Abstract
The IMD pathway signaling plays a pivotal role in the Drosophila defense against bacteria. During the last two decades, significant progress has been made in identifying the components and deciphering the molecular mechanisms underlying this pathway, including the means of bacterial sensing and signal transduction. While these findings have contributed to the understanding of the immune signaling in insects, they have also provided new insights in studying the mammalian NF-κB signaling pathways. Here, we summarize the current view of the IMD pathway focusing on how it regulates the humoral immune response of Drosophila.
Keywords: Immune response, NF-κB signaling, Drosophila
1. Introduction
Like many insects, the fruit fly, Drosophila melanogaster, is highly resistant to microbes (Hoffmann et al., 1993). Drosophila live in an environment rich with fungi, viruses and bacteria, but do not seem to succumb in infections, thanks to their well-adjusted immune responses. Unlike mammals, insects have no adaptive immunity, no somatic recombination or major histocompatibility complex (MHC) -mediated antigen presentation, and they rely primarily on innate immune responses for host defense. These responses include cellular responses, such as phagocytosis and encapsulation, as well as humoral defense responses. The study of the humoral responses, such as the secretion of potent antimicrobial peptides (AMPs) from the insect equivalent of liver, the fat body, started in the 1970s. Hans Boman and colleagues reported that fruit flies that were vaccinated (or primed) with a preparation of attenuated Enterobacter cloacae preparation before an exposure to pathogenic Pseudomonas aeruginosa, survived better from an otherwise lethal dose of bacteria (Boman et al., 1972). This was one of the first publications to demonstrate the existence of the inducible, cell-free antibacterial defense in Drosophila. It still took eight more years before the first antimicrobial peptide, Cecropin, was characterized from Cecropia moths (Hultmark et al., 1980). Other antimicrobial peptides were soon discovered in vertebrates, such as defensins in human neutrophils (Ganz et al., 1985) and magainins in frog skin (Zasloff, 1987), but also in invertebrates. In Drosophila, we now have seven well-characterized AMP families, which include Attacins, Cecropins, Diptericins, Drosomycins, Defensins, Drosocins, and Metchnikowins (Imler and Bulet, 2005). In addition to these, Drosophila have several immune inducible genes encoding small proteins or peptides, which likely have antimicrobial properties (De Gregorio et al., 2001). Mass spectrometry has identified other immune inducible proteins in the hemolymph, which may also be antimicrobial (Uttenweiler-Joseph et al., 1998). Another antimicrobial peptide, Andropin, is linked to a specific organ and expressed in the male reproductive tract and seminal fluid (Samakovlis et al., 1991). The production of AMPs is highly inducible following septic infection, the levels of AMPs ranging from very low and mostly undetectable in uninfected animals to hundreds of micromolars in circulation during infection (Imler and Bulet, 2005). The recognition and signaling events that control the production and secretion of antimicrobial peptides are a focus of this review.
In 1995, Lemaitre et al. reported a new mutant that failed to induce the expression of most AMP genes following septic bacterial infection (Lemaitre et al., 1995). This phenotype was found in the mutant line known as Black cells (Bc) due to the circulating melanized crystal cells that were visible through the larval cuticle in these animals. However, careful genetic analysis revealed that the immune deficient phenotype was not only linked to Bc, but also to a nearby locus termed immunodeficiency (imd). Adult flies carrying this mutation alone had impaired production of most AMPs following a mixed infection with Escherichia coli and Micrococcus luteus, however the antifungal Drosomycin remained inducible (Lemaitre et al., 1995; Lemaitre et al., 1996). In a later study, it was shown that Drosomycin induction, following M. luteus or fungal infection, was regulated by the Toll pathway (Lemaitre et al., 1996), while the response to most gram-negative bacteria and certain Gram-positives was blocked by the imd mutation.
Several groups identified other members of the pathway shortly after the discovery of IMD. These include the nuclear factor κB (NF-κB) transcription factor Relish (Dushay et al., 1996; Hedengren et al., 1999), the Drosophila inhibitor of κB kinase (IKK) complex (Lu et al., 2001; Rutschmann et al., 2000; Silverman et al., 2000), the caspase-8 homolog Death-related ced-3/Nedd2-like protein (DREDD) (Leulier et al., 2000), Drosophila Fas-Associated protein with Death Domain (dFADD) (Leulier et al., 2002; Naitza et al., 2002), TGF-β activated kinase 1 (TAK1) (Silverman et al., 2003; Vidal et al., 2001), and finally the receptor, peptidoglycan recognition protein-LC (PGRP-LC) (Choe et al., 2002; Gottar et al., 2002; Rämet et al., 2002). Later, the pathway was supplemented with another receptor, PGRP-LE (Takehana et al., 2002; Takehana et al., 2004), TAK1-associated binding protein (TAB2) (Gesellchen et al., 2005; Kleino et al., 2005), the ubiquitination machinery components inhibitor of apoptosis 2 (IAP2) (Gesellchen et al., 2005; Huh et al., 2007; Kleino et al., 2005; Leulier et al., 2006a), Bendless (Ubc13), Uev1a (Zhou et al., 2005), and Effete (Ubc5) (Paquette et al., 2010), and the transcription cofactor Akirin (Goto et al., 2008). Furthermore, several negative regulators, which function at various steps in the pathway, have also been characterized (see section 5).
The IMD pathway is triggered by meso-diaminopimelic acid (DAP)-type peptidoglycan (PGN) (Kaneko et al., 2004; Leulier et al., 2003; Stenbak et al., 2004; Werner et al., 2003), which comprises the cell wall of most Gram-negative bacteria, as well as some Gram-positive bacteria, such as Bacillus and Listeria species. This discovery is consistent with the early characterization of which microbes trigger the IMD pathway, including most Gram-negatives as well as Bacillus subtilis (Lemaitre et al., 1997; Stenbak et al., 2004). Both receptors implicated in the IMD pathway, PGRP-LC on the plasma membrane, and the intracellular PGRP-LE, bind specifically to DAP-type PGN (Kaneko et al., 2006). Once bound to PGN, these receptors likely dimerize or multimerize (Mellroth et al., 2005) and intracellular signal is transmitted to the adaptor protein IMD (Choe et al., 2005).
IMD recruits dFADD (Leulier et al., 2002) and the caspase DREDD (Leulier et al., 2000). Dredd cleaves IMD, which is then further activated by K63-ubiquitination (Paquette et al., 2010)(see section 3.1. for more details). According to the current model, these K63-polyubiquitin chains recruit and activate TAK1 via the ubiquitin-binding domain of its regulatory partner TAB2. TAK1 is responsible for activating both the JNK and IKK/Relish branches of the IMD pathway. Relish becomes activated by IKK-mediated phosphorylation and cleavage by DREDD, after which the Relish NF-κB domain translocates to the nucleus and initiates the transcription of target genes.
Activation of the IMD pathway in response to bacterial challenge is rapid. Signal transduction, as monitored by cleavage and K63-ubiquitination of IMD, or cleavage, phosphorylation, and nuclear translocation of Relish, occurs within minutes (Paquette et al., 2010). Transcription of target genes, especially AMPs, peaks within hours (Lemaitre et al., 1997; Valanne et al., 2007; Vodovar et al., 2005). In comparison, another important immune signaling pathway, the Toll pathway, which is thought to mainly respond to fungi and to some extent to Gram-positive bacteria, is activated within hours, and the transcription of target genes, such as the antifungal Drosomycin, persists for days (Lemaitre et al., 1997). Therefore, quickly-responding IMD pathway response is likely more effective against fast-replicating pathogens, such as bacteria.
