartung T, Rovida C. Chemical regulators have overreached. Nature. 2009;460:1080–1081. doi: 10.1038/4601080a. [DOI] [PubMed] [Google Scholar]
- Hartung T. Food for thought … on alternative methods for cosmetics safety testing. ALTEX. 2008;25:147–162. doi: 10.14573/altex.2008.3.147. [DOI] [PubMed] [Google Scholar]
- Hartung T. Food for thought … on validation. ALTEX. 2007;24:67–72. doi: 10.14573/altex.2007.2.67. [DOI] [PubMed] [Google Scholar]
- Hartung T, Bremer S, Casati S, Coecke S, Corvi R, Fortaner S, Gribaldo L, Halder M, Hoffmann S, Roi AJ, Prieto P, Sabbioni E, Scott L, Worth A, Zuang V. A Modular Approach to the ECVAM principles on test validity. ATLA – Altern. Lab. Anim. 2004;32:467–472. doi: 10.1177/026119290403200503. [DOI] [PubMed] [Google Scholar]
- Heidema AG, Boer JMA, Nagelkerke N, Mariman ECM, Van Der A, Feskens EJM. The challenge for genetic epidemiologists: how to analyze large numbers of SNPs in relation to complex diseases. BMC Genet. 2006;7:23. doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijne WH, Kienhuis AS, van Ommen B, Stierum RH, Groten JP. Systems toxicology: applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteomics. 2005a;2:767–780. doi: 10.1586/14789450.2.5.767. [DOI] [PubMed] [Google Scholar]
- Heijne WHM, Lamers R-JAN, van Bladeren PJ, Groten JP, van Nesselrooij JH, van Ommen B. Profiles of metabolites and gene expression in rats with chemically induced hepatic necrosis. Toxicol. Pathol. 2005b;33:425–433. doi: 10.1080/01926230590958146. [DOI] [PubMed] [Google Scholar]
- Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICH. Validation of Analytical Procedures. Geneva: International Conference on Harmonization (ICH) of Technical Requirements for the Registration of Pharmaceuticals for Human Use; 1995. [Google Scholar]
- ICH. Validation of analytical procedures: Methodology. Geneva: International Conference on Harmonization (ICH) of Technical Requirements for the Registration of Pharmaceuticals for Human Use; 1996. [Google Scholar]
- Issaq HJ, Fox SD, Chan KC, Veenstra TD. Global proteomics and metabolomics in cancer biomarker discovery. J. Sep. Sci. 2011;34:3484–3492. doi: 10.1002/jssc.201100528. [DOI] [PubMed] [Google Scholar]
- Iyer VV, Ovacik MA, Androulakis IP, Roth CM, Ierapetritou MG. Transcriptional and metabolic flux profiling of triadimefon effects on cultured hepatocytes. Toxicol. Appl. Pharmacol. 2010;248:165–177. doi: 10.1016/j.taap.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Jackson JE. A User’s Guide to Principal Components. Hoboken, N.J: Wiley-Interscience; 2003. [Google Scholar]
- Jenkins H, Hardy N, Beckmann M, Draper J, Smith AR, Taylor J, Fiehn O, Goodacre R, Bino RJ, Hall R, Kopka J, Lane GA, Lange BM, Liu JR, Mendes P, Nikolau BJ, Oliver SG, Paton NW, Rhee S, Roessner-Tunali U, Saito K, Smedsgaard J, Sumner LW, Wang T, Walsh S, Wurtele ES, Kell DB. A proposed framework for the description of plant metabolomics experiments and their results. Nat. Biotechnol. 2004;22:1601–1606. doi: 10.1038/nbt1041. [DOI] [PubMed] [Google Scholar]
- Jobu K, Sun C, Yoshioka S, Yokota J, Onogawa M, Kawada C, Inoue K, Shuin T, Sendo T, Miyamura M. Metabolomics study on the biochemical profiles of odor elements in urine of human with bladder cancer. Biol. Pharm. Bull. 2012;35:639–642. doi: 10.1248/bpb.35.639. [DOI] [PubMed] [Google Scholar]
- Johansen K, Wang L, Aasly JO, White LJ, Matson WR, Henchcliffe C, Flint Beal M, Bogdanov M. Metabolomic profiling in LRRK2-related Parkinson’s disease. PLoS One. 2009;4:e7551. doi: 10.1371/journal.pone.0007551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P, Johansson AI, Gullberg J, Trygg J, Jiye A, Grung B, Marklund S, Sjostrom M, Antti H, Moritz T. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Kalantzi OI, Martin FL, Thomas GO, Alcock RE, Tang HR, Drury SC, Carmichael PL, Nicholson JK, Jones KC. Different levels of polybrominated diphenyl ethers (PBDEs) and chlorinated compounds in breast milk from two U.K. Regions.". Environ. Health Perspect. 2004;112:1085–1091. doi: 10.1289/ehp.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, GotoS HM, Hirakawa M, Itoh M, KatayamaT KS, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita K-F, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Data-base issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Gopalacharyulu P, Holm L, Oresic M. MPEA--metabolite pathway enrichment analysis. Bioinformatics. 2011;27:1878–1879. doi: 10.1093/bioinformatics/btr278. [DOI] [PubMed] [Google Scholar]
