Abstract
Fragmented QRS (f-QRS) has been proven to be an efficient biomarker for several diseases, including remote and acute myocardial infarction, cardiac sarcoidosis, non-ischaemic cardiomyopathy, etc. It has also been shown to have higher sensitivity and/or specificity values than the conventional markers (e.g. Q-wave, ST-elevation, etc.) which may even regress or disappear with time. Patients with such diseases have to undergo expensive and sometimes invasive tests for diagnosis. Automated detection of f-QRS followed by identification of its various morphologies in addition to the conventional ECG feature (e.g. P, QRS, T amplitude and duration, etc.) extraction will lead to a more reliable diagnosis, therapy and disease prognosis than the state-of-the-art approaches and thereby will be of significant clinical importance for both hospital-based and emerging remote health monitoring environments as well as for implanted ICD devices. An automated algorithm for detection of f-QRS from the ECG and identification of its various morphologies is proposed in this work which, to the best of our knowledge, is the first work of its kind. Using our recently proposed time–domain morphology and gradient-based ECG feature extraction algorithm, the QRS complex is extracted and discrete wavelet transform (DWT) with one level of decomposition, using the ‘Haar’ wavelet, is applied on it to detect the presence of fragmentation. Detailed DWT coefficients were observed to hypothesize the postulates of detection of all types of morphologies as reported in the literature. To model and verify the algorithm, PhysioNet's PTB database was used. Forty patients were randomly selected from the database and their ECG were examined by two experienced cardiologists and the results were compared with those obtained from the algorithm. Out of 40 patients, 31 were considered appropriate for comparison by two cardiologists, and it is shown that 334 out of 372 (89.8%) leads from the chosen 31 patients complied favourably with our proposed algorithm. The sensitivity and specificity values obtained for the detection of f-QRS were 0.897 and 0.899, respectively. Automation will speed up the detection of fragmentation, reducing the human error involved and will allow it to be implemented for hospital-based remote monitoring and ICD devices.
Keywords: electrocardiography, fragmented QRS, wavelet transform
1. Introduction
Recently in last 5 years, fragmented QRS (f-QRS) has gained clinical significance in the diagnosis of various cardiologic disorders, including remote and acute myocardial infarction, cardiac sarcoidosis, non-ST-elevation myocardial infarction, ventricular aneurysm, etc. [1–15]. These studies have shown that f-QRS complexes can be an important biomarker for detection of several diseases and has resulted in higher sensitivity and/or specificity than other conventional markers e.g. Q-wave, ST-elevation, etc. [1–15]. However, despite its enormous diagnosis significance cardiologists often disregard or do not report fragmentation in most of the cases except those of bundle branch block (BBB). Therefore, automatic detection and identification of morphologies of f-QRS will precisely report on all the cases and may help in finding correlations of potential clinical significance by analysing hundreds of tracings, and hence will facilitate its widespread clinical acceptance, adaptation and application.
In general, the diagnostic information is obtained from standard 12-lead ECG using the conventional biomarkers, e.g. P, Q, R, S waves, their duration and peaks, ST elevation/depression, PT interval and (non-)inverted T wave [16]. However, there are certain diseases, e.g. cardiac sarcoidosis, myocardial infarction, etc. [2–5,9], which cannot be detected by these conventional biomarkers, e.g. Q-wave, ST elevation/depression, PT interval, etc. Patients suffering from these diseases may have to undergo several invasive or non-invasive tests for reliable diagnosis which may be unaffordable [2–5,9]. f-QRS in this context has been found to be a marker for aforementioned diseases and its occurrence in standard 12-lead ECG makes it an inexpensive and easily available tool for diagnosis. Table 1 provides information on diseases which have been found to have high sensitivity and/or specificity values for diagnosis using f-QRS [2–5,8,9] along with the conventional markers/tests that are used to diagnose these diseases and their limitations. Computerized ECG interpretation and feature extraction are being successfully used in a hospital-based environment [17]. Furthermore, significant prevalence of cardiovascular diseases throughout the world resulted in emerging remote health monitoring systems, which also demand automated, low-complexity feature extraction algorithms ported into sophisticated mobile devices or within on-board sensors processing modules ([16,18], http://www.chiron-project.eu/). An automated algorithm can find direct applications in remote health monitoring. It has been found that f-QRS can be used as a selection criterion for implantation of ICD devices [19,20]. Long-term monitoring is required for this purpose and an automated system will considerably reduce the effort required for verification of the signals. However, to the best of our knowledge f-QRS detection and identification of its various morphologies have not been automated and implemented in practice. Here, we propose an automated algorithm for f-QRS detection and morphology identification.
Table 1.
Cardiologic disorders along with their conventional marker and limitations.
| cardiologic disorder | conventional marker | remarks |
|---|---|---|
| acute and remote myocardial infarction with coronary artery disease | acute ST-elevation, Q-wave | after an acute MI resolves repolarization, abnormalities stabilize and only Q-wave then remains as a marker of MI. Q-waves regress or even disappears with time and there is no specific sign of a non-Q-wave MI non-ST elevation MI [2,4]. |
| BBB, premature ventricular complexes and paced rhythm | QRS ≥ 120 ms | no ECG diagnosis prior MI scar without the presence of Q-wave has been defined. Patients have to undergo expensive diagnosis test e.g. SPECT test, echocardiography, etc [5]. |
| left ventricular aneurysm with QRS ≤ 120 ms | ST elevation with the presence of prominent R wave in aVR | low specificity as ST-segment elevation is present in many cardiac diseases and Goldberger's sign of prominent R also has low sensitivity [3]. |
| cardiac sarcoidosis | no current marker | diagnosed by myocardial biopsy, cardiac magnetic resonance imaging with gadolinium-delayed enhancement images, echocardiography. Myocardial biopsy is invasive and has low sensitivity. There is no single diagnostic test but a combination of clinical data and investigation of CMR with GDE are used for diagnosis [9]. |
| non-ST-elevation myocardial infarction | ischaemic T-waves, ST-segment depression, microvolt T-wave alternans, late potentials on the signal-averaged ECG, pathologic Q-waves | sensitivity and specificity of f-QRS have been found to be higher than ischaemic T-waves. For other biomarkers, their correlation with the exact anatomic location of the culprit lesion is not very high [8]. |
2. Material and methods
Figure 1 presents the procedure followed in the proposed method. The raw ECG signal is passed through a preprocessing module comprising baseline wandering removal and denoising, which will be discussed in §2.2. Then, out of existing ECG feature extraction techniques [16,18,21–24] our recently proposed time–domain morphology and gradient (TDMG)-based algorithm [16] is applied to extract the QRS complex which is interpolated next and fed as an input to the fragmentation detection and identification (FDMI) module. We skip here the detailed discussion on the TDMG algorithm. Interested readers may consult Mazomenos et al. [16] for the details. If fragmentation is detected, its morphology is then identified. This FDMI module is discussed next. Online QRS detection has been investigated by several researchers [21–24], and hence the availability of an accurately detected QRS can be safely assumed. In this work, DWT has been applied using the ‘Haar’ wavelet. As the Haar wavelet is discontinuous and anti-symmetric [25], it is suitable for discontinuity and edge detection [25]. Moreover, owing to its low computational complexity and power consumption, it is suitable for remote health monitoring applications [18]. Hence, Haar is used in this investigation. Detailed discussion on the ‘Haar’ wavelet-based transform beyond the scope of this paper. However, interested readers are requested to consult section III of reference [18] for more detail.
Figure 1.
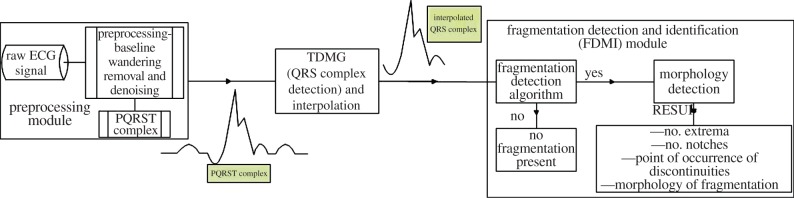
Procedure followed for the detection and identification of fragmentation in QRS of an ECG signal. (Online version in colour.)
2.1. Fragmentation detection and identification module
This section has been further subdivided into two subsections viz. fragmentation detection and morphology identification.
