Abstract
Sponges (phylum: Porifera) react to external light or mechanical signals with contractile or metabolic reactions and are devoid of any nervous or muscular system. Furthermore, elements of a photoreception/phototransduction system exist in those animals. Recently, a cryptochrome-based photoreceptor system has been discovered in the demosponge. The assumption that in sponges the siliceous skeleton acts as a substitution for the lack of a nervous system and allows light signals to be transmitted through its glass fiber network is supported by the findings that the first spicules are efficient light waveguides and the second sponges have the enzymatic machinery for the generation of light. Now, we have identified/cloned in Suberites domuncula two additional potential molecules of the sponge cryptochrome photoreception system, the guanine nucleotide-binding protein β subunit, related to β-transducin, and the nitric oxide synthase (NOS)–interacting protein. Cryptochrome and NOSIP are light-inducible genes. The studies show that the NOS inhibitor L-NMMA impairs both morphogenesis and motility of the cells. Finally, we report that the function of primmorphs to produce reactive nitrogen species can be abolished by a NOS inhibitor. We propose that the sponge cryptochrome-based photoreception system, through which photon signals are converted into radicals, is coupled to the NOS apparatus.
Keywords: sponges, Suberites domuncula, photoreception, phototransduction, cryptochrome, nitric oxide synthase-interacting protein, beta-transducin
Introduction
It is an inherent property of any living entity to respond to signals, stimuli, and cues from the environment with an adaptation of its internal metabolism and reactions. This circle is seen in unicellular systems (Luria and Delbrück 1943; Demerec 1950) as well as in multicellular organisms (Fregly and Blatteis 1996). Focusing on animals, already the earliest descendants of the hypothetical ancestor of all metazoans, the urmetazoan, the sponges (Müller et al. 2004), are provided with the main structural and functional molecules to maintain multicellularity. Sponges are lacking any extracellular muscles or any neuronal nexuses (Hyman 1940) but show, especially the larvae, the competence to react to light (Leys et al. 2002) and other cues (reviewed in Rivera et al. 2012). Before understanding light signal recognition in sponges, on a molecular level, the first neuronal receptor was cloned in the demosponge Geodia cydonium (Perović et al. 1999). The metabotropic glutamate/GABA-like receptor has been found to undergo sensitization to the excitatory amino acid glutamate, resulting in an increase in the intracellular calcium concentration [Ca2+]i. As a first molecule involved in light recognition in sponges, the cryptochrome has been cloned and functionally analyzed in the demosponge Suberites domuncula (Müller et al. 2010). This protein is different from the DNA photolyase that has been identified and characterized in the hexactinellid Aphrocallistes vastus (Schröder et al. 2003). Searches in sequence databases, including expressed sequence tags (ESTs) from S. domuncula (SpongeBase 2010) or genomic tags from the demosponge Amphimedon queenslandica (Srivastava et al. 2010), revealed that the opsin-based light sensory apparatus is missing in sponges, even though the covalently bound cofactor retinal is synthesized in S. domuncula (Müller, Binder, et al. 2011). Likewise, the master control gene, Pax6, for eye development in bilateria (Gehring and Seimiya 2010), has not been found in sponges. In turn, we proposed that, in sponges, the cryptochrome represents the (major) photoreceptive system (Müller et al. 2010), a finding that has been corroborated recently in the sponge A. queenslandica (Rivera et al. 2012). The experimental data gathered indicate that it is blue light that is most sensitively perceived by the cryptochrome system; this light spectral range is generated by the sponge luciferase system (Müller et al. 2009) and also exists in the marine twilight zone, where sponges live. The bioluminescence emission spectrum of the S. domuncula luciferase (at pH 8.0) ranges between 480 and 620 nm. The spicules from siliceous sponges allow the transmission of light within the wavelength ranges from 600 to 1300 nm (Müller, Wendt, et al. 2006); hence, the proposed coupling of luciferase-generated light to the spicules occurs within the “white light spectrum.” The expression of the cryptochrome gene is correlated with the light-dark cycle and is highest during the light phase (Müller et al. 2010). In the S. domuncula and also the A. queenslandica systems (Müller et al. 2010; Rivera et al. 2012), cryptochrome, with its flavin-based cofactor, is coupled to the siliceous spicular system. In S. domuncula, the skeletal elements, the monaxonial tylostyles, comprise dimensions of about 200 µm in length and 5 to 10 µm in diameter. These siliceous spicules have been proven to act as light waveguides (Cattaneo-Vietti et al. 1996; Aizenberg et al. 2005; Müller, Wendt, et al. 2006), allowing blue light to pass through. In sponges, the individual spicules are connected either by spongin-like fibrous molecules or DUF protein (Wang et al. 2010) (as in demosponges) or are even fused together, like in hexactinellids (see Wang et al. 2012). Our recent studies revealed that calcareous spicules, fabricated with silicatein from siliceous sponges, are characterized by exceptionally stabile and flexible properties (Natalio et al. 2013). These observations suggest that sponges can react to both exogenous light (environmental day light) and endogenous light (via the luciferase system; Müller et al. 2009). The latter reactivity was deduced from the finding that, in complete darkness, the sponge S. domuncula is flashing light, emanating from luciferase reactions (Müller et al. 2009), in a characteristic wave pattern (Wiens et al. 2010).
The available studies suggest that the cryptochrome not only exists intracellularly but also in the extracellular matrix (Müller et al. 2010). Studies with animals in vivo revealed that in the sponge Tethya aurantium, light is transmitted via the spicules into the central part of the animals (Brümmer et al. 2008). Under the supposition that sponge cryptochrome molecules are directly involved in photon harvesting, after passing of the photons through the spicules, a membrane-associated form must be postulated. Therefore, we approached this question by asking which molecules/proteins become associated with cryptochrome. Co-immunoprecipitation experiments presented here show that the S. domuncula cryptochrome becomes associated with a G protein β subunit (β-transducin) and one regulatory molecule of the nitric oxide synthase (NOS) pathways (the NOS-interacting protein).
The published data with S. domuncula (Müller et al. 2010) and A. queenslandica (Rivera et al. 2012) suggest that cryptochrome is the signal/photon receiver (photoreceptor), especially for blue light in sponges, like in plants and perhaps also in other, higher animal species (Lian et al. 2011). The subsequent question arises: By which mechanism do the light-absorbing cryptochromes, with their cofactor(s), transmit the primary environmental cues to the effector cells in sponges (phototransduction)? Photoreception/phototransduction should be fast and temporary to become effective. Surely, the reactions of the sponges to environmental stimuli are comparatively slow with respect to the signal transmission along the neuronal system. In sponges (e.g., Tethya wilhelma), mechanical stimuli spread over the surfaces with a velocity of 12.5 µm s−1 (Nickel 2004). In comparison, nerve conduction velocity, along some myelinated neurons, is much faster at speeds up to 120 m s−1 (Andrew and Part 1972). In turn, it can be assumed that the conversion of a photon signal of the light into a first step of the light transduction cascade might not involve electrochemical signals but rather hormone-like molecules or radicals (nitric oxide [NO]). These two latter signal transmission systems are substantially slower than neural impulses coupled to synaptic transmission (Pribyl et al. 2003; Utsumi et al. 2003). Cell biological evidence has been presented that both neurohormones (e.g., serotonin) (Weyrer et al. 1999) and also NO•-radical generating systems (Müller, Ushijima, et al. 2006) exist in sponges. We obtained the latter result with primmorphs, a special three-dimensional (3D) cell culture system, prepared from the demosponge S. domuncula (Müller et al. 1999).