Bacteria and other microbes typically enter the body of a fly either through an open wound in the cuticle or through the barrier epithelia lining the respiratory, reproductive, and especially the digestive tract. Epithelial cells in these tissues are capable of producing AMPs, and this local immune response is IMD pathway dependent (Ferrandon et al., 1998; Tzou et al., 2000). However, the regulation of the IMD pathway signaling during local immune response somewhat differs from that of the systemic immune response. For example, the AMP production in the trachea is induced by the IMD pathway, but antagonized by constitutively active signaling through Toll-8/Tollo, Spätzle 2/DNT1 and Ectoderm-expressed 4 (Ect-4), which is the fly homolog of sterile alpha- and armadillo-motif-containing protein (SARM)(Akhouayri et al., 2011). In the intestinal epithelia, the homeobox protein Caudal specifically suppresses the IMD pathway signaling, preventing deleterious effects of immune signaling in response to harmless commensal microbes (Ryu et al., 2008) (see also the review elsewhere in this special issue). In contrast, a POU transcription factor known as Drifter or Ventral veins lacking drives constitutive AMP production in the male ejaculatory duct, bypassing the requirement for Relish and GATA factors in the absence of infection (Junell et al., 2010). Furthermore, nitric oxide (NO) signaling has been reported to induce Diptericin expression, likely via the IMD pathway (Foley and O’Farrell, 2003). Feeding or injecting larvae with an NO source was sufficient to trigger Diptericin expression, while pharmacological inhibition of nitric oxide synthase (NOS) sensitized the animals to Erwinia carotovora carotovora (Ecc) infections (Foley and O’Farrell, 2003; Nappi et al., 2000). NO signaling has also been reported to drive Diptericin expression in malpighian tubules of adult flies. Moreover, targeted expression of NOS in malpighian tubules enhances the fly survival from systemic E. coli infection (McGettigan et al., 2005). However, the link between NO signaling and the local immune response is not clearly defined, as NOS null mutants do not display defects in Diptericin expression during systemic or natural Ecc infection (Chakrabarti et al., 2012).
In addition to responding to bacteria, the IMD pathway has also been implicated in defense against some RNA viruses. When analyzing the transcription of UAS/GAL4-driven Sindbis virus replicon in flies, it was found that mutations in relish, imd, dFADD, dredd, ird5, kenny, and Tab2 resulted in increased replication of the Sindbis virus construct. These findings were further validated in Sindbis virus-infected RelishE20 (null) mutants, which showed increased virus titers compared to wild-type flies (Avadhanula et al., 2009). Others have reported that PGRP-LC, Tak1, ird5, kenny, and relish mutants are all more susceptible to Cricket paralysis virus infections. However, unlike in bacterial infections, the IMD pathway response against these viruses did not involve strong induction of AMP expression. Curiously, PGRP-LC and PGRP-LE were not required for limiting the transcription of the Sindbis virus replicon, while IMD and dFADD (but not PGRP-LC) were dispensable for survival after Cricket paralysis virus-infection (Avadhanula et al., 2009; Costa et al., 2009). It therefore remains unclear how viruses trigger the IMD signaling, and how IMD pathway activation translates to improved resistance to viral pathogens.
Next, we will focus on these signaling events in the classic bacterial-triggered IMD signaling pathway.
2. Recognition of PGN by PGRPs
Peptidoglycan is a large polymer essential to the structure of the bacterial cell wall, which is formed by carbohydrate chains of alternating N-acetylmuramic acid and N-acetylglucosamine residues cross-linked together by chains of 4 to 5 amino acid residues. In DAP-type PGN, the third residue in these chains is meso-diaminopimelic acid, while in Lys-type PGN, which is more commonly found in Gram-positive bacteria, the third residue is L-lysine. DAP-type PGN is commonly found in Gram-negative bacteria, but also in some Gram-positive bacterial species, such as in Listeria and in Bacillus.
Drosophila genome encodes 13 PGRPs, producing 17 proteins via alternative splicing (Dziarski, 2004; Werner et al., 2000). The PGRPs are structurally related to a class of enzymes known as N-acetylmuramyl-L-alanine amidases (NAMLAAs), or type 2 amidases, which cleave the lactyl bond between the carbohydrate backbone and stem-peptides of PGN. The domain similar to type 2 amidase is often referred to the PGRP domain. Five of the PGRPs retain all key catalytic residues and function as PGN degrading enzymes. Of these, PGRP-SB1, -SB2, -SC1, -SC2, and -LB are able to process DAP-type PGN. Interestingly, the other seven of the Drosophila PGRPs lack a critical catalytic cysteine in the type 2 amidase domain and lack amidase activity but most of them still bind PGN. In particular, DAP-type PGN is specifically recognized by PGRP-LC and PGRP-LE, two non-catalytic PGRPs (Kaneko et al., 2006; Takehana et al., 2002; Takehana et al., 2004).
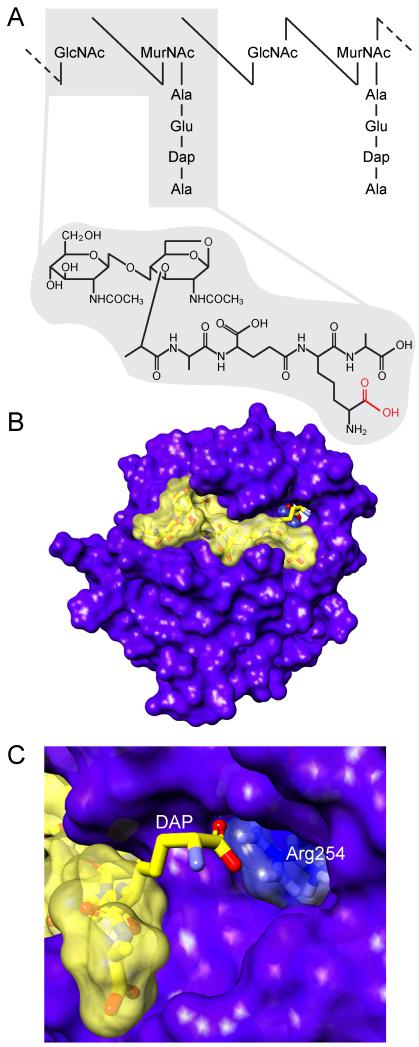
PGRP-LC encodes three alternative splice variants, PGRP-LCa, PGRP-LCx, and PGRP-LCy (referred to as PGRP-LC-RB, PGRP-LC-RA, and PGRP-LC-RC, respectively, in Flybase) (Werner et al., 2003). All the proteins encoded by these transcripts have identical transmembrane and cytoplasmic parts but different PGRP domains (Stenbak et al., 2004; Werner et al., 2000). In addition, PGRP-LCy has a longer linker region between the transmembrane and peptidoglycan recognition domain but the function of this isoform is still mainly unknown, although it may play a minor role in antagonizing the IMD pathway response (Neyen et al., 2012; Werner et al., 2003). PGRP-LCx, and PGRP-LCa are characterized better, and they are known to have differential binding specificities for different forms of DAP-PGN. PGRP-LCx binds polymeric DAP-PGN in a deep binding cleft typical of this family of receptors/enzymes, and probably this polyvalent ligand will cluster multiple PGRP-LCx receptors; we propose that these clusters of PGRP-LCx will trigger downstream signaling events. On the other hand, the deep PGN binding cleft is occluded in PGRP-LCa and is not capable of binding polymeric PGN in this typical manner (Chang et al., 2005). Instead, it participates in the recognition of monomeric fragment of DAP-PGN, a disaccharide tetrapeptide known as tracheal cytotoxin (TCT) (Fig. 1A). Structural and biochemical studies demonstrate that TCT binds in the deep binding cleft of PGRP-LCx, and PGRP-LCa interacts with this complex through both protein:protein and protein:PGN interactions to form a ligand-induced heterodimer (Chang et al., 2006; Chang et al., 2005; Kaneko et al., 2005; Mellroth et al., 2005).
Figure 1.
The principle of DAP-PGN recognition by PGRP-LE. (A) Schematic illustration of polymeric DAP-PGN and TCT. The carboxylate group of DAP that interacts with Arg254 of PGRP-LE is highlighted with red color. (B) Structure of the PGRP domain of PGRP-LE bound to TCT. (C) Atomic-level interactions of Arg254 from PGRP-LE and the carboxylic acid specific to DAP from TCT.
