Abstract
Several members of the Ly-6/uPAR (LU)-protein domain family are differentially expressed in human squamous epithelia. In some cases, they even play important roles in maintaining skin homeostasis, as exemplified by the secreted single domain member, SLURP-1, the deficiency of which is associated with the development of palmoplantar hyperkeratosis in the congenital skin disorder Mal de Meleda. In the present study, we have characterized a new member of the LU-protein domain family, which we find to be predominantly expressed in the stratum granulosum of human skin, thus resembling the expression of SLURP-1. In accordance with its expression pattern, we denote this protein product, which is encoded by the LYPD5 gene, as Haldisin (human antigen with LU-domains expressed in skin). Two of the five human glycolipid-anchored membrane proteins with multiple LU-domains characterized so far are predominantly confined to squamous epithelia (i.e., C4.4A), to stratum spinosum, and Haldisin to stratum granulosum under normal homeostatic conditions. Whether Haldisin is a prognostic biomarker for certain epithelial malignancies, like C4.4A and SLURP-1, remains to be explored.
Keywords: LU, Ly-6/uPAR protein domain family, LYPD5, PRO4356, Haldisin, uPAR, C4.4A, squamous epithelium, stratum granulosum, differentiation marker
Introduction
The Ly-6/uPAR (LU) protein domain superfamily is a well-defined group of functionally diverse proteins involved in a multitude of biological processes, as exemplified by their role in the toxic insult by snake venom α-neurotoxins (Kini and Doley 2010), maintenance of squamous epithelial integrity by SLURP-1 (Favre et al. 2007), regulation of complement activation by CD59 (Nevo et al. 2013), translocation of lipoprotein lipase across capillaries by GPIHBP-1 (Davies et al. 2010; Young and Zechner 2013), cytokine signaling by TGF-β receptors (Hinck 2012), and focalizing cell-surface associated proteolysis by urokinase-type plasminogen receptor (uPAR) (Kriegbaum, Persson, et al. 2011). The individual members of the LU-protein domain family share a relatively low overall sequence similarity as they are primarily defined by a consensus sequence motif encompassing eight cysteines arranged in a characteristic disulfide-bonding pattern. This disulfide configuration translates into the canonical three-fingered toxin fold, where three loops project from a globular cysteine-rich core and are assembled on a five- to six-stranded central β-sheet (Ploug and Ellis 1994). In the genome of Homo sapiens, at least 45 genes encode proteins with LU-domains, and these are scattered throughout the genome involving at least nine different chromosomes (Galat 2008).
The far majority of the members of the LU-domain family contain only a single copy of the LU-domain, which can exist as a soluble secreted version (e.g., SLURP-1), as tethered to the cell membrane by a glycosylphosphatidylinositol (GPI) anchor (e.g., GPIHBP-1 and CD59), or as the extracellular ligand binding domain in type-1 transmembrane receptors (activin and TGF-β receptors). A small subset of these genes, nonetheless, encodes proteins with two to four LU-domains and these genes are intriguingly all confined to a small gene cluster on the human chromosome 19q13 (Supplemental Fig. S1, panel A). The best characterized of these multidomain members of the LU-protein domain family is the GPI-anchored uPAR, where the functional importance of having a multidomain architecture is particularly evident as the inherent flexibility between its three extracellular LU-domains drives the allosteric regulation of this multifunctional receptor (Llinas et al. 2005; Huai et al. 2006; Gårdsvoll, Jacobsen, et al. 2011; Gårdsvoll, Kjaergaard, et al. 2011; Mertens et al. 2012). The other multidomain members of this family are (1) C4.4A, which is a strong biomarker for poor prognosis in non-small cell lung adenocarcinoma patients (Hansen et al. 2007; Jacobsen et al. 2013), although its normal expression is predominantly confined to the stratum spinosum of squamous epithelia (Kriegbaum, Jacobsen, et al. 2011); (2) TEX101, which is a marker of testicular germ cells and essential for male fertility (Kurita et al. 2001; Fujihara et al. 2013); and (3) CD177, which is a receptor for neutrophil proteinase 3 and involved in the pathogenesis of the autoimmune disease Wegener’s granulomatosis (Korkmaz et al. 2008). The remaining two genes, LYPD4 and LYPD5, encode protein products that have so far not been characterized.
In the present study, we have expressed and purified the protein encoded by LYPD5, which is predicted to contain two archetypical LU-domains tethered to the cell surface via a GPI-anchor. We have subsequently used this protein preparation to generate a specific polyclonal rabbit antibody suitable for immunohistochemistry in both mouse and human paraffin-embedded tissue. Our global immunohistochemical analysis of resected mouse organs reveals that the protein product of LYPD5 is primarily expressed in the stratum granulosum of squamous epithelium. This unique expression pattern is recapitulated in human skin, and we have consequently termed this protein Haldisin (human antigen with LU-domains expressed in skin). Our studies demonstrate that two homologous multidomain members of the LU-protein domain family intriguingly are differentiation markers for squamous epithelium, but they clearly define different stages of development: C4.4A is a marker of stratum spinosum (Kriegbaum, Jacobsen, et al. 2011) and Haldisin of stratum granulosum (this study).
Materials & Methods
Materials
The plasmid (pBluescript-Haldisin) with cDNA encoding full-length human Haldisin was obtained from American Type Culture Collection, ATCC No. 10435601 (LGC/ATCC; Borås, Sweden). HPLC-purified DNA oligos were purchased from DNA Technology A/S (Aarhus, Denmark). Restriction enzymes were from New England Biolabs (Hertfordshire, UK). Accuprime Pfx DNA polymerase, pMT/BiP/V5-His A, pCoHYGRO, pcDNA5/FRT/TO, CellFectin, Drosophila melanogaster Schneider 2 (S2) cells, Schneider’s Drosophila medium (SDM), Express Five serum free medium (SFM), SDS gels, Flp-In T-REx System, goat anti-rabbit Alexa Fluor-488 conjugated F(ab’)2 fragment, and ProLong Gold antifade reagent with DAPI were all obtained from Invitrogen/Life Technologies (Groningen, The Netherlands). Human Factor Xa (FXa) was from Enzyme Research Laboratories (HFXa 1011; South Bend, IN). ECL reagents, films for immunoblotting, Hi TRAP Protein G, and MidiTrap G-25 were obtained from GE Healthcare (Brøndby, Denmark). Freunds Complete and Incomplete Adjuvant were from Statens Serum Institut (Copenhagen, Denmark). Purified rabbit immunoglobulin from non-immunized healthy rabbits (product code no. x0903), Antibody Diluent (product code no. S3022), and horseradish peroxidase-conjugated EnVision rabbit reagent (product code no. K4003) were all purchased from Dako (Copenhagen, Denmark). Shandon racks for mounting of tissue sections were from Thermo Shandon (Pittsburgh, PA), and NovaRed chromogene was from Vector Laboratories (Burlingame, CA).
Human and Animal Tissue
Fresh, normal human skin from mammary gland surgery was received from Rigshospitalet (Copenhagen, Denmark). Animal tissue was obtained as follows: 12-week-old FVB/N mice and 8-week-old Sprague Dawley rats were anesthetized using 0.1 ml/10 g of a 1:1 mixture of Hypnorm (fluanisone 5 mg/ml and fentanyl 0.1 mg/ml) and Dormicum (midazolam 5 mg/ml) prior to perfusion with PBS followed by 4% (v/v) buffered formaldehyde. Human skin as well as resected mouse and rat organs were paraformaldehyde fixed for 24 hr at 4C before they were mounted in paraffin, sectioned, and developed by immunohistochemistry. The experiments performed on human skin were approved by the Regional Scientific Ethics Committee (H-1-2012-141). The animals were housed in a certified facility and the institutional guidelines for animal welfare and experimental conduct were followed.