In our previous report, we showed that, in primmorphs, NO• is synthesized by UV-B/H2O2-treated cells with the consequence of the induction of apoptosis (Müller, Ushijima, et al. 2006). This effect could be abolished by the NO-specific scavenger PTIO (2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide) (Lakshmi and Zenser 2007) and ethylene (Krasko et al. 1999). Nitric oxide is synthesized in metazoans by nitric oxide synthases (NOSes), which are divided into three isoforms (reviewed in Förstermann and Münzel 2006; Benarroch 2011): the NOS neuronal NOS (nNOS, NOS I), inducible NOS (iNOS, NOS II), and endothelial NOS (eNOS, NOS III). Although nNOS and eNOS are constitutively expressed and produce only low levels of NO, iNOS is inducible by cytokines and generates high NO levels. Besides being regulated at the gene expression level, NOS activity, especially eNOS and nNOS, is regulated via the nitric oxide synthase–interacting protein (NOSIP) (Dedio et al. 2001; Dreyer et al. 2004). NOSIP is a 34-kDa protein that interacts with eNOS and subsequently translocates eNOS from the cell membrane to the cell interior (Dedio et al. 2001). Through a similar molecular mechanism, NOSIP translocates nNOS from the cell membrane to the cytoplasm with functional consequences (Dreyer et al. 2004). In turn, NOSIP modulates both nNOS and eNOS activity (Dreyer et al. 2004; Yu et al. 2012). The NOSIP contains a RING finger domain through which it binds via zinc atoms to proteins (Borden and Freemont 1996), among them also to nNOS (Dreyer et al. 2004). Furthermore, NOSIP is an inducible protein (Yu et al. 2012).
The studies reported here show that, after light exposure, the primmorphs respond with an increased production of NO•. Parallel to this process, the membrane-bound cryptochrome is associated with NOSIP, suggesting that the cryptochrome pathway is coupled to the NO synthase signaling cascade. As initially described by us (Müller et al. 2010) and subsequently confirmed (Rivera et al. 2012), light response of the sponge cryptochrome results in a photoreduction, the photoresponsiveness of the flavoprotein, under formation of FADH2 (reduced flavin adenine dinucleotide) from flavin adenine dinucleotide (FAD). The reoxidation process of FADH2 to FAD is not yet understood in animals. In plants (Arabidopsis thaliana), experimental evidence has been presented that suggests that the reoxidation of the flavin involves molecular oxygen and an intermediate spin-correlated FADH-superoxide radical pair (FADH• and O2•−) (Weber et al. 2010; Müller and Ahmad 2011). Essentially, this reaction also takes place in the modular eukaryotic NOS isoforms that are composed of (1) the C-terminal reductase domain that binds reduced nicotinamide adenine dinucleotide phosphate (NADPH), flavin mononucleotide (FMN), and FAD and (2) the N-terminal oxygenase domain (see Förstermann and Münzel 2006). The latter domain carries a prosthetic heme group (reviewed in Förstermann and Münzel 2006; Ghosh et al. 2006) that provides an electron for reducing O2. On the basis of this evidence, we propose that the cryptochrome photoreduction/photoresponsiveness is intimately coupled to the NOS system via the FADH-superoxide radical in the S. domuncula system.
In the present study, we provide new evidence that, in S. domuncula, the cryptochrome system is coupled not only to the NOS pathway but also to G protein–coupled signal transduction. Transducin is the linker molecule between the photochemical reaction, cis-trans retinal isomerization by light, and the downstream signaling cascade. In mammalian systems, activation of the trimeric G protein transducin is initiated by the receptor-stimulated replacement by GTP of GDP bound to the α subunit in concert with the βγ heterodimer (Rondard et al. 2001). In addition to its function to dissociate dynamically from the inner cell membrane, the β subunit of transducin contributes not only to the activation of GTP hydrolysis but is also involved in the binding of the heterotrimeric G protein to the cell membrane (Gospe et al. 2011). In an earlier contribution, we showed that the trimeric G proteins are highly conserved within the metazoan kingdom from Porifera to mammals (Seack et al. 1998).
Materials and Methods
Chemicals, Materials, and Enzymes
The sources of the materials used for this study were provided previously (Müller, Wang, et al. 2011; Wang et al. 2013).
Sponges and Primmorphs
Specimens of the marine sponge S. domuncula (Porifera, Demospongiae, Hadromerida) were collected in the Northern Adriatic near Rovinj (Croatia). They were kept in aquaria in Mainz (Germany) at a temperature of 17C for more than 12 months (LePennec et al. 2003).
Primmorphs, the 3D cell aggregates, were prepared from single cells of S. domuncula (Müller et al. 1999). They were cultivated (at 19C) for up to 6 days in natural seawater (Sigma; Taufkirchen, Germany) supplemented with organic nutrients (0.2% RPMI 1640 medium; Gibco BRL, Eggenstein, Germany). After 2 days in culture, the cells formed 0.8- to 1-mm aggregates; they increased in size over the following 4 days. If not mentioned otherwise, the experiments described were performed with 6-day-old primmorphs.
Where indicated, the primmorphs remained in complete darkness or were illuminated with 30 lux, using a 15-watt Cool White fluorescent light (spectrum: 420–740 nm; Westinghouse Electric, Hamburg, Germany).
Total extracts of primmorphs were obtained by adding an equal amount (v/w) of 20 mM Tris-HCl buffer (pH 7.5; containing 100 mM KCl, 5 mM MgCl2, 5% v/v glycerol, and 10 µl/ml of the protease inhibitor mixture cOmplete ULTRA [Roche, Mannheim, Germany; 1 tablet/10 ml]) to the primmorphs. After two cycles of freezing and thawing, followed by homogenization (Downs homogenizer), the suspension was centrifuged (10,000 × g, 4C; 5 min) and the supernatant was collected (1.5 mg protein/ml−1).
Permissions: No specific permits were required for the described animal studies (locations/activities). The location of the collection site is not privately owned or protected in any way. The studies did not involve endangered or protected species.
NOS Activity
NOS activity in a living system was determined as described (Chiou et al. 1997) by applying the fluorimetric detection system FCANOS-1 (Sigma). Primmorphs were prepared and remained 6 days in the dark. Then the aggregates were incubated for 24 hr in the dark or exposed to 30 lux of white light. Samples of primmorphs were taken at time zero or after 3, 12, or 24 hr. The FCANOS-1 detection system was used to assay the NOS enzymatic activity in the cells according to the instructions of the manufacturer (Sigma). After incubation, the fluorescence intensity (caused by triazolofluorescein) was determined at the wavelength of excitation/emission (485 nm/530 nm). Parallel samples were used for determination of the DNA content of the cells in the assays. The data are given in relative fluorescence units (RFU) and were normalized to 1 µg of DNA. The compound NG-methyl-l-arginine (L-NMMA) acetate salt (M7033; Sigma), a competitive inhibitor of all three isoforms of NOS (eNOS, iNOS, and nNOS; Achike and Kwan 2003), at a concentration of 10 µM, was used as an inhibitor of the NOS activity (Akarid et al. 1995).
