Abstract
Many biological tissues are viscoelastic, behaving as elastic solids on short timescales and fluids on long timescales. This collective mechanical behaviour enables and helps to guide pattern formation and tissue layering. Here, we investigate the mechanical properties of three-dimensional tissue explants from zebrafish embryos by analysing individual cell tracks and macroscopic mechanical response. We find that the cell dynamics inside the tissue exhibit features of supercooled fluids, including subdiffusive trajectories and signatures of caging behaviour. We develop a minimal, three-parameter mechanical model for these dynamics, which we calibrate using only information about cell tracks. This model generates predictions about the macroscopic bulk response of the tissue (with no fit parameters) that are verified experimentally, providing a strong validation of the model. The best-fit model parameters indicate that although the tissue is fluid-like, it is close to a glass transition, suggesting that small changes to single-cell parameters could generate a significant change in the viscoelastic properties of the tissue. These results provide a robust framework for quantifying and modelling mechanically driven pattern formation in tissues.
Keywords: embryonic development, tissue modelling, glassy dynamics, supercooled fluid, tissue viscoelasticity, active matter
1. Introduction
A quantitative description of the mechanical behaviour of groups of cells in biological tissues is critical for an understanding of many fundamental biological processes, including embryogenesis [1–7], wound healing [8,9], stem cell dynamics, regeneration [10,11] and tumourigenesis [12–15]. Previous work demonstrates that the macroscopic response of many tissues is viscoelastic [1], where the tissue behaves as an elastic solid over short timescales and a viscous fluid over long timescales. The relaxation timescale, a material parameter that is different for different tissue types, characterizes how much time it takes for a tissue to crossover from elastic to viscous behaviour.
Recently, researchers have discovered that some two-dimensional tissues exhibit glassy behaviour [16,17]. In these confluent tissues, individual cells have difficulty moving past one another or exchanging neighbours, resulting in a ‘frozen’ system with a macroscopic response that is solid-like even on long timescales. Solid tissues support stresses and provide structure to living organisms.
A wide variety of materials exhibit glass transitions, which generally occur when the individual agents that comprise the material (i.e. atoms, polymers, droplets and cells) approach high densities or low individual motilities (i.e. atoms at low temperature). Glasses display universal features, including long-range spatial correlations in velocity fields and slowing down of individual agents [18–20].
By contrast, embryonic explants are fluid-like on long timescales, allowing cells in the embryo to rearrange and flow in order to form new patterns and structures. Therefore, they are not glasses. However, embryonic tissues share several properties with another class of materials called supercooled fluids—they are tightly packed in disordered structures and display elastic behaviour on short timescales and fluid behaviour on long timescales. Supercooled fluids have a viscoelastic relaxation timescale that is controlled by their proximity to a glass transition [18,21] and display glassy dynamics such as anomalous slowing of individual motions and ‘caging’ behaviour [22,23]. Caging behaviour occurs when the motion of a single element (particle, atom or cell) is trapped and impeded from motion by a surrounding ‘cage’ of its nearest neighbours due to the dense packing of the system; only rare, high-energy fluctuations allow the element to be released from the cage and permitted to move a new location.
To test whether the viscoelasticity of embryonic tissues is similarly controlled by proximity to a glass transition, we must first determine whether cell trajectories in embryonic tissues display signatures of glassy dynamics, and then we must develop a framework for determining how ‘close’ the system is to a glass transition phase boundary. Such a description has the potential to transform existing definitions for adaptability and robustness in biology. Tissues near a phase boundary could significantly change their behaviour with only small changes to single-cell properties and are therefore highly adaptable, while those far from phase boundaries are robust to perturbations. This might also give insights into disease. For example, if a mutation causes the tissue to cross a phase boundary and transition from fluid-like to solid-like behaviour, it would significantly interfere with pattern formation in embryonic development. Finally, we can use information from phase diagrams to make quantitative predictions about macroscopic tissue viscoelasticity.
The remainder of this paper is organized as follows: first, we track individual cells in three-dimensional zebrafish explants and demonstrate that cell trajectories exhibit anomalous diffusion and caging behaviour, which are both signatures of glassy dynamics.
Second, we develop a novel minimal model with three dimensionless parameters that can be used to quantify how close the material is to a glass transition. The parameters have simple biophysical interpretations: (i) the ratio between the adhesion and the cortical tension, (ii) the magnitude of forces actively generated by cells exerting tension on their neighbours and (iii) the typical timescale over which those forces act. As a function of the three parameters, we find that the model does have a ‘jamming’ or glass transition and demonstrate that the best-fit model parameters describe a supercooled fluid that is controlled by this nearby transition.
Third, we use this calibrated model with no fit parameters to make predictions about macroscopic viscoelastic response as measured by tissue compression and tissue fusion assays. We demonstrate that our minimal model accurately predicts the viscoelastic relaxation timescale seen in both types of experiments, providing a strong validation of the model. The model also reproduces qualitative observations of surface properties, such as the existence of a tissue surface tension, but it does not correctly reproduce the magnitude of the tissue surface tension. This is consistent with previous work [24,25] suggesting that steady-state surface properties are sensitive to the detailed shapes and tensions of individual cells, which are not included in our model.
We conclude with a discussion of how these results can be extended to better understand more complicated biological processes. Now that we have a good handle on the mechanical behaviour in simple environments, we can systematically add additional degrees of freedom to the model to capture biochemical signalling near tissue boundaries, coupling to extracellular matrix or chemotaxis that occurs in signalling gradients.
2. Material and methods
2.1. Experimental methods
2.1.1. Explant preparation
Maternal zygotic one-eyed pinhead (MZoep) and 50–100 pg of cyclops mRNA-injected wild-type zebrafish aggregates, corresponding to ectoderm and mesendoderm, respectively, were generated and fluorescently labelled as previously described [1,24,26].
2.1.2. Explant fusion
Fusion experiments were carried out on a Zeiss Axiovert 200M microscope (Zeiss, Göttingen, Germany), equipped with a SPOT camera (Diagnostic Instruments, Inc., MI, USA) and Metamorph v. 4.6 (Molecular Devices, LLC, CA, USA) software and on a Olympus DSU microscope (Tokyo, Japan), equipped with a Hamamatsu camera (Hamamatsu City, Japan) and Slidebook 5.1 software. The aspect ratio (AR) of fusing tissue aggregates was determined by finding the object's convex hull using a built-in MATLAB (MathWorks; Natick, MA, USA) function, and extracting its major and minor axes (see the electronic supplementary material). The AR as a function of time was plotted and fitted with an exponential + constant [27].
2.1.3. Tissue surface tension measurements
The tissue surface tensiometer (TST) was constructed as previously described [28,29], without the water circulation and with a digital CAHN electrobalance D-200 (Cerritos, CA, USA) and the lower compression plate connected to a Newport NewStep NSA 12 actuator (Irvine, CA, USA), allowing for computer-controlled motion through a custom LabVIEW program (National Instruments, Austin, TX, USA). Aggregates were imaged using a Basler A601f camera (Ahrensburg, Germany) attached to a Leica S8APO stereo microscope (Wetzlar, Germany). The tensiometer was calibrated as described in the electronic supplementary material text. Tissue surface tension and Young's modulus were calculated as described in earlier studies [26,30].
2.1.4. Two-photon imaging and cell tracking
For time-lapse in vivo imaging, explants were embedded in 1% low-melting point agarose (Cat. no. 15517-022, Invitrogen) in E2-medium [1] to minimize motion. Imaging was carried out on a custom-built two-photon laser scanning microscope, constructed on an upright BX51 Olympus microscope (Tokyo, Japan). A tunable Ti : sapphire pulse laser (Mira 900, Coherent, 100-fs pulses at 80 MHz) was used to excite the sample with approximately 920 nm pulses. The emitted light was collected simultaneously through an Olympus LUMPlan Fl/IR water-immersion objective (NA = 0.8) and an oil-immersion condenser (NA = 1.4) using GaAS photomultiplier tubes (Hamamatsu Photonics K.K; Hamamatsu City, Japan). MATLAB ScanImage software was used for image acquisition [31]. Fluorescently labelled nuclei were identified in Z-stacks of two-photon images, with one Z-stack captured every 2 min, and resolution of 0.86 μm/pixel in X and Y and 4 μm/pixel in Z. The images were analysed using a bandpass filter and three-dimensional feature finding algorithms [32]. Features were binned according to position and volume and spurious features were identified as those with volumes less than 20% of average or located outside the spherical aggregate shape. Features identified at each time point are linked into nuclei tracks according to a standard tracking algorithm [32], and automated tracks are compared to manual tracks to ensure accuracy. Because images for the ectoderm explants are slightly noisier, our tracking algorithm occasionally wrongly identifies noise as nuclei ‘features’, but these mislabelled features have very short tracks (two to three frames) and do not affect results at timescales longer than 5 min.