2.1.1. Fragmentation detection
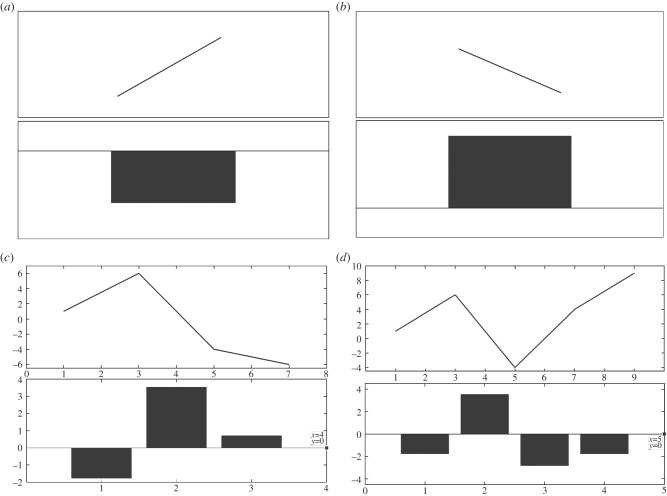
The detailed DWT coefficients behave in a particular manner when a discontinuity1 is encountered. A peak or nadir in any signal can be detected by the zero crossing of the wavelet transform. This technique has been previously used to detect QRS complexes of ECG [22]. In this section, we explain this behaviour with the help of figure 2. Consider a line segment with positive slope joining two points and the corresponding bar plot of the detailed DWT coefficient as shown in figure 2a, it can be seen that the coefficient is negative for an increasing segment. Figure 2b shows a line segment with negative slope and the corresponding bar plot of the detailed DWT coefficient which in this case is positive. In figure 2c, a set of points have drawn along with the bar plot of their detailed DWT coefficients. It can be inferred from figure 2a–c that the increasing part of the curve results in negative and the decreasing part of the curve results in positive detailed DWT coefficients, respectively. It should also be noted that the magnitude of the coefficients depends on the slope of the tangent at that point. The greater the slope, the greater the magnitude of the coefficient. Whenever a local extremum appears, there is a transition in the sign of the detailed coefficient depending on the presence of a maxima or minima. We confirm this hypothesis with figure 2d in which both the extrema are identified, i.e. first the maxima and then the minima. This phenomenon has been used in modelling and designing of the proposed fragmentation detection algorithm (FDA). The notches [1–15] that occur in the QRS complex are identified as frequent changes in the sign of detailed coefficients and a peak is identified as a sudden change in sign with a constant follow up of the coefficients with the same sign.
Figure 2.
Explanation of patterns observed on occurrence of a discontinuity.
In figure 2, bar plots of detailed DWT coefficients of interpolated QRS complex are plotted and rules for identifying extrema and notches are formulated by observing and correlating the patterns occurring in the QRS complex and detailed coefficients. It is to be noted that the bars may not seem to align because the scales of the x-axis of the wave and the bar plot of detailed discrete wavelet transform (DWT) coefficients are different. When DWT is applied the number of detailed coefficients obtained is half the number of samples on which the DWT was applied. The main purpose of using figure 2 is to show the behaviour of detailed coefficients when an increasing or decreasing part of the wave is encountered. In figure 2c, the wave has seven samples and the total number of detailed coefficients is 4, however, the 4th coefficient is zero. Similarly, in figure 2d, the number of samples is 9 and coefficients are 5, however, the last coefficient is zero, which is indicated in figure 2. Detailed discussion of the DWT can be found in [18]. With the help of few patients, the criteria in table 2 were hypothesized and were then iteratively refined by applying them on leads of 40 subjects such that the mentioned criteria accurately and reliably captured all sorts of discontinuities occurring in the QRS complexes. Table 2 presents the rules framed for detection of local extrema and notches. The algorithm starts evaluating the QRS complex from the left-most side of the bar plot and proceeds to the right. While traversing through the coefficients, patterns are recognized and if any pattern matches to those mentioned in table 2 the corresponding discontinuity2 is realized and noted. Algorithm pointer (k) pointing at a particular point of the QRS complex is incremented according to the presence and type of discontinuity encountered. If a discontinuity is spotted, it is identified and the pointer increments as per the mentioned rule (table 2) otherwise it increments by one. For example, in pattern A1 as shown in table 2 there are two consecutive sign changes in the values of detailed coefficients which can be attributed to the occurrence of a local extremum pair in close proximity, and hence can assumed to be a notch. Similarly, in pattern A2, there are three consecutive sign changes implying that there are three extrema and the one that is identified as a notch depends on the magnitude of the detailed coefficient, the other is identified as an extremum. Other patterns for the identification of notch and extremum can be interpreted in a similar fashion as mentioned in table 2.
Table 2.
Rules for identification of discontinuities.
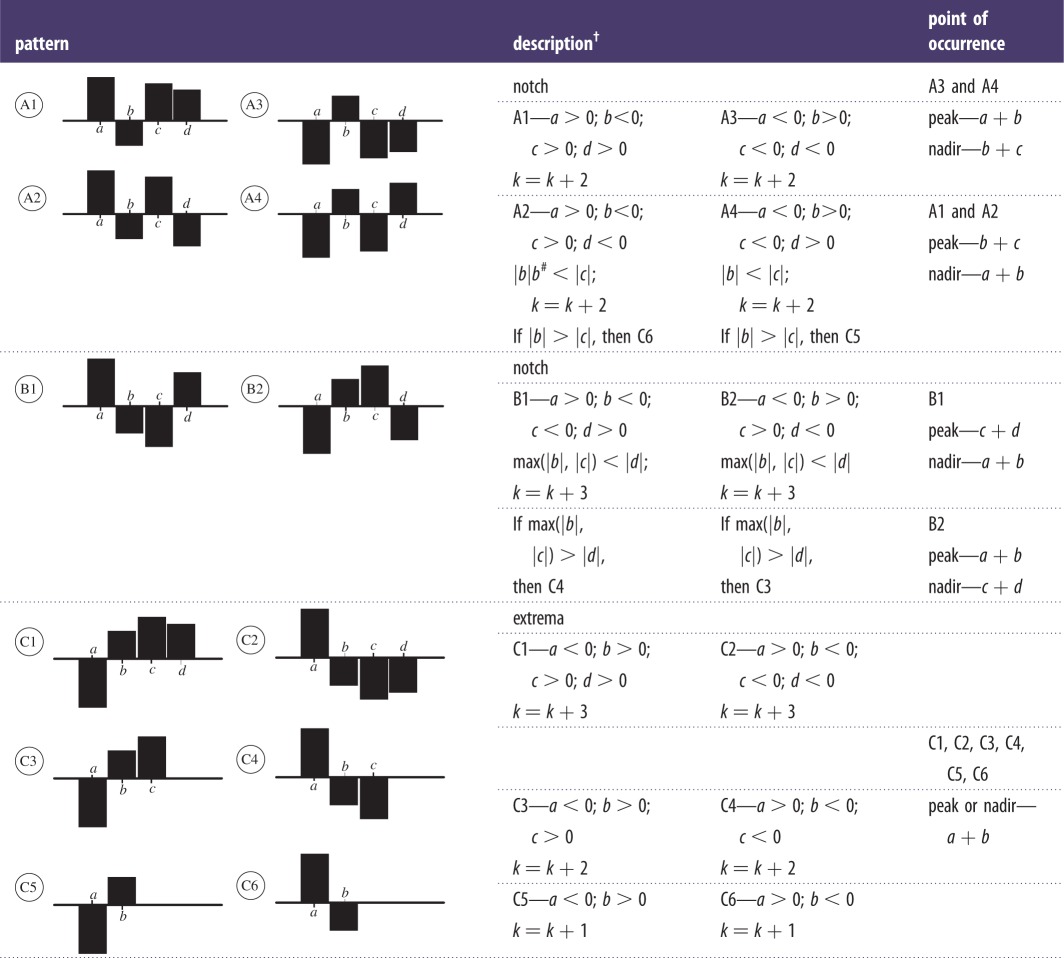 |
†Pointer ‘k’ initially starts at ‘a’. Here ‘a’, ‘b’, ‘c’ and ‘d’ are consecutive points on the bar plot of discrete coefficients and denote the corresponding boxes. Incrementing ‘k’ shifts it from box ‘a’ to box ‘b’.
#|·|, denotes magnitude of the detailed coefficient at a particular point.
2.1.2. Morphology identification
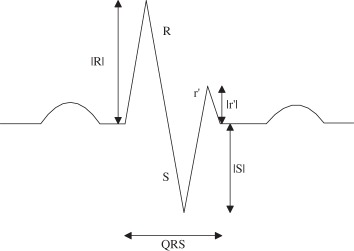
There exist six fundamental morphologies of f-QRS and several other variations of RSR’ patterns which were exemplified in the literature [1–15]. Apart from those mentioned, for the sake of completion we have attempted to encompass all the possible variations in RSR’ patterns, for example, some morphologies were found to have a Q-wave that was not reported in the six fundamental morphology as shown in B2, C1, etc., in tables 3 and 4. However, all the originally mentioned morphologies identified as fragmented QRS have been included. Tables 3 and 4 summarize and state the criteria for identification of the corresponding morphologies. Table 3 presents all the 10 morphologies that were quantified for QRS ≤ 120 ms, and table 4 presents the criterion for the identification of morphologies with QRS ≥ 120 ms which are generally encountered in practice and appeared in literature. The criterion of identification preferably starts once the number of maxima, minima and notches, the point of occurrence, i.e. positive or negative side of the reference axis, sequence of occurrence and height and depth of R and S waves, respectively, have been obtained from the fragmentation detection step. We have attempted to maintain a clear difference between an extremum pair and a notch. Morphologies with two R waves (R’) viz. A, C, E and I have been considered to dominate over the presence of notches so as to prevent such morphologies to be identified as f-QRS. Notched S morphology where the notch is being identified as an extremum pair was encountered, this case has also been taken into account. Similar morphologies for R waves exist and has been identified as RsR’ patterns. These criteria have been formulated based on the appearance of the QRS complex and have been assigned to morphologies such that each one can be identified distinctly, i.e. criteria assigned to morphologies can distinctively identify them without any conflict.