- Kanoh S, Kobayashi H, Motoyoshi K. Exhaled ethane: an in vivo bio-marker of lipid peroxidation in interstitial lung diseases. Chest. 2005;128:2387–2392. doi: 10.1378/chest.128.4.2387. [DOI] [PubMed] [Google Scholar]
- Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley: Hoboken, NJ; 2005. [Google Scholar]
- Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40(Database issue):D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilk K, Mahlapuu R, Soomets U, Langel U. Analysis of in vitro toxicity of five cell-penetrating peptides by metabolic profiling. Toxicology. 2009;265:87–95. doi: 10.1016/j.tox.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Kim HK, Choi YH, Verpoorte R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol. 2011;29:267–275. doi: 10.1016/j.tibtech.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Klawitter J, J Klawitter, et al. "Bezielle (BZL101)-induced oxidative stress damage followed by redistribution of metabolic fluxes in breast cancer cells: a combined proteomic and metabolomic study.". Int. J. Cancer. 2011;129:2945–2957. doi: 10.1002/ijc.25965. [DOI] [PubMed] [Google Scholar]
- Kleensang A, Franke D, Alcaïs A, Abel L, Müller-Myhsok B, Ziegler A. An extensive comparison of quantitative trait Loci mapping methods. Hum. Hered. 2010;69:202–211. doi: 10.1159/000289596. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Smith AM, West PR, Conard K, Fontaine B, Weir-Hauptman A, Palmer J, Knudsen TB, Dix DJ, Donley ELR, Cezar GG. Identifying developmental toxicity pathways for a subset of ToxCast chemicals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 2011;257:111–121. doi: 10.1016/j.taap.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Korman A, Oh A, Raskind A, Banks D. Statistical methods in metabolomics. Methods Mol. Biol. 2012;856:381–413. doi: 10.1007/978-1-61779-585-5_16. [DOI] [PubMed] [Google Scholar]
- Krull IS, Swartz M. Analytical method development and validation for the academic researcher. Analyt. Lett. 1999;32:1067–1080. [Google Scholar]
- Kutyshenko VP, Molchanov M, Beskaravayny P, Uversky VN, Timchenko MA. Analyzing and mapping sweat metabolomics by high-resolution NMR spectroscopy. PLoS One. 2011;6:e28824. doi: 10.1371/journal.pone.0028824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One. 2010;5:e10538. doi: 10.1371/journal.pone.0010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Hasiwa M, Daneshian M, Hartung T. Validation and quality control of replacement alternatives – current status and future challenges. Toxicol. Res. 2012;1:8. [Google Scholar]
- Li F, Lu J, Ma X. Profiling the reactive metabolites of xenobiotics using metabolomic technologies. Chem. Res. Toxicol. 2011;24:744–751. doi: 10.1021/tx200033v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shrestha B, Vertes A. Atmospheric pressure infrared MALDI imaging mass spectrometry for plant metabolomics. Anal. Chem. 2008;80:407–420. doi: 10.1021/ac701703f. [DOI] [PubMed] [Google Scholar]
- Lin L, Huang Z, Gao Y, Yan X, Xing J, Hang W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 2011a;10:1396–1405. doi: 10.1021/pr101161u. [DOI] [PubMed] [Google Scholar]