3. Results
3.1. Statistics of individual cell trajectories
We first analyse the structure and dynamics of cells in ectoderm and mesendoderm zebrafish explants. We reconstruct the three-dimensional static positions for a subset of the nuclei in the explant at each timepoint and estimate cell shapes by taking a three-dimensional Voronoi tessellation [33] of the nuclei positions. In both tissue types, the structure of the tissue is disordered; the cell nuclei are not arranged in a crystalline pattern, and the cell shapes are irregular polyhedra with roughly similar volumes. A two-dimensional slice through the tissue therefore appears as curved polygons with widely varying areas (figure 1a).
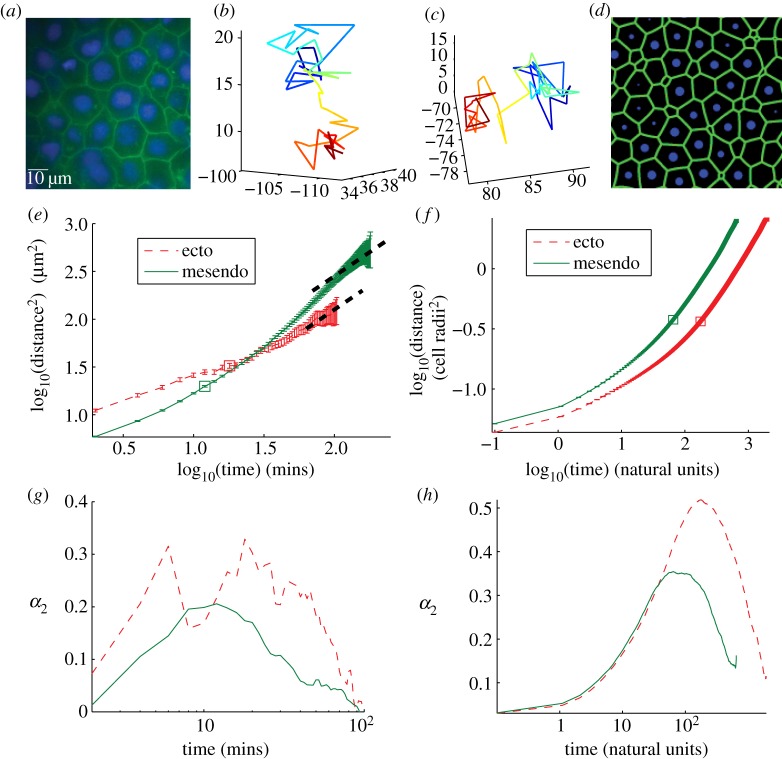
Figure 1.
(a) Two-dimensional cross section of three-dimensional experimental tissue showing packing fraction unity. Cell membranes are labelled using Gap43GFP, cell nuclei using Hoechst. (b,c) Two sample cell tracks extracted from experimental data illustrating ‘caging’ behaviour as described in the text. (d) Two-dimensional cross section of three-dimensional tissue simulation, interfaces generated using a Voronoi tessellation and the surface evolver computer program [34]. (e) MSD data for experimental ectoderm (thin dashed) and mesendoderm (solid) explants. Thick dashed lines are slope 1, drawn to guide the eye. (f) MSD data for best-fit simulation parameters shown in natural units as discussed in the electronic supplementary material text. (g,h) Non-Gaussian parameter (described in text) for experimental (g) and best-fit simulation (h) data achieve a maximum at a timescale tc discussed in the main text. Secondary peak at very short timescales for ectoderm data is likely caused by misidentified features. (Online version in colour.)
A second observation is that the tissue is confluent, where there are no visible extracellular gaps in membrane-labelled images. One way to quantify a cellular structure is the dimensionless packing fraction ϕ, which is the ratio of the sum of the volumes of all the individual cells compared to the total volume taken up by the aggregate. For tissues with extracellular gaps, the packing fraction is less than one, but for completely confluent tissues the packing fraction is unity. This value can be directly compared with results from simulations.
To non-dimensionalize other observables, we define the average effective radius R of cells by calculating the average distance between nuclei in the middle of the aggregate, which is 15±2 μm. Since the overlap between soft disordered spheres at packing fraction unity is approximately 15% [35], we find that twice the effective radius averages 17 μm and the average effective radius is R = 8 ± 1 μm.
By combining the static three-dimensional positions of nuclei from different timepoints, we can track them over time and analyse their dynamics inside the tissue explants [32].
A standard metric for studying the motion of particles is the mean-squared displacement (MSD), which is the square of the net distance an individual particle moves as a function of time, averaged over all particles. The motion of the nuclei is diffusive if the MSD scales linearly with time and super (sub)-diffusive if the MSD increases with the time to a power greater (less) than one. For diffusive tissues in three dimensions, the diffusion constant D is one-sixth the long time limit of the ratio between the MSD and time.
Figure 1e shows the log of the MSD as a function of log of the time for ectoderm and mesendoderm tissues. We find that D = 0.22 ± 0.05 μm2 min–1 for ectoderm and D = 0.60 ± 0.05 μm2 min–1 for mesendoderm. Ectoderm and mesendoderm explants have previously been shown [1] to differ by a factor of 2 in their tissue surface tensions, thus the observed difference in the diffusion constant is not surprising as we would expect cells in more adhesive tissues to move less. The slope of this plot, which is a second dimensionless observable α, is nearly unity at long times for both tissues indicating that the nuclei movement is diffusive. Furthermore, we observe that both tissues exhibit a subdiffusive regime at short timescales where the slope is significantly smaller than unity (α < 1) on a log-log plot.
While there are many instances in biology where a material exhibits subdiffusive behaviour, these MSD curves are reminiscent of those seen in supercooled colloids that are just above the glass transition temperature [23]. In those materials, individual colloidal particles are trapped in a ‘cage’ of neighbours, and they must wait for a rare, high-energy temperature fluctuation to escape that cage and continue diffusing. Therefore, a signature of caging behaviour is a trajectory that displays long periods where particle displacements are small compared with the particle diameter, punctuated by a period of directed travel that traverses roughly a particle diameter.
In particulate matter, caging becomes more pronounced as the packing fraction increases. Because our tissues are at packing fraction unity, we expect that a significant amount of mechanical energy is likely required for one cell to move past another, and therefore the subdiffusive regime in tissues could be generated by a similar mechanism. A schematic illustrating this caging effect is shown in figure 2. A sample cage-breaking event that occurred inside a zebrafish explant is shown in the electronic supplementary material, movie S5.
Figure 2.
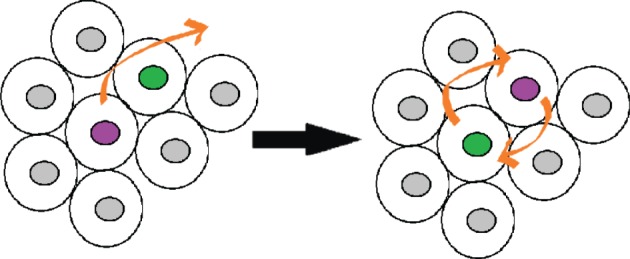
Schematic illustrating the caging effect. In confluent or nearly confluent tissues, cells are surrounded by a cage of neighbours and only a high-energy collective rearrangment allows them to escape the cage. (Online version in colour.)
To test this hypothesis, we analysed images of the real-space trajectories of individual nuclei inside ectoderm explants. We find many trajectories that exhibit signatures of caging behaviour—long periods where cell displacements are small punctuated by a quick, large displacement of roughly a cell diameter. Two examples are shown in figure 1b,c, and a large set of trajectories is shown in the electronic supplementary material, figure S6.