Time-Lapse Analysis of Aggregate Formation
The dynamics of primmorph formation was studied in rectangular well plates. The dissociated cells were suspended at a density of 2 × 106 cells/ml in 2-ml assays; the assays were exposed to 30 lux. After 1 day in culture, one series remained in the absence and the parallel series in the presence of the NOS inhibitor L-NMMA (10 µM). Time-lapse microscopy was conducted using a Wild binocular microscope M11-59316 (Wild, Heerbrugg, Switzerland), equipped with a Nikon D4 camera (Nikon; Tokyo, Japan). Photomicrographs were taken every 20 min.
NO Quantification
The method applied uses the Griess detection system (Chiou et al. 1997; Miranda et al. 2001). The primmorphs (6 days) were incubated in 96-well plates (Greiner; Frickenhausen, Germany) in 150 µl culture medium in the dark or exposed to light. After the indicated period (0 hr or after 3, 12, or 24 hr), 100-µl aliquots were removed and added to the Griess reagent (G441; Sigma) and the absorbance was measured at 540 nm in a microplate reader. NO production was calculated from a standard calibration curve (Amano and Noda 1995).
S. domuncula NOSIP Protein
The S. domuncula sponge database (SpongeBase 2010) contains several ESTs, comprising almost the complete coding region for the NOSIP, the NOS interacting protein. The complete complementary DNA (cDNA) spanning the putative NOSIP was obtained by application of the 3′- and 5′-racing technique using the CapFishing Full-length cDNA Premix Kit (Seegene; Rockville, MD) as described (Simkin et al. 2004). The 1074-nt-long cDNA without the poly(A) tail, termed SDNOSIP-r, comprised between nt16-18 and nt928-930(stop) the complete open reading frame (ORF) for the deduced polypeptide, termed NOSIP-r_SUBDO.
S. domuncula TBLR-Related Protein
The cDNA, encoding the sponge transducin β-like protein (TBLR protein), was likewise extracted from the sponge database (SpongeBase 2010). The EST tags were again completed by the racing technique. The nt sequence comprising the complete ORF for the TBLR protein, a guanine nucleotide-binding protein subunit β (GNB), was termed SDGNB-Trans. This cDNA is 1599 nt long. The ORF of the deduced TBLR protein, termed GNB-Trans_SUBDO, spans nt280 to nt1233-1235 within the cDNA.
Sequence Analyses
Similarity searches were conducted through servers at the European Bioinformatics Institute (Hinxton, UK) and the National Center for Biotechnology Information (NCBI; Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/). The multiple alignments were run with ClustalW version 1.6 (Thompson et al. 1994), and the resulting phylogenetic trees were constructed by applying the neighbor-joining method to distance matrices with the Dayhoff PAM matrix model (Dayhoff et al. 1978; Saitou and Nei 1987). The degree of support for internal branches was assessed by bootstrapping (Felsenstein 1993). The graphical output of the bootstrap figures was produced through the “Treeview” software and GeneDoc (Nicholas and Nicholas 1997).
Preparation of Recombinant NOSIP and TBLR Protein and Antibodies
The recombinant proteins were obtained by expression of the complete ORF in the Gateway System from Invitrogen (Karlsruhe; Germany). The cDNAs, SDNOSIP-r and SDGNB-Trans, were amplified by standard polymerase chain reaction (PCR) as described (Natalio et al. 2010). The attB PCR products, together with the attb recombination sites, were purified from the gel using the “NucleoSpin Extract II” (Macherey-Nagel; Düren, Germany) according to the instruction of the manufacturer. The BP recombination reaction was performed with the obtained attB PCR products (including the respective cDNA fragment) and using the pDONR 221 entry vector (Invitrogen). This construct was used to transform TOP10 cells (Invitrogen). Positive clones were transformed into competent Escherichia coli BL21 AI One Shot cells (Invitrogen). The cells were incubated in LB medium with 50 µg/ml carbenicillin (Roth; Karlsruhe, Germany) at 37C, and the recombinant protein was produced (Müller et al. 2012). Analyses of the recombinant proteins, r-NOSIP-r_SUBDO and r-GNB-Trans_SUBDO, were performed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant proteins had a purity of >95%, as checked by SDS-PAGE.
Polyclonal antibodies (PoAb) were raised in female rabbits (White New Zealand) as described (Müller et al. 2005; Wiens et al. 2007) and using r-NOSIP-r_SUBDO or r-GNB-Trans_SUBDO as antigens. The titer of the two produced PoAb sera (termed PoAb-r-NOSIP-r_SUBDO and PoAb-r-GNB-Trans_SUBDO) was determined and found to be >1:5000. As controls for the western blot analyses and the immunoprecipitation studies, adsorbed antibodies were used. Those samples were prepared as described (Schröder et al. 2006). PoAb sera (100 µl), both r-NOSIP-r_SUBDO and r-GNB-Trans_SUBDO, were incubated with 20 µg of the respective recombinant antigen (r-NOSIP-r_SUBDO or r-GNB-Trans_SUBDO) for 1 hr (4C). Then the samples were centrifuged (10,000 × g, 4C; 5 min), and the adsorbed antibodies were collected. These preparations failed to recognize the antigen as analyzed by western blotting or by immunofluorescence studies (not shown). As further controls, preimmune sera were used and showed no antibody-antigen signals.
Antibodies against S. domuncula Cryptochrome
Recombinant S. domuncula cryptochrome was prepared in E. coli and used as antigen to raise PoAb (PoAb-r-CRY_SUBDO) as described (Müller et al. 2010).
Immunohistology
Tissue slices were prepared as described (Müller et al. 2005). Slices of 8 µm were fixed in paraformaldehyde. After washing with phosphate-buffered saline (PBS), unspecific binding was blocked with 2% (v/v) goat serum (Invitrogen). Subsequently, the specimens were incubated with the primary antibodies, rabbit anti-cryptochrome (PoAb-r-CRY_SUBDO), anti-NOSIP (PoAb-r-NOSIP-r_SUBDO), or anti-GNB-Trans (PoAb-r-GNB-Trans_SUBDO), diluted by 1:1500 in blocking solution and incubated while shaking at 4C overnight. As controls, the adsorbed sera were used. Unbound antibodies were removed by washing four times with PBS prior to the incubation with fluorescently labeled (Cy5 [red] or Cy3 [green] fluorescent) (Dianova; Hamburg, Germany) secondary antibodies (dilution 1:3000). Parallel slices were stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma). Then, the slices were inspected with an Olympus AHBT3 microscope (Olympus Europa, Hamburg, Germany), where indicated specimens were inspected under immunofluorescent light at an excitation light wavelength of 546 nm (Cy3-stained structures) or 490 nm (DAPI).