Table 3.
QRS ≤ 120 ms (f-QRS).
| morphology | name | criteria | conflict |
|---|---|---|---|
 |
(A) rSr’ |
(A) Nmax = 2 Nmin = 1 Pmaxv(1,2) > 0 Pminv(1) < 0 Nnotch = NFa |
when this morphology is encountered, the presence of notches on R, R’ or S wave will not lead it to be termed as f-QRS |
 |
(B) notched R (rsR’) |
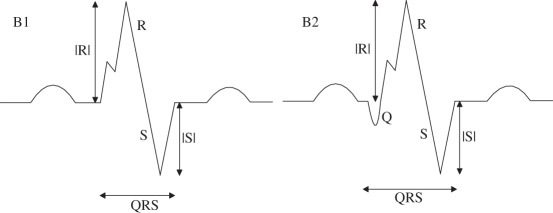
Nmax = 1; Nnotch = 1 Pmaxv(1) > 0; Pnotchv(1) > 0 Pnotchh(1) < Pmaxh(1) |
|
| (B1) without Q |
(B1) Nmin = 1; Pminv(1) < 0 |
when Nnotch > 1 then the morphology resembles f-QRS | |
| (B2) with Q |
(B2) Nmin = 2; Pminv(1,2) < 0 |
||
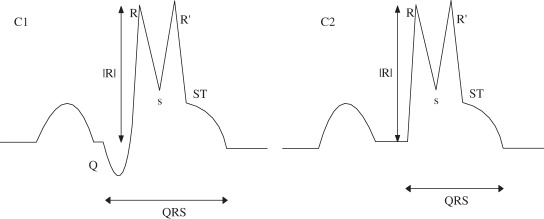 |
(C) RsR’ with ST elevation |
Nmax = 2; Nnotch = 0; Pmaxv(1,2) > 0; End of QRS complex must lie above horizontal axis. |
case may arise when sR’ is identified as a notch instead of a minima–maxima pair. Then all the cases arising are captured by morphology (F). For an extremum to be R, it has to be identified as a maximum |
| (C1) with Q |
(C1) Nmin = 2; Pminv(1) < 0; Pminv(2) > 0 |
||
| (C2) without Q |
(C2) Nmin = 1; Pminv(1) > 0 |
||
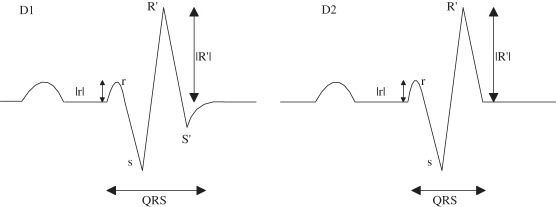 |
(D) rSR’ |
Nmax = 2; Pmaxv(1,2) > 0; Pmaxh(1) < Pminh(1) |
the presence of notches will not affect the morphology |
| (D1) with S’ |
(D1) Nmin = 2; Pminv(1,2) < 0 |
||
| (D2) without S’ |
(D2) Nmin = 1; Pminv(1) < 0; Pmaxv(1) < Pminv(1); Pminv(1) < Pmaxv(2); |
||
 |
(E) RsR’ without ST elevation |
Nmax = 2; Nnotch = 0; Pmaxv(1,2) > 0; |
if Nnotch ≥ 1, then the morphology resembles f-QRS. R’ or R must be detected as a maxima and not a notch for its morphology to be RsR’ |
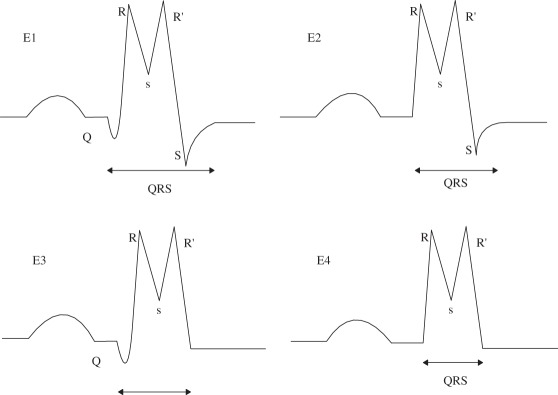
| (E1) with Q and S |
(E1) Nmin = 3; Pminv(1,3) < 0; Pminv(2) > 0 |
||
| (E2) S without Q |
(E2) Nmin = 2; Pminv(1) > 0; Pminv(2) < 0 |
||
| (E3) Q without S |
(E3) Nmin = 2; Pminv(1) < 0; Pminv(2) > 0; Pminh(1) < Pmaxh(1) |
||
| (E4) without Q and S |
(E4) Nmin = 1; Pminv(1) > 0; |
||
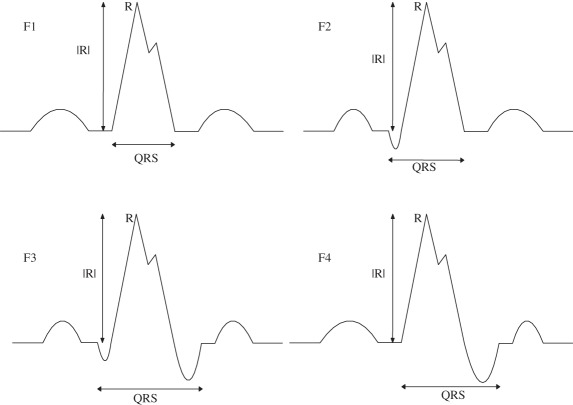 |
(F) Rsr’ |
Nmax = 1; Nnotch = 1; Pmaxv(1) > 0; Pnotchh(1) > Pmaxh(1); Pnotchv > 0 |
sr’ must be identified as a notch and not an extremum pair. If more than one notch occurs the morphology resembles f-QRS |
| (F1) without Q and S wave |
(F1) Nmin = 0 |
||
| (F2) only Q |
(F2) Nmin = 1; Pminv(1) < 0; Pminh(1) < Pmaxh(1) |
||
| (F3) both Q and S present |
(F3) Nmin = 2; Pminv(1,2) < 0; Pminh(1) < Pmaxh(1); Pminh(2) > Pmaxh(1) |
||
| (F4) Only S |
(F4) Nmin = 1; Pminv(1) < 0; Pminh(1) > Pmaxh(1) |
||
 |
(G) RSr’ |
Nmax = 2; Nnotch = UD Nmin = 1; Pmaxv(1,2) > 0 Pminv(1) < 0 Pmaxv(1) > |Pminv(1)| Pmaxv(2) < |Pminv(1)| |
the presence of notches will not affect the morphology |
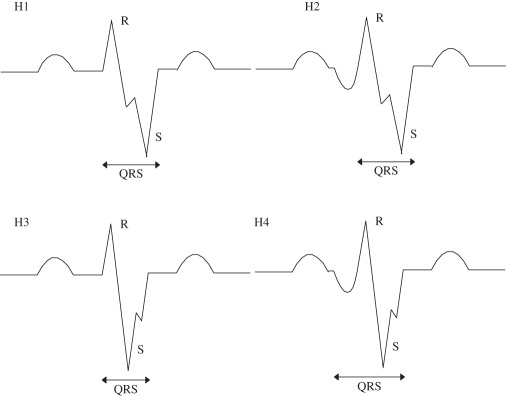 |
(H) notched S |
if more than one notch is present, then the morphology will be termed as f-QRS. Notch may be discovered as a extremum pair but still the morphology resembles notched S | |
| (H1 and H3) notch in downstroke and upstroke of S wave. Q not present |
(H1 and H3) Nmax = 1; Nnotch = 1; Nmin = 1; Pminv(1) < 0 Pmaxv(1) > 0; Pnotchv < 0 Pnotchh(1) > Pmaxh(1); |
||
| (H2 and H4) notch in downstroke and upstroke of S wave. Q present. |
(H2 and H4) Nmax = 1; Nnotch = 1; Nmin = 2; Pminv(1,2) < 0 Pmaxv(1) > 0; Pnotchv < 0 Pnotchh(1) > Pmaxh(1); |
||
| (H5 and H6) if notch is detected as extremum pair in any of the cases |
(H5) Nmax = 2; Nmin = 2; Nnotch = 1; Pmaxv(1) > 0; Pmaxv(2) < 0 Pminv(1,2) < 0 (H6) Nmax = 2; Nmin = 3 Nnotch = 1; Pmaxv(1) > 0; Pmaxv(2) < 0 Pminv(1,2,3) < 0 |
||
 |
(I) RSR’ |
(I)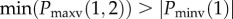 Nmax = 2; Nmin = 1 Pmaxv(1,2) > 0 Pminv(1) < 0 Nnotch = NF |
the presence of notch does not affect the morphology |
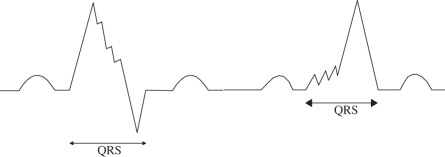 |
(J) f-QRS |
(J) if none of the aforementioned morphologies are detected and Nnotch ≥ 1; or Nmax ≥ 2; Nmin ≥ 2 |
visually identified notch may be detected as an extremum pair. If so, then such morphology will be termed as f-QRS. Notches may occur in R and/or S wave |
aNF, number and the presence of notches not fixed. Nmax, number of maxima; Nmin, number of minima; Nnotch, number of notch; Pmaxv(i), position at which ith maxima occurs on the vertical axis (magnitude along with sign); Pmaxh(i), position at which ith maxima occurs on the horizontal axis; Pminv(i), position at which ith minima occurs on the vertical axis; Pminh(i), position at which ith minima occurs on the horizontal axis; Pnotchv(i), position at which ith notch occurs on the vertical axis; Pnotchh(i), position at which ith notch occurs on the horizontal axis.