To systematically quantify these caging effects, we calculate the average non-Gaussian parameter for all cell trajectories [22], which quantifies the directedness of cell trajectories as a function of timescale. We expect cell displacements to be random and roughly Gaussian-distributed at short timescales when they are caged by their neighbours, and then again at very long timescales after they have exchanged many neighbours. However, at intermediate timescales when they break out of their cages, we expect the non-Gaussian parameter to be large because the trajectories are more directed and less random. Therefore, a peak in the non-Gaussian parameter as a function of timescale is a quantitative demonstration of caging behaviour in a statistically large set of cell trajectories.
Figure 1g shows the non-Gaussian parameter α2 as a function of timescale and demonstrates that there is a peak in the non-Gaussian parameter. We denote the timescale at which this peak occurs as tc ∼ 20 min for ectoderm and tc ∼ 10 min for mesendoderm tissues, and it occurs at roughly the same timescale as the crossover from subdiffusive to diffusive behaviour in the MSD trajectories. This strongly supports our hypothesis that cell trajectories are supercooled or caged, and allows us to define a third dimensionless crossover time observable τ* = Dtc/(R2). For the ectoderm aggregates, a spurious peak occurs at very short timescales (3–5 min) because of mislabelled features in our tracking algorithm, but as discussed above this does not affect the results for longer tracks.
Biologically, we can understand caging as follows: owing to the dense packing of cells in the tissue, they can only move larger distances when another cell allows them to pass, similar to a person trying to move around in a very crowded room. Thus, cells are jiggling at one location for most of the time, and occasionally they can escape and move larger distances.
3.2. A minimal model for cell dynamics
As discussed in the electronic supplementary material text, existing models for tissue mechanics [4,6,36–45] are not suitable for describing our observations of zebrafish explants. We now develop a new model with the minimal ingredients necessary to explain the macroscopic bulk viscoelastic response of simple embryonic tissues. Instead of allowing many degrees of freedom per cell, corresponding to the viscoelasticity and activity of the actin–myosin cytoskeleton, we allow one degree of freedom per cell, the centre of mass (COM) and introduce several types of interactions between cells to capture single-cell viscoelasticity, adhesion and active force generation.
We hypothesize that the emergent mechanical behaviour of a large group of cells does not depend on detailed cell shapes and activities, but instead on a small set of variables that govern the rates at which cells can squeeze past one another in this tightly packed, disordered structure. Therefore, we focus on determining the correct length and timescales for typical cell–cell interactions, and later verify that the exact forms for these interactions are not important for determining the emergent behaviour. We identify four general classes of interactions that occur between cells: resistance to shape changes and adhesion (captured by an interaction term Fint), damping (Fdamp) and active cell motility (Fa). A detailed description of each of these terms as well as their mathematical representation is given in the electronic supplementary material, and figure 3 is a schematic illustrating several ingredients in the model.
Figure 3.
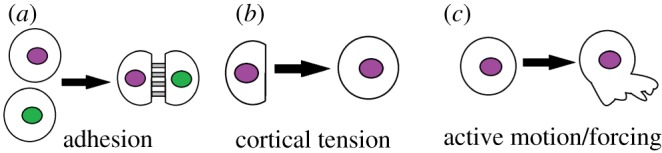
Schematic illustrating ingredients in our minimal model: (a) adhesion causes cells to stick together, (b) cortical tension resists changes to a cell's shape, making an isolated cell round and (c) active forcing describes the additional forces generated as cells actively develop protrusions and make new contacts. (Online version in colour.)
The most important difference between this model and others in the literature is that the active forcing term is not random white noise [41,44–46]. Instead, it is structured to be more biologically realistic; it enforces that cells can only move by exerting tension on adhesive contacts with other cells, and incorporates a timescale pt that characterizes how much time a cell typically spends moving in the direction of one adhesive contact before switching directions to move towards a different adhesive contact.
Assuming that the cell dynamics are overdamped the equation of motion for each cell i is
| 3.1 |
We non-dimensionalize the equations with units of length equal to the average cell radius R, units of force equal to the product K R, where K is an effective spring constant that characterizes the cortical tension, and time τ = b/K, where b is a damping coefficient. The equation of motion for the position of the COM ri for cell i is
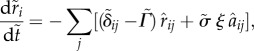 |
3.2 |
where 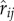 is the unit vector connecting the two cell centres, and
is the unit vector connecting the two cell centres, and 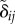 is the cell overlap (how close two cells centres are compared to their radii). Here
is the cell overlap (how close two cells centres are compared to their radii). Here 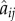 is a unit vector in the direction of the active force calculated as described in the electronic supplementary material, and ξ is a unit variance χ-distributed random variable with k = 3 and persistence time
is a unit vector in the direction of the active force calculated as described in the electronic supplementary material, and ξ is a unit variance χ-distributed random variable with k = 3 and persistence time 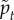 . There are three dimensionless parameters
. There are three dimensionless parameters 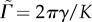 , which is the ratio between the adhesion energy and cortical tension,
, which is the ratio between the adhesion energy and cortical tension, 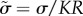 , which is the ratio of the magnitude of the active forces to the cortical tension and
, which is the ratio of the magnitude of the active forces to the cortical tension and 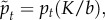 which characterizes the persistence time for active forces. Therefore, we must identify three dimensionless observables in the simulations and experiments to calibrate the model.
which characterizes the persistence time for active forces. Therefore, we must identify three dimensionless observables in the simulations and experiments to calibrate the model.
We first perform a simulation of a rounded droplet that mimics the experiments in which we studied individual cell tracks. We integrate the equations of motion (equation (3.2)) for slightly polydisperse cells from the droplet initial conditions with non-periodic boundary conditions. Simulation initialization is described in the electronic supplementary material text. The tissues in the simulations reach a steady-state droplet volume after approximately 10–20 natural time units, and then we continue to run the simulations for approximately 500 natural time units.
Figure 1d is a reconstructed image of a two-dimensional slice through the three-dimensional tissue simulation. The COM is denoted by a sphere of radius 0.5 R and the green lines (generated using the program Surface Evolver in two dimensions [34]) minimize the surface tension between cells. In the best-fit parameter regime, the simulations exhibit a disordered structure, as confirmed by an analysis of the pair correlation function (see electronic supplementary material, figure S4).
The simulated aggregates also show liquid-like dynamical behaviour similar to that seen in the experimental cell aggregates. One of these behaviours is the rounding up of tissue fragments into a spherical shape [1,26] as shown in electronic supplementary material, figure S4 and movie S1, which suggests that surface tension governs the final shape of aggregates.
We observe that the qualitative behaviour of the MSD and the non-Gaussian parameter in the simulations is similar to those seen in the experiments, as shown in figure 1f,h. In addition, all cells remained part of a single connected cluster throughout the simulation, which is also generally seen in healthy experimental explants.
Now that we have shown that the model is qualitatively similar in structure and dynamics to the experimental data, we calibrate it by varying the three dimensionless parameters (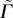 ,
, 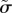 ,
, 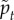 ) and identifying the best match with three dimensionless observables: the product of the diffusion constant and the crossover timescale τ* = Dtc/R2, the scaling of the power law relationship between the MSD and time α and the packing fraction ϕ. Figure 4a,b illustrate the results of hundreds of simulations with varying model parameters. Hatched regions correspond to parameter ranges where the simulation matches the experimentally observed ϕ and α, respectively, while the circle and cross pinpoint the exact parameter values which match τ* for the ectoderm and mesendoderm, respectively. Details are described in the electronic supplementary material. Figure 4c demonstrates that the functional form of the MSD for the best-fit ectoderm simulation is very similar to that for the experimental data. Table 1 summarizes the best-fit parameters and conversion factors from our simulation model to both tissue types.
) and identifying the best match with three dimensionless observables: the product of the diffusion constant and the crossover timescale τ* = Dtc/R2, the scaling of the power law relationship between the MSD and time α and the packing fraction ϕ. Figure 4a,b illustrate the results of hundreds of simulations with varying model parameters. Hatched regions correspond to parameter ranges where the simulation matches the experimentally observed ϕ and α, respectively, while the circle and cross pinpoint the exact parameter values which match τ* for the ectoderm and mesendoderm, respectively. Details are described in the electronic supplementary material. Figure 4c demonstrates that the functional form of the MSD for the best-fit ectoderm simulation is very similar to that for the experimental data. Table 1 summarizes the best-fit parameters and conversion factors from our simulation model to both tissue types.