In addition to PGRP-LC, PGRP-LE also participates in DAP-PGN recognition (Kaneko et al., 2006; Lim et al., 2006; Takehana et al., 2002; Takehana et al., 2004; Yano et al., 2008). PGRP-LE does not harbor a recognizable transmembrane domain and instead localizes to the cytoplasm, where it recognizes DAP-type PGN that enters the cytosol through unknown mechanisms (Kaneko and Silverman, 2005; Yano and Kurata, 2011) or PGN introduced by cytosolic bacteria, such as Listeria monocytogenes (Yano et al., 2008). Also, it is thought that PGRP-LE is processed and released from cells, by unknown mechanisms, and then collaborates with the extracellular domain of PGRP-LC in the recognition of extracellular DAP-type PGN (Kaneko et al., 2006; Takehana et al., 2002). Upon TCT binding, PGRP-LE is known to form homopolymers, with atomic interactions nearly identical to that observed in the LCx:LCa heterodimer (Lim et al., 2006), see Figure 1.
Currently, the mechanism how DAP-type PGN binding to its receptors triggers the downstream signaling events is unclear. The structure of the PGRP-domain and its PGN binding pocket in PGRP-LC and PGRP-LE has been characterized at the atomic level (Chang et al., 2006; Chang et al., 2005; Lim et al., 2006), while much less is known about the function of the relatively long cytoplasmic/signaling N-terminal region of these receptors. Interestingly, truncated PGRP-LC lacking the cytoplasmic tail can function as dominant negative (Maillet et al., 2008), while a version lacking the ectodomain is constitutively active (Choe et al., 2005). Although the N-terminal domains of PGRP-LC and PGRP-LE are clearly important for signal transduction, their structure and interaction mechanisms are poorly known. Therefore one can only speculate that interaction with IMD (and possibly other downstream components) is triggered by multimerization or clustering of PGRP-LC or –LE, or that a PGN-induced conformational change is transmitted to the signaling domains, or both.
3. Receptor-proximal signaling events
Deletion analysis of both PGRP-LC and PGRP-LE signaling domains identified a conserved region critical for signal transduction in both receptors. The N-terminal signaling domains of PGRP-LC and PGRP-LE are largely dissimilar, except for this short motif involved in IMD signaling. Interestingly, this motif resembles the RIP Homotypic Interaction Motif (RHIM-motif) found in receptor-interacting protein 1 (RIP1) and RIP3 as well as TIR-domain-containing adapter-inducing interferon-β (TRIF) and DNA-dependent activator of IRFs/Z-DNA binding protein 1 (DAI/ZBP1) (Kaneko et al., 2004). These mammalian proteins are involved in the innate immune signaling, and the RHIM motifs of RIP1 and RIP3 are critical for their homotypic interaction and their role in programmed necrosis (Cho et al., 2009). Recently, it has been argued that these RHIM motifs form amyloid fibrils in the context of the programmed necrosis (Li et al., 2012). However, the molecular mechanism of the RHIM-like motif in PGRP-LC and –LE -mediated signaling is not properly understood.
Genetic studies suggest that the IMD protein functions most proximal to these receptors and, in fact, IMD interacts with both PGRP-LC and PGRP-LE (Kaneko et al., 2006). While the PGRP-LE-IMD interaction requires an intact RHIM-like motif, the PGRP-LC interaction does not (Kaneko et al., 2006). IMD has no ortholog in mammals, but its death domain shares striking homology with the death domain of the mammalian RIP1 (Georgel et al., 2001), suggesting some common functions for these two proteins that are both involved in NF-κB signaling pathways, in mammals and insects.
IMD interacts via its death domain with the Drosophila homolog of FADD called dFADD or BG4 (Naitza et al., 2002). dFADD in turn recruits the caspase-8 homolog DREDD to the signaling complex via a homotypic Death-effector domain (DED) interaction (Hu and Yang, 2000). The subcellular localization of these interactions or whether or not they change upon immune stimulation has not been thoroughly characterized. Curiously, PGRP-LC resides at the plasma membrane, yet IMD is mostly localized in the nucleus and can only be detected at the plasma membrane when the signaling pathway is activated (Boyer et al., 2011). In addition to IMD, mammalian FADD has also been reported to be mostly nuclear, and to harbor strong nuclear localization and nuclear export signals (Gomez-Angelats and Cidlowski, 2003; Screaton et al., 2003). It is not known if this is also true for dFADD, but it raises the questions how IMD, dFADD and DREDD are recruited to the signaling complex, and which additional proteins or cellular structures are involved in their presumed transport to the plasma membrane.
Regardless of the exact nature and localization of the receptor proximal signaling complex, the immediate result of PGN stimulation through either PGRP-LC or –LE is thought to be the activation of DREDD. DREDD is an inititiator caspase very similar to mammalian caspase-8. The molecular mechanism of DREDD activation is not clear, yet DREDD is critical for the cleavage of two proteins in the IMD pathway. First, it cleaves IMD and promotes its further activation by exposing a binding site for the ubiquitin E3 ligase IAP2 (Paquette et al., 2010). Second, DREDD is also required for, and likely directly mediates, the cleavage of the NF-κB precursor Relish (Ertürk-Hasdemir et al., 2009; Leulier et al., 2000; Stöven et al., 2000; Stöven et al., 2003). To date it is not thoroughly understood how these sequential steps are regulated.
3.1. K63-polyubiquitination of IMD and Dredd
Ubiquitination of proteins can lead to different outcomes depending on the nature of the ubiquitin linkage. The best-known example of poly-ubiquitination is conjugation with K48-linked chains, which targets the proteins to the proteasome for destruction. More recently, many other types of poly-ubiquitin chains have been identified, including K63 ubiquitin chains. This type of linkage often results in activation of target proteins and/or downstream pathways, rather than proteasomal degredation (Komander, 2009).
Interestingly, the IMD pathway is likely regulated by both K48 and K63 polyubiquitination, but to opposing purposes.
A major target of ubiquitination in the IMD pathway is the IMD protein itself. Upon activation of the signaling pathway, DREDD cleaves IMD just after aspartate 30, removing the N-terminus and exposing a highly conserved interaction site for the E3 ubiquitin ligase IAP2. This newly exposed N-terminus is often referred to as an IAP binding motif (IBM), and is known to interact with BIR domains, found in all IAP proteins. Interestingly, IBMs are often located at the N-terminus of processed caspases and pro-apoptotic proteins, such as Reaper/Hid/Grim (RHG proteins) and Smac (Wu et al., 2000), and apoptosis is controlled by the mutually exclusive binding of either caspases or RHG and is thus a key regulator of programmed cell death (Wilson et al., 2002). On the other hand, Drosophila IAP2 is a central component of the IMD pathway, interacting with and ubiquitinating cleaved IMD, but also as a minor contribution to programmed cell death (Leulier et al., 2006b).
The key attributes of the IBM are an exposed alanine in the neo-N-terminus and a proline in position 3. In IMD, these residues promote the interaction with BIR2 and BIR3 domains of IAP2 (Paquette et al., 2010). In addition, IAP2 has a C-terminal RING domain, which is indispensable for its function in immune response (Huh et al., 2007). Further, the RING domain is a signature motif of E3 ubiquitin ligases. Genetic evidence argues that IAP2 is the E3 ligase required for the immune-induced ubiquitination of IMD (Paquette et al., 2010), but this has not yet been confirmed in vitro.
RNAi studies suggest that the IAP2 E3 ligase functions with E2 ubiquitin conjugating enzymes Uev1a and Bendless (Ubc13), as well as Effete (Ubc5) to drive IMD polyubiqitination (Paquette et al., 2010; Zhou et al., 2005). In this respect the IMD pathway signaling events resemble the mammalian tumor necrosis factor receptor (TNFR) signaling pathway, where Uev1a and Ubc13 have been reported to participate in the activation of TNF receptor-associated factor 2 (TRAF2) via K63-linked ubiquitination of RIP (Wang et al., 2001), while cellular IAP1 (cIAP1), cIAP2, and Ubc5 likely catalyze polyubiquitination of RIP1 (Bertrand et al., 2008; Liu and Chen, 2011; Mahoney et al., 2008; Xu et al., 2009).
Curiously, activation of the caspase DREDD is also thought to be dependent on K63-polyubiquitination mediated by IAP2 (Meinander et al., 2012). IAP2 interacts with the death-effector domain (DED) of DREDD via its BIR2/3 domains, which supports the K63-ubiquitination of DREDD. Dredd44 flies carry a single amino acid substitution (G120R) in the DED domain of DREDD, which prevents its ubiquitination, although DREDDG120R can still interact with IAP2. Un-ubiquitinated DREDD cannot cleave Relish or IMD, which suppresses the IMD pathway signaling and renders the flies as susceptible to microbial challenge as a null allele, DreddB118 (Meinander et al., 2012). Curiously, Drosophila IAP2 also auto-ubiquitinates following immune activation (Paquette et al., 2010).