Design and Construction of Haldisin Expression Vectors
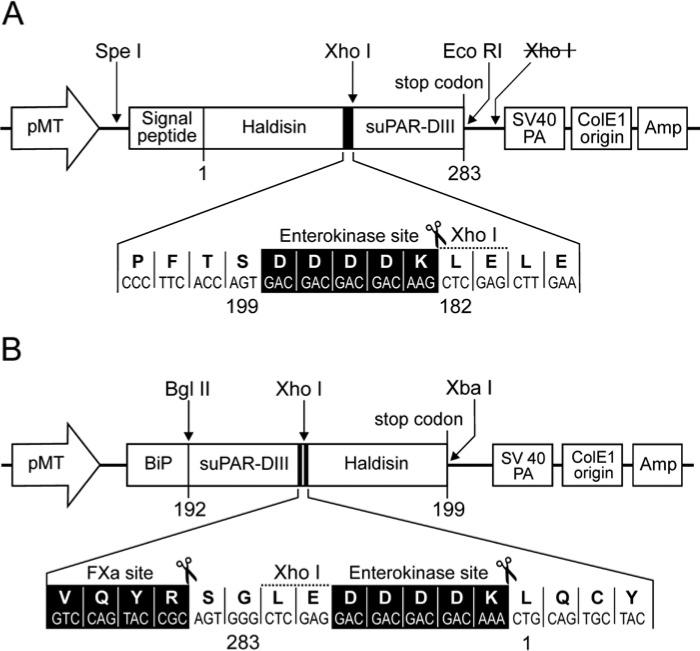
To express recombinant soluble Haldisin by Drosophila melanogaster S2 cells, two fusion protein constructs were generated, each containing a soluble version of the third domain of uPAR (suPAR-DIII) as a purification tag located either at the C- or N-terminal of Haldisin (see Fig. 1). The endogenous signal sequences for GPI-anchoring are deleted in these Haldisin constructs to enable secretion and facilitate subsequent purification of the fusion proteins from the conditioned media by immunoaffinity chromatography (Gårdsvoll et al. 2007). Expression of Haldisin containing a C-terminal purification tag is driven by its native signal sequence, whereas secretion for the N-terminal tagged version depends on the Drosophila BiP signal sequence present in the original pMT/BiP/V5-His vector. Both vectors contain the metallothionein promoter, which enables a strong, inducible expression of heterologous proteins by CuSO4. These two new expression vectors were denoted pMTC-X/Haldisin/ent/suPAR-DIII and pMTBiP/suPAR-DIII/ent/Haldisin, respectively (see Figs. 1A and 1B), and were constructed as briefly described: The vector pBluescript-Haldisin containing the full-length human sequence was used as a template in a PCR reaction using upstream primer: 5-TATACTAGTCCAGCTCAGCAATGGCAATGGGGGTCC-3’ and downstream primer: 5-ATTCTCGAG CTTGTCGTCGTCGTCACTGGTGAAGGGCTGGGTCATGGATTTCC-3’, which contain a sequence encoding the enterokinase recognition site (bold). Flanking this PCR product is a SpeI site and a XhoI site (underlined sequences) enabling insertion into the pMT-X/suPAR-DIII tagging vector constructed previously (Gårdsvoll et al. 2007). To create a new vector for use in N-terminal suPAR-DIII tagging the plasmid pMT-X/suPAR-DIII was in this case used as a template in a PCR reaction using upstream primer: 5’-TATAGATCTCGCCAGTGTTACAGCTGCAAGGGGAAC-3’ and downstream primer: 5’-ATACTCGAGCCCACTGCGGTACTGGACATCCAGGTC-3’. Flanking this PCR product is a BglII site and a XhoI site (underlined sequences) enabling insertion into the commercially available vector pMT/BiP/V5-HisA in fusion with the BiP signal sequence contained within this vector, thus creating the plasmid pMTBiP/suPAR-DIII/V5-HisA. The Haldisin insert for this vector was prepared in a new PCR reaction with pBluescript-Haldisin as a template using upstream primer: 5-TATCTCGAG GACGACGACGACAAACTGCAGTGCTACAGCTTTGAGCACACCTAC-3’ (sequence of enterokinase recognition site indicated in bold) and downstream primer: 5’-ATTTCTAGA TTAACTGGTGAAGGGCTGGGTCATGGATTTCC-3’ (stop codon indicated in bold). This PCR product is flanked by XhoI and XbaI sites (underlined sequences) for use in cloning into pMTBiP/suPAR-DIII/V5-His A.
Figure 1.
Schematic representation of the S2 cell expression vectors used for suPAR-DIII tagging of Haldisin. The vectors used for expression of Haldisin with a suPAR-DIII tag positioned at the C- or N-terminal are shown in panel A and panel B, respectively. pMT, Drosophila metallothionein promoter; Signal peptide, DNA sequence encoding the signal sequence of Haldisin; BiP, Drosophila secretion signal; Haldisin, DNA sequence encoding amino acid residues 1–199 hereby omitting the residues encoding GPI-anchoring; suPAR-DIII, soluble version of uPAR domain III (residues 182–283 for C-terminal tagging, and residues 192–283 for N-terminal tagging). Unique restriction enzyme sites are indicated above, and the recognition sites for enterokinase (DDDDK) and FXa (VQYR) are indicated below as inserts in black with indication of actual cleavage points (scissors).
To enable expression of full-length GPI-anchored Haldisin in HEK293 cells, pBluescript-Haldisin was used as a template in a PCR reaction using upstream primer: 5-ATAGGATCCAGGTGGCCCAGCTCAGCAATGGCAATG-3’ and downstream primer: 5-AATGCGGCCGCTGAAGCAGCAAGAATGAGGTGTGTGAG-3’. Flanking this PCR product are a BamHI site and a NotI site (underlined sequences) enabling insertion into the pcDNA/FRT/TO vector creating pcDNA5/FRT/TO/Haldisin-GPI.
Expression and Purification of Haldisin Fusion Proteins
Stably transfected S2 cell lines capable of producing secreted Haldisin/ent/suPAR-DIII and suPAR-DIII/ent/Haldisin fusion proteins were established, and the corresponding recombinant proteins were produced as described in detail previously (Gårdsvoll et al. 2007), with the only exception being the use of Express Five SFM during the 7-day production period in shaker flasks.
The secreted fusion proteins were purified from the conditioned media by immunoaffinity chromatography, and the suPAR-DIII tag was removed by enterokinase cleavage as described previously (Gårdsvoll et al. 2007). The enterokinase cleavage of Haldisin/ent/suPAR-DIII will leave the enterokinase recognition sequence attached to the C-terminus of the released Haldisin, which is accordingly denoted Haldisin/ent. Enterokinase cleavage of suPAR-DIII/ent/Haldisin releases a tag-free recombinant Haldisin. Factor Xa (FXa) cleavage of suPAR-DIII/ent/Haldisin leaves an extension consisting of nine amino acid residues (SGLEDDDDK) on the N-terminus of the released Haldisin, which accordingly is denoted *Haldisin. The FXa digestion was performed in an enzyme:substrate ratio of 1:50 (w/w) at room temperature overnight and was followed by the addition of PMSF to a final dose of 1 mM. After concentration on an Amicon filter, the *Haldisin product was further purified by gel filtration on a Superdex 75 column operated at 1 ml/min in 20 mM NaH2PO4, 150 mM NaCl, pH 7.2. Collected fractions were analyzed by SDS-PAGE and silver staining, and pooled fractions of *Haldisin were given additional supplementation of PMSF to a final dose of 1 mM before the proteins were concentrated on Amicon filtration using an Ultracell-3K filter having a 3 kDa cut-off.