- Lin S, Yang Z, Liu H, Cai Z. Metabolomic analysis of liver and skeletal muscle tissues in C57BL/6J and DBA/2J mice exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Biosyst. 2011b;7:1956–1965. doi: 10.1039/c1mb05057e. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Nicholson JK, Holmes E, Keun HC, Craig A, Pearce JT, Bruce SJ, Hardy N, Sansone SA, Antti H, Jonsson P, Daykin C, Navarange M, Beger RD, Verheij ER, Amberg A, Baunsgaard D, Cantor GH, Lehman-McKeeman L, Earll M, Wold S, Johansson E, Haselden JN, Kramer K, Thomas C, Lindberg J, Schuppe-Koistinen I, Wilson ID, Reily MD, Robertson DG, Senn H, Krotzky A, Kochhar S, Powell J, van der Ouderaa F, Plumb R, Schaefer H, Spraul M. Summary recommendations for standardization and reporting of metabolic analyses. Nat. Biotechnol. 2005a;23:833–838. doi: 10.1038/nbt0705-833. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Keun HC, Ebbels TM, Pearce JM, Holmes E, Nicholson JK. The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005b;6:691–699. doi: 10.2217/14622416.6.7.691. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang L, You L, Cong M, Zhao J, Wu H, Li C, Liu D, Yu J. Toxicological responses to acute mercury exposure for three species of Manila clam Ruditapes philippinarum by NMR-based metabolomics . Environ. Toxicol. Pharmacol. 2011;31:323–332. doi: 10.1016/j.etap.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- Lucio M, Fekete A, Weigert C, Wagele B, Zhao X, Chen J, Fritsche A, Haring HU, Schleicher ED, Xu G, Schmitt-Kopplin G, Lehmann R. Insulin sensitivity is reflected by characteristic metabolic fingerprints--a Fourier transform mass spectrometric non-targeted metabolomics approach. PLoS One. 2010;5:e13317. doi: 10.1371/journal.pone.0013317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malley JD, Malley KG, Pajevic S. Cambridge, UK: Cambridge University Press; 2011. Statistical Learning for Biomedical Data (Practical Guides to Biostatistics and Epidemiology) p. 298. [Google Scholar]
- Mathur-De VR. The scope and limitations of a QA system in research. Accred. Quality Assur. 2000;5:3–10. [Google Scholar]
- Meyer H, Liebeke M, Lalk M. A protocol for the investigation of the intracellular Staphylococcus aureus metabolome. Anal. Biochem. 2010;401:250–259. doi: 10.1016/j.ab.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Monton MR, Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry . J. Chromatogr. A. 2007;1168:237–246. doi: 10.1016/j.chroma.2007.02.065. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Neerathilingam M, Volk DE, Sarkar S, Alam TM, Alam NK, Ansari GA, Luxon BA. 1H NMR-based metabonomic investigation of tributyl phosphate exposure in rats. Toxicol. Lett. 2010;199:10–16. doi: 10.1016/j.toxlet.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Neuweger H, Albaum SP, Dondrup M, Persicke M, Watt T, Niehaus K, Stoye J, Goesmann A. MeltDB: a software platform for the analysis and integration of metabolomics experiment data. Bioinfor-matics. 2008;24:2726–2732. doi: 10.1093/bioinformatics/btn452. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Everett JR, Lindon JC. Longitudinal pharmacometabonomics for predicting patient responses to therapy: drug metabolism, toxicity and efficacy. Expert Opin. Drug Metab. Toxicol. 2012;8:135–139. doi: 10.1517/17425255.2012.646987. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, Azuma T, Yoshida M. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7:e40459. doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC. Toxicity testing in the 21st century: a vision and a strategy. Washington, DC: National Academy Press; 2007. [Google Scholar]
- OECD. Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment. Paris: Environmental Health and Safety Monograph Series on Testing and Assessment No. 34. OECD; 2005. [Google Scholar]
- Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. Concordance of the Toxicity of Pharmaceuticals in Humans and in Animals. Reg. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Parman T, Bunin DI, Ng HH, McDunn JE, Wulff JE, Wang A, Swezey R, Rasay L, Fairchild DG, Kapetanovic IM, Green CE. Toxicogenomics and metabolomics of pentamethylchromanol (PMCol)-induced hepatotoxicity. Toxicol. Sci. 2011;124:487–501. doi: 10.1093/toxsci/kfr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Lanz C, Gonzalez FJ, Idle JR. The role of mass spectrometry-based metabolomics in medical countermeasures against radiation. Mass Spectrom. Rev. 2010;29:503–521. doi: 10.1002/mas.