SDS-PAGE and Western Blot Analysis
SDS-PAGE was performed as described (Laemmli 1970; Schröder et al. 2006). Samples of 5 to 8 µg of protein were mixed with loading buffer (Roti-Load; Roth), boiled for 8 min, and subjected to SDS-PAGE (12% acrylamide and 0.1% SDS). The gels were stained with Coomassie brilliant blue. The protein size standard “Dual Color” (Roth) was used to estimate protein sizes. For western blot analyses, size-separated proteins were transferred to PVDF-Immobilon membranes (Wiens et al. 1998). Subsequently, the membranes were blocked at room temperature with blocking solution (Roche; 1% [v/v] in TBS-T buffer; 20 mM Tris-HCl [pH 7.6], 137 mM NaCl, and 0.1% [v/v] Tween-20). Then, the blots were incubated with PoAb-r-NOSIP-r_SUBDO or PoAb-r-GNB-Trans_SUBDO (1:5000 dilution in TBS-T, supplemented with 0.1% [v/v] blocking solution), followed by alkaline phosphatase (AP)–conjugated species-specific secondary antibodies (1:4000 dilution) and 4-nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Invitrogen).
Co-immunoprecipitation
The co-immunoprecipitation experiments were performed as described (Wang and Elion 2003; Schröder et al. 2006). Primmorph samples were homogenized (1:1 w/v) in 20 mM Tris-HCl buffer, containing the protease inhibitor mixture cOmplete ULTRA. The extract was centrifuged at low speed (1300 × g, 4C; 3 min) and the supernatant was collected. The particle-free supernatant (100 µl) was subjected to 10 µl PoAb-r-GNB-Trans_SUBDO coupled to Protein A/G Plus agarose beads (Pierce-Thermo Fisher Scientific; Rockford, IL). These beads were prepared by incubating (2 hr; 4C) them in undiluted PoAb-r-GNB-Trans_SUBDO serum. The suspension of the anti–GNB-Trans antibodies, coupled to Protein A/G beads, was incubated for 2 hr (4C) and then centrifuged (2000 × g; 5 min; 20 sec); the bound proteins were eluted from the beads with 4× SDS-PAGE sample buffer and analyzed by immunoblot.
Cell Membranes
Cell membranes were prepared from primmorphs following a described procedure (Warren et al. 1966; Leyhausen et al. 1983). Six-day-old primmorphs were used for the experiments; they remained in the dark the last 3 days prior to the experiment. Then these 3D aggregates were exposed to light for up to 8 hr. After gently breaking the cells, the membranes were centrifuged (60,000 × g, 2 hr; Beckman Allegra 64R; Beckman Coulter, Brea, CA) through a sucrose step-gradient and collected on the 50% sucrose layer. Membranes were suspended in 20 mM Tris-HCl buffer; the samples had a protein content of approximately 150 µg/ml. The membrane-free, soluble fraction was taken from the surface of the 10% sucrose upper layer (≈100 µg/ml of protein). Aliquots (10 µl) of those samples were then analyzed by 12% SDS-PAGE, with the exception that samples were not boiled prior to gel loading. Proteins were visualized with Coomassie blue staining (Rath et al. 2009). Then the proteins were transferred onto the membrane and subjected to western blot analysis.
Quantitative Real-Time RT-PCR (qRT-PCR)
The (absolute) steady-state expression levels of the genes encoding for GNB-Trans (SDGNB-Trans), NOSIP (SDNOSIP-r), and cryptochrome (SDCRYPTO) were determined by quantitative real-time RT-PCR (qRT-PCR) as described (Müller et al. 2010; Wiens et al. 2010). In short, after extraction of RNA and its treatment with DNAse to eliminate genomic DNA, first-strand cDNA synthesis was performed using SuperScript III Reverse Transcriptase in a reaction mixture containing 5 µg of total RNA, dNTPs, oligo(dT)18, and reverse transcriptase buffer (Invitrogen) at 42C for 1 hr. Then the reverse transcriptase was inactivated (65C, 15 min) and the reactions were diluted; 2 µl of those dilutions was used as a template for the 30-µl reaction for qPCR. They were run in an iCycler (Bio-Rad; München, Germany) using 1/10 serial dilutions in triplicate as described (Livak and Schmittgen 2001). Each reaction contained “Absolute Blue SYBR Green” master mixture (ABgene; Hamburg, Germany) and 5 pmol of each primer pair. All reactions were run with an initial denaturation at 95C for 10 min, followed by 40 cycles each at 95C for 20 sec, then 60C for 20 sec, and 72C for 35 sec. The following primers were used for amplification:
SDGNB-Trans: forward, 5′-GCTTTGTGGCTTGTGGTGGTCTG-3′ (nt541 to nt563), and reverse, 5′-ACAAGACGAGAGGTAGCCAGTGTG-3′ (nt659 to nt636), with a PCR product size of 119 bp
SDNOSIP-r: forward, 5′-CATCGTGTCCAAGCCAATCAACCC-3′ (nt375 to nt398), and reverse, 5′-AGATCGTTGATCTTCTGCCGACCC-3′ (nt492 to nt469), with a PCR product size of 118 bp
SDCRYPTO (accession number FN421335): forward, 5′-GGATGACCCGACTAGAAAAGCA-3′ (nt963 to nt942), and reverse, 5′-CATTGTTGCTCTCCCAAGGTAG-3′ (nt1030 to nt1009), with a PCR product size of 68 bp
As a reference housekeeping gene, we have used theS. domuncula glycerol 3-phosphate dehydrogenase (SDGAPDH; AM902265.1) and the following primers: forward, 5′-TCCAAACCAGCCAAGTACGATG-3′ (nt816 to nt837), and reverse, 5′-AGTGAGTGTCTCCCCTGAAGTC-3′ (nt945 to nt924), with a fragment size of 130 bp. After qRT-PCR, the threshold position was set to 50.0 RFU above the PCR-subtracted baseline for all runs. Mean Ct values and efficiencies were calculated by the iCycler (Bio-Rad) software. Relative expression level was calculated by EGAPDHCtGAPDH / ESDGNB-Trans(SDNOSIP-r,SDCRYPTO)CtSDGNB-Trans(SDNOSIP-r,SDCRYPTO), where E is the PCR efficiency and Ct is the threshold cycle.
Further Methods
For quantification of protein the Bradford method (Compton and Jones 1985), Roti-Quant solution (Roth) was used. Statistical analysis was performed with the paired Student’s t-test (Sachs 1984).
Results
S. domuncula and Its Primmorphs
The in vitro 3D cell culture system (primmorphs) was used to determine the effect of light on the metabolic response in sponges. The primmorphs were prepared from single cells (Fig. 1A). Three days after starting aggregate formation, the primmorphs grew to ~1 mm (Fig. 1B) and increased to ~2 to 3 mm during a total incubation period of 6 days (Fig. 1C).
Figure 1.
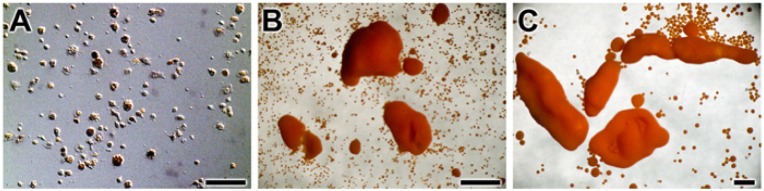
Primmorph formation from single cells of the sponge Suberites domuncula. Single sponge cells (A; scale bar = 50 µm) form after 3 days irregularly shaped primmorphs (B; scale bar = 1 mm) that round to more spherical aggregates after a total incubation period of 6 days (C; scale bar = 1 mm).