HEK293 cells stably transfected with glycolipid-anchored Haldisin
GPI-anchored Haldisin was expressed in transfected HEK293 cells using the Flp-In T-REx system. The transfection and selection as well as growth of cells were performed as outlined previously (Gårdsvoll, Kjaergaard, et al. 2011). We generated three separate monoclonal cell lines of the HEK293 Haldisin-transfected cells by traditional protocols using cloning rings.
Production and Purification of Polyclonal Antibodies Directed against Haldisin
Three New Zealand white rabbits were immunized according to a previously described immunization schedule (Rasch et al. 2010). In brief, rabbits were injected subcutaneously with 20 µg purified Haldisin/ent in PBS per injection mixed 1:1 (v/v) with either Freunds Complete Adjuvant (first injection) or Freunds Incomplete Adjuvant (subsequent nine injections). The rabbits were bled before the first immunization to generate pre-immune serum and were subsequently primed with antigen every 4th week until a total bleeding was performed after 6 months. The specificity of the collected sera was tested by immunoblotting analysis using conditioned medium from S2 cells producing recombinant Haldisin/ent/suPAR-DIII. All three rabbits mounted a comparable immune response (results not shown). Finally, IgG fractions were purified from the immune sera by Hi TRAP Protein G affinity chromatography.
Immunoblotting of Cell Lysates and Media after PI-PLC Treatment
Adherent HEK293 cells with stable expression of GPI-anchored Haldisin were washed with PBS and subsequently treated with 0.2 units Phosphatidylinositol-specific phospholipase C (PI-PLC)/ml in 400 µl PBS at 20C. Aliquots were withdrawn at indicated time intervals. In a separate setup, untreated HEK293 cells were harvested from confluent cultures grown in six-well plates and lysed directly in SDS-PAGE sample buffer. After centrifugation, supernatants from either cell lysates or PI-PLC treatments were subjected to SDS-PAGE using a Bis-Tris 4% to 12% gradient gel followed by electroblotting onto a PVDF membrane. The blot was incubated with 0.25 µg/ml of our polyclonal rabbit anti-Haldisin antibody overnight at 4C followed by incubation with peroxidase-conjugated swine anti-rabbit antibodies diluted 1:5000 for 1 hr. The immunoblot was developed with an ECL chemoluminescence detection system.
Immunofluorescence Analysis
Gelatin-coated glass coverslips were placed in a 24-well plate and Haldisin-transfected HEK293 cells were seeded and grown for 2 to 3 days at 37C in a 5% CO2 incubator until they were subconfluent. Cells were washed gently once in PBS before fixation using 4% paraformaldehyde (PFA) for 10 min followed by washing twice in PBS. Blocking of unspecific binding was performed by incubation for 1 hr in PBS containing 10% (v/v) normal goat serum (NGS). Incubation with pre-immune serum or our polyclonal rabbit anti-Haldisin antibody diluted to 1.7 µg/ml in PBS containing 1% (w/v) BSA and 1% (v/v) NGS proceeded for 1 hr and was followed by four washes in PBS. Specificity of our polyclonal anti-Haldisin antibody (pAb) was verified by pre-absorption using a 50-fold molar excess of either Haldisin/ent/suPAR-DIII or C4.4A/ent/suPAR-DIII (Gårdsvoll et al. 2007) at room temperature for 30 min prior to its addition to the coverslips. These coverslips were subsequently incubated with goat anti-rabbit Alexa Fluor-488 conjugated F(ab’)2 fragment diluted to 10 µg/ml in the above dilution buffer for 1 hr in the dark. Finally, after repeated washings in PBS and once in water, the coverslips were mounted in ProLong Gold antifade reagent with DAPI. Images of stained cells were recorded using an Axio Imager 2 microscope equipped with an LSM 700 confocal scanning module and processed with the Zen 2011 light software (all from Carl Zeiss; Thornwood, NY). The exposure conditions for each image within an experiment were comparable.
Immunoperoxidase Staining
Five-µm thick paraffin-embedded tissue sections were deparaffinized in xylene and hydrated in decreasing concentrations of ethanol in water. Antigen retrieval was achieved by pre-treatment with 5 µg/ml Proteinase K for 10 min (mouse tissue) or 15 min (human and rat tissues) at 37C. Subsequently, endogenous peroxidase activity was blocked by incubation with 1% (v/v) H2O2 for 15 min. After the tissue sections were mounted in Shandon racks, they were briefly washed in Tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 7.6) containing 0.5% (v/v) Triton X-100 and incubated overnight at 4C with primary antibodies diluted to 1 µg/ml in Antibody Diluent. The specificity of our rabbit anti-Haldisin pAb was verified by pre-absorption with 60-fold molar excess of Haldisin/ent/suPAR-DIII at room temperature for 2 hr prior to its addition to the sections. The primary antibodies were detected using the Envision reagent containing anti-rabbit IgG horseradish peroxidase-labeled polymers and visualized with NovaRed chromogene according to the manufacturer’s instructions. The sections were counterstained with Mayer’s haematoxylin, dehydrated in ethanol solutions, and mounted in Pertex.
Results
Expression and Purification of Recombinant Soluble Haldisin
Inspection of the cDNA-derived sequence for Haldisin reveals that the nascent protein is composed of a 25 amino acid (aa) signal peptide, a 199 aa extracellular protein part consisting of two LU-domains, and a 27 aa C-terminal signal sequence responsible for a putative GPI-anchoring (Supplemental Fig. S2, panel A). Accordingly, we used two different strategies to produce soluble secreted versions of Haldisin in Drosophila S2 cells. The first version complies with our original protocol for expression of tagged LU-domain proteins (Gårdsvoll et al. 2007) exploiting the endogenous signal sequence of the LYPD5 gene and placing the suPAR-DIII tag at the C-terminus of the fusion protein (Haldisin/ent/suPAR-DIII), as depicted in Fig. 1A. Our second approach positioned the suPAR-DIII tag at the N-terminus of Haldisin and used the endogenous Drosophila BiP signal sequence (suPAR-DIII/ent/Haldisin), which is illustrated in Fig. 1B. Both constructs were produced well by the S2 cells, yielding between 2 and 4 mg pure Haldisin fusion protein per liter of culture medium after immunoaffinity purification using the monoclonal anti-uPAR antibody R2. As judged by silver staining after SDS-PAGE, the purity of the final preparations was very high (>95%) with no obvious sign of degradation (lane 1 in Figs. 2A and 2B).
Figure 2.