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persicke M, Rückert C, Plassmeier J, Stutz L, Kessler N, Kalinowski J, Goesmann A, Neuweger H. MSEA: metabolite set enrichment analysis in the MeltDB metabolomics software platform: metabolic profiling of Corynebacterium glutamicum as an example. Metabolomics. 2012;8:310–322. [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li T, Sun J, Liu X, Ren J, Fei J. Metabolomic analysis of normal (C57BL/6J, 129S1/SvImJ) mice by gas chromatography-mass spectrometry: detection of strain and gender differences. Talanta. 2011;85:718–724. doi: 10.1016/j.talanta.2011.04.060. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- Ramautar R, Somsen GW, de Jong GJ. CE-MS in metabolomics. Electrophoresis. 2009;30:276–291. doi: 10.1002/elps.200800512. [DOI] [PubMed] [Google Scholar]
- Ramautar R, Mayboroda OA, Somsen GW, de Jong GJ. CE-MS for metabolomics: developments and applications in the period 2008-2010. Electrophoresis. 2011;32:52–65. doi: 10.1002/elps.201000378. [DOI] [PubMed] [Google Scholar]
- Robertson DG. Metabonomics in toxicology: a review. Toxicol. Sci. 2005;85:809–822. doi: 10.1093/toxsci/kfi102. [DOI] [PubMed] [Google Scholar]
- Robertson DG, Watkins PB, Reily MD. Metabolomics in Toxicology: Preclinical and Clinical Applications. Toxicol. Sci. 2011;120:S146–S170. doi: 10.1093/toxsci/kfq358. [DOI] [PubMed] [Google Scholar]
- Robertson DG, Bulera SJ. High-throughput toxicology: practical considerations. Curr. Opin. Drug Discov. Devel. 2000;3:42–47. [PubMed] [Google Scholar]
- Robertson DG, Lindon JC. Metabonomics in Toxicity Assessment. Boca Raton, FL: Taylor & Francis; 2005. p. 536. [Google Scholar]
- Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, Kennedy MA. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn. Reson. Chem. 2009;47(Suppl 1):S36–S46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- Roux A, Lison D, Junot C, Heilier J-F. Applications of liquid chroma-tography coupled to mass spectrometry-based metabolomics in clinical chemistry and toxicology: A review. Clin. Biochem. 2011;44:119–135. doi: 10.1016/j.clinbiochem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Ruepp SU, Tonge RP, Shaw J, Wallis N, Pognan F. Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol. Sci. 2002;65:135–150. doi: 10.1093/toxsci/65.1.135. [DOI] [PubMed] [Google Scholar]
- Ruiz-Aracama A, Peijnenburg A, Kleinjans J, Jennen D, van Delft J, Hellfrisch C, Lommen A. An untargeted multi-technique metabolomics approach to studying intracellular metabolites of HepG2 cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics. 2011;12:251. doi: 10.1186/1471-2164-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos EM, Ball JS, Williams TD, Wu H, Ortega F, van Aerle R, Katsiadaki I, Falciani F, Viant MR, Chipman JK, Tyler CR. Identifying health impacts of exposure to copper using transcriptomics and metabolomics in a fish model. Environ. Sci. Technol. 2010;44:820–826. doi: 10.1021/es902558k. [DOI] [PubMed] [Google Scholar]
- Sato E, Kohno M, Yamamoto M, Fujisawa T, Fujiwara K, Tanaka N. Metabolomic analysis of human plasma from haemodialysis patients. Eur. J. Clin. Invest. 2011;41:241–255. doi: 10.1111/j.1365-2362.2010.02398.x. [DOI] [PubMed] [Google Scholar]
- Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spec-trometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg LK, Jones RC, Thyparambil S, Taylor JT, Han T, Tong W, Hansen DK, Fuscoe JC, Edmondson RD, Beger RD, Dragan YP. An integrated study of acute effects of valproic acid in the liver using metabonomics, proteomics, and transcriptomics platforms. OMICS. 2006;10:1–14. doi: 10.1089/omi.2006.10.1. [DOI] [PubMed] [Google Scholar]
- Schnackenberg LK, J Sun, et al. Metabolomics in Systems Toxicology: Towards Personalized Medicine. General, Applied and Systems Toxicology, John Wiley & Sons, Ltd; 2009. [Google Scholar]
- Seattle Proteome Centre. [accessed 26 July 2012];Formats:mzXML. 2012 http://tools.proteomecenter.org/wiki/index.php?title=Formats:mzXML.