Table 4.
QRS ≥ 120 ms (f-wQRS).
| name | definition | criteria |
|---|---|---|
| f-BBB | various RSR’ patterns with or without Q-wave with greater than two R waves (R’) or greater than two notches in the R wave or greater than two notches in downstroke or upstroke of S wave | Nmax > 2 or at least three notches in the positive half plane or at least three notches in the negative half plane |
| fragmented premature ventricular complex | greater than two R waves (R’) or greater than two notches in the downstroke or the upstroke of S wave or only two notches in the R with greater than 40 ms separation | Nmax > 2 or at least three notches in the negative half plane or two notches in the positive half plane with > 40 ms delay |
| fragmented paced rhythms | greater than two R waves (R’) or greater than two notches in the downstroke or the upstroke of S wave | Nmax > 2 or at least three notches in the negative half plane |
2.2. Preprocessing and feature extraction
Wavelet-based techniques have been implemented for low-power and low-complexity applications [18]. Several wavelet transform-based artefact removal algorithms have been proposed in the literature [26–29]. However, there exists no clear demarcation upon the performance of these denoising techniques for comparison. Hence, all these techniques were employed and the denoised signals were visually observed. DWT has been used to remove the baseline wandering and have tested four different variants of wavelet transform to denoise the raw ECG signal. Baseline wandering removal using DWT [26] involved decomposition down to level 9 and the wavelet filter used was Symlet 10. The denoising approaches are as follows:
Approach 1. Denoising and artefact removal using DWT [27]. Decomposition down to level 3. Wavelet filter used was Symlet 4.
Approach 2. Denoising and artefact removal using stationary wavelet transform (SWT) [28]. Decomposition down to level 4. Wavelet filter used was Symlet 4. Empirical Bayes posterior median thresholding was used.
Approach 3. Denoising and artefact removal using undecimated wavelet transform (UWT) [29]. Wavelet filters were Daubechies 6. The level of decomposition was self-determined by the code [30]. We used classical standard deviation type of variance estimator and hard thresholding.
Approach 4. Denoising and artefact removal using translation invariant wavelet transform (TIWT) [26]. Wavelet filters used were Symlet 8. Hard thresholding was used and level of decomposition was self-determined by the code [31].
It was found that Approach 2 and in very rare cases Approach 1 were tampering with the QRS complex. Figure 3 shows two instances taken from two different subjects, where the tampering with the original signal was found while using Approach 2 (SWT), the denoised signal is plotted against the baseline wandering-removed signal which was initially verified as not tampering with the signal. It can be seen that notches have been introduced even though the original complex did not have one; other denoising techniques have smoothly retraced the original QRS complex. This visual inspection was done for more than 40 subjects and in rare instances DWT was also found to tamper the complex but none such cases were encountered for Approaches 3 and 4. Approach 3 (UWT) and Approach 4 (TIWT) were satisfactorily denoising the signals. For designing and verification of the algorithm, Approach 4 was adopted as this denoising technique was applied and verified with signals obtained at higher sampling frequencies [32] and satisfactory results were obtained.
Figure 3.
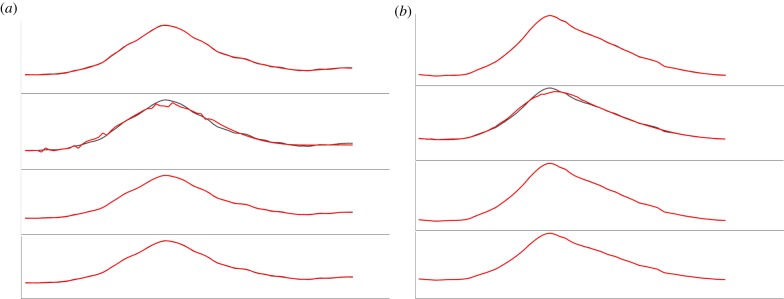
(a,b) Two instances in which Approach 2 was found to be tampering the QRS complex.
The proposed algorithm was implemented on MATLAB (v. 7.10.0-2010a). Appendix A provides the MATLAB code snippet for the implementation of the baseline wandering and denoising techniques.
3. Experiments and results
This section has been divided into the following subsections. Section 3.1 presents the experimental set-up, §3.2 discusses the case studies to understand the working principle of the algorithm, §3.3 presents the evaluation methods used to measure the performance of the algorithm in terms of accuracy, and §3.4 presents the results.
3.1. Experimental set-up
The PTB database (PTBDB) [33,34] from PhysioNet has been used for the designing and verification of the proposed algorithm. PTBDB is an unprocessed or raw 15-lead database comprising conventional 12 leads and three orthogonal Frank leads digitized simultaneously at a sampling frequency of 1 kHz and captured at the standard speed of 25 mm s−1 and 10 mm mV−1 with grid intervals being 0.2 s and 0.5 mV. The database was categorized on the basis of cardiac disorders reported and ECGs of patients from various categories were used for designing and modelling the algorithm. PTBDB consists of patients belonging to various diagnostic classes viz. myocardial infarction, cardiomyopathy/heart failure, BBB, dysrthymia, myocardial hypertrophy, Valvular heart disease, myocarditis, healthy controls and other miscellaneous conditions [33,34]. A high sampling rate is desired to capture the occurrence of high-frequency notches in QRS complexes. A simple linear interpolation is then applied on the preprocessed signal to further increase the number of samples so that the detailed coefficients obtained after applying DWT can accurately detect all discontinuities (local extrema and notches), as DWT diminishes the time resolution by a factor of 2. Equation (3.1) shows the methodology adopted for interpolation
| 3.1 |
A mean was calculated for every two consecutive samples and was inserted in between them thus increasing the total number of samples. Interpolation in this manner is extremely simple and does not affect the points of extrema. Requirement of interpolation is based on observational interpretations during the designing of the algorithm. Upon interpolation, it was observed that the detailed coefficients were found to capture all the discontinuities without fail. The MATLAB code snippet for the implementation of linear interpolation and DWT can be found in appendix A.