The cleavage and robust ubiquitination of IMD, as well as the weaker ubiquitination of DREDD and IAP2, all occur within a minute of DAP-type PGN-stimulation of Drosophila cells. The amount of K63-polyubiquitinated IMD peaks in 10 minutes, after which the levels start to decrease until they are almost undetectable at 30 min. (Paquette et al., 2010). The rapid and transient nature of IMD ubiquitination suggests it is tightly regulated, and de-ubiquitinating proteases have been implicated in regulating this aspect of the IMD pathway (see section 5.4.).
4. Kinases and the activation of Relish
Genetic studies suggest that the next signaling components downstream of IMD are TAK1 and its partner protein TAB2 (Lu et al., 2001; Rutschmann et al., 2000; Silverman et al., 2003; Vidal et al., 2001). Once K63-polyubiquitinated, IMD likely recruits the TAB2/TAK1 complex via the highly conserved Npl4 zinc finger (NZF) domain of TAB2. Studies with mammalian TAB2 homologs have shown this domain to interact specifically with K63-linked ubiquitin (Kulathu et al., 2009). Mutations in the NZF domain of mammalian TAB2 abolished this binding ability, which suppressed the activation of TAK1 and IKK (Kanayama et al., 2004). Drosophila TAB2 harbors another motif that is predicted to interact with ubiquitin conjugating enzymes, known as CUE domain, but neither the CUE nor NZF domains from Drosophila TAB2 have been biochemically characterized for their ubiquitin binding properties. Once activated, TAB2/TAK1 likely phosphorylates and activates the IKK complex as well as the JNK pathway through Hemipterous (MKK7), similar to mammalian NF-κB pathways (Aggarwal, 2003). Interestingly, activation of the JNK pathway is in turn thought to destabilize TAK1 in a feedback loop involving upregulation of the E3 ubiquitin ligase Plenty of SH3 (POSH) (Tsuda et al., 2005).
The Drosophila IKK complex consists of two subunits, the catalytic subunit called Immune response deficient 5 (IRD5 or IKKβ) and the regulatory subunit Kenny or IKKγ, which are homologous to mammalian IKKβ and IKKγ, respectively (Rutschmann et al., 2000; Silverman et al., 2000). The IKK complex is required for the activation of Relish.
Relish, one of the three Drosophila NF-κB proteins, is the critical transcription factor in the IMD pathway and AMP gene induction. Unlike the other two NF-κB proteins DIF and Dorsal, which are involved in Toll pathway signaling, Relish consists of both an N-terminal Rel homology (or NF-κB) domain and a C-terminal ankyrin-repeat/IκB-like domain, thus resembling the mammalian NF-κB precursors p100 and p105. The N-terminal transcription factor domain (RelN or Rel68) is released by endoproteolytic cleavage, after which it translocates to the nucleus and initiates the transcription of its target genes. The IκB part (Rel49) remains in the cytoplasm, where its function(s) are unclear (Stöven et al., 2000; Stöven et al., 2003; Wiklund et al., 2009).
The IKK complex controls Relish activation through at least two distinct mechanisms, one involving the catalytic (kinase) activity and the other independent of kinase activity. On the one hand, both IKK subunits are essential for robust AMP gene induction and both are required for Relish cleavage (Silverman et al., 2000; Stöven et al., 2003). However, Relish cleavage occurs normally in transgenic flies carrying a catalytically inactive ird5, yet AMP induction is dramatically reduced. In addition, the Drosophila IKK has been shown to directly phosphorylate serine residues 528 and 529 of Relish, which are part of the mature Rel68 cleavage product (Ertürk-Hasdemir et al., 2009). These phosphorylations are argued to be required for the proper activation of Relish and AMP gene induction. This would fully explain the role of kinase activity of the Drosophila IKK complex in the IMD pathway, but requires further validation in vivo.
The phenotype of kinase-inactive IRD5 (IKKβ) flies resembles the phenotype of the TAK1 mutants. While TAK1 mutants exhibit dramatically reduced AMP gene induction, this mitogen activated protein kinase kinase kinase (MAP3K) is not required for Relish cleavage. The function of TAK1 in driving AMP expression remains controversial. One report argued that the activation of JNK signaling by TAK1 is essential for AMP induction (Delaney et al., 2006), while others have argued that JNK signaling is likely to down-modulate AMP expression (Kim et al., 2007) (see section 5.5 for more details). The alternative builds upon the finding that TAK1 is required for IKK activation and Relish phosphorylation (Silverman et al., 2003), and Relish phosphorylation is required for the induction of AMP expression (Ertürk-Hasdemir et al., 2009). These results are consistent with the very similar phenotypes reported for TAK1 mutant and catalytically inactive IKKβ flies.
As alluded to above, the activation is complex and involves several discrete points of regulation. In addition to a non-catalytic role for the IKK complex, Relish cleavage also requires the PGN receptor PGRP-LC, IMD, and DREDD, both in cells and in adult flies. Using RNAi in immune inducible Drosophila cell lines, dFADD is also implicated in Relish cleavage. The role of IAP2 and ubiquitination has only been examined in cell lines, where a partial role in Relish cleavage is suggested. As mentioned above, TAK1 and the IKK kinase activity are not involved in Relish cleavage. Likewise, in cell lines TAB2 RNAi is reported to have either no role, or only a partial role in Relish cleavage, but was found critical for the nuclear translocation of Relish (Ertürk-Hasdemir et al., 2009; Kleino et al., 2005). Altogether, these data suggest that the IMD pathway regulates Relish by modulating its cleavage, phosphorylation and transcriptional activity, and nuclear translocation, yet still more study is required to fully understand the molecular mechanisms underlying all these events.
One additional factor implicated in Relish dependent AMP gene induction is Akirin, a nuclear protein with no other recognizable motifs (Goto et al., 2008). In vivo RNAi of Akirin reduced AMP induction and sensitized the flies to Gram-negative bacterial infections by an unknown mechanism. In mammals, Akirin2 seems to be essential for development, since homozygous knock-out mice die before embryonic day 9.5. However, the embryonic fibroblasts derived from these animals show defects in their ability to induce NF-κB dependent cytokines (Goto et al., 2008). Unlike mammals, which have two Akirins, the Drosophila genome encodes only one Akirin. The null mutation of Akirin is lethal, suggesting that Akirin may possess functions beyond the IMD signaling. More recent studies have suggested that the Akirins may play a more general role in transcriptional regulation through their chromatin activities (Clemons et al., 2013; Nowak et al., 2012).
5. Negative regulators
Uncontrolled immune responses can have detrimental outcomes and seriously affect the health, fitness and survival of an organism. Therefore, the localization, intensity, and duration of the immune response must be carefully regulated. As described above, activation of Relish is under multiple layers of control, perhaps to limit the damage of an inappropriate immune response, and the IMD pathway in general is modulated through multiple negative feedback loops to ensure a properly timed and adjusted response. Figures 2 and 3 summarize both the signaling events underlying IMD signaling, and the modulatory factors that keep this response in check.
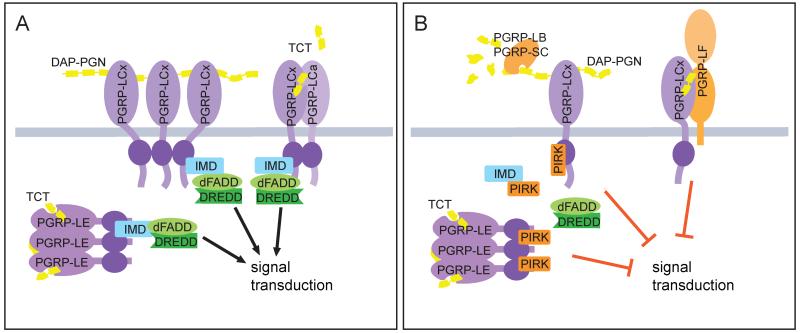
Figure 2.