Total NOS Activity in Cells of Primmorphs Dependent on Light Exposure
Primmorphs were allowed to be formed during a 6-day incubation period in the dark. Then samples remained in the dark for the subsequent 0 to 24 hr of incubation or were exposed to light (30 lux, white light) for the same periods. The NOS activity in these aggregates was determined by application of the fluorimetric detection system. The primmorphs were incubated with the cell-permeable diacetate derivative of 4,5-diaminofluorescein, and the fluorescence was measured. The relative values obtained were subsequently correlated to the DNA content in the samples to obtain absolute values. The data revealed that during the selected incubation period of 24 hr, no statistically significant change of the intracellular NOS activity was seen; values around 0.14 RFU/µg DNA were measured (Fig. 2). The intracellular NOS activity was abolished by 70% after incubating the aggregates for 24 hr with 10 µM of the NOS inhibitor L-NMMA (Fig. 2).
Figure 2.
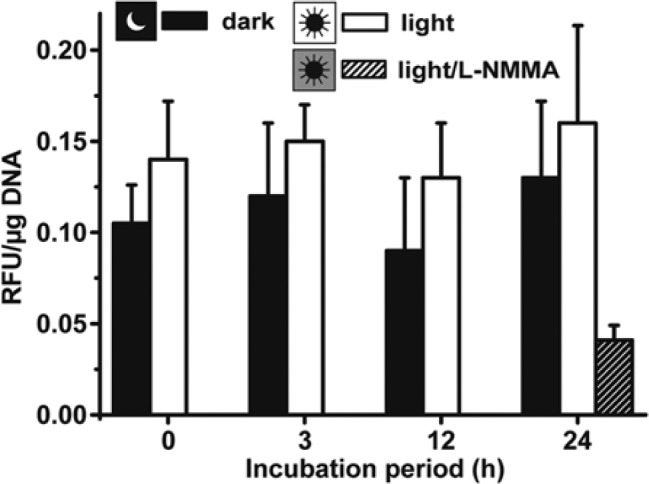
Determination of total nitric oxide synthase (NOS) activity in primmorphs, kept under dark (dark) or in the light (light). Six-day-old primmorphs that were formed in the dark remained for the subsequent 24 hr in the dark or were transferred to light. Then the total intracellular NOS activity was determined, as described in the Materials and Methods. In one control series, primmorphs were kept for 24 hr in the light and additionally exposed to 10 µM of the NOS inhibitor L-NMMA (light/L-NMMA). Results are means (± SD) from six separate experiments. RFU, relative fluorescent units.
Increased NO Production by Primmorphs during Incubation in the Light
The accumulation of NO in the culture medium of 6-day-old primmorphs kept in the dark was low compared with those that had been exposed to light. In the dark (time: 0 hr), the NO level increased from <1 nmol/µg DNA only to 2.2 ± 0.4 nmol/µg at 3 hr and finally to 4.7 ± 0.9 nmol/µg after the 24-hr incubation period (Fig. 3). However, if the aggregates were kept under light, a significantly higher NO level was seen even after an incubation period of 3 hr with 4.1 ± 0.9 nmol/µg, a value that strongly and significantly further increased to 24.3 ± 4.8 nmol/µg (Fig. 3).
Figure 3.
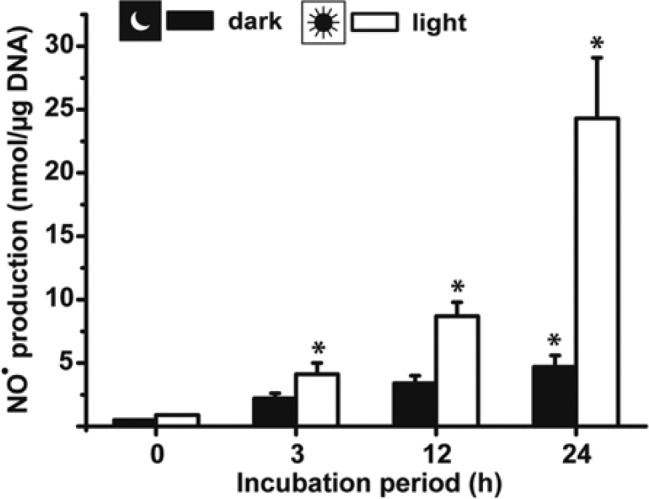
Time-dependent increase in NO• in supernatants of primmorph culture. The 6-day-old aggregates were incubated for 0, 3, 12, or 24 hr either in the dark or in the light. Subsequently, the culture supernatants were collected and nitric oxide (NO) was quantified by the Griess detection system. Results are means (± SD) from six separate experiments; *p<0.05.
Effect of L-NMMA on the Motility of Cells: Time-Lapse Analysis of Aggregate (Primmorph) Formation
NO• is known to display a modulating effect on morphogenetic processes (Diwan et al. 2010). During sponge primmorph formation, the cells migrate to each other, then adhere to each other to form functionally active aggregates (Müller et al. 2000). To elucidate if the NO• signaling molecule has a related function in the sponge primmorph system, the aggregating cells were exposed to the NOS inhibitor L-NMMA (10 µM) one day after starting the experiments.
In the experiment shown in Fig. 4, dissociated cells were allowed to form aggregates during a 24-hr period. Then one series remained in the absence of the NOS inhibitor for the subsequent 200 min, while the parallel aggregates were supplemented with 10 µM L-NMMA. The photomicrographs show that, in the absence of the inhibitor, the cell aggregates migrated to each other, forming up to 0.6-mm primmorphs (Fig. 4A–J, left panel). In contrast, if the aggregates were treated with L-NMMA, a distinct retardation of primmorph formation was seen (Fig. 4A–J, right panel). Only infrequently were primmorphs seen larger than 0.3 mm during the incubation period.
Figure 4.