3.2. Case studies
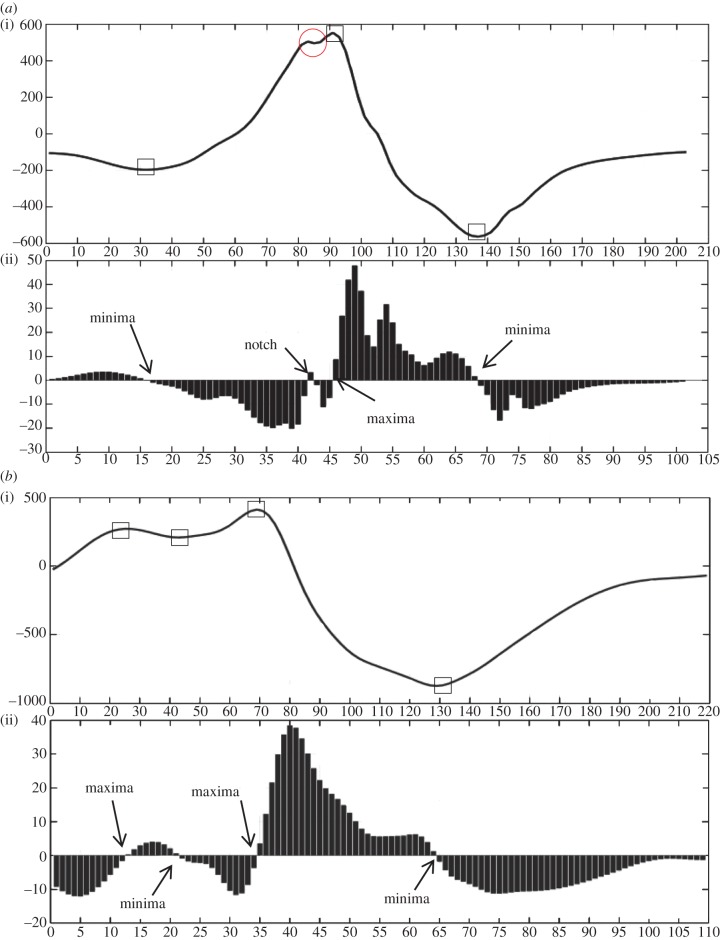
The outputs obtained from the algorithm are the number of extrema points (maxima and minima), notches, time instants at which they occur, magnitude of extrema, the occurrence on the lower or upper half of the magnitude–time plane and the morphology of fragmentation encountered. In this section, we discuss eight different cases and the output obtained from the algorithm for them which are intended to show the working principle of our proposed algorithm. These complexes were randomly selected from the large database avoiding the selections where similar morphologies were selected. Figure 4a–h shows the plot of these cases with part ‘(i)’ of the plot showing the interpolated QRS complex and part ‘(ii)’ of the plot showing the bar plot of the detailed coefficients obtained after applying DWT. Notches and extrema are denoted by circle and rectangle, respectively. The following cases correspond to the QRS complexes shown in figure 4. This case study provides insight into the working of the algorithm and will help in the reproduction of the work. Rules mentioned in table 2 have been used to detect the discontinuities, and criteria mentioned in tables 3 and 4 have been used to identify the morphology.
Case 1. This case is an example of notched R (rsR’) morphology. Number of maxima, minima and notches are 1, 2 and 1, respectively. On examining figure 4a(i), it can be seen that a minima (<0) is encountered first followed by a notch (>0), maxima (>0) and a second minima (<0).
Case 2. This case is an example of RsR’ without ST elevation. Number of maxima, minima and notches are 2, 2 and 0, respectively. In figure 4b(ii), a maxima (>0) is encountered first followed by minima (>0), maxima (>0) and minima (<0).
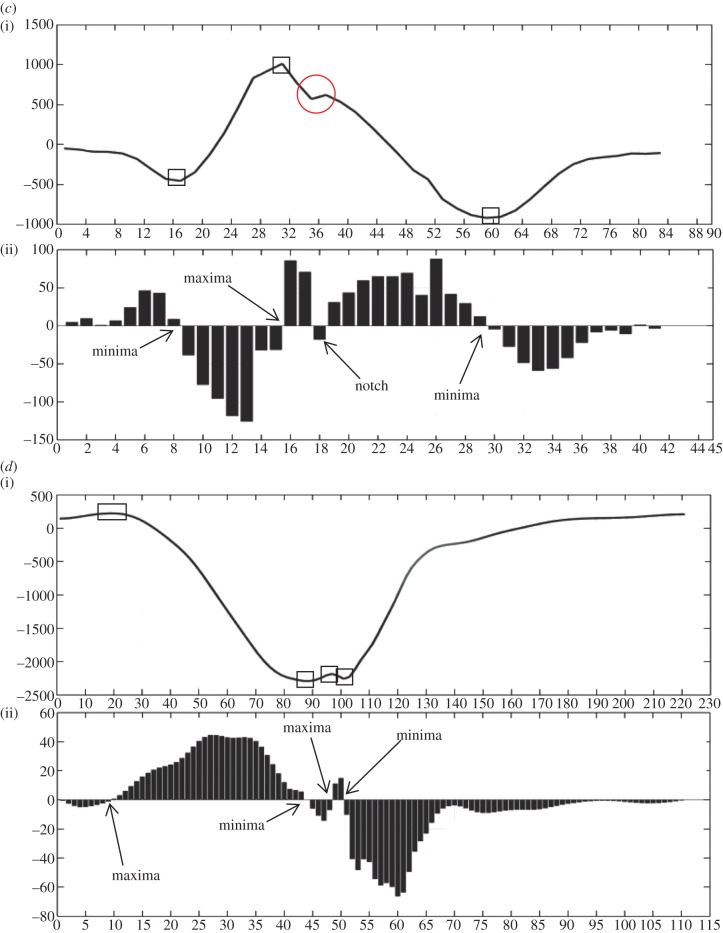
Case 3. This case is an example of Rsr’. Number of maxima, minima and notches are 1, 2 and 1, respectively. In figure 4c(ii), a minima (<0) is encountered first followed by maxima (>0), notch (>0) and minima (<0).
Case 4. This case is an example of notched S. Number of maxima, minima and notches are 2, 2 and 0, respectively. In figure 4d(ii), a maxima (>0) is encountered first followed by minima (<0), maxima (<0) and minima (<0).
Case 5. This case is an example of rSr’. Number of maxima, minima and notches are 2, 1 and 1, respectively. In figure 4e(ii), a maxima (>0) is encountered first followed by notch (>0), minima (<0) and maxima (<0).
Case 6. This case is an example of f-QRS. Number of maxima, minima and notches are 2, 3 and 0, respectively. In figure 4f(ii), a minima (<0) is encountered first followed by maxima (>0), minima (<0), maxima (>0) and minima (<0).
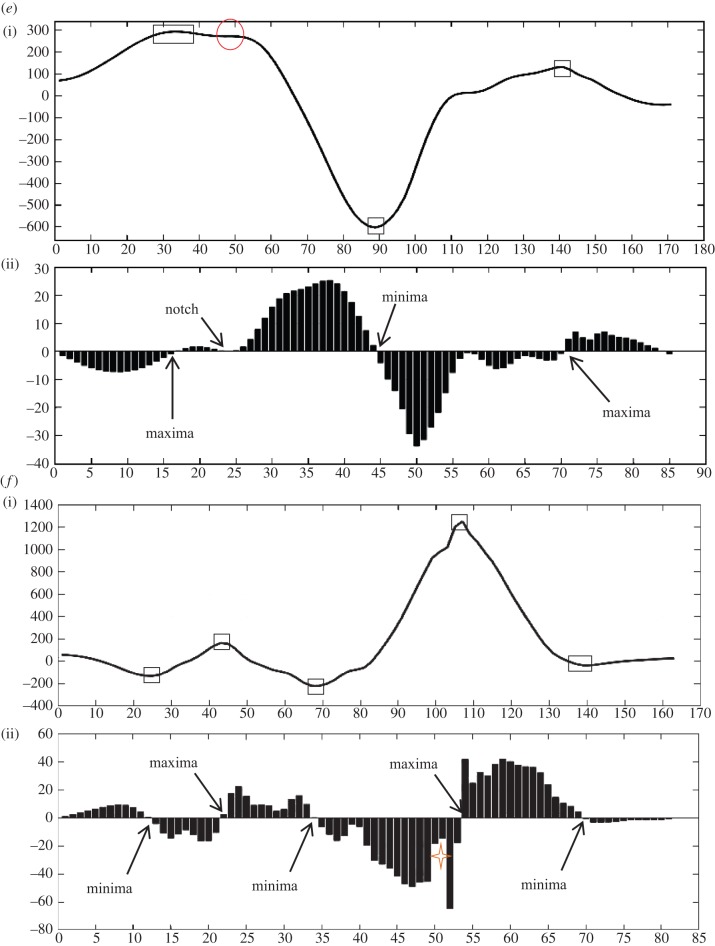
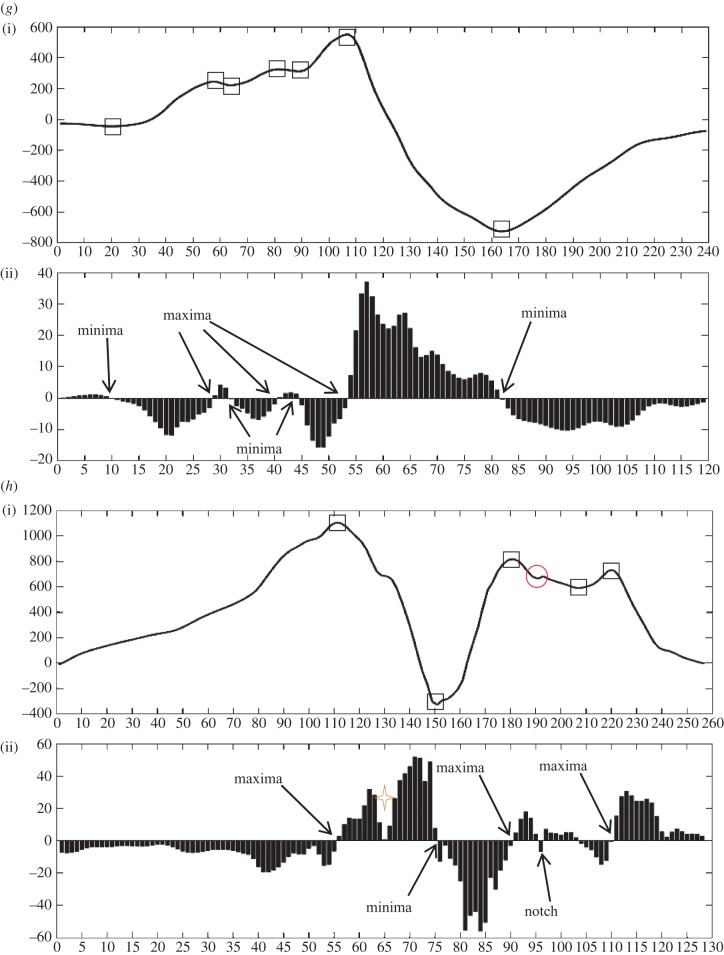
Case 7. This case is an example of f-QRS. Number of maxima, minima and notches are 3, 4 and 0, respectively. In figure 4g(ii), a minima (<0) is encountered first followed by maxima (>0), minima (>0), maxima (>0), minima (>0), maxima (>0) and minima (<0).