Positive and negative regulation of the receptor-proximal signaling events in the IMD pathway. (A) Activation of the IMD pathway signaling by polymeric and monomeric DAP-PGN through recognition by PGRP-LC on the plasma membrane, or PGRP-LE in the cytoplasm. TCT likely multimerizes intracellular PGRP-LE, dimerizes cell surface PGRP-LCx and –LCa, while polymeric DAP-type peptidoglycan is thought to cluster PGRP-LCx. Ligand binding is believed to lead to the recruitment of IMD, dFADD, and the caspase-8 homolog DREDD to the signaling complex. (B) Negative regulation of receptor activation. Active amidases PGRP-LB and PGRP-SC digest polymeric PGN to small subunits that do not activate PGRP-LC. PGRP-LF binds PGRP-LC and blocks signaling by competitive inhibition. PIRK binds both IMD and the RHIM-like motif found in PGRP-LC and PGRP-LE, which suppresses the receptor-IMD interaction and downstream signaling events.
5.1.Catalytic PGRP proteins
The IMD pathway is activated by binding of DAP-type peptidoglycan to the receptor PGRP-LC or -LE (Choe et al., 2005; Kaneko et al., 2004; Leulier et al., 2003). Therefore, the first level of negative regulation in this pathway targets the interaction of the PGN ligand with its receptor. This is achieved through enzymatic degradation of PGN into smaller fragments with greatly diminished immunostimulatory activity, and thereby attenuating pathway activation (Bischoff et al., 2006; Mellroth et al., 2003; Zaidman-Remy et al., 2006). Five of the Drosophila PGRPs, namely PGRP-LB, PGRP-SB1, PGRP-SB2, PGRP-SC1, and PGRP-SC2, are known or predicted amidases (Mellroth et al., 2003; Werner et al., 2000). PGRP-SC1/2 has been shown to be able to degrade both DAP-and Lys-type peptidoglycan, while PGRP-LB appears to be specifically degrading DAP-type peptidoglycan (Mellroth et al., 2005; Zaidman-Remy et al., 2006). With genetic studies, both of these amidases have been shown to downregulate the IMD pathway activation in vivo (Bischoff et al., 2006; Mellroth et al., 2003; Zaidman-Remy et al., 2006). Moreover, both PGRP-LB and PGRP-SC2 are themselves targets of IMD signaling, as part of a regulatory feedback loop (De Gregorio et al., 2002; Valanne et al., 2007).
PGRP-LB is expressed in the fat body, from where it is secreted to the hemolymph (Zaidman-Remy et al., 2006). In addition, both PGRP-LB and PGRP-SC1/2 are expressed in the gut epithelium and released into the lumen, suggesting that the regulation of IMD signaling by dampening the initial stimulus may be particularly important to the gut, which has to tolerate the presence of commensal bacteria and the fragments of PGN they produce. Curiously, in the absence of PGRP-LB or PGRP-SC, flies express elevated levels of antimicrobial peptide genes during infection (Bischoff et al., 2006; Zaidman-Remy et al., 2006) but also under normal rearing conditions (Zaidman-Remy et al., 2006), consistent with the idea that constitutively active immune response is harmful to the host.
Another member of the PGRP protein family, PRGP-LF, has been shown to downregulate IMD but is not a PGN-degrading amidase (Basbous et al., 2011; Maillet et al., 2008; Persson et al., 2007). PGRP-LF is a transmembrane protein, which has a very short cytoplasmic part likely not to have a signaling function. Moreover, unlike other PGRPs, PGRP-LF has two extracellular PGRP domains. The mechanism of its inhibitory action is somewhat controversial. On the one hand, Persson et al. find that PGRP-LF binds PGN and could therefore serve as a decoy receptor (Persson et al., 2007). On the other hand, Basbous et al. solved the crystal structure of both PGRP domains of LF and conclude that neither PGRP-LFz nor -LFw can bind PGN in the typical manner due to an obstructed binding cleft, inconsistent with a decoy receptor model. In addition, Basbous et al. further show that PGRP-LF strongly interacts with the ectodomain of PGRP-LCx in the presence of TCT, competing with the multimers or clusters of PGRP-LCx involved in signal transduction, thereby blocking signaling (Basbous et al., 2011).
5.2. PIRK
PIRK, also referred to as PIMS and Rudra, was originally identified in two independent microarray analyses as a target of the IMD pathway (De Gregorio et al., 2001; Kallio et al., 2005). However, the expression kinetics of pirk differ from the other highly induced genes. The expression of pirk peaks much earlier than AMPs, around an hour after the immune stimulation, while the transcript levels of AMPs typically increase for several hours. In addition, the transcription of pirk is dependent on IMD pathway activation and Relish (Aggarwal et al., 2008; Kleino et al., 2008; Lhocine et al., 2008; Valanne et al., 2007). The upstream/regulatory region of pirk contains four putative NF-κB binding sites, one of which is identical to the DNA-binding motif of Relish (Lhocine et al., 2008). Moreover, PIRK is a potent negative regulator of IMD signaling. Hence, the induction of pirk upon infection creates a negative feedback loop that efficiently suppresses the IMD pathway signal transduction (Aggarwal et al., 2008; Kleino et al., 2008; Lhocine et al., 2008).
Pirk encodes a 21 kDa protein with no obvious motifs. However, the central portion includes a series of repeats. Orthologs containing conserved middle and C-terminal regions of PIRK are readily identified within other Diptera, but no obvious mammalian homologs of PIRK have been identified, and the structure is unknown (Kleino et al., 2008). Biochemically, PIRK is known to interact with both PGRP-LC and PGRP-LE, as well as with IMD itself (Aggarwal et al., 2008; Kleino et al., 2008; Lhocine et al., 2008). The interaction with PGRPs is at least to some extent mediated by the RHIM-like motif of PGRP-LC and PGRP-LE (Aggarwal et al., 2008). Since Pirk interacts both with the receptor and the immediate downstream component of the pathway in a mutually exclusive manner, it has been proposed that PIRK might act by interfering with the formation of the PGRP-LC-IMD signaling complex (Aggarwal et al., 2008; Kleino et al., 2008). According to another hypothesis, PIRK mediates the trafficking of PGRP-LC from the plasma membrane to lysosomes, where the receptor would be degraded (Lhocine et al., 2008). More experimental evidence is needed before either or both of these hypotheses can be confirmed.
5.3. Caspase inhibition
The caspase-8 like DREDD is the central player in IMD signaling, responsible for the cleavage and activation of both IMD and Relish. It is not yet understood how DREDD is activated and modulated to coordinate both of these events. However, it is clear that DREDD is tightly controlled and the target of negative regulation.
Defense repressor 1 (DNR1) has been proposed to suppress the activity of DREDD (Foley and O’Farrell, 2004; Guntermann et al., 2009). dnr1 encodes an evolutionarily conserved protein containing a C-terminal RING finger domain, which suggests that DNR1 might function as an E3 ligase. Both in cells and in flies, depletion of DNR1 results in enhanced signaling through the IMD pathway, with elevated AMP expression both in the absence of or following infection (Foley and O’Farrell, 2004; Guntermann et al., 2009). DNR1 has been shown to physically interact with DREDD but the mechanism of the inhibition still remains unknown. The RING finger domain of DNR1 is required for the suppression of the IMD pathway signaling (Guntermann et al., 2009), suggesting that DNR1 functions through a ubiquitin-dependent mechanism. Besides DREDD, DNR1 has been reported to affect DRONC-dependent regulation of apoptosis (Primrose et al., 2007), suggesting a more general function of DNR1 as a repressor of initiator caspases. Interestingly, a recent report also associates DNR1 with neurodegeneration by demonstrating that loss-of-function mutations in dnr1 lead to IMD pathway activation, and increased, Relish-dependent, expression of AMPs in the fly brain, which eventually results in neuropathology. Furthermore, bacterial infection in the fly brain, or neuronal overexpression of AMPs, was sufficient to induce neurodegeneration (Cao et al., 2013). This highlights the importance of downregulation in the immune signaling to avoid tissue damage and pathology, and suggests a novel role for DNR1 in the brain.