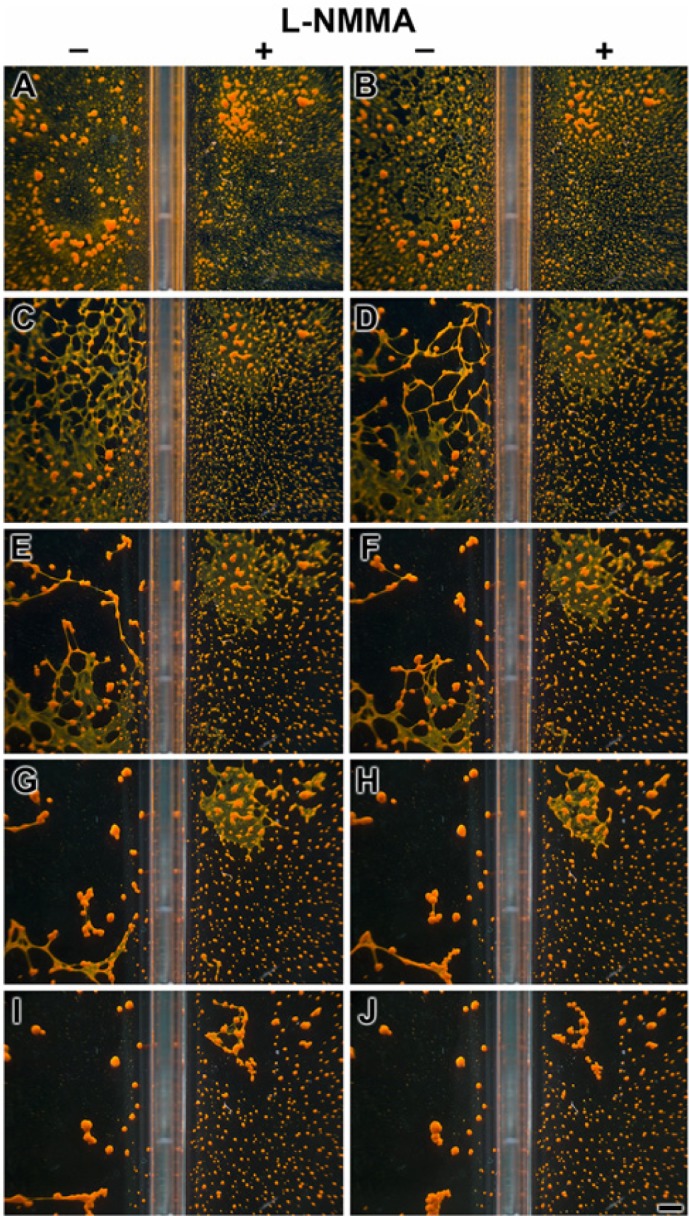
Delayed formation of primmorphs in the presence of the nitric oxide synthase (NOS) inhibitor L-NMMA (10 µM). One day after starting the reaggregation of dissociated cells (performed under 30 lux of light), the assays, performed in parallel, were started in rectangular well plates; one series remained in the absence of the inhibitor, while the second one was supplemented with 10 µM L-NMMA, as indicated. Photomicrographs were taken every 20 min. Scale bar = 1 mm.
Cloning of the S. domuncula NOSIP Protein
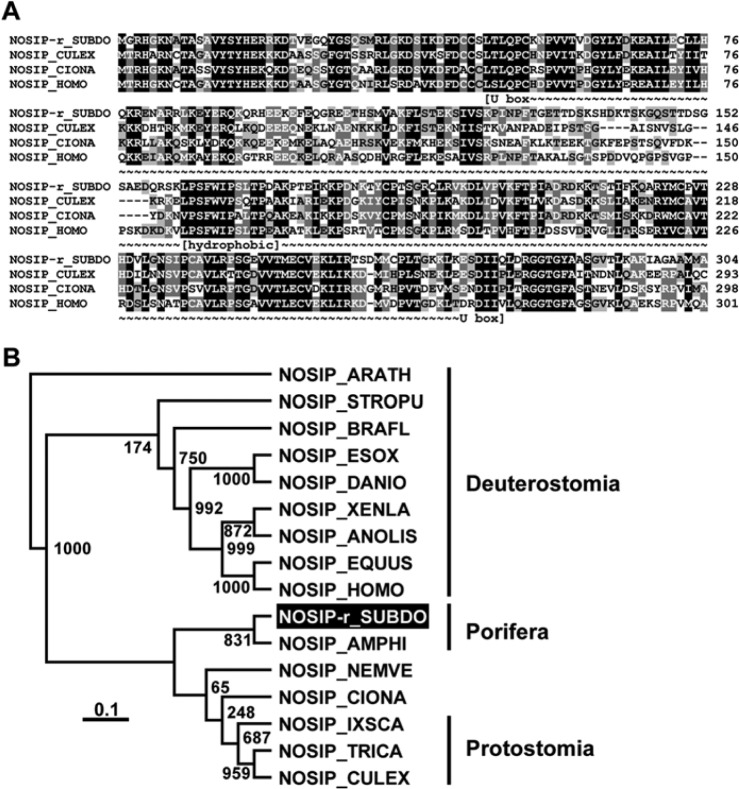
The complete polypeptide for the NOSIP protein was deduced from the corresponding S. domuncula cDNA SDNOSIP-r. The 304–amino acid (aa)–long deduced protein (NOSIP-r_SUBDO) has a calculated size of 33.9 kDa and a theoretical pI (isoelectric point) of 9.06. Between aa 48 and aa 278, the conserved “modified RING finger domain” (U box) exists, which shows a high similarity to the RING consensus sequences (Aravind and Koonin 2000) with an “expect value” E = 4.80e−08 (Coligan et al. 2000). Members of the U-box family of proteins have been reported to interact with chaperones, suggesting that the function of those proteins is to control degradation of unfolded/misfolded proteins (Hatakeyama et al. 2004). At least one hydrophobic cluster exists within the conserved parts of the protein (aa 161 to aa 173), which is conserved in the metazoan NOSIP sequences and is likewise predictive for docking to membrane protein structures (Bordner 2009) (Fig. 5A).
Figure 5.
The Suberites domuncula nitric oxide synthase–interacting protein (NOSIP) (NOSIP-r_SUBDO). (A) The deduced sponge NOSIP is aligned with related sequences from Culex quinquefasciatus (NOSIP_CULEX; accession number XP_001865022.1), Ciona intestinalis (NOSIP_CIONA; XP_002122091.1), and human (NOSIP_HOMO; NP_057037.1). The U box consensus (~~~) and the predicted membrane docking segment of the protein (hydrophobic) are marked. Residues conserved (similar or related with respect to their physicochemical properties) in all sequences are shown in white on black, and those in at least three sequences in black on gray. (B) These four sequences are compared with the related sequences from Equus caballus (NOSIP_EQUUS; XP_001492183.1), the bird Anolis carolinensis (NOSIP _ANOLIS; XP_003222746.1), the fishes Esox lucius (NOSIP_ESOX; ACO13473.1), and Danio rerio (NOSIP_DANIO; NP_001007435.1), as well as to NOSIP from Xenopus laevis (NOSIP_XENLA; NP_001084604.1), then Branchiostoma floridae (NOSIP_BRAFL; XP_002608258.1) and Strongylocentrotus purpuratus (NOSIP_STROPU; XP_790354.2), the insects Ixodes scapularis (NOSIP_IXSCA; XP_002434038.1) and Tribolium castaneum (NOSIP_TRICA; XP_966634.1), and the sequences from Nematostella vectensis (NOSIP_NEMVE; XP_001632123.1) and Amphimedon queenslandica (NOSIP_AMPHI; XP_003385646.1). The plant sequence from Arabidopsis thaliana (NOSIP_ARATH; NP_564781.1) was used as an out-group to root the tree. The scale bar indicates an evolutionary distance of 0.1 amino acid substitutions per position in the sequence.
High sequence similarity of the sponge NOSIP-r_SUBDO exists for the related sequences from all metazoans listed in the databases. Some selected sequences have been included in the alignment and the tree. The phylogenetic tree separates the sequences of the Protostomia (e.g., Culex quinquefasciatus, which has identical/similar aa of 49%/66%) from the Deuterostomia (e.g., a human sequence of 51%/66%). From the protostomians, the two poriferan sequences branch off (Fig. 5B).