Figure 4.
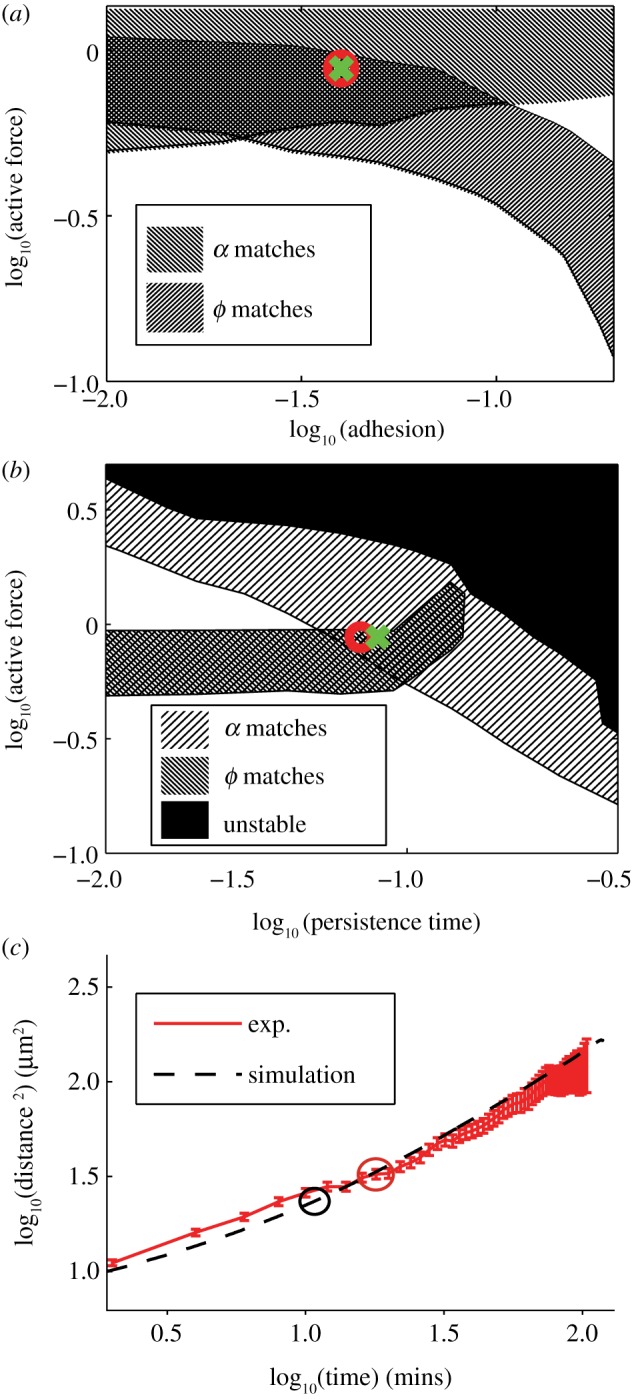
Calibrating the model. (a) Simulation phase diagram for active force and adhesion parameters. Circle and cross show best-fit parameters for ectoderm and mesendoderm, respectively. (b) Phase diagram for active force and persistence time model parameters. (c) Comparison of MSD for ectoderm explants compared to best-fit parameter from simulations. (Online version in colour.)
Table 1.
Tissue simulation parameters.
| parameter | value |
|---|---|
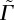 |
0.04 |
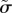 |
0.88 |
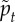 |
0.07 (ecto) 0.08 (meso) |
| τ | 4 s |
| R | 8 µm |
We also calculate how sensitive the observables (i.e. the diffusion constant and packing fraction) are to changes in the model parameters (see electronic supplementary material, table S1) near the best-fit parameter values. The diffusion constant is very sensitive to changes in all three parameters, and most sensitive to changes in 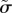 , which means that the model parameters are strongly constrained by the diffusion data. For example, changing the active force magnitude by 15% changes the diffusion constant by 100%. By contrast, the packing fraction ϕ is less sensitive to changes in model parameters.
, which means that the model parameters are strongly constrained by the diffusion data. For example, changing the active force magnitude by 15% changes the diffusion constant by 100%. By contrast, the packing fraction ϕ is less sensitive to changes in model parameters.
In addition, the model exhibits a jamming or glassy phase transition when the active forcing magnitude and persistence time are smaller than the best-fit values for ectoderm and mesendoderm. As discussed in the electronic supplementary material text, we define the glass transition as the point at which the diffusion constant D < 1 × 10−4 in natural units, because we find that this coincides with the onset of dynamical arrest. Although the best-fit model parameters are in a regime that is not jammed, the transition is nearby. For example, the model predicts that reducing either the active forcing magnitude or the persistence time by 20% would result in a glassy tissue where cells cannot migrate. This suggests that the viscoelasticity observed in these tissues might be controlled by the nearby glass transition.
3.3. Predictions for macroscopic tissue response
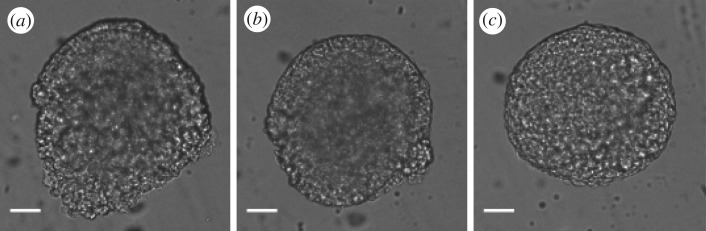
As a test of the predictive powers of this model, we keep the model parameters fixed at the values in table 1, and simulate the response of ectoderm tissues to large-scale mechanical perturbations such as compression and fusion with no adjustable parameters, and find qualitatively similar behaviour. We then quantitatively compare the emergent mechanical responses and timescales to those observed in the experimental data, finding reasonable agreement for bulk properties such as viscoelasticty, but disagreement for surface properties such as tissue surface tension. Tissue surface tension is a well-studied property of tissues [1,24,47,48], which quantifies how much a macroscopic tissue resists changes to its surface area. Tissue surface tension causes zebrafish explants to round-up as shown in figure 5 and governs cell sorting in embryonic tissues [1,24].
Figure 5.
Brightfield still images showing a piece of zebrafish embryonic tissue round-up. Scale bar, 30 μm. Electronic supplementary material, movie S1 shows the entire sequence.
The first set of simulations for a quantitative comparison are tissue surface tension parallel plate compression tests, where we seek to replicate the experiment in which a cellular aggregate is compressed between two parallel plates. In our model, the walls are represented by a two-dimensional triangular crystalline array of particles lying in a plane, as discussed in the electronic supplementary material text. Because we represent the wall with a single layer of particles, the necessarily stiff interaction potential makes the wall artificially sensitive to small changes in the positions of cells. Therefore, while the average value of the force on the wall is physically meaningful, the fluctuations in the forces on the wall are larger than those seen in the experiments.