Another modulator of IMD signaling, Caspar, shares homology with the human Fas associated factor 1 (FAF1). FAF1 has been reported to associate with several factors involved in NF-κB and/or cell death signaling, including FADD, FAS, RelA, IKKβ, and caspase-8, which is the mammalian homolog of DREDD (Chu et al., 1995; Park et al., 2004; Park et al., 2007; Ryu et al., 2003). Two mechanisms have been suggested for the inhibitory action of FAF1. First, it has been shown to physically interact with RelA and interfere with its nuclear localization (Park et al., 2004). Second, FAF1 binding to IKKβ has been suggested to disrupt the IKK complex formation (Park et al., 2007). In Drosophila, depletion of caspar results in elevated transcription of antimicrobial peptide genes both with and without infection (Kim et al., 2006). This suggests that Caspar is required for suppressing constitutive IMD pathway activation. Caspar has been shown to inhibit DREDD-dependent cleavage of Relish in vivo (Kim et al., 2006), but the mechanism of suppression, as well as the target(s) of Caspar are currently unknown.
5.4. More ubiquitin: the ubiquitin-proteasome pathway
Besides DNR1 and Caspar, a number of other ubiquitin-proteasome related proteins have been implicated in the negative regulation of IMD signaling. While IMD is activated by conjugation with K63-polyubiquitin chains by the E3 ligase IAP2 (Paquette et al., 2010), it also seems to be deubiquitinated in order to suppress the IMD pathway signaling. A ubiquitin-specific protease called Scrawny, or dUSP36, has been associated with this function in vivo (Thevenon et al., 2009). The authors reported that the levels of activated, K63-polyubiquitinated IMD were reduced by dUSP36, which in turn increased the proteasomal degradation of IMD (Thevenon et al., 2009), possibly by K48-linked ubiquitination of IMD by a yet unknown E3 ligase. K63-ubiquitination, deubiquitination, and possible K48-conjugation of IMD likely provides a very fine-tuned temporal control over the IMD pathway activity.
Another deubiquitinase reported to downregulate the IMD response is Cylindromatosis (CYLD) (Tsichritzis et al., 2007). Mammalian CYLD is a tumor suppressor gene that suppresses NF-κB signaling by removing K63-linked ubiquitin chains from TRAF2, TRAF6, and NEMO/IKKγ (Kovalenko et al., 2003; Trompouki et al., 2003). In flies, CYLD was shown to interact with Kenny, the fly ortholog of IKKγ. Flies deficient for CYLD exhibit increased AMP gene expression, especially prior infection, but showed decreased survival upon E. coli infection (Tsichritzis et al., 2007). This poor survival may be due to altered morphology and function of the fly fat body in CYLD mutants, since CYLD also regulates the fat homeostasis in flies by unknown mechanisms (Tsichritzis et al., 2007). In addition, CYLD is required for Eiger (Drosophila TNF homolog) -mediated JNK signaling and cell death, probably through deubiquitinating dTRAF2 (Xue et al., 2007). The target of CYLD deubiquitination in the IMD pathway is still uncharacterized.
Other approaches have further implicated the ubiquitin-proteasome pathway in regulating the IMD pathway. A forward genetic screen identified skpA as a negative regulator of the IMD pathway, and found that skpA mutation resulted in the constitutive expression of Diptericin in larvae and adults (Khush et al., 2002). SkpA is homologous to the mammalian and yeast Skp1 proteins, which are components of Skp1/Cullin/F-box protein (SCF)-E3 ubiquitin ligases (Cardozo and Pagano, 2004). The SCF complex is involved in the K48-linked ubiquitination and proteasomal degradation of substrate proteins. In addition to SkpA, depletion of other components of the SCF complex, such as dCullin or Slimb, resulted in the expression of Diptericin without infection. In addition, RNAi silencing of skpA or slimb was shown to increase the levels of both full-length and cleaved Relish suggesting that the SCF complex might regulate the stability of Relish and thereby modulate the IMD pathway activity (Khush et al., 2002). However, the direct target of the SCF complex in IMD signaling has not been identified.
5.5. Negative regulation in the nucleus
Transcription factors of the JAK/STAT (Janus kinase/signal transducer and activator of transcription) and the JNK pathways have been implicated in the negative regulation of the transcriptional activity of Relish. In particular, Drosophila activator protein 1 (dAP-1) and Stat92E have been suggested to form a repressosome complex with a Drosophila High mobility group (HMG) protein called Dorsal switch protein 1 (DSP1) in response to continuous immune signaling (Kim et al., 2007). In cell lines, this complex functions by replacing Relish at overlapping cis-regulatory elements, and recruiting a histone deacetylase to close chromatin and inhibit transcription of the target genes. Reducing the activity of either dAP-1, Stat92E or DSP1 by mutation or RNAi increased transcription of AMP genes in vivo, in a Relish-dependent manner. Furthermore, these mutant flies were more susceptible to E. coli infections compared to wild-type flies, but could be rescued by reducing the copy number of Relish. These results suggest that the survival following infection, observed in the repressosome complex mutants, was in fact due to the elevated immune responses, perhaps even hyperactivated AMP production (Kim et al., 2007). These studies are contradictory to the conclusions from Delaney et al. (2006), using clonal analysis in the JNK pathway components in fat body to argue that JNK signaling is essential for AMP gene induction (Delaney et al., 2006). Additional studies are required to resolve this apparent contradiction.
Recently, Zinc finger homeodomain 1 (Zfh1) was also identified as a negative regulator of the IMD pathway signaling (Kleino et al. 2005, Myllymäki and Rämet, 2012). ZFH1 includes 9 C2H2 zinc fingers, a homeodomain as well as a nuclear localization signal that targets it into the nucleus. RNAi knock-down of ZFH1 dramatically hyperactivates the IMD response in immune responsive cell lines, but curiously in vivo RNAi targeting of this gene caused elevated AMP gene expression only for Cecropin B. The mechanism of ZFH1-mediated IMD pathway suppression remains obscure, as well as if the other ZFH in flies, ZFH2, is partially redundant with ZFH1 function. Interestingly, the human zfh1 homolog, ZEB1, appears to function as a positive regulator of TNF signaling, because it is required for NF-κB dependent responses in HeLa cells (Myllymäki and Rämet, 2013).
6. Hormonal regulation of the IMD pathway
The effects of hormones, especially glucocorticoids, on the mammalian immune responses are well known (Baschant and Tuckermann, 2010). In Drosophila, at least two different hormones participate in the regulation of immune responses. The steroid hormone 20-hydroxyecdysone (20E), and the sesquiterpenoid juvenile hormone (JH) are both involved in the regulation of various aspects of the fly physiology, especially development and metamorphosis, but also in the cellular and humoral arms of the immune response.
In plasmatocytes and plasmatocyte-like cultured cells 20E promotes differentiation of the cells. They flatten, spread, and become more adherent and macrophage-like, while their ability to phagocytose also increases (Dimarcq et al., 1997; Lanot et al., 2001). Furthermore, 20E regulates the inducibility of AMP genes; Diptericin induction is especially 20E-dependent (Dimarcq et al., 1997; Meister and Richards, 1996; Silverman et al., 2000), which suggests a role for 20E in the regulation of the IMD pathway. 20E-supported humoral immune responses can be suppressed by JH (or its analogs) through unknown mechanisms (Flatt et al., 2008), suggesting a network of counteracting hormonal controls that can modulate IMD responses throughout the Drosophila lifecycle.
The mechanism of 20E-mediated hormonal control over the IMD pathway activation is not yet fully understood. 20E-induced humoral responses require two nuclear receptors, Ecdysone Receptor (ER) and Ultraspiracle (USP), which are orthologs of the mammalian liver X receptor (LXR) and retinoid x receptor (RXR), respectively (King-Jones and Thummel, 2005). Recently, seven transcription factors (BR-C, Eip93F, Eip74EF, Eip78C, HR46, Serpent and Pannier), which are all induced by 20E, were found to be critical for IMD-dependent responses in cell lines and adult flies, (Rus et al., 2013). Furthermore, another early 20E-induced factor, Eip75B, was found to negatively regulate the immune response, which is in line with a previous report (Kleino et al., 2005). Eip75B is a nuclear hormone receptor, which has been shown to heterodimerize and interfere with HR46, suggesting a mechanism for the Eip75B function (Thummel, 1997; White et al., 1997; Yamanaka and O’Connor, 2011). Most importantly, the expression of the IMD pathway receptor, PGRP-LC, was found to be highly dependent on 20E, which naturally affects all aspects of the PGRP-LC-dependent IMD signaling. Forced expression of PGRP-LC can bypass the requirement for 20E pretreatment of cells for PGN-induced IMD signal transduction and the activation of some AMP genes, but other AMP genes, i.e. Diptericin, still require hormonal input to be immune inducible (Rus et al., 2013).