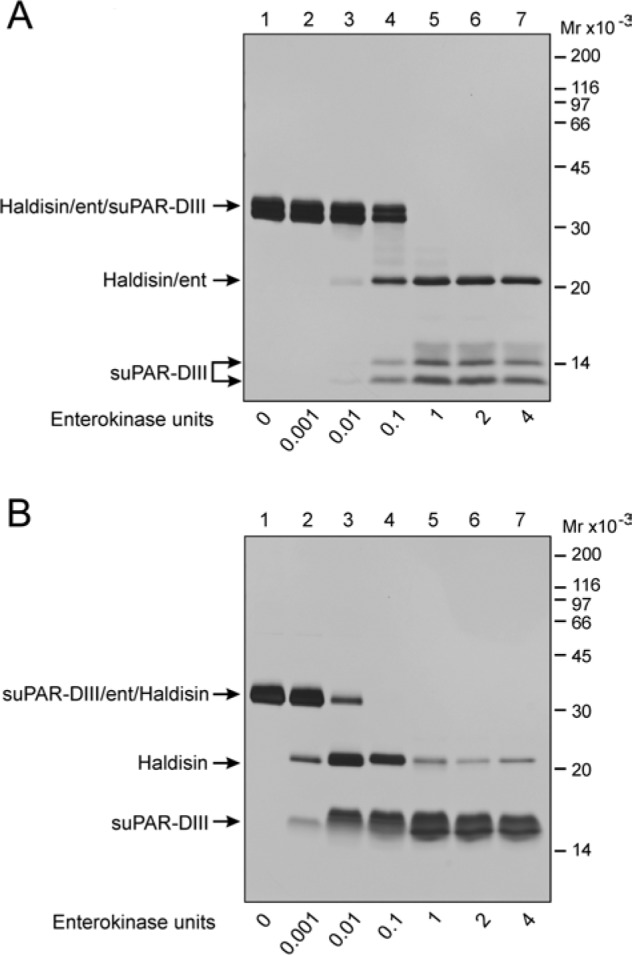
Efficacy of enterokinase cleavage of C- and N-terminal suPAR-DIII-tagged recombinant Haldisin. The immunoaffinity purified Haldisin/ent/suPAR-DIII (panel A) and suPAR-DIII/ent/Haldisin (panel B) were analyzed by SDS-PAGE and silver staining before and after treatment with various amounts of enterokinase. Digestion was performed at 37C for 16 hr using EKMax from Invitrogen. Samples were all non-reduced. Note that 1 unit of enterokinase per. 20 µg fusion protein is needed to obtain 100% cleavage with C-terminal tagging (panel A), whereas the same cleavage efficiency is achieved for the N-terminal tagging using 10-fold less enzyme (panel B).
To remove the purification tag from these purified Haldisin hybrid proteins, we incubated them with a different concentration of enterokinase for 16 hr at room temperature and analyzed them by SDS-PAGE followed by silver staining (Figs. 2A and 2B). The engineered cleavage site for enterokinase is readily accessible in both of these hybrid proteins, as the enzyme efficiently catalyzes the release of the suPAR-DIII tag and Haldisin, with the N-terminally tagged version being particularly sensitive to the enterokinase-mediated release of suPAR-DIII. Nonetheless, it was clear that additional internal cleavage occurred in this particular suPAR-DIII/ent/Haldisin construct (Fig. 2B), whereas Haldisin released by enterokinase from the Haldisin/ent/suPAR-DIII fusion construct appeared to be more stable, as evaluated by SDS-PAGE of the cleavage mixture under non-reducing conditions (Fig. 2A). When the corresponding purified Haldisin/ent was analyzed under reducing conditions, it was, however, evident that approximately 50% of the purified product unfortunately carries an internal cleavage within one of its two LU-domains, and the fragments generated remain covalently linked by disulfide bonds (Fig. 3A). Analysis by MALDI mass spectrometry of the purified Haldisin/ent preparation yielded an average molecular mass of 23,346 Da, which is in fair agreement with the mass calculated for Haldisin/ent (23,349 Da), including one of the biantennary N-linked glycosylations typical of recombinant proteins expressed by S2 cells (Gårdsvoll et al. 2004). Reduction of the disulfide bonds in this Haldisin/ent preparation produced two fragments with molecular masses of 18,477 Da and 4,887.6 Da, where the larger fragment was reduced to 17,442 Da after N-glycanase treatment (Table 1). This clearly shows that the recombinant Haldisin contains only one N-linked carbohydrate despite the presence of two Asn-X-Thr/Ser motifs in the primary sequence. The internal cleavage in Haldisin/ent occurs between GYR42↓APV in the first LU-domain in the exposed loop 2 connecting βIC and βID, with the scissile bond being located close to the intron-exon boundary between exons 2 and 3 (Supplemental Fig. S2A).
Figure 3.
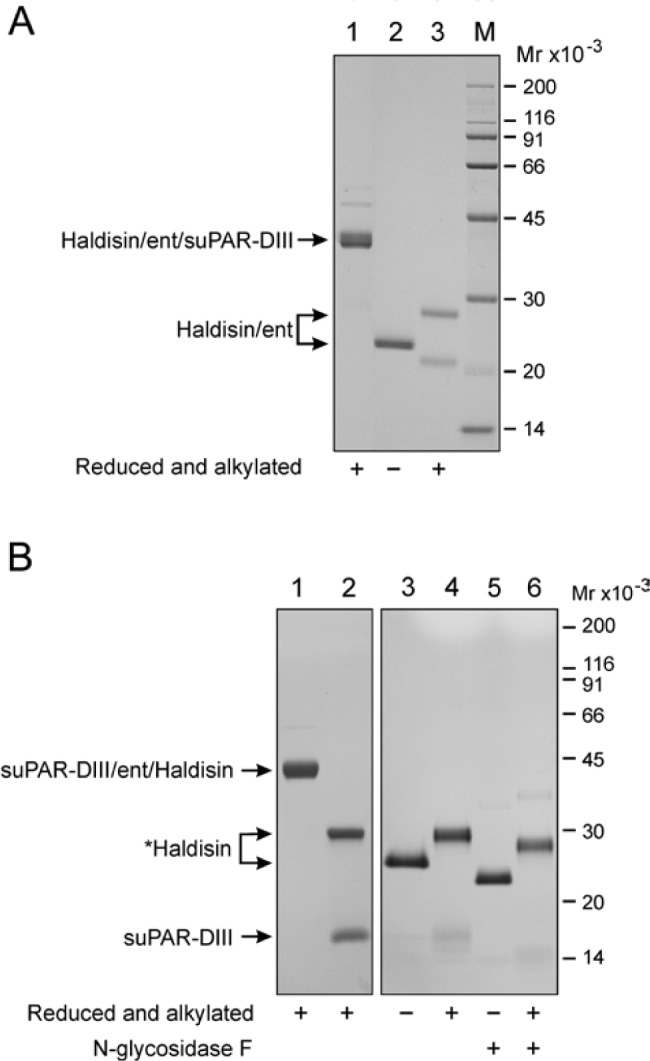
Purification of recombinant soluble Haldisin released from C- or N-terminal suPAR-DIII-tagged fusion proteins. Panel A: Immunoaffinity purified Haldisin/ent/suPAR-DIII was analyzed by SDS-PAGE and Coomassie staining (lane 1). Likewise, the isolated Haldisin/ent product is shown after removal of the suPAR-DIII tag (lanes 2 and 3). Note that the purified Haldisin/ent preparation contains a significant proportion of molecules with an internal cleavage as revealed after reduction and alkylation (compare lanes 2 and 3). A total of 1 to 3 µg was applied in each lane. Panel B: Purified suPAR-DIII/ent/Haldisin (lane 1) was subjected to FXa cleavage to liberate *Haldisin from the suPAR-DIII tag (lane 2), and the corresponding isolated *Haldisin product was analyzed by SDS-PAGE and silver staining (lanes 3 and 4). The (*) indicates that the recombinant product *Haldisin contains an N-terminal extension consisting of the residues SGLEDDDDK (see Fig. 1, panel B). N-glycosidase F treatment of *Haldisin is also shown (lanes 5 and 6). Note that liberation of *Haldisin by FXa proceeds without any internal cleavage, as demonstrated by the single band in SDS-PAGE of reduced and alkylated samples (lanes 4 and 6).