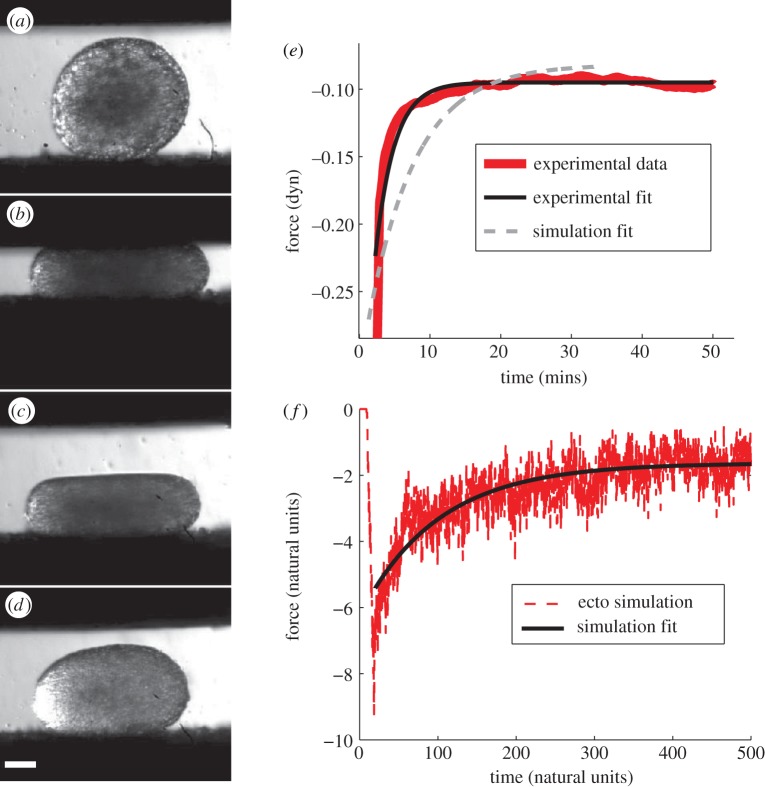
In both simulations and experiments, we measure the net force exerted on the explant by the upper compression plate. Images of an explant in a typical compression experiment are shown in figure 6a–d. All of the experimental TST data demonstrates the same qualitative response shown in figure 6e; as the plates are quickly brought together, there is a sharp decrease in the net force. This indicates a large force downward on the aggregate generated by the short-time elastic response of the aggregate. After the initial response, we observe a slow relaxation towards a non-zero equilibrium force as the cells rearrange and relax stress, just as a molecular fluid does. The non-zero force at long times is generated by the effective surface tension of the tissue. Previous work has established that the extracted surface tension does not depend on amount of deformation, demonstrating that this is a surface effect and not a bulk effect [26]. We fit the relaxation process to a constant + exponential. The exponential relaxation time of (4.5±0.8) min (mean±s.e.; n = 7) for ectoderm explants is a robust material property that describes the viscous tissue rheology.
Figure 6.
(a–d) Image series of an ectoderm explant compression in the TST: (a) precompression, (b) compressed, (c) right after release and (d) more than 10 min after compression. Scale bar, 100 mm. (e) TST experimental ectoderm force-balance curve (thick solid line) and fit (thin solid line). For comparison, the normalized ectoderm simulation fit is shown by a dashed line. (f) Simulation force-balance curves for ectoderm (dashed line). Solid line is a fit to ectoderm data. (Online version in colour.)
The tissue surface tension γ is obtained from the TST data using a modified Laplace's law, which is more robust to fitting procedures for experimental data [30]
| 3.3 |
where Feq is the steady-state force in the equatorial plane (of radius R1) at long times, and R2 is the local radius of curvature at the equator, extracted by fitting circles to brightfield images of the aggregate edge. Finally, we can also calculate the effective Young's modulus Y for ectoderm aggregates for small compressions by assuming that the initial response of the tissue is elastic and applying a Hertzian model for the spherical object [26]. We find that the Young's modulus for ectoderm is 44±11 Pa (n = 5), in agreement with previously published results of Y = 48±9 Pa [26].
The force–time response for a parallel plate compression of simulated ectoderm aggregates are shown in figure 6f. A first observation is that the mechanical response is qualitatively identical, capturing the features related to elastic, viscous and surface tension effects. In analogy to the experimental data, we fit the simulated data to a single exponential plus a constant, and find a relaxation time of 98 ± 14 natural time units, which corresponds to (6.7 ± 1.0) min. The fact that with no fit parameters the simulated ectoderm has an emergent relaxation time that is similar to that for the experimental tissue is a strong validation of the predictive powers of the model.
To analyse whether the surface tension in the simulations matches experiments, we need an independent estimate of the force scale. If we assume that Young's modulus for a single cell is the same as Young's modulus for the entire aggregate (which is reasonable since the tissue is confluent) then Y ∼ 40 − 50 Pa. This means that the natural force units in our simulation should be approximately 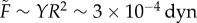 . An alternative path to the same result is to note that pipette aspiration experiments indicate that a typical effective cortical tension for tissues is γc = 1 dyn cm−1 [49], which corresponds to a natural force unit for our simulation model of
. An alternative path to the same result is to note that pipette aspiration experiments indicate that a typical effective cortical tension for tissues is γc = 1 dyn cm−1 [49], which corresponds to a natural force unit for our simulation model of 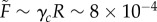 [50]. For the remainder of this paper, we use the value derived from TST data:
[50]. For the remainder of this paper, we use the value derived from TST data: 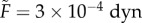 .
.
Because the observables that are most robust to fits in the simulations are different from those in the experiments, we fit to a different form of Laplace's law that involves the radius of contact between the plate and the aggregate, R3, as discussed in the electronic supplementary material. We find that Fwall = 1.45, R1 = 8.9, R2 = 10.1 and R3 = 5.3 in simulation units, which leads to a calculated value for the surface tension of γ ∼ 0.034 dyn cm−1. This is about 25 times smaller than that seen in experiments, where γ = 0.8 ± 0.2 dyn cm−1 (n = 6) and indicates that our model is not quantitatively capturing the surface tension effects in real tissues.
In hindsight, this is perhaps not surprising, as we have shown in previous work that the surface tension depends sensitively on individual cell shapes at the surface of the cell aggregate, as well as temporary mechanical polarization of those cells [24,25]. As our simulation explicitly disregards cell shapes and polarizations, it does not quantitatively capture behaviours governed by surface tension, although it does capture qualitative features such as rounding up of aggregates. In the discussion, we will address how our model might be augmented to better capture surface tension effects.
A second test for the predictive powers of the model is tissue fusion experiments. For tissue fusion simulations, we initialize the system with two droplets of 1000 cells each and position them so that the average radii of the two droplets overlap by half of a single-cell radius. We then evolve all of the COMs according to equation (3.2).
Figure 7a,b are snapshots from an experiment demonstrating that two rounded ectoderm explants join together to form a single rounded tissue (see electronic supplementary material, movie S2). To quantify this behaviour, we first identify the convex hull of the two-dimensional images of the aggregate using standard image analysis techniques (see electronic supplementary material, text movie S3), and the resulting hull is illustrated by lines in figure 7a,b. Using a method similar to Brangwynne et al. [27], we define the AR to be the ratio between major and minor axes, and figure 7e illustrates that it varies from approximately two at the beginning of a fusion experiment to approximately unity when the fusion has completed for the example shown in figure 7a,b. We fit the decay of the AR and find a characteristic decay timescale of (207 ± 23) min (mean ± s.e.; n = 6) for ectoderm explants.
Figure 7.
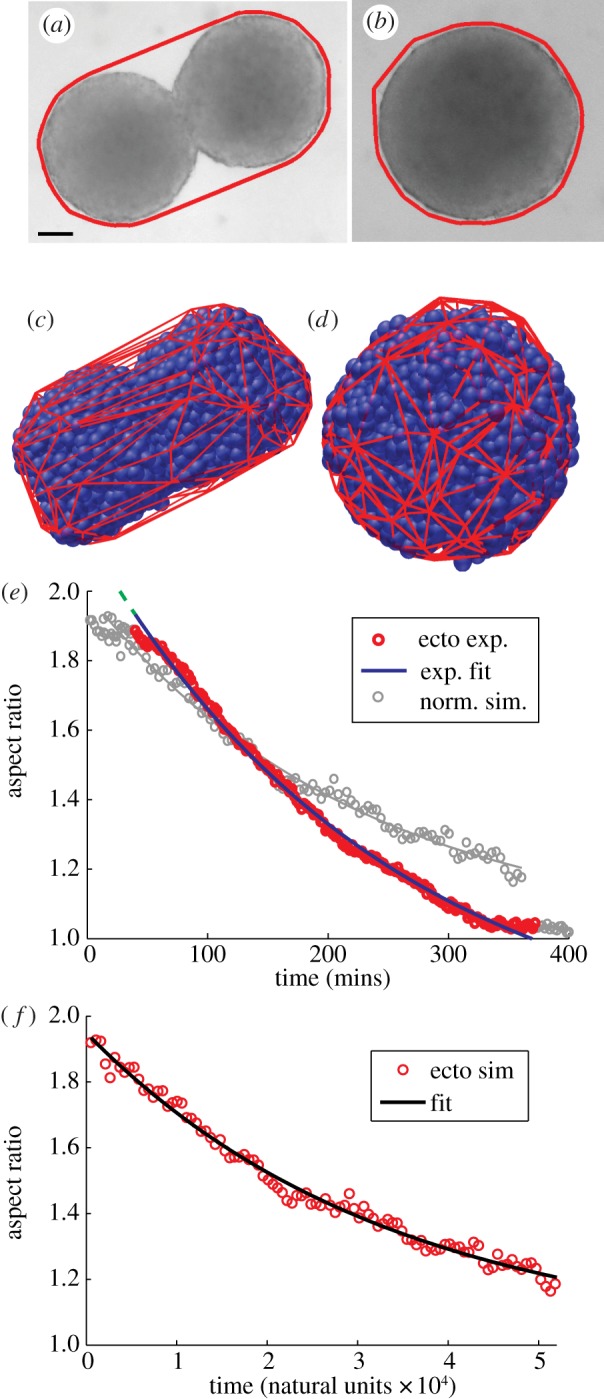
(a,b) Images from the beginning and end of experimental aggregate fusion. Thick line indicates convex hull. Scale bar, 100 mm. (c,d) Images from beginning and end of simulation of aggregate fusion. (e) Experimental data for AR as a function of time (dark circles) and fit (dark line). Light circles and line correspond to simulation data that have been normalized to account for different aggregate size and surface tension as measured by the TST simulations. (f) Simulation data for AR as a function of time (circles) and fit (solid line). (Online version in colour.)