7. Conclusions and future perspectives
Since the discovery of the imd in 1995, great progress has been made on understanding the physiological role of this immune response pathway, the underlying molecular mechanisms that contribute to the remarkably rapid and robust induction of AMP genes, and the myriad of feedback and physiogical regulators that modulate and tune the outputs of this immune response. These regulatory activities, whether they are hormonal, feedback regulators, or tissue specific modulators, are likely crucial to the overall health and fitness of the animal. It is well known that hyperactive or inappropriately activated immune responses can be pathological, in mammals as well as in insects. In addition, the fat body, the major site of systemic AMP production, is also crucial for reproduction in insects. In fact, experimental activation of the IMD pathway has been shown to reduce egg production (Zerofsky et al., 2005), and likely has strong effect on overall fitness. The hormonal control of IMD signaling further suggests that the immune response may be adjusted, through hormonal inputs, depending on the stage of development and may also indicate that steroid signal may have roles, during stress or aging, to modulate immune output.
Although much progress has been made in identifying the components of the IMD signaling pathway and the basic roles of these factors in signal transduction have been elucidated, many fundamental questions about the underlying biochemical mechanisms of signaling and AMP gene induction remain. As highlighted above, Relish activation is tightly controlled through several regulatory checkpoints, which are not fully understood. The mechanisms whereby PGRP-receptor ligation by DAP-type PGN leads to the activation of intracellular signaling also remains obscure. The role of the kinases, TAK1 and IKK, in IMD signaling and in AMP production also requires more in depth biochemical and genetic study. The role of the IMD pathway in protecting the gut epithelia from infection, yet supporting the gut microflora, is also a major area for further study, and the subject of a separate review in this issue. Furthermore, the complex regulatory network of ubiquitin signaling still requires further clarification. And finally, how the interplay of the IMD pathway and other signaling pathways regulates the overall health and fitness of the fly. Future research will continue to elucidate these aspects and increase our understanding of the fine-tuned fly immunity, but also the conserved mechanisms of the human immune response.
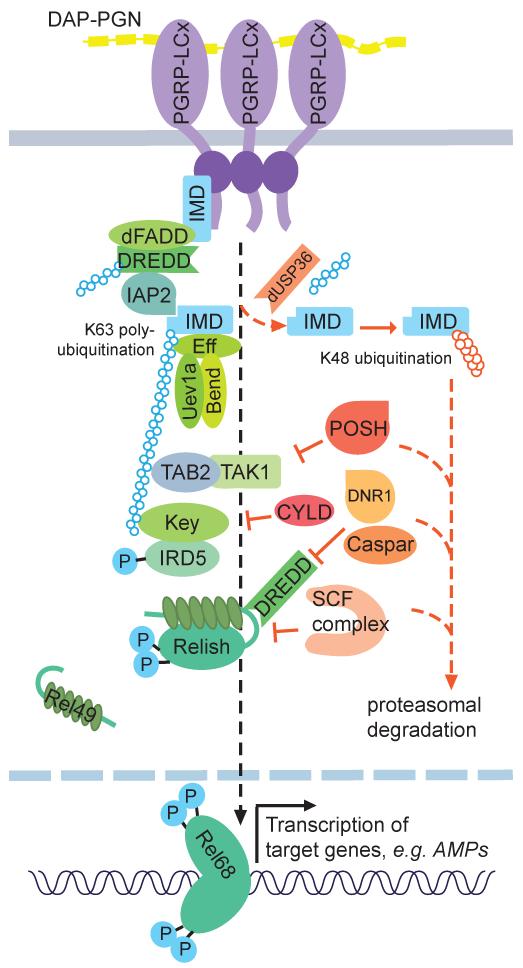
Figure 3.
Ubiquitin signaling in the activation and deactivation of the IMD pathway signaling. Following PGN recognition and the assembly of a receptor proximal signaling complex, IMD signaling proceeds through at least two distinct paths. Both paths require the proteolytic activity of DREDD, which presumably acts as part of this receptor proximal complex. Once activated, DREDD cleaves Relish, which is necessary for its activation, nuclear translocation and induction of target genes. DREDD also likely cleaves IMD, exposing an IBM at position 31. Cleaved IMD then interacts with IAP2 and is robustly and transiently conjugated with K63-polyubiquitin chains. These chains are thought to serve as a scaffold for both the TAK1/TAB2 complex, through NZF domain of TAB2, and the IKK complex, through the IKKγ subunit. The IKK complex then phosphorylates Relish, which is important for its activation, and contributes in a non-catalytic manner to Relish cleavage. The IMD pathway is also subject to numerous negative regulatory controls, including deubiquitination of IMD by dUSP36. IMD may then be re-ubiquitinated with K48-chains and degraded. Likewise, the ubiquitin-proteasome system is implicated in the control of TAK1, DREDD and Relish by POSH, DNR1 and the SCF complex respectively.
8. Acknowledgements
We are grateful to Dr. Iivari Kleino for technical help in the preparation of illustrations for this article, and the members of the Silverman lab for insightful discussions. This work was supported by National Institutes of Health (NIH) grants AI060025 and AI099708 to NS, and the EMBO Long-Term Fellowship, Sigrid Juselius Foundation, and Maud Kuistila Memorial Foundation to AK.
Glossary
Abbreviations
- 20E
20-hydroxyecdysone
- AMP
antimicrobial peptide
- DAP
meso-diaminopimelic acid
- DREDD
Death-related ced-3/Nedd2-like protein
- FADD
Fas-Associated protein with Death Domain
- IAP
Inhibitor of apoptosis
- IκB
Inhibitor of κB
- IKK
IκB kinase
- imd
immune deficiency
- JNK
c-Jun N-terminal kinase
- PGN
peptidoglycan
- PGRP
peptidoglycan recognition protein
- RHIM
RIP homotypic interaction motif
- RIP
Receptor-interacting protein
- RNAi
RNA interference
- TAB
TAK1-associated binding protein 2
- TAK1
TGF-β activated kinase 1
- TCT
tracheal cytotoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9. References
- Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Aggarwal K, Rus F, Vriesema-Magnuson C, Ertürk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbous N, Coste F, Leone P, Vincentelli R, Royet J, Kellenberger C, Roussel A. The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 2011;12:327–333. doi: 10.1038/embor.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschant U, Tuckermann J. The role of the glucocorticoid receptor in inflammation and immunity. J Steroid Biochem Mol Biol. 2010;120:69–75. doi: 10.1016/j.jsbmb.2010.03.058. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman HG, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere GM, Ip WK, Fracchia S, Hennessy E, Ertürk-Hasdemir D, Reichhart JM, Silverman N, Lacy-Hulbert A, Stuart LM. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- Chang CI, Ihara K, Chelliah Y, Mengin-Lecreulx D, Wakatsuki S, Deisenhofer J. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc Natl Acad Sci U S A. 2005;102:10279–10284. doi: 10.1073/pnas.0504547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- Chu K, Niu X, Williams LT. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci U S A. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons AM, Brockway HM, Yin Y, Kasinathan B, Butterfield YS, Jones SJ, Colaiacovo MP, Smolikove S. akirin is required for diakinesis bivalent structure and synaptonemal complex disassembly at meiotic prophase I. Mol Biol Cell. 2013;24:1053–1067. doi: 10.1091/mbc.E12-11-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SA, Dow JA. Modulation of epithelial innate immunity by autocrine production of nitric oxide. Gen Comp Endocrinol. 2009;162:113–121. doi: 10.1016/j.ygcen.2008.09.012. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Stöven S, Uvell H, Anderson KV, Engström Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 2006;25:3068–3077. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarcq JL, Imler JL, Lanot R, Ezekowitz RA, Hoffmann JA, Janeway CA, Lagueux M. Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem Mol Biol. 1997;27:877–886. doi: 10.1016/s0965-1748(97)00072-6. [DOI] [PubMed] [Google Scholar]
- Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci U S A. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stöven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, Garbuzov A, Palli SR, Tatar M, Silverman N. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol. 2008;211:2712–2724. doi: 10.1242/jeb.014878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O’Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 2003;17:115–125. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O’Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Angelats M, Cidlowski JA. Molecular evidence for the nuclear localization of FADD. Cell Death Differ. 2003;10:791–797. doi: 10.1038/sj.cdd.4401237. [DOI] [PubMed] [Google Scholar]
- Goto A, Matsushita K, Gesellchen V, El Chamy L, Kuttenkeuler D, Takeuchi O, Hoffmann JA, Akira S, Boutros M, Reichhart JM. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat Immunol. 2008;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Guntermann S, Primrose DA, Foley E. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev Comp Immunol. 2009;33:127–134. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Hetru C, Reichhart JM. The humoral antibacterial response of Drosophila. FEBS Lett. 1993;325:63–66. doi: 10.1016/0014-5793(93)81414-u. [DOI] [PubMed] [Google Scholar]