Sponge GNB-Trans/TBLR-Related Protein
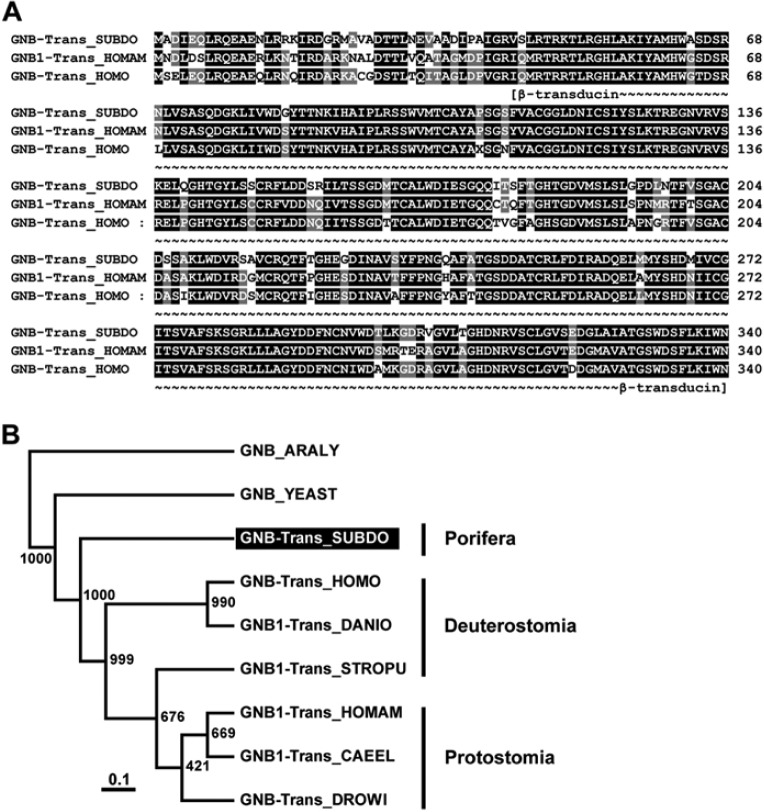
The complete β subunit of transducin, GNB-Trans_SUBDO, was deduced from the isolated cDNA SDGNB-Trans. The 340-aa-long polypeptide has a size of 37.1 kDa and a pI of 5.75. The highest sequence similarity was found for the metazoan transducin β-like proteins (TBLR proteins), with an identical/similarity score of >78%/89%. This high sequence relationship is demonstrated by the alignment of the sponge polypeptide with the related proteins from humans and the American lobster Homarus americanus (Fig. 6A). The WD or β-transducin repeat within the sponge sequence is found between aa 45 and aa 340 with an E value = 2.8e−10; as expected, the six Trp-Asp (WD) repeat profile segments are interspersed within the β-transducin repeat (Gilman 1987; Seack et al. 1998).
Figure 6.
The Suberites domuncula putative transducin β subunit (GNB-trans_SUBDO). (A) The deduced sponge polypeptide GNB-Trans_SUBDO was aligned with the related Gβ-subunit from human (GNB-Trans_HOMO; AAA35922.1) and the American lobster Homarus americanus (GNB1-Trans_HOMAM; O45040.1). Residues conserved (identical or similar with respect to their physicochemical properties) in all three sequences are shown in white on black; those that share similarity to at least two residues are shown in white on gray. The regions spanning the β-transducin repeats (β-transducin) are marked. (B) These three proteins were compared with the related Gβ-subunits from the fish Danio rerio (GNB1-Trans_DANIO; NP_997774), insect Drosophila willistoni (GNB-Trans_DROWI; XP_002075419.1), nematode Caenorhabditis elegans (GNB1-Trans_CAEEL; NP_001254312.1), sea urchin Strongylocentrotus purpuratus (GNB1-Trans_STROPU; XP_001176793.1), and yeast Saccharomyces cerevisiae (GNB_YEAST; AAA35114.1). The protein, from the plant Arabidopsis lyrata (GNB_ARALY; XP_002867119.1), was used as an out-group to root the tree. Scale bar indicates an evolutionary distance of 0.1 amino acid substitutions per position in the sequence.
The phylogenetic analysis was performed with selected metazoan sequences and the yeast (Saccharomyces cerevisiae) and plant (Arabidopsis lyrata) GNB-Trans-related polypeptides (Fig. 6B). Again, a distinct separation of the S. domuncula branch from the protostomian and deuterostomian GNB sequences was resolved.
Recombinant Proteins and Antibodies
Recombinant sponge proteins were obtained by heterologous expression of the complete ORF of the respective cDNA (SDNOSIP-r and SDGNB-Trans) in E. coli, as described in the Materials and Methods. The proteins (r-NOSIP-r_SUBDO and r-GNB-Trans_SUBDO) were isolated and purified to >95%, as assessed by SDS-PAGE. Then polyclonal antibodies were raised in rabbits; the two PoAb sera samples, PoAb-r-NOSIP-r_SUBDO and PoAb-r-GNB-Trans_SUBDO, had a titer of >1:5000. Primmorph extracts were prepared and subjected to western blot analysis to identify the wild-type NOSIP and TBLR protein. The extracts were size separated (Fig. 7A, lane b), the proteins were blotted, and the membranes were probed with either PoAb-r-NOSIP-r_SUBDO (Fig. 7B, lane a) or PoAb-r-GNB-Trans_SUBDO (Fig. 7C, lane a). The results show that the anti-NOSIP-r_SUBDO antibodies recognized a 34-kDa polypeptide, matching the size of the protein deduced from the respective cDNA. The antiserum against GNB-Trans_SUBDO recognized the 37.5-kDa polypeptide, again as expected from the cDNA SDGNB-Trans. The adsorbed antisera did now show any signal after blotting. Likewise, the preimmune sera showed no signals on the blots (data not shown).
Figure 7.
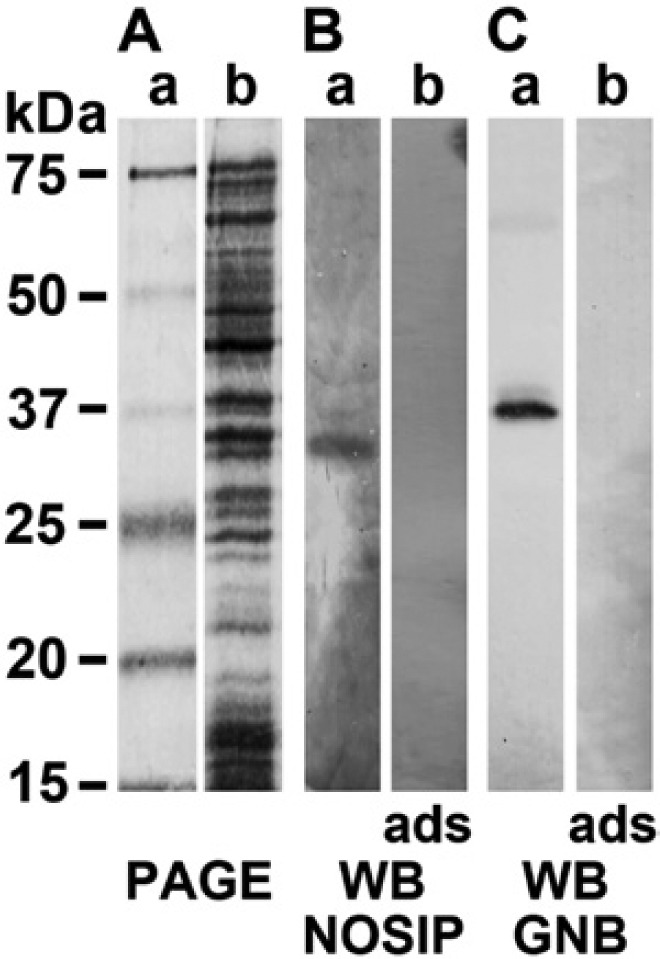
Detection of wild-type nitric oxide synthase–interacting protein (NOSIP) and TBLR protein in extracts, prepared from primmorphs. (A) Proteins were extracted from 6-day-old primmorphs and were size-separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE); the gels were stained with Coomassie brilliant blue (lane a). (B) The wild-type NOSIP protein was detected by western blotting (WB) using PoAb-r-NOSIP-r_SUBDO antibodies. The 34-kDa polypeptide was detected (lane a); no antigen-antibody signals are seen if the membranes are incubated with adsorbed (ads) antibodies (lane b). (C) The transducin β-subunit (GNB) was identified in the extract as a 37.5-kDa polypeptide using the PoAb-r-GNB-Trans_SUBDO antiserum (lane a); the adsorbed sample showed no reaction (lane b). In A, lane a, the size marker used is shown.