We also perform the same test on simulated ectoderm explants. Figure 7c,d are snapshots from the three-dimensional simulations, with the cells denoted by spheres and the mesh denoting the convex hull. A full-time sequence is available in electronic supplementary material, movie S4. We again calculate the AR for each timepoint using the same definitions as for the experiments (scaled from two to three dimensions). The resulting AR evolution, which is qualitatively very similar to the experimental observation, is shown in figure 7f. The AR does not quite asymptote to unity because of the difference in the way the major and minor chords are calculated in two versus three dimensions. We fit this time evolution to the same exponential + constant function, and find a decay time of 33 500 in natural units or 2250 mins. This is more than an order of magnitude larger than the experimental observation, but there are two important differences between the simulated aggregate and the experimental aggregate. First, the experimental aggregate has about eight times as many cells as the simulated aggregate; for consistency we used a simulated droplet of the same size as those in the simulations in figure 4a,b. Second, the surface tension of the simulated aggregate as measured by the TST simulation is about 20 times less than that for the experimental aggregate. In 1939, Young [51] derived an analytical expression for the change in the AR (ra) of an ellipsoidal droplet as it approaches a spherical droplet under the influence of surface tension
| 3.4 |
where γ is the surface tension, η is the viscosity, V is the volume and f(ra) is a complicated function that depends on whether the ellipsoid is oblate or prolate. This equation indicates that we can account for the known differences between the simulated aggregate and the experimental aggregate by multiplying the timescale by Rexp/Rsim ∼ 2 and dividing the timescale by the fit from the TST data γexp/γsim ∼ 23. The resulting curve for the ‘effective’ AR has a time decay constant of 195 min, which compares remarkably well with the experimental timescale of 207 min, given there are no fit parameters. We also plot the effective simulated AR in light grey in figure 7e, illustrating the similarity between the two. Therefore, the simulated tissue fusion data are consistent with the experimental fusion data under the assumption that the simulated aggregate has the same bulk properties but a reduced surface tension.
4. Discussion
We have shown that a minimal model for cell interactions with three dimensionless parameters predicts several types of emergent, collective mechanical responses in embryonic tissues. It disregards a significant amount of information about cell shapes and small-scale details of the mechanical interactions between cells. It is simple enough that it can be calibrated from experimental data using only single-cell trajectories and static nuclei imaging. Despite this simplicity, it captures complex features of the experimental data, such as caging behaviour and crossover timescales. When we additionally calibrate the force scale using TST data, the model can also predict the fusion behaviour of two explants.
Our model indicates that macroscopic tissue behaviour is most sensitive to the two model parameters that describe forces actively generated by cells. Tissues do not change their structure (i.e. they are solid-like) when cells generate weak forces or change their direction rapidly, and they do change their structure (i.e. explants round-up and behave like a liquid) when cells generate larger forces or change their direction slowly. This is analogous to commuters trying to exit a crowded subway train: a person must push persistently in one direction or push harder to move others out of the way and make a path to the exit. In addition, tissues become more solid-like when the ratio between the adhesion and the cortical tension is high. The more strongly cells stick to their neighbours, the harder it is to get past them.
Calculating a phase diagram as in figure 4 and estimating how close a tissue is to the phase boundary between liquid and solid has important biological implications. Tissues that are near the boundary are much more adaptable: small changes to the single-cell mechanical properties via mutations or knockdowns can drastically change the macroscopic tissue response. For example, our observations of caging and subdiffusivity in zebrafish embryonic explants suggests that they are very close to the boundary, and therefore our model predicts that if individual cells became slightly more adhesive or slightly less active, the tissue will transition to a solid. Since the fluid-like behaviour of embryonic tissues is critical for pattern formation in early development, our model will be useful for determining what types of motility defects are likely to lead to pathologies. More generically, it will be useful for predicting which embryonic tissues are fluid-like and therefore governed by tissue surface tension, and which tissues are solid-like and governed by elasticity, which will enable better predictions about structure formation and organogenesis in embryonic development.
If the specific details of single-cell mechanics are important for the large-scale mechanics of the tissue, we would expect our model to fail. The fact that the model succeeds does not mean that single-cell mechanics are not important for bulk properties, but suggests that details such as cell motility, directionality, elastic modulus, etc., can be successfully coarse-grained into a small number of properly chosen parameters.
Furthermore, we show that the motion of cells past one another, which must be generated by cells actively exerting tension on contacts with other cells, is best modelled by a special type of structured noise (both multiplicative and coloured), instead of positional or angular white noise [41,44–46]. While white noise is uncorrelated in time and thus describes random fluctuations (and would allow a cell to at any instant randomly switch its direction of motion), coloured noise has a non-zero autocorrelation for non-zero time lags, thus enforces that cells can exhibit persistent motion for a certain time in the direction of an adhesive contact before switching contacts. This choice for the noise is motivated both by experimental observations at the single-cell scale and the fact that our simple model cannot reproduce the macroscopic observations without it.
Why does such a simple model work so well? As we and others [16,52] have noted, active tissues display glassy dynamics: the motion of individual cells is constrained because they are surrounded by tightly packed neighbouring cells that impede their progress. It has been shown that the dynamics in non-biological glassy or jammed materials display universality: dynamical features do not depend on the details of the interactions [53,54]. Our work suggests that models with only a few parameters could be adequate for describing active tissues near this glass transition.
An important observation about our experimental zebrafish explants is that the tissue boundary remains remarkably coherent during all mechanical perturbations, including hanging drop, TST compression and tissue fusion experiments. No cells exit the aggregate, even though cells move over large distances on the inside and together behave collectively as a fluid. This remarkable property is not observed in typical non-biological materials with short-ranged interactions; if particles behave as a liquid inside the droplet, they also necessarily leave the droplet and generate a steady-state vapour pressure in a closed system. We note that vapour pressures also occur in many models for biological tissues. For example, the one introduced by Ranft et al. [41] does possess a regime, where the bulk is fluid-like, but unless the model also has artificially long-ranged interactions, it will generate a significant vapour pressure in the fluid regime. By contrast, the model introduced here has a self-generated boundary that reproduces the experimentally observed absence of a vapour pressure, because the direction of the noise acting on one cell depends on the location of the cell's neighbours. A cell does not push off another cell, but instead is biased to move in a direction where it can make new contacts.
Although there is little or no vapour pressure in the three-dimensional system, recent experiments [52] have shown that cell aggregates can wet adhesive substrates and generate a two-dimensional vapour pressure on the surface. By replacing the non-adhesive walls in our TST experiments with adhesive passive particles, we would be able to model those behaviours. We expect that our model would very naturally recapitulate the presence of a two-dimensional surface vapour pressure and lack of a three-dimensional vapour pressure.