- Hu S, Yang X. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J Biol Chem. 2000;275:30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, Guo M, Park JM, Hay BA. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- Imler JL, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy. 2005;86:1–21. doi: 10.1159/000086648. [DOI] [PubMed] [Google Scholar]
- Junell A, Uvell H, Davis MM, Edlundh-Rose E, Antonsson A, Pick L, Engström Y. The POU transcription factor Drifter/Ventral veinless regulates expression of Drosophila immune defense genes. Mol Cell Biol. 2010;30:3672–3684. doi: 10.1128/MCB.00223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Rämet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Golenbock D, Silverman N. Peptidoglycan recognition by the Drosophila Imd pathway. J Endotoxin Res. 2005;11:383–389. doi: 10.1179/096805105X76823. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell Microbiol. 2005;7:461–469. doi: 10.1111/j.1462-5822.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Ertürk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Khush RS, Cornwell WD, Uram JN, Lemaitre B. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr Biol. 2002;12:1728–1737. doi: 10.1016/s0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, Kim J, Jeong K, Shim J, Kim-Ha J, Kim YJ. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5:e238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci U S A. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kleino A, kiMyllymä H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Rämet M. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymäki H, Enwald H, Stöven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Rämet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol Cell Biol. 2006a;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Lecreulx D, Lemaitre B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, Zachariou A, Pichaud F, Ueda R, Meier P. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 2006b;13:1663–1674. doi: 10.1038/sj.cdd.4401868. [DOI] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, Lemaitre B, Gstaiger M, Meier P, Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wu LP, Anderson KV. The antibacterial arm of the drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15:104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, Arora V, Mak TW, Lacasse EC, Waring J, Korneluk RG. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
- McGettigan J, McLennan RK, Broderick KE, Kean L, Allan AK, Cabrero P, Regulski MR, Pollock VP, Gould GW, Davies SA, Dow JA. Insect renal tubules constitute a cell-autonomous immune system that protects the organism against bacterial infection. Insect Biochem Mol Biol. 2005;35:741–754. doi: 10.1016/j.ibmb.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, Broemer M, Leulier F, Zvelebil M, Silverman N, Meier P. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M, Richards G. Ecdysone and insect immunity: the maturation of the inducibility of the diptericin gene in Drosophila larvae. Insect Biochem Mol Biol. 1996;26:155–160. doi: 10.1016/0965-1748(95)00076-3. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Hakansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci U S A. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Myllymäki H, Rämet M. Transcription factor zfh1 downregulates Drosophila Imd pathway. Dev Comp Immunol. 2013;39:188–197. doi: 10.1016/j.dci.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Naitza S, Rosse C, Kappler C, Georgel P, Belvin M, Gubb D, Camonis J, Hoffmann JA, Reichhart JM. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- Nappi AJ, Vass E, Frey F, Carton Y. Nitric oxide involvement in Drosophila immunity. Nitric Oxide. 2000;4:423–430. doi: 10.1006/niox.2000.0294. [DOI] [PubMed] [Google Scholar]
- Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue-and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. 2012;189:1886–1897. doi: 10.4049/jimmunol.1201022. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Aihara H, Gonzalez K, Nibu Y, Baylies MK. Akirin links twist-regulated transcription with the Brahma chromatin remodeling complex during embryogenesis. PLoS Genet. 2012;8:e1002547. doi: 10.1371/journal.pgen.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Ertürk-Hasdemir D, Reichhart JM, Meier P, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Jang HD, Lee SY, Lee KJ, Kim E. Fas-associated factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J Biol Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- Park MY, Moon JH, Lee KS, Choi HI, Chung J, Hong HJ, Kim E. FAF1 suppresses IkappaB kinase (IKK) activation by disrupting the IKK complex assembly. J Biol Chem. 2007;282:27572–27577. doi: 10.1074/jbc.C700106200. [DOI] [PubMed] [Google Scholar]
- Persson C, Oldenvi S, Steiner H. Peptidoglycan recognition protein LF: a negative regulator of Drosophila immunity. Insect Biochem Mol Biol. 2007;37:1309–1316. doi: 10.1016/j.ibmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Primrose DA, Chaudhry S, Johnson AG, Hrdlicka A, Schindler A, Tran D, Foley E. Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster. J Cell Sci. 2007;120:1189–1199. doi: 10.1242/jcs.03417. [DOI] [PubMed] [Google Scholar]
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, Yates E, Tatar M, Silverman N. Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J. 2013 doi: 10.1038/emboj.2013.100. doi: 10.1038/emboj.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Ryu SW, Lee SJ, Park MY, Jun JI, Jung YK, Kim E. Fas-associated factor 1, FAF1, is a member of Fas death-inducing signaling complex. J Biol Chem. 2003;278:24003–24010. doi: 10.1074/jbc.M302200200. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Kylsten P, Kimbrell DA, Engström A, Hultmark D. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. EMBO J. 1991;10:163–169. doi: 10.1002/j.1460-2075.1991.tb07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Kiessling S, Sansom OJ, Millar CB, Maddison K, Bird A, Clarke AR, Frisch SM. Fas-associated death domain protein interacts with methyl-CpG binding domain protein 4: a potential link between genome surveillance and apoptosis. Proc Natl Acad Sci U S A. 2003;100:5211–5216. doi: 10.1073/pnas.0431215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Erlich RL, Hunter M, Bernstein E, Schneider D, Maniatis T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, Herve M, Chaput C, Boneca IG, Lee WJ, Lemaitre B, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S, Silverman N, Junell A, Hedengren-Olcott M, Ertürk D, Engström Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci U S A. 2002;99:13705–13710. doi: 10.1073/pnas.212301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 2004;23:4690–4700. doi: 10.1038/sj.emboj.7600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Benaud C, Baudier J, Taillebourg E, Fauvarque MO. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6:309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Dueling orphans--interacting nuclear receptors coordinate Drosophila metamorphosis. Bioessays. 1997;19:669–672. doi: 10.1002/bies.950190806. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, Mosialos G. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134:2605–2614. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci U S A. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Kleino A, Myllymäki H, Vuoristo J, Rämet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Dev Comp Immunol. 2007;31:991–1001. doi: 10.1016/j.dci.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci U S A. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem. 2003;278:26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Wiklund ML, Steinert S, Junell A, Hultmark D, Stöven S. The N-terminal half of the Drosophila Rel/NF-kappaB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev Comp Immunol. 2009;33:690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Igaki T, Kuranaga E, Kanda H, Miura M, Xu T. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13:446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, O’Connor MB. Nitric oxide directly regulates gene expression during Drosophila development: need some gas to drive into metamorphosis? Genes Dev. 2011;25:1459–1463. doi: 10.1101/gad.2080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem. 2011;150:143–149. doi: 10.1093/jb/mvr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–108. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Zhou R, Silverman N, Hong M, Liao DS, Chung Y, Chen ZJ, Maniatis T. The role of ubiquitination in Drosophila innate immunity. J Biol Chem. 2005;280:34048–34055. doi: 10.1074/jbc.M506655200. [DOI] [PubMed] [Google Scholar]