Case 8. The length of QRS is more than 120 ms (QRS ≥ 120 ms). This case resulted in fragmented BBB (f-BBB) with 3 or more R waves (R’). Number of maxima, minima and notches are 3, 2 and 1, respectively. In figure 4h(ii), a maxima (>0) is encountered first followed by minima (<0), maxima (>0), notch (>0), minima (>0), maxima (>0).
Figure 4.
Eight different morphologies (a–h) comprising an interpolated plot along with bar plot of its detailed coefficients obtained after applying DWT. Square boxes denote extrema and circle denotes notch. Four-point star shows the sudden changes in gradient of the wave, however, these do not lead to discontinuity. This star has been used to demonstrate the sensitivity of the algorithm in capturing gradients of the wave encountered.
3.3. Evaluation criteria
Forty patients were selected at random from the database, and the QRS complexes were extracted using the TDMG feature extraction algorithm and were examined by the cardiologists. Out of 40 patients, nine patients were removed from the study owing to the poor quality of their ECGs as per the suggestion of the cardiologists. The remaining QRS complexes were then independently examined by the two cardiologists in a blind fashion, and finally a consensus was reached to produce the final result which will be hereby referred as cardiologist's status (CS). The measurements obtained from the cardiologists are assumed to be the gold standard. The QRS complexes were then input to the FDA and the results obtained were compared with CS, and sensitivity and specificity values were calculated. The following will discuss the test results and test nature:
True positive. The cardiologists detected fragmentation in a particular lead of the patient and the algorithm reported correctly.
False positive. The cardiologists did not detect fragmentation but the algorithm reported the presence of fragmentation in a particular lead.
True negative. The cardiologists did not detect fragmentation and the algorithm reported correctly.
False negative. The cardiologists detected fragmentation in a particular lead but the algorithm could not detect.
The sensitivity and specificity were calculated using the following equations:
| 3.2 |
and
| 3.3 |
3.4. Experimental results
The QRS complexes obtained from TDMG were evaluated by two experienced cardiologists and were simultaneously used to obtain results from the algorithm. From 372 leads (31 patients) selected, i.e. only 12-lead ECG of 31 patients and not the Frank leads, the results of 89.8%, i.e. 334 leads from FDA complied with that of CS. The sensitivity and specificity values obtained were 0.897 and 0.899, respectively. As cardiologists are often interested in fragmentation in BBB subjects, out of randomly selected 31 patients five patients had BBB and the sensitivity and specificity were found to be 0.932 and 0.933, respectively. It is to be noted that in this paper we have evaluated the presence or the absence of fragmentation at a lead level rather than patient level which is usually the case in the literature. It is important that the algorithm detects fragmentation correctly in every lead of the patient as well identifies its various morphologies. The sensitivity and specificity values were obtained for the presence or the absence of fragmentation and not for the morphology obtained by the algorithm and cardiologists. We have not found any work that standardizes the morphologies of fragmentation for the common agreement, and hence we have avoided the evaluation of the morphological results obtained from the algorithm.
4. Discussion
From figure 4a–h, we can see that whenever the gradient of the wave is high the magnitude of the detailed coefficients is more. Sensitivity of the algorithm can be estimated from the fact that it not only captures the formation of an extremum pair but also sudden changes in gradient which did not result in an extremum pair, e.g. in figure 4f,h a sudden gradient change before the occurrence of final maxima (figure 4f(ii)) and after the first maxima (figure 4h(ii)) can be seen (denoted by a four-point orange star) with the magnitude of detailed coefficients suddenly decreasing and then increasing. This highlights the sensitivity of the detailed coefficients to sudden changes in the ECG, and hence it is important to denoise the ECG before applying the FDA algorithm otherwise a noisy part in the QRS complex may be detected as a notch, which upon denoising may not be present. Hence, baseline wandering removal and denoising is required for the correct detection of morphology of f-QRS.
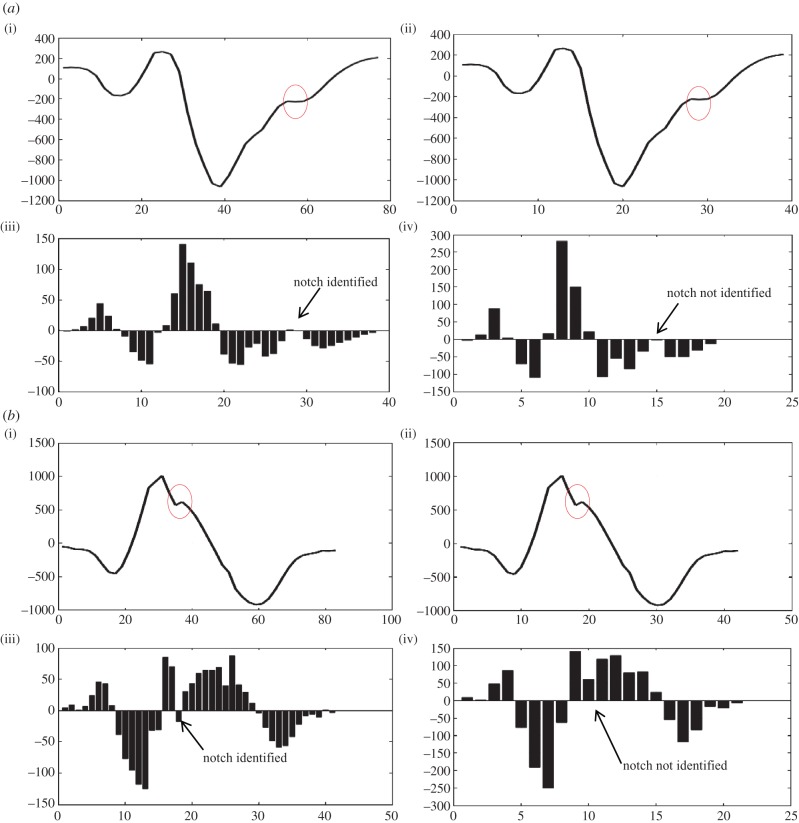
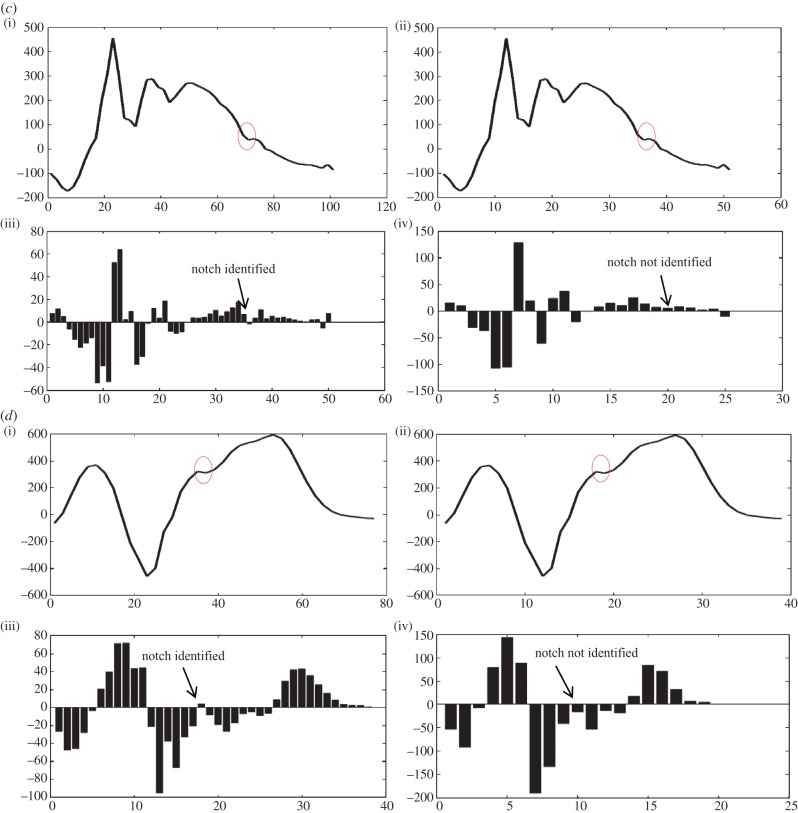
Sampling rate plays an important role in capturing fragmentation. We have observed that a sampling rate of 2 kHz will be appropriate for algorithm implementation. This can be deduced from the role played by interpolation to double the number of samples as the sampling rate of the PTB database is 1 kHz. The significance of number of samples can be seen from figure 5a–d (notches have been denoted by a red circle). Figure 5a–d(i)(iii) shows a plot of interpolated QRS along with a bar plot of detailed coefficients. Figure 5a–d(ii)(iv) of original QRS along with the bar plot of its detailed coefficients. All four figures show that on increasing the number of samples, undetectable notches can also be identified. This has been our main motivation behind interpolating. A notch that is detectable will only become elongated or may become converted to an extremum pair; this can be resolved with the help of criteria and postulates for morphology realization and identification, respectively, but an undetectable notch will result in an incorrect determination of the number of notches, and hence the morphology, which may lead to an incorrect diagnosis. Figure 5a was identified by the algorithm as notched S, 5B as Rsr’, 5C as f-QRS and 5D as rSR’.