Another interesting direction is to investigate modifications to our model that could help explain the much larger surface tensions seen in experiments compared to simulations. We believe our model fails to correctly capture the magnitude of tissue surface tension because it likely depends on cell shapes and a strong feedback between adhesion and cortical tension that occurs at tissue interfaces, as discussed in other work [24]. We could augment our model to account for this by replacing the isotropic interaction given by electronic supplementary material, equation S2, by a non-isotropic interaction for surface cells.
While the model was inspired by and calibrated using data from embryonic zebrafish explants, we expect that variations of this model will be applicable to a broad range of biological tissues. Because it is simple, it can be effectively calibrated for different tissue types. It bridges microscopic mechanical information (cell elastic modulus, cell surface tension, adhesion and rate of protrusions) and macroscopic tissue mechanics (tissue viscosity, tissue surface tension and tissue elasticity). To expand the model to capture more complicated tissues than the ones studied here, one could group several spheres together to allow shape changes and cell divisions. Another possibility is to allow for higher order interactions between multiple cells; for cells that are very soft, the surface area in contact and the elastic interaction do not depend only on the two-body overlap, but also on the direction of the contact and on the locations of neighbouring cells. A cluster expansion borrowed from statistical physics might be able to address such effects.
5. Conclusion
We have studied the trajectories of individual cells in embryonic explants and find that although the tissue is fluid-like on long timescales, it displays features of glassy or supercooled dynamics, including subdiffusive MSDs on short timescales and caging behaviour. This suggests that the tissue might act as a viscoelastic material where the viscoelastic response is governed by a nearby glass transition.
To explore this hypothesis, we have developed a three-parameter model that makes predictions for the emergent properties of simple embryonic tissues. Two key observations that we incorporate into our model are (i) that cells are biased to move in the direction of their neighbours and (ii) that this motion occurs over a time window that is not infinitesimally small compared with other timescales in the tissue. The model exhibits a glass transition between solid-like behaviour and dynamical arrest (when active forces are small and occur over short time windows), and liquid-like behaviour (when active forces are large and persistent).
We calibrated the model using only the tracking and structural data from experiments on zebrafish embryonic explants. The best-fit parameters suggest that embryonic tissues are liquid-like but close to the glass transition, and therefore our model can be used to help explain how small single-cell motility defects might lead to large differences in tissue response during development.
We verify that the calibrated model makes accurate quantitative and qualitative predictions about the macroscopic tissue response in parallel plate compressions and tissue fusion assays, including a coherent self-generated boundary. Because this simple model can explain the bulk tissue response, the exact details about individual cell shapes and mechanics are not critical for this experimental system; instead, we can effectively coarse-grain those details into a few parameters that can easily be extracted from experiments. The fact that the model fails to explain surface properties suggests that cell shapes and mechanical polarizations play an important role in those processes.
From a practical standpoint, this model can easily be expanded to make predictions about other tissues in development and disease. From an intellectual standpoint, it explains how disordered tissue structures and high cell densities generate a viscoelastic response. It also provides an explanation for the timescale for the crossover from elastic to viscous behaviour as a caging timescale. Because it is simple and yet different from existing models for active matter, it provides a framework for thinking about the types of phase transitions (such as jamming or flocking) that are possible in biological tissues.
Acknowledgements
The authors thanks T. Bacarian for initial cell tracking, S. Thiberge and the Lewis-Sigler Institute imaging core facility for two-photon usage, and J. Shaevitz, S. Henkes and J. Posakony for comments on the manuscript.
Funding statement
E.-M.S. is funded by the Lewis-Sigler Fellowship and the Burroughs Wellcome Fund CASI award. M.L.M. acknowledges computational support from the Princeton Center for Theoretical Science and the College of Arts and Sciences at Syracuse University.
References
- 1.Schötz E, Burdine RD, Jülicher F, Steinberg MS, Heisenberg C, Foty RA. 2008. Quantitative differences in tissue surface tension influence zebrafish germ layer positioning. HFSP J. 2, 42–56. ( 10.2976/1.2834817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller R, Shook D, Skoglund P. 2008. The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys. Biol. 5, 015007 ( 10.1088/1478-3975/5/1/015007) [DOI] [PubMed] [Google Scholar]
- 3.Farge E. 2003. Mechanical Induction of twist in the Drosophila foregut/stomodeal primordium. Curr. Biol. 13, 1365–1377. ( 10.1016/S0960-9822(03)00576-1) [DOI] [PubMed] [Google Scholar]
- 4.Farhadifar R, Roper J-C, Aigouy B, Eaton S, Julicher F. 2007. The influence of cell mechanics, cell–cell interactions, and proliferation on epithelial packing. Curr. Biol. 17, 2095–2104. ( 10.1016/j.cub.2007.11.049) [DOI] [PubMed] [Google Scholar]
- 5.McMahon A, Supatto W, Fraser SE, Stathopoulos A. 2008. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science 322, 1546–1550. ( 10.1126/science.1167094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandersius SA, Chuai M, Weijer CJ, Newman TJ. 2011. A ‘chemotactic dipole’ mechanism for large-scale vortex motion during primitive streak formation in the chick embryo. Phys. Biol. 8, 045008 ( 10.1088/1478-3975/8/4/045008) [DOI] [PubMed] [Google Scholar]
- 7.Weijer CJ. 2009. Collective cell migration in development. J. Cell Sci. 122, 3215–3223. ( 10.1242/jcs.036517) [DOI] [PubMed] [Google Scholar]
- 8.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. 2007. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl Acad. Sci. USA 104, 15 988–15 993. ( 10.1073/pnas.0705062104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider L. 2010. Directional cell migration and chemotaxis in wound healing response to PDGF-AA are coordinated by the primary cilium in fibroblasts. Cell Physiol. Biochem. 25, 279–292. ( 10.1159/000276562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano J, Rudiger S, Pullarkat P, Ott A. 2009. Mechanogenetic coupling of Hydra symmetry breaking and driven turing instability model. Biophys. J. 96, 1649–1660. ( 10.1016/j.bpj.2008.09.062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laird DJ, von Andrian UH, Wagers AJ. 2008. Stem cell trafficking in tissue development, growth, and disease. Cell 132, 612–630. ( 10.1016/j.cell.2008.01.041) [DOI] [PubMed] [Google Scholar]
- 12.Christiansen JJ, Rajasekaran AK. 2006. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 66, 8319–8326. ( 10.1158/0008-5472.CAN-06-0410) [DOI] [PubMed] [Google Scholar]
- 13.Friedl P, Gilmour D. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445–457. ( 10.1038/nrm2720) [DOI] [PubMed] [Google Scholar]