- Senn T, Hazen SL, Wilson WH, Tang WH. Translating metabolomics to cardiovascular biomarkers. Prog. Cardiovasc. Dis. 2012;55:70–76. doi: 10.1016/j.pcad.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serkova NJ, Van Rheen Z, Tobias M, Pitzer JE, Wilkinson JE, Stringer KA. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;295:L152–L161. doi: 10.1152/ajplung.00515.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wahlang B, Wei X, Yin X, Falkner KC, Prough RA, Kim SH, Mueller EG, McClain CJ, Cave M, Zhang X. Metabolomic analysis of the effects of polychlorinated biphenyls in nonalcoholic fatty liver disease. J. Proteome Res. 2012;11:3805–3815. doi: 10.1021/pr300297z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Miyawaki I, Bando K, Horie H, Zaitsu K, Katagi M, Bamba T, Tsuchihashi H, Fukusaki E. "Influences of methamphetamine-induced acute intoxication on urinary and plasma metabolic profiles in the rat.". Toxicol. 2011;287:29–37. doi: 10.1016/j.tox.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Shintu L, Baudoin R, Navratil V, Prot J-M, Pontoizeau C, Defernez M, Blaise BJ, Domange C, Péry ARR, Toulhoat P, Legallais C, Brochot C, Leclerc E, Dumas M-E. Metabolomics-on-a-chip and predictive systems toxicology in microfluidic bioartificial organs. Anal. Chem. 2012;84:1840–1848. doi: 10.1021/ac2011075. [DOI] [PubMed] [Google Scholar]
- Shulaev V. Metabolomics technology and bioinformatics. Brief. Bioinform. 2006;7:128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- Shurubor YI, Matson WR, Martin RJ, Kristal BS. Relative contribution of specific sources of systematic errors and analytical imprecision to metabolite analysis by HPLC-ECD. Metabolomics. 2005;1:159–168. [Google Scholar]
- Sieber M, Wagner S, Rached E, Amberg A, Mally A, Dekant W. Metabonomic study of ochratoxin a toxicity in rats after repeated administration: phenotypic anchoring enhances the ability for biomarker discovery. Chem. Res. Toxicol. 2009;22:1221–1231. doi: 10.1021/tx800459q. [DOI] [PubMed] [Google Scholar]
- Singh OV. Proteomics and metabolomics: the molecular make-up of toxic aromatic pollutant bioremediation. Proteomics. 2006;6:5481–5492. doi: 10.1002/pmic.200600200. [DOI] [PubMed] [Google Scholar]
- Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, Faught W, Sawyer MB. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010;16:5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Smolinska A, Blanchet L, Coulier L, Ampt KA, Luider T, Hintzen RQ, Wijmenga SS, Buydens LMC. Interpretation and visualization of non-linear data fusion in kernel space: study on metabolomic characterization of progression of multiple sclerosis. PLoS One. 2012;7:e38163. doi: 10.1371/journal.pone.0038163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- Southam AD, Lange A, Hines A, Hill EM, Katsu Y, Iguchi T, Tyler CR, Viant MR. Metabolomics reveals target and off-target toxicities of a model organophosphate pesticide to roach (Rutilus rutilus): implications for biomonitoring. Environ. Sci. Technol. 2011;45:3759–3767. doi: 10.1021/es103814d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagou K, Wilson ID, Masson P, Theodoridis G, Raikos N, Coen M, Holmes E, Lindon JC, Plumb RS, Nicholson JK, Want EJ. HILIC-UPLC-MS for exploratory urinary metabolic profiling in toxicological studies. Anal. Chem. 2011;83:382–390. doi: 10.1021/ac102523q. [DOI] [PubMed] [Google Scholar]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin. Cancer Res. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–1167. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- Stein SE. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectr. 1999;10:770–781. [Google Scholar]
- Steinfath M, Groth D, Lisec J, Selbig J. Metabolite profile analysis: from raw data to regression and classification. Physiol. Plant. 2008;132:150–161. doi: 10.1111/j.1399-3054.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- Stephens NS, Siffledeen J, Su X, Murdoch TB, Fedorak RN, Slupsky CM. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J. Crohns Colitis, ahead of print. 2012 doi: 10.1016/j.crohns.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis. Curr. Bioinform. 2012;7:96–108. doi: 10.2174/157489312799304431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y, Setou M. Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: toward in situ pharmacometabolomes. J. Neuroimmune Pharmacol. 2010;5:31–43. doi: 10.1007/s11481-009-9162-6. [DOI] [PubMed] [Google Scholar]