Figure 5.
Significance of interpolation and magnitude criteria used in table 2.
When patterns similar to table 2, A1 and A3 are encountered, it unfailingly denotes the presence of a notch but when patterns like A2 and A4 are encountered it becomes difficult to interpret whether such a pattern should be treated as a notch or an extremum pair. Figure 5b(i)(iii) shows a similar case. In the absence of magnitude criterion, the maxima would have been identified as a notch and the peak of the notch would have been identified as a maxima.
Properties of the detailed coefficients as explained in figure 2 can be used to explain and understand the occurrence of such patterns. Similarly, if the magnitude criteria stated in tables 3 and 4, B1 and B2 are spared then on implementation of the algorithm in figure 5c(ii)(iv) would have resulted in detection of the first maxima and second minima as two notches which is inaccurate. On iterative refinement of the algorithm and evaluating it on more than 40 patients, we deemed it necessary to incorporate the magnitude criterion in the postulates. In the designing and modelling phase, the method of QRS detection was adopted from the appendix of [25]. To obtain accurate results from the algorithm, it is necessary to input an accurately detected QRS complex. Any extra discontinuity will lead to the identification of some other morphology. In the worst-case scenarios, the algorithm will identify the morphology as f-QRS, leaving no f-QRS undetected.
5. Conclusion
We have introduced a novel approach for detection of discontinuities in the QRS complexes and have verified it using PTBDB. A less complex ‘Haar’ wavelet was used for low-power consumption which can be used in battery-operated devices viz. mobile phone/PDA/tablets [18] or battery-operated ICD devices. This approach is not signal specific and the method can be applied to any other kind of biomedical signal for detection of its specific important aspects and features. For ECG-specific applications, we have formulated the postulates for detection of notches and extrema and have proposed criteria for identification of various morphologies. The significance of denoising techniques and all types of discrepancies encountered have been discussed.
Funding statement
This work is partly supported by the DIT, India under the ‘Cyber Physical Systems Innovation Hub’ under grant no.: 13(6)/2010-CC&BT, dated 1 March 2011 and ‘IOT for Smarter Healthcare’ under grant no.: 13(7)/2012-CC&BT, dated 25 February 2013.
Appendix A
The appendix provides a snippet of the Matlab codes which will be helpful in reproduction of the work. The part of the code considered important and necessary has been provided. Figure 6 provides the Matlab code for implementation of denoising techniques, and figure 7 provides the code for implementation of interpolation and DWT.
Figure 6.
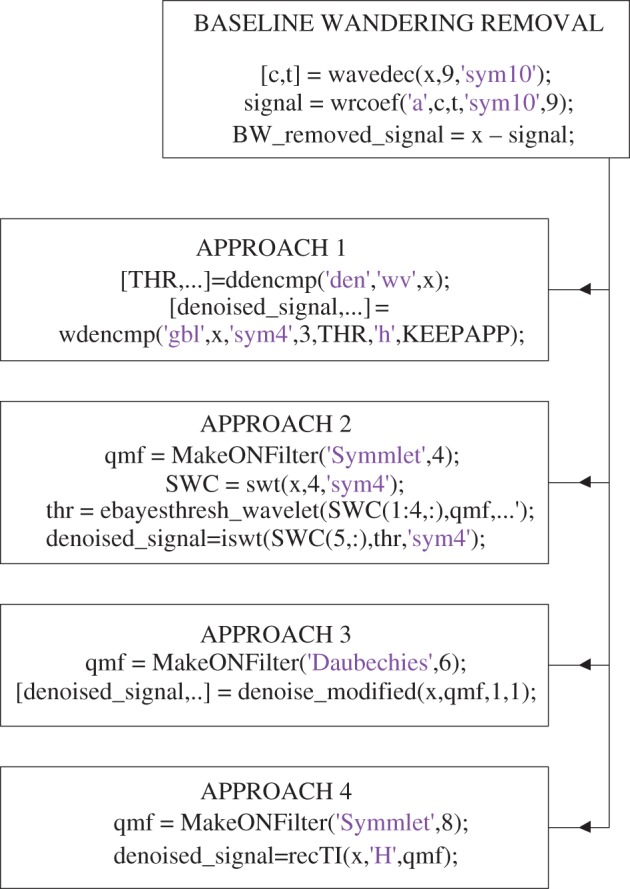
MATLAB code snippet for baseline wandering removal, denoising, interpolation and DWT. (Online version in colour.)
Figure 7.
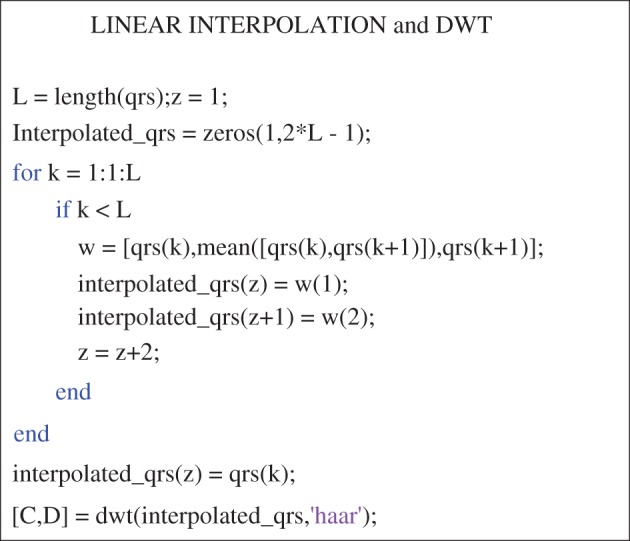
MATLAB code snippet for linear interpolation and DWT. (Online version in colour.)
Endnotes
Discontinuity is discussed later in this subsection.
Discontinuity in general refers to any local extrema considered, i.e. notch, maxima or minima.