Table 1.
MALDI-MS Analysis of Recombinant Haldisin.
| Sample | Recorded Masses | Calculated Masses | Comment |
|---|---|---|---|
| Enterokinase cleaved suPAR-DIII/ent/Haldisin | |||
| Haldisin1-199-DDDDK | 23,346 Da | 23,349 Da | + 1 CHO (1039 Da) |
| + N-glycanase | 22,308 Da | 22,311 Da | corrected for Asn to Asp conversion |
| After reduction of disulfides: | |||
| Haldisin43-199-DDDDK | 18,477 Da | 18,496 Da | Haldisin43-199-DDDDK + 1 CHO |
| Haldisin43-199-DDDDK + N-glycanase | 17,442 Da | 17,458 Da | corrected for Asn to Asp conversion |
| Haldisin1-42 | 4,887.3 Da | 4,887.6 Da | Haldisin1-42 |
| Factor Xa cleaved suPAR-DIII/ent/Haldisin | |||
| SGLEDDDDK-Haldisin1-199 | 23,731 Da | 23,735 Da | + 1 CHO (1039 Da) |
| + N-glycanase | 22,680 Da | 22,697 Da | corrected for Asn to Asp conversion |
Haldisin was deposited on the MS target as a sandwich using sinipinic acid as matrix. Reduction of disulfides was accomplished by incubation in 20 mM DTT for 30 min at 56C. The surface was washed extensively with 0.1% (v/v) TFA before recording the mass spectra on an Autoflex II MS/MS operated in the linear mode (Bruker, Germany).
In an attempt to find an alternate strategy for the release of intact Haldisin from the fusion proteins, we exposed both constructs to purified thrombin and FXa. It is important that none of these proteases catalyzed internal cleavage in any of the fusion proteins (data not shown), but it was unexpected that FXa caused an efficient release of Haldisin from the suPAR-DIII tag, but only in suPAR-DIII/ent/Haldisin (Fig. 3B). Based on the mass of the released *Haldisin, determined by MALDI-MS before and after N-glycanase treatment (Table 1), it is clear that the scissile bond hydrolyzed by FXa is between Arg-Ser located internally in the uPAR-DIII linker (VQYR↓SGLE), as depicted in Fig. 1B.
Immunofluorescence Analysis of Cellular Haldisin Expressed by Transfected HEK293 Cells
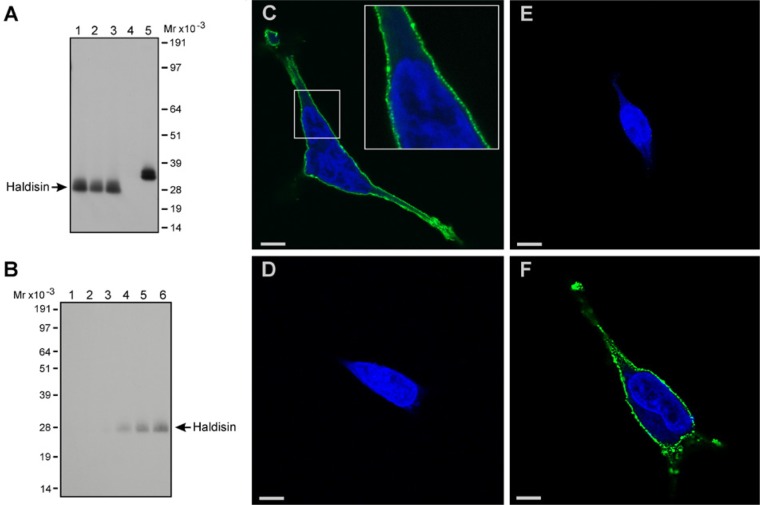
To enable future studies on Haldisin expression in cells and tissues, we generated polyclonal rabbit anti-human Haldisin antibodies by immunizing three rabbits with the purified recombinant Haldisin/ent expressed by Drosophila S2 cells, as shown in Fig. 3A. The specificity of one of the purified anti-Haldisin pAbs was validated by immunoblotting analysis on western blots of total cell lysates from three different HEK293 cell lines stably transfected with full-length Haldisin cDNA (Fig. 4, panel A). As expected, the pAb detected only a single band in the lysates with an electrophoretic mobility of approximately 30 kDa, and, accordingly, no reaction was detected in the mock-transfected cells. When the cells were treated with PI-PLC, a soluble, shed version of this protein was detected in the media by our pAb, thus demonstrating that Haldisin is indeed a GPI-anchored protein (Fig. 4B). We therefore continued to explore if this specific reactivity could be extended to GPI-anchored Haldisin expressed on the membrane of transfected HEK293 cells plated on gelatin-coated coverslips. As shown in Fig. 4C, the antibody recognized a component that was localized topographically in discrete and punctuate foci on the cell surface. This reactivity was specific for Haldisin, as it was absent if the anti-Haldisin pAb was replaced with pre-immune serum (Fig. 4D) or if the anti-Haldisin pAb was pre-absorbed with recombinant Haldisin purified from S2 cells (Fig. 4E). Along the same line of evidence, pre-absorption of the anti-Haldisin pAb with the structural homolog C4.4A purified from S2 cells (Hansen et al. 2004) did not attenuate the membrane staining with the pAb (Fig. 4F).
Figure 4.
Immunoblotting and immunofluorescence of HEK293 cells transfected with GPI-anchored Haldisin. In panel A, our rabbit pAb directed against recombinant Haldisin was tested at 0.25 µg/ml in immunoblotting analysis on total cell lysates derived from three different HEK293 cell lines stably transfected with GPI-anchored Haldisin (lanes 1–3). Mock-transfected HEK293 cell lysate was used as a negative control (lane 4), and the purified Haldisin/ent/suPAR-DIII served as a positive control (lane 5). In panel B, HEK293 cells expressing GPI-anchored Haldisin were incubated for 1, 2, or 4 hr in PBS in the absence (lanes 1–3) or presence of 0.2 units PI-PLC/ml (lanes 4–6). Released, soluble Haldisin in the media was revealed by immunoblotting as in panel A. Panels C–F show the immunofluorescence staining of a Haldisin-transfected HEK293 cell line. Cells grown on gelatin-coated coverslips were fixed and incubated with 1.7 µg/ml of the anti-Haldisin pAb, either alone (C) or after pre-incubation with a molar excess of purified recombinant Haldisin (E) or C4.4A (F). Pre-immune serum with an IgG concentration corresponding to 1.7 µg/ml was used as a negative control (D). Immunodetection was performed with Alexa Fluor 488-conjugated antibodies, whereas nuclei were stained by DAPI. Bars C, D, E = 10 µm; bar F = 5 µm.