- Sun J, Shannon M, Ando Y, Schnackenberg LK, Khan NA, Portilla D, Beger RD. Serum metabolomic profiles from patients with acute kidney injury: a pilot study. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012;893-894:107–113. doi: 10.1016/j.jchromb.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- Tang M, Zhang T, Xue Y, Wang S, Huang M, Yang Y, Lu M, Lei H, Kong L, Yuepu P. Dose dependent in vivo metabolic characteristics of titanium dioxide nanoparticles. J. Nanosci. Nanotechnol. 2010;10:8575–8583. doi: 10.1166/jnn.2010.2482. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenori L, Oakman C, Claudino WM, Bernini P, Cappadona S, Nepi S, Biganzoli L, Arbushites MC, Luchinat C, Bertini I, Di Leo A. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: a pilot study. Mol. Oncol. 2012;6:437–444. doi: 10.1016/j.molonc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ravenzwaay B, Coelho-Palermo Cunha G, Leibold E, Looser R, Mellert W, Prokoudine A, Walk T, Wiemer J. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol. Lett. 2007;172:21–28. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- van Ravenzwaay B, Herold M, Kamp H, Kapp MD, Fabian E, Looser R, Krennrich G, Mellert W, Prokoudine A, Strauss V, Walk T, Wiemer J. Metabolomics: a tool for early detection of toxicological effects and an opportunity for biology based grouping of chemicals-from QSAR to QBAR. Mutat. Res. 2012;746:144–150. doi: 10.1016/j.mrgentox.2012.01.006. [DOI] [PubMed] [Google Scholar]
- van Vliet E. Current standing and future prospects for the technologies proposed to transform toxicity testing in the 21st century. ALTEX. 2011;28:17–44. doi: 10.14573/altex.2011.1.017. [DOI] [PubMed] [Google Scholar]
- van Vliet E, Morath S, Eskes C, Linge J, Rappsilber J, Honegger P, Hartung T, Coecke S. A novel in vitro metabolomics approach for neurotoxicity testing, proof of principle for methyl mercury chloride and caffeine. Neurotoxicol. 2008;29:1–12. doi: 10.1016/j.neuro.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Vulimiri SV, Misra M, Hamm JT, Mitchell M, Berger A. Effects of mainstream cigarette smoke on the global metabolome of human lung epithelial cells. Chem. Res. Toxicol. 2009;22:492–503. doi: 10.1021/tx8003246. [DOI] [PubMed] [Google Scholar]
- Wang J, Reijmers T, Chen L, Van Der Heijden R, Wang M, Peng S, Hankemeier T, Xu G, Van Der Greef J. Systems toxicology study of doxorubicin on rats using ultra performance liquid chromatography coupled with mass spectrometry based metabolomics. Metabolomics. 2009;5:407–418. doi: 10.1007/s11306-009-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang A, Han Y, Wang P, Sun H, Song G, Dong T, Yuan Y, Yuan X, Zhang M, Ning X, Zhang H, Dong H, Dong W. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol. Cell. Proteomics. 2012;11:370–380. doi: 10.1074/mcp.M111.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, Heilier JF, Ducruix C, Ezan E, Junot C, Tabet JC. Mass spectrometry for the identification of the discriminating signals from metabolomics: current status and future trends. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;871:143–163. doi: 10.1016/j.jchromb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- West PR, Weir AM, Smith AM, Donley EL, Cezar GG. Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 2010;247:18–27. doi: 10.1016/j.taap.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Wikipedia. [accessed 26 July 2012];Mass spectrometry data format. 2012 http://en.wikipedia.org/wiki/Mass_spectrometry_data_format.
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava P, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS. Quantitative metabolomics using NMR. TrAC - Trends Analyt. Chem. 2008;27:228–237. [Google Scholar]
- Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometr. Intellig. Lab. Syst. 2001;58:109–130. [Google Scholar]
- Woo HM, Kim KM, Choi MH, Jung BH, Lee J, Kong G, Nam SJ, Kim S, Bai SW, Chung BC. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta. 2009;400:63–69. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(suppl 2):W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010a;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010b;38(Web Server issue):W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J. Biomed. Biotechnol. 2012 doi: 10.1155/2012/805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, He K, Raghavan N, Wang L, Mitroka J, Maxwell BD, Knabb RM, Frost C, Schuster A, Hao F, Gu Z, Humphreys WG, Grossman SJ. Comparative metabolism of 14C-labeled apixaban in mice, rats, rabbits, dogs, and humans. Drug Metab. Dispos. 2009a;37:1738–1748. doi: 10.1124/dmd.108.025981. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu H, Wu J, Zhang X, Liu M, Wang Y. Metabonomic alterations in hippocampus, temporal and prefrontal cortex with age in rats. Neurochem. Int. 2009b;54:481–487. doi: 10.1016/j.neuint.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol. Biosyst. 2012;8:470–481. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]