In Situ Localization of Cryptochrome, GNB-Trans, and NOSIP
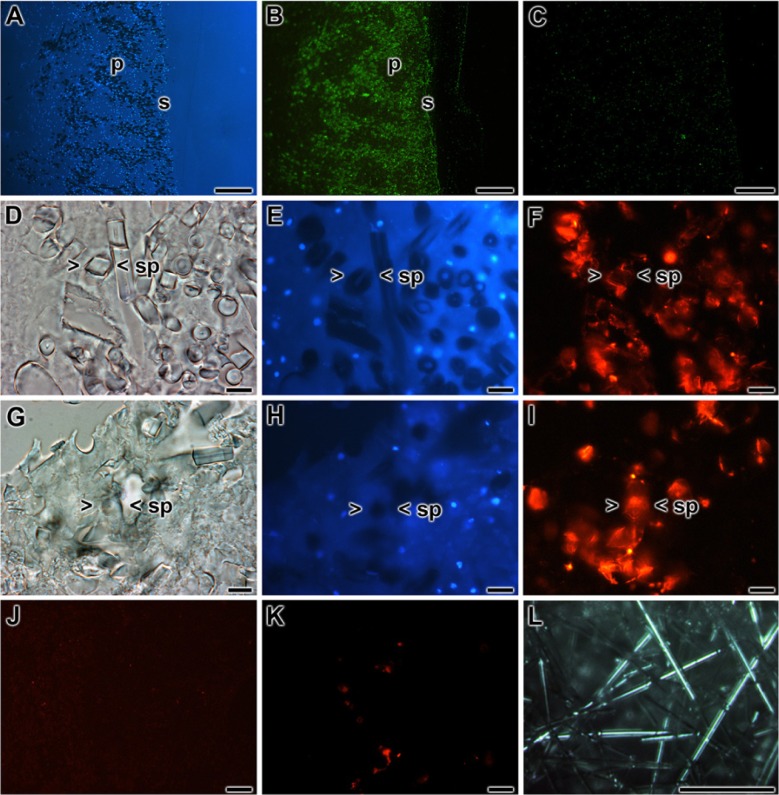
Using the antibodies developed here, together with those raised earlier, the localization of the molecules under discussion within tissue or primmorphs was determined. In accordance with the data given earlier (Müller et al. 2010), cryptochrome (using the PoAb-r-CRY_SUBDO as a tool) was localized around the spicules and especially around those localized in the surface pinacoderm region within the sponge tissue (Fig. 8A,B). Adsorbed antibodies failed to show a signal (Fig. 8C). The reactivity of the antibodies, raised in the present study against GNB-Trans and NOSIP, was determined in primmorphs. According to the immunofluorescence images, the highest antigen levels were found around the spicules (Fig. 8F, I); those images were compared with a bright-field (Fig. 8D, G) and DAPI-stained (Fig. 8E, H) photomicrograph to discriminate between spicules and cells. In controls, using adsorbed antibodies, it was established that the antibody-antigen reactions were specific for the anti-GNB-Trans (Fig. 8J) and anti-NOSIP antibodies (Fig. 8H) used.
Figure 8.
Localization of cryptochrome, GNB-Trans, and nitric oxide synthase–interacting protein (NOSIP) in tissue of Suberites domuncula in the area around the spicules. The sections through the tissue or the primmorphs are stained with antibodies or reacted with DAPI. (A, B) The highest levels of cryptochrome within the sponge tissue are found in the surface (s) region of sponge tissue, the pinacoderm (p). (A) Slices through this region show the (B) highest immune signals coming from the reaction between the cell’s antigen and anti-cryptochrome antibodies. The antibody-antigen complexes are stained with Cy3-labeled secondary antibodies. (C) Immune staining with adsorbed antibodies did not result in a significant lighting up. (D–F) Reaction of slices through primmorphs. (D) Bright-field image through a primmorph; (E) the same region is stained with DAPI to highlight the nuclei. (F) Distinct staining of the areas around the spicules after immune staining with antibodies against GNB-Trans (a Cy5-labed secondary antibody was used). The same region around a spicule (> <sp) is marked. (G–I) Reactivity within primmorph toward anti-NOSIP (detection with Cy5-labed secondary antibodies). (G) Slices (bright-field image) were incubated with (H) DAPI or (I) anti-NOSIP antibodies. Again, the areas framing spicules are highlighted (one spicule is marked: > <sp). (J, K) Controls were performed with adsorbed antibodies against GNB-Trans (J) or NOSIP (K). (L) Light transmission guided through isolated tylostyles, monaxonal spicules from S. domuncula; they had been illuminated with a white light beam. (A–C) Scale bars = 100 µm; (D–K) scale bars = 10 µm; (L) scale bar = 100 µm.
Spicules as Light Waveguide
The monaxonial tylostyles, the skeletal elements of the siliceous sponge S. domuncula, comprised a length of 150 to 320 µm and an average diameter of 6 µm. After orienting a white light beam toward the end of the spicules, the light was guided through the longitudinal axis of the spicules (Fig. 8L).
Influence of Light on the Expression of GNB-Trans, Cryptochrome, and NOSIP
Dark-adapted primmorphs were exposed to light. After 0, 1, 2, and 4 hr of light exposure, the primmorphs were collected and RNA was extracted. Using these samples, the steady-state expression of GNB-Trans, cryptochrome, and NOSIP was determined by qRT-PCR (Fig. 9). The expression values were normalized for the expression of the transcripts of the housekeeping gene GAPDH. As shown in Fig. 9, the expression level of GNB-Trans did not change significantly during the 0 to 4 hr of light exposure. In contrast, the expression levels of the cryptochrome and NOSIP genes were low immediately after termination of the dark period; for cryptochrome, the level was determined to be 0.019 ± 0.005 with respect to GAPDH, and the level for NOSIP was 0.005 ± 0.001. These levels significantly increased after the short incubation period of 1 hr and reached expression levels of 0.031 ± 0.007 for cryptochrome and 0.012 ± 0.003 for NOSIP. These values increased further during the 4-hr light exposure period (Fig. 9).
Figure 9.