Immunohistochemical Localization of Haldisin in Human and Rodent Tissues
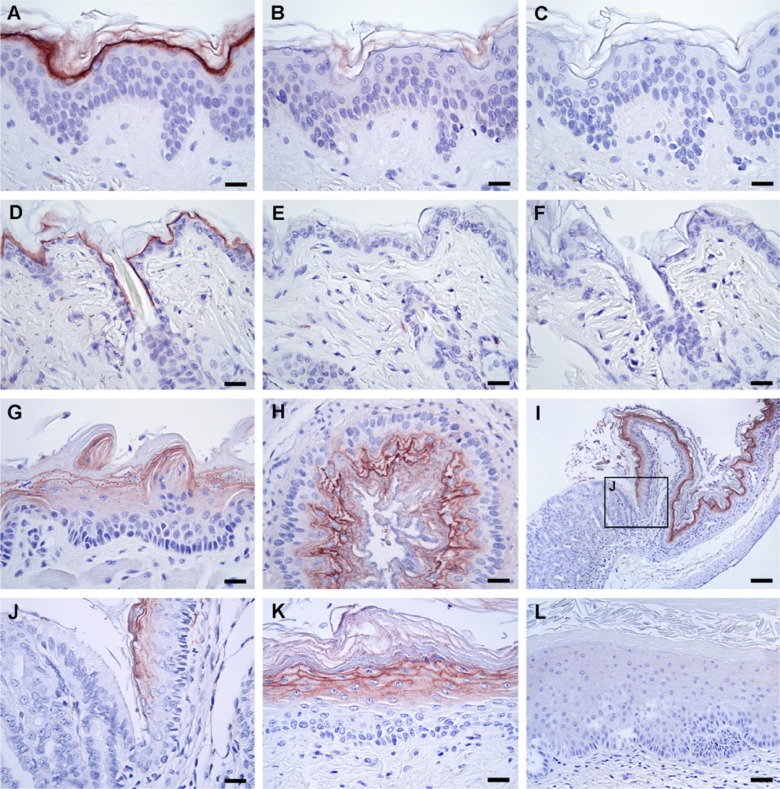
To characterize the expression profile of Haldisin in vivo, we explored the aptness of our anti-Haldisin pAb for specific detection of endogenously expressed Haldisin in paraformaldehyde-fixed and paraffin-embedded tissues by immunohistochemistry. It is interesting that these studies clearly showed that Haldisin exhibits a distinct expression in human epidermis where it is primarily localized to the highly differentiated keratinocytes of the stratum granulosum, leaving most of the stratum corneum, stratum spinosum, and stratum basale negative for staining (Fig. 5, panel A). In the epidermis, Haldisin expression was also noted in keratinocytes lining hair follicles, in the ducts of sebaceous glands, and in the cuboidal epithelium of sweat glands (results not shown). The staining appears highly specific because of its distinct demarcation and confinement to the cell membrane. To further validate the specificity of the staining experimentally, we performed pre-absorption of the anti-Haldisin pAb with recombinant Haldisin prior to staining and tested the staining potential of rabbit IgG of irrelevant specificity (Fig. 5, panels B, C). As expected, no specific recognition could be observed in either of these two control experiments. Pre-absorption of our anti-Haldisin pAb with equivalent amounts of purified C4.4A/ent/suPAR-DIII, human suPAR, or the purified ent/suPAR-DIII tag did not attenuate the immunostaining of the stratum granulosum in samples of human breast skin (Supplemental Figs. S3A–S3D), whereas both purified Haldisin/ent/suPAR-DIII and Haldisin/ent did remove this immunoreactivity completely (Supplemental Figs. S3E and S3F).
Figure 5.
Haldisin expression in human skin and mouse tissues determined by immunohistochemistry. Immunohistochemistry was performed on serial sections of paraformaldehyde-fixed and paraffin-embedded human (A–C) and murine skin (D–F) using 1 µg/ml of our rabbit anti-Haldisin pAb either alone (A, D) or after pre-incubation (B, E) of the antibody with a molar excess of purified recombinant human Haldisin prior to staining. Incubation of the tissue sections with 1 µg/ml control rabbit IgG of irrelevant specificity did not result in any staining (C, F). Moreover, the staining of Haldisin was evaluated in several other tissues from the adult mouse and rat as shown in Table 2, and a selected number of these stainings are presented in panels G–L. Shown is Haldisin expression in the stratified squamous epithelium of the mouse tongue (G), esophagus (H), and non-glandular part of the stomach from the upper gastrointestinal tract (I, J). Note that Haldisin is expressed in the squamous epithelium of the caudal end of vagina (K), but only weakly or absent in the cranial vaginal portion near the transition zone to the uterus (L). Bars A–H, J, K = 25 µm; bar L = 50 µm; and bar I = 100 µm.
We next evaluated if any cross-reactivity of the anti-Haldisin pAb with murine Haldisin (75% sequence conservation; see Supplemental Fig. S4) could be exploited for immunohistochemistry in mouse tissues—in analogy with previous findings for our anti-C4.4A pAb (Kriegbaum, Jacobsen, et al. 2011). Using the same criteria for specificity as we used for human Haldisin, it is clear that the cross-reactivity of the pAb raised against human Haldisin is sufficient to visualize expression of murine Haldisin in paraffin-embedded tissues and that the staining pattern observed in human skin is recapitulated in mouse skin (Fig. 5, panels D–F). This observation allows an extensive survey of the global Haldisin expression in the normal adult mouse and rat (Table 2).
Table 2.
Protein Expression of Haldisin in the Adult Mouse and Rat.
| Tissue | Haldisin Expression in Adult Mouse | Haldisin Expression in Adult Rat | Tissue | Haldisin Expression in Adult Mouse | Haldisin Expression in Adult Rat |
|---|---|---|---|---|---|
| Tongue | + (+) | + (+) | Ovaries | - (-) | - (-) |
| Esophagus | + (+) | + (+) | Uterus | - (-) | - (-) |
| Stomach | Vagina | ||||
| Glandular | - (-) | - (-) | Cranial | - (+) | - (+) |
| Non-glandular | + (+) | + (+) | Caudal | + (+) | + (+) |
| Duodenum | - (-) | - (-) | Adipose tissue | - (-) | - (-) |
| Jejunum | - (-) | - (-) | Brain | - (-) | ND |
| Ileum | - (-) | - (-) | Thymus | - (+) | - (+) |
| Ascending colon | - (-) | - (-) | Spleen | - (-) | - (-) |
| Sigmoid colon | - (-) | - (-) | Liver | - (-) | - (-) |
| Rectum | - (-) | - (-) | Pancreas | - (-) | - (-) |
| Anorectal junction | + (+) | ND | Mammary gland | - (-) | - (-) |
| Bone | - (-) | ND | Kidney | - (-) | - (-) |
| Bone marrow | - (-) | ND | Bladder | - (-) | - (-) |
| Eye | - (+) | - (+) | Heart | - (-) | - (-) |
| Testicle | - (-) | - (-) | Striated muscle | - (-) | - (-) |
| Epididymis | - (-) | - (-) | Smooth muscle | - (-) | - (-) |
| Seminal vesicle | - (-) | ND | Nasal cavity | + (+) | ND |
| Skin | + (+) | + (+) | Trachea | - (-) | - (-) |
| Hair follicle | + (+) | + (+) | Lung | - (-) | - (-) |
Haldisin expression in squamous epithelium; -, absence of Haldisin expression; ND, not determined. C4.4A expression in the corresponding organs is shown in brackets (Kriegbaum, Jacobsen, et al 2011).