References
- 1.Maehara K, Kokubun T, Awano N, Taira K, Ono M, Furukawa T, Shimizu Y, Maruyama Y. 1999. Detection of abnormal high-frequency components in the QRS complex by the wavelet transform in patients with idiopathic dilated cardiomyopathy. Jpn Circ. J. 63, 25–32. ( 10.1253/jcj.63.25) [DOI] [PubMed] [Google Scholar]
- 2.Das MK, Khan B, Jacob S, Kumar A, Mahenthiran J. 2006. Significance of a fragmented QRS complex versus a Q wave in patients with coronary artery disease. Circulation 113, 2495–2501. ( 10.1161/CIRCULATIONAHA.105.595892) [DOI] [PubMed] [Google Scholar]
- 3.Reddy CVR, Cheriparambill K, Saul B, Makan M, Kassotis J, Kumar A, Das MK. 2006. Fragmented left sided QRS in absence of bundle branch block: sign of left ventricular aneurysm. Ann. Noninvas. Electro. 11, 132–138. ( 10.1111/j.1542-474X.2006.00094.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michael MA, Masry HE, Khan BR, Das MK. 2007. Electrocardiographic signs of remote myocardial infarction. Prog. Cardiovasc. Dis. 50, 198–208. ( 10.1016/j.pcad.2007.05.003) [DOI] [PubMed] [Google Scholar]
- 5.Das MK, Suradi H, Maskoun W, Michael MA, Shen C, Peng J, Dandamudi G, Mahenthiran J. 2008. Fragmented wide QRS on a 12-lead ECG : a sign of myocardial scar and poor prognosis. Circ. Arrhythm. Electrophysiol. 1, 258–268. ( 10.1161/CIRCEP.107.763284) [DOI] [PubMed] [Google Scholar]
- 6.Das MK, Zipes DP. 2009. Fragmented QRS: a predictor of mortality and sudden cardiac death. Heart Rhythm 6(Suppl. 3), S8–S14. ( 10.1016/j.hrthm.2008.10.019) [DOI] [PubMed] [Google Scholar]
- 7.Pietrasik G, Goldenberg I, Zdzienicka J, Moss AJ, Zareba W. 2007. Prognostic significance of fragmented QRS complex for predicting the risk of recurrent cardiac events in patients with Q-wave myocardial infarction. Am. J. Cardiol. 100, 583–586. ( 10.1016/j.amjcard.2007.03.063) [DOI] [PubMed] [Google Scholar]
- 8.Guo R, Li Y, Tang K, Li W. 2012. Significance of fragmented QRS complexes for identifying culprit lesions in patients with non-ST-elevation myocardial infarction: a single-center, retrospective analysis of 183 cases. BMC Cardiovasc. Disord. 12, 44 ( 10.1186/1471-2261-12-44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homsi M, Alsayed L, Safadi B, Mahenthiran J, Das MK. 2009. Fragmented QRS complexes on 12-lead ECG: a marker of cardiac sarcoidosis as detected by gadolinium cardiac magnetic resonance imaging. Ann. Noninvasive Electrocardiol. 14, 319–326. ( 10.1111/j.1542-474X.2009.00320.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita H, Kusano KF, Miura D, Nagase S, Nakamura K, Morita ST, Ohe T, Zipes DP, Wu J. 2008. Fragmented QRS as a marker of conduction abnormality and a predictor of prognosis of Brugada syndrome. Circulation 118, 1697–1704. ( 10.1161/CIRCULATIONAHA.108.770917) [DOI] [PubMed] [Google Scholar]
- 11.Nagao S, Watanabe H, Kodama M, Tanaka J, Suzuki E, Narita I, Aizawa Y. 2011. Fragmented QRS complexes, conduction disorder, and ST segment abnormality on twelve-lead electrocardiography predict severe cardiac manifestation in sarcoidosis. Circulation 124, A9485. [Google Scholar]
- 12.Vanga SR, Koerber S, DiBiase L, Verma A, Bommana S, Swarup V, Natale A, Dawn B, Lakkireddy D. 2010. Prevalence of fragmented QRS complexes in non-ischemic dilated cardiomyopathy patients and long term prognosis. Circulation 122, A20836. [Google Scholar]
- 13.Korhonen P, Husa T, Konttila T, Tierala I, Mäkijärvi M, Väänänen H, Ojanen J, Vehtari A, Toivonen L. 2010. Fragmented QRS in prediction of cardiac deaths and heart failure hospitalizations after myocardial infarction. Ann. Noninvas Electrocardiol. 15, 130–137. ( 10.1111/j.1542-474X.2010.00353.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Refaat MM, et al. 2011. Fragmented wide QRS on a 12-lead electrocardiogram increases the risk of ventricular arrhythmias in patients with severe systolic dysfunction. Circulation 124, A15244. [Google Scholar]
- 15.Michael MA, Das MK. 2006. Fragmented QRS on a 12-lead ECG is a sign of acute or recent myocardial infarction. Circulation 114, II_512. [Google Scholar]
- 16.Mazomenos EB, Chen T, Acharyya A, Bhattacharya A, Rosengarten J, Maharatna K. 2012. A time-domain morphology and gradient based algorithm for ECG feature extraction. In 2012 IEEE Int. Conf. on. Ind. Technol. (ICIT), 117–122. ( 10.1109/ICIT.2012.6209924) [DOI] [Google Scholar]
- 17.Gao GQ. 2003. Computerised detection and classification of five cardiac conditions. Master thesis, AUT, Auckland, New Zealand. [Google Scholar]
- 18.Mazomenos EB, Biswas D, Acharyya A, Chen T, Maharatna K, Rosengarten J, Morgan J, Curzen N. 2013. A low-complexity ECG feature extraction algorithm for mobile healthcare applications. IEEE Trans. Inform. Technol. Biomed. 17, 459–469. ( 10.1109/TITB.2012.2231312) [DOI] [PubMed] [Google Scholar]
- 19.Goernig M, DiPietroPaolo D, Haueisen J, Erné SE. 2009. Biomagnetic risk stratification by QRS fragmentation in patients with Implanted cardioverter defibrillators. In 4th European Conf. of the Int. Federation for Medical and Biological Engineering 2009, IFMBE Proc. 22, 5–7. ( 10.1007/978-3-540-89208-3_2) [DOI] [Google Scholar]
- 20.Chatterjee S, Changawala N. 2010. Fragmented QRS complex: a novel marker of cardiovascular disease. Clin. Cardiol. 33, 68–71. ( 10.1002/clc.20709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan J, Tompkins WJ. 1985. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 32, 230–236. ( 10.1109/TBME.1985.325532) [DOI] [PubMed] [Google Scholar]
- 22.Sahambi JS, Tandonz SN, Bhatt RKP. 1997. Using wavelet transforms for ECG characterization: an on-line digital signal processing system. IEEE. Eng. Med. Biol. Mag. 16, 77–83. ( 10.1109/51.566158) [DOI] [PubMed] [Google Scholar]
- 23.Martínez JP, Almeida R, Olmos S, Rocha AP, Laguna P. 2004. A wavelet-based ECG delineator: evaluation on standard databases. IEEE Trans. Biomed. Eng. 51, 570–581. ( 10.1109/TBME.2003.821031) [DOI] [PubMed] [Google Scholar]
- 24.Rinćon F, Recas J, Khaled N, Atienza D. 2011. Development and evaluation of multilead wavelet-based ECG delineation algorithms for embedded wireless sensor nodes. IEEE Trans. Inform. Technol. Biomed. 15, 854–863. ( 10.1109/TITB.2011.2163943) [DOI] [PubMed] [Google Scholar]
- 25.Loring Z, Chelliah S, Selvester RH, Wagner G, Strauss DG. 2011. A detailed guide for quantification of myocardial scar with the Selvester QRS score in the presence of electrocardiogram confounders. J. Electrocardiol. 44, 544–554. ( 10.1016/j.jelectrocard.2011.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D. 2005. Wavelet approach for ECG baseline wander correction and noise reduction. In 27th Ann. Int. Conf. of the Engineering in Medicine and Biology Society, IEEE-EMBS 2005, 27, 1212–1215. ( 10.1109/IEMBS.2005.1616642) [DOI] [PubMed] [Google Scholar]
- 27.Hostalkov E, Prochazka A. 1905. Wavelet signal and image denoising. Institute of Chemical Technology, Prague Department of Computing and Control Engineering Technicḱa 166 28 Prague 6.
- 28.Li S, Lin J. 2009. The optimal de-noising algorithm for ECG using stationary wavelet transform. In 2009 WRI World Congress on Computer Science and Information Engineering. 6, 469–473. ( 10.1109/CSIE.2009.999) [DOI] [Google Scholar]
- 29.Hernandez O, Olvera E. 2009. Noise cancellation on ECG and heart rate signals using the undecimated wavelet transform. In Int. Conf. on eHealth, Telemedicine, and Social Medicine, 2009. eTELEMED '09, 145–150. ( 10.1109/eTELEMED.2009.49) [DOI] [Google Scholar]
- 30.Baraniuk R, et al. 2002. Rice wavelet toolbox See http://www.dsp.rice.edu/software/rwt.shtml.
- 31.Antoniadis A, Bigot J, Sapatinas T. 2001. Wavelet estimators in nonparametric regression: a comparative simulation study. J. Stat. Softw. 6, 1–83. [Google Scholar]
- 32.Fugal DL. 2009. Conceptual wavelets in digital signal processing. San Diego, CA: Space and Signals Technical Publishing. [Google Scholar]
- 33.Goldberger AL, et al. 2000. PhysioBank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 101, e215–e220. ( 10.1161/01.CIR.101.23.e215) [DOI] [PubMed] [Google Scholar]
- 34.Bousseljot R, Kreiseler D, Schnabel A. 1995. Nutzungder EKG signaldatenbank CARDIODAT der PTB ber das Internet. Bio- medizinischeTechnik 1995, 40, S317–S318.