- 14.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL. 2012. Mechanical compression drives cancer cells toward invasive phenotype. Proc. Natl Acad. Sci. USA 109, 911–916. ( 10.1073/pnas.1118910109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegedus B, Marga F, Jakab K, Sharpe-Timms KL, Forgacs G. 2006. The interplay of cell–cell and cell–matrix interactions in the invasive properties of brain tumors. Biophys. J. 91, 2708–2716. ( 10.1529/biophysj.105.077834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelini TE, Hannezo E, Trepat X, Marquez M, Fredberg JJ, Weitz DA. 2011. Glass-like dynamics of collective cell migration. Proc. Natl Acad. Sci. USA 108, 4714–4719. ( 10.1073/pnas.1010059108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nnetu KD, Knorr M, Ks J, Zink M. 2012. The impact of jamming on boundaries of collectively moving weak-interacting cells. New J. Phys. 14, 115012 ( 10.1088/1367-2630/14/11/115012) [DOI] [Google Scholar]
- 18.Debenedetti PG, Stillinger FH. 2001. Supercooled liquids and the glass transition. Nature 410, 259–267. ( 10.1038/35065704) [DOI] [PubMed] [Google Scholar]
- 19.Ediger M. 2000. Spatially heterogeneous dynamics in supercooled liquids. Annu. Rev. Phys. Chem. 51, 99–128. ( 10.1146/annurev.physchem.51.1.99) [DOI] [PubMed] [Google Scholar]
- 20.Abate AR, Durian DJ. 2007. Topological persistence and dynamical heterogeneities near jamming. Phys. Rev. E 76, 021306 ( 10.1103/PhysRevE.76.021306) [DOI] [PubMed] [Google Scholar]
- 21.Richert R, Angell CA. 1998. Dynamics of glass-forming liquids. V. On the link between molecular dynamics and configurational entropy. J. Chem. Phys. 121, 9016 ( 10.1063/1.476348) [DOI] [Google Scholar]
- 22.Kob W, Donati C, Plimpton SJ, Poole PH, Glotzer SC. 1997. Dynamical heterogeneities in a supercooled Lennard-Jones liquid. Phys. Rev. Lett. 79, 2827–2830. ( 10.1103/PhysRevLett.79.2827) [DOI] [Google Scholar]
- 23.Weeks ER, Weitz DA. 2002. Properties of cage rearrangements observed near the colloidal glass transition. Phys. Rev. Lett. 89, 095704 ( 10.1103/PhysRevLett.89.095704) [DOI] [PubMed] [Google Scholar]
- 24.Manning ML, Foty RA, Steinberg MS, Schoetz E-M. 2010. Coaction of intercellular adhesion and cortical tension specifies tissue surface tension. Proc. Natl Acad. Sci. USA 107, 12 517–12 522. ( 10.1073/pnas.1003743107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amack J, Manning M. 2012. Knowing the boundaries: extending the differential adhesion hypothesis in embryonic cell sorting. Science 338, 212–215. ( 10.1126/science.1223953) [DOI] [PubMed] [Google Scholar]
- 26.Schoetz E-M. 2008. Dynamics and mechanics of zebrafish embryonic tissues—a study of the physical properties of zebrafish germlayer cells and tissues and cell dynamics during early embryogenesis. Saarbrücken, Germany: VDM publishing group. [Google Scholar]
- 27.Brangwynne CP, Mitchison TJ, Hyman AA. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339. ( 10.1073/pnas.1017150108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foty RA, Forgacs G, Pfleger CM, Steinberg MS. 1994. Liquid properties of embryonic tissues: measurement of interfacial tensions. Phys. Rev. Lett. 72, 2298–2301. ( 10.1103/PhysRevLett.72.2298) [DOI] [PubMed] [Google Scholar]
- 29.Foty RA, Pfleger CM, Forgacs G, Steinberg MS. 1996. Surface tensions of embryonic tissues predict their mutual envelopment behavior. Development 122, 1611–1620. [DOI] [PubMed] [Google Scholar]
- 30.Mgharbel A, Delanoe-Ayari H, Rieu J-P. 2009. Measuring accurately liquid and tissue surface tension with a compression plate tensiometer. HFSP J. 3, 213–221. ( 10.2976/1.3116822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pologruto T, Sabatini B, Svoboda K. 2003. ScanImage: flexible software for operating laser scanning microscopes. BioMed. Eng. OnLine 2, 13 ( 10.1186/1475-925X-2-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crocker JC, Grier DG. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 298–310. ( 10.1006/jcis.1996.0217) [DOI] [Google Scholar]
- 33.Voronoi G. 1908. Nouvelles applications des paramètres continus à la théorie des formes quadratiques. Premier Mémoire: Sur quelques propriétés des formes quadratiques positives parfaites. J. Reine Angew. Mathematik 133, 97–178. [Google Scholar]
- 34.O'Hern CS, Langer SA, Liu AJ, Nagel SR. 2002. Random packings of frictionless particles. Phys. Rev. Lett. 88, 075507 ( 10.1103/PhysRevLett.88.075507) [DOI] [PubMed] [Google Scholar]
- 35.Brakke KA. 1992. The surface evolver. Exp. Math. 1, 141–165. ( 10.1080/10586458.1992.10504253) [DOI] [Google Scholar]
- 36.Mombach J, Glazier J, Raphael R, Zajac M. 1995. Quantitative comparison between differential adhesion models and cell sorting in the presence and absence of fluctuations. Phys. Rev. Lett. 75, 2244–2247. ( 10.1103/PhysRevLett.75.2244) [DOI] [PubMed] [Google Scholar]
- 37.Graner F, Glazier JA. 1992. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys. Rev. Lett. 69, 2013–2016. ( 10.1103/PhysRevLett.69.2013) [DOI] [PubMed] [Google Scholar]
- 38.Shirinifard A, Gens J, Zaitlenand B, Poplawski N, Swat M, Glazier J. 2009. Multi-cell simulation of tumor growth and angiogenesis. PLoS ONE 4, e7190 ( 10.1371/journal.pone.0007190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N, Glazier JA, Izaguirre JA, Alber MS. 2007. A parallel implementation of the cellular Potts model for simulation of cell-based morphogenesis. Comp. Phys. Commun. 176, 670–681. ( 10.1016/j.cpc.2007.03.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brodland GW, Yang J, Sweny J. 2009. Cellular interfacial and surface tensions determined from aggregate compression tests using a finite element model. HFSP J. 4, 273–281. ( 10.2976/1.3175812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranft J, Basan M, Elgeti J, Joanny J, Prost J, Jülicher F. 2010. Fluidization of tissues by cell division and apoptosis. Proc. Natl Acad. Sci. USA 107, 20 863–20 868. ( 10.1073/pnas.1011086107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staple DB, Farhadifar R, Röper JC, Aigouy B, Eaton S, Jülicher F. 2010. Mechanics and remodelling of cell packings in epithelia. Eur. Phys. J. E 33, 117–127. ( 10.1140/epje/i2010-10677-0) [DOI] [PubMed] [Google Scholar]
- 43.Vicsek T, Czirók A, Ben-Jacob E, Cohen I, Shochet O. 1995. Novel type of phase transition in a system of self-driven particles. Phys. Rev. Lett. 75, 1226–1229. ( 10.1103/PhysRevLett.75.1226) [DOI] [PubMed] [Google Scholar]
- 44.Gregoire G, Chate H. 2004. Onset of collective and cohesive motion. Phys. Rev. Lett. 92, 025702 ( 10.1103/PhysRevLett.92.025702) [DOI] [PubMed] [Google Scholar]
- 45.Henkes S, Fily Y, Marchetti MC. 2011. Active jamming: self-propelled soft particles at high density. Phys. Rev. E 84, 040301 ( 10.1103/PhysRevE.84.040301) [DOI] [PubMed] [Google Scholar]
- 46.Palsson E. 2001. A three-dimensional model of cell movement in multicellular systems. Future Gen. Comp. Syst. 17, 835–852. ( 10.1016/S0167-739X(00)00062-5) [DOI] [Google Scholar]
- 47.Steinberg MS. 1963. Reconstruction of tissues by dissociated cells. Science 141, 401–408. ( 10.1126/science.141.3579.401) [DOI] [PubMed] [Google Scholar]
- 48.Foty RA, Steinberg MS. 2005. The differential adhesion hypothesis: a direct evaluation. Dev. Biol. 278, 255–263. ( 10.1016/j.ydbio.2004.11.012) [DOI] [PubMed] [Google Scholar]
- 49.Evans E, Yeung A. 1989. Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration. Biophys. J. 56, 151–160. ( 10.1016/S0006-3495(89)82660-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durian DJ. 1997. Bubble-scale model of foam mechanics:mMelting, nonlinear behavior, and avalanches. Phys. Rev. E 55, 1739–1751. ( 10.1103/PhysRevE.55.1739) [DOI] [Google Scholar]
- 51.Young G. 1939. On the mechanics of viscous bodies and elongation in ellipsoidal cells. Bull. Math. Biophys. 1, 31–46. ( 10.1007/BF02478011) [DOI] [Google Scholar]
- 52.Douezan S, Guevorkian K, Naouar R, Dufour S, Cuvelier D, Brochard-Wyart F. 2011. Spreading dynamics and wetting transition of cellular aggregates. Proc. Natl Acad. Sci. USA 108, 7315–7320. ( 10.1073/pnas.1018057108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haxton T, Schmiedeberg M, Liu A. 2011. Universal jamming phase diagram in the hard-sphere limit. Phys. Rev. E 83, 031503 ( 10.1103/PhysRevE.83.031503) [DOI] [PubMed] [Google Scholar]
- 54.Schmiedeberg M, Haxton T, Nagel S, Liu A. 2011. Mapping the glassy dynamics of soft spheres onto hard-sphere behavior. Europhys. Lett. 96, 36010 ( 10.1209/0295-5075/96/36010) [DOI] [Google Scholar]