A distinct Haldisin expression was generally confined to most of the keratinized squamous epithelia examined. In the gastrointestinal tract, expression was thus found in the epithelium of the tongue, esophagus, non-glandular part of the rodent stomach (Fig. 5, panels G–J), and anorectal junction (not shown). In the latter two tissues, the expression terminated abruptly with the transition from squamous to columnar epithelium at the squamo-columnar junctions. A notable deviation from this robust expression pattern in squamous epithelia was found in the female reproductive system. In this particular organ, the squamous epithelium did not present a continuum of Haldisin positive cells, in contrast to C4.4A (Kriegbaum, Jacobsen, et al. 2011). Although Haldisin, as expected, is highly expressed in the squamous epithelium of the caudal vagina (Fig. 5, panel K), it is absent or only very weakly expressed in the cranial vagina near the transition zone to the columnar epithelium of the uterus (Fig. 5, panel L). The absence of Haldisin expression in the non-keratinized stratified squamous epithelium of the cornea of the eye (data not shown) furthermore emphasizes the primary confinement of Haldisin to stratum granulosum giving rise to stratum corneum.
In conclusion, all Haldisin-positive stratified squamous epithelia revealed a progression of Haldisin expression toward the epithelial surface with a negative stratum basale and a peak expression in the stratum granulosum. Thus, the expression of Haldisin seems to reflect the late differentiation of the corresponding stratified epithelia.
Discussion
In the past decade, bioinformatics have identified a vast array of genes encoding single domain proteins belonging to the LU-domain protein family, where the corresponding proteins adopt the archetypal three-fingered folding topology found in snake venom α-neurotoxins (Galat 2008). From a functional point of view, this protein fold has proven highly versatile, as evolution has created single domain paralogs with such diverse functions as bona fide cytotoxins that inhibit acetylcholine esterases (fasciculins), ion channels (calciseptine) or nicotinic acetylcholine receptors (e.g., α-neurotoxins), inhibitors of autologous complement activation (CD59), cofactors of lipolysis enabling trans-capillary trafficking of lipoprotein lipase (GPIHBP-1), and maintenance of paracellular diffusion barriers in Drosophila by trafficking and assembly of septate junction components (crooked, crimpled, coiled, and boudin) (Kini and Doley 2010; Nilton et al. 2010; Nevo et al. 2013; Young and Zechner 2013). Nevertheless, the biochemical function of a large proportion of these single domain LU-proteins still remains enigmatic, as illustrated by the >20 murine Ly-6 antigens encoded on chromosome 15 and predominantly expressed by neutrophils (Lee et al. 2013).
A specialized subgroup of the LU-protein family is encoded by a small gene cluster located on human chromosome 19, which comprises six multidomain membrane proteins (Supplemental Fig. S1). The founding member of this subgroup is uPAR, which is by far the best characterized member, both structurally and functionally (Kriegbaum, Persson, et al. 2011; Ploug 2013). Recent studies have clearly demonstrated that the interdomain flexibility of its three LU-domains enables an allosteric regulation of uPAR, where receptor occupancy with its high-affinity serine protease ligand, uPA, affects the subsequent interaction of uPAR with the extracellular matrix component vitronectin (Gårdsvoll, Jacobsen, et al. 2011; Gårdsvoll, Kjaergaard, et al. 2011; Mertens et al. 2012). The flexible multidomain architecture of uPAR may thus provide a conformational switch enabling a functional rendezvous between matrix remodeling and matrix-assisted adhesion and migration. Whether a similar regulatory mechanism is operational and enabled by such interdomain flexibility in the other multidomain members remains, at present, highly speculative. Nevertheless, the occurrence of a unique structural abnormality in the N-terminal LU-domain (i.e., a missing consensus disulfide), which is present in all six multidomain members (Supplemental Fig. S1) but disallowed for the correct folding of the single domain members, may argue for a shared interdomain flexibility in these proteins (Kjaergaard et al. 2008).
In the present study, we have expressed and characterized the LU-domain-containing protein encoded by LYPD5, which we now term Haldisin because of its prevalent expression in human skin. Like uPAR, Haldisin undergoes posttranslational modifications including attachment of one N-linked glycosylation on either Asn95 or Asn149 as well as addition of a C-terminal glycolipid membrane anchor (GPI) responsible for the tethering of Haldisin to the outer external leaflet of the plasma membrane. Concordant with this biochemical property, cytological and histological staining using our polyclonal anti-Haldisin antibody reveal its pronounced expression on the cell membrane (Figs. 4 and 5).
It is surprising that this distinct expression pattern of Haldisin, which is confined to the stratified squamous epithelia, is shared by another of the glycolipid-anchored members of the multidomain LU-protein family denoted C4.4A (Table 2). A notable difference in their expression patterns is, nonetheless, immediately obvious, as C4.4A is predominantly confined to stratum spinosum (Hansen et al. 2004; Kriegbaum, Jacobsen, et al. 2011), whereas Haldisin expression is most pronounced in stratum granulosum. The function of these proteins in the normal stratified epithelium remains at present elusive. Nevertheless, due to their high expression levels on the keratinocyte plasma membrane, one might speculate that a possible function could relate to skin barrier function or to intraepithelial adhesion. Such a function is not completely without precedence within the LU-protein domain family, since the glycolipid-anchored single LU-domain proteins crooked, crimpled, coiled, and boudin are involved in trafficking and assembly of septate junction components important in the establishment of a functional blood–brain barrier in Drosophila (Nilton et al. 2010; Syed et al. 2011).
Several studies have demonstrated that expression of C4.4A is a strong biomarker for poor prognosis of non-small cell lung cancer patients with adenocarcinomas (Hansen et al. 2007; Jacobsen et al. 2013). The normal lung epithelium is devoid of C4.4A, but early, non-neoplastic changes in the bronchial epithelium (e.g., basal cell hyperplasia and metaplasia) are generally associated with the emergence of C4.4A (Jacobsen et al. 2012). In this histological subtype, the levels of C4.4A are of no prognostic significance as it merely reflects the non-malignant transformation into squamous epithelia (Hansen et al. 2007; Jacobsen et al. 2013). However, pre-malignant reactive changes of the alveolar epithelium (atypical adenomatous hyperplasia) also display C4.4A expression in approximately 25% of the cases (Jacobsen et al. 2012). It is interesting that a similar fraction of lung adenocarcinomas also express C4.4A, which prompts the intriguing possibility that C4.4A expression in early lesions and the development of particularly aggressive adenocarcinomas may be correlated (Jacobsen et al. 2013). Other studies have shown an additional prognostic impact of C4.4A expression in colorectal adenocarcinomas (Paret et al. 2007; Konishi et al. 2010). Whether Haldisin expression contains similar prognostic information, however, remains to be studied.
Supplementary Material
Acknowledgments
We thank Haldis Egholm Mønsted, Gitte Juhl Funch, Lotte Frederiksen, and John Post for excellent technical assistance.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Danish National Research Foundation (Danish-Chinese Centre for Proteases and Cancer); Siemenfondens legat and Novo Scholarship (EPH); and The Danish Cancer Society (MCK and WA).
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
References
- Davies BS, Beigneux AP, Barnes RH, 2nd, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyren R, Goldberg I, Olivecrona G, et al. 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Plantard L, Aeschbach L, Brakch N, Christen-Zaech S, de Viragh PA, Sergeant A, Huber M, Hohl D. 2007. SLURP1 is a late marker of epidermal differentiation and is absent in Mal de Meleda. J Invest Dermatol. 127:301–308 [DOI] [PubMed] [Google Scholar]
- Fujihara Y, Tokuhiro K, Muro Y, Kondoh G, Araki Y, Ikawa M, Okabe M. 2013. Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc Natl Acad Sci U S A. 110:8111–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A. 2008. The three-fingered protein domain of the human genome. Cell Mol Life Sci. 65:3481–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gårdsvoll H, Hansen LV, Jørgensen TJ, Ploug M. 2007. A new tagging system for production of recombinant proteins in Drosophila S2 cells using the third domain of the urokinase receptor. Protein Expr Purif. 52:384–394 [DOI] [PubMed] [Google Scholar]
- Gårdsvoll H, Jacobsen B, Kriegbaum MC, Behrendt N, Engelholm L, Østergaard S, Ploug M. 2011. Conformational regulation of urokinase receptor function: impact of receptor occupancy and epitope-mapped monoclonal antibodies on lamellipodia induction. J Biol Chem. 286:33544–33556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gårdsvoll H, Kjaergaard M, Jacobsen B, Kriegbaum MC, Huang M, Ploug M. 2011. Mimicry of the regulatory role of urokinase in lamellipodia formation by introduction of a non-native interdomain disulfide bond in its receptor. J Biol Chem. 286:43515–43526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gårdsvoll H, Werner F, Søndergaard L, Danø K, Ploug M. 2004. Characterization of low-glycosylated forms of soluble human urokinase receptor expressed in Drosophila Schneider 2 cells after deletion of glycosylation-sites. Protein Expr Purif. 34:284–295 [DOI] [PubMed] [Google Scholar]
- Hansen LV, Gårdsvoll H, Nielsen BS, Lund LR, Danø K, Jensen ON, Ploug M. 2004. Structural analysis and tissue localization of human C4.4A: a protein homologue of the urokinase receptor. Biochem J. 380:845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LV, Skov BG, Ploug M, Pappot H. 2007. Tumour cell expression of C4.4A, a structural homologue of the urokinase receptor, correlates with poor prognosis in non-small cell lung cancer. Lung Cancer. 58:260–266 [DOI] [PubMed] [Google Scholar]
- Hinck AP. 2012. Structural studies of the TGF-betas and their receptors—insights into evolution of the TGF-beta superfamily. FEBS Lett. 586:1860–1870 [DOI] [PubMed] [Google Scholar]
- Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li Y, Yuan C, Bian C, Chen L, et al. 2006. Structure of human urokinase plasminogen activator in complex with its receptor. Science. 311:656–659 [DOI] [PubMed] [Google Scholar]
- Jacobsen B, Muley T, Meister M, Dienemann H, Christensen IJ, Santoni-Rugiu E, Laerum OD, Ploug M. 2013. Ly6/uPAR-related protein C4.4A as a marker of solid growth pattern and poor prognosis in lung adenocarcinoma. J Thorac Oncol. 8:152–160 [DOI] [PubMed] [Google Scholar]
- Jacobsen B, Santoni-Rugiu E, Illemann M, Kriegbaum MC, Laerum OD, Ploug M. 2012. Expression of C4.4A in precursor lesions of pulmonary adenocarcinoma and squamous cell carcinoma. Int J Cancer. 130:2734–2739 [DOI] [PubMed] [Google Scholar]
- Kini RM, Doley R. 2010. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon. 56:855–867 [DOI] [PubMed] [Google Scholar]
- Kjaergaard M, Hansen LV, Jacobsen B, Gårdsvoll H, Ploug M. 2008. Structure and ligand interactions of the urokinase receptor (uPAR). Front Biosci. 13:5441–5461 [DOI] [PubMed] [Google Scholar]
- Konishi K, Yamamoto H, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Takao T, Doki Y, et al. 2010. Expression of C4.4A at the invasive front is a novel prognostic marker for disease recurrence of colorectal cancer. Cancer Sci. 101:2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz B, Kuhl A, Bayat B, Santoso S, Jenne DE. 2008. A hydrophobic patch on proteinase 3, the target of autoantibodies in Wegener granulomatosis, mediates membrane binding via NB1 receptors. J Biol Chem. 283:35976–35982 [DOI] [PubMed] [Google Scholar]
- Kriegbaum MC, Jacobsen B, Hald A, Ploug M. 2011. Expression of C4.4A, a structural uPAR homolog, reflects squamous epithelial differentiation in the adult mouse and during embryogenesis. J Histochem Cytochem. 59:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegbaum MC, Persson M, Haldager L, Alpízar-Alpízar W, Jacobsen B, Gårdsvoll H, Kjaer A, Ploug M. 2011. Rational targeting of the urokinase receptor (uPAR): development of antagonists and non-invasive imaging probes. Curr Drug Targets. 12:1711–1728 [DOI] [PubMed] [Google Scholar]
- Kurita A, Takizawa T, Takayama T, Totsukawa K, Matsubara S, Shibahara H, Orgebin-Crist MC, Sendo F, Shinkai Y, Araki Y. 2001. Identification, cloning, and initial characterization of a novel mouse testicular germ cell-specific antigen. Biol Reprod. 64:935–945 [DOI] [PubMed] [Google Scholar]
- Lee PY, Wang JX, Parisini E, Dascher CC, Nigrovic PA. 2013. Ly6 family proteins in neutrophil biology. J Leukoc Biol. 10.1189/jlb.0113014 [DOI] [PubMed] [Google Scholar]
- Llinas P, Le Du MH, Gårdsvoll H, Danø K, Ploug M, Gilquin B, Stura EA, Menez A. 2005. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 24:1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M. 2012. A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR). J Biol Chem. 287:34304–34315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo Y, Ben-Zeev B, Tabib A, Straussberg R, Anikster Y, Shorer Z, Fattal-Valevski A, Ta-Shma A, Aharoni S, Rabie M, et al. 2013. CD59 deficiency is associated with chronic hemolysis and childhood relapsing immune-mediated polyneuropathy. Blood. 121:129–135 [DOI] [PubMed] [Google Scholar]
- Nilton A, Oshima K, Zare F, Byri S, Nannmark U, Nyberg KG, Fehon RG, Uv AE. 2010. Crooked, coiled and crimpled are three Ly6-like proteins required for proper localization of septate junction components. Development. 137:2427–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret C, Hildebrand D, Weitz J, Kopp-Schneider A, Kuhn A, Beer A, Hautmann R, Zöller M. 2007. C4.4A as a candidate marker in the diagnosis of colorectal cancer. Br J Cancer. 97:1146–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug M. 2013. Structure-driven design of radionuclide tracers for non-invasive imaging of uPAR and targeted radiotherapy. The tale of a synthetic peptide antagonist. Theranostics. 3:467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug M, Ellis V. 1994. Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins. FEBS Lett. 349:163–168 [DOI] [PubMed] [Google Scholar]
- Rasch MG, Lund IK, Illemann M, Høyer-Hansen G, Gårdsvoll H. 2010. Purification and characterization of recombinant full-length and protease domain of murine MMP-9 expressed in Drosophila S2 cells. Protein Expr Purif. 72:87–94 [DOI] [PubMed] [Google Scholar]
- Syed MH, Krudewig A, Engelen D, Stork T, Klämbt C. 2011. The CD59 family member Leaky/Coiled is required for the establishment of the blood-brain barrier in Drosophila. J Neurosci. 31:7876–7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Zechner R. 2013. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 27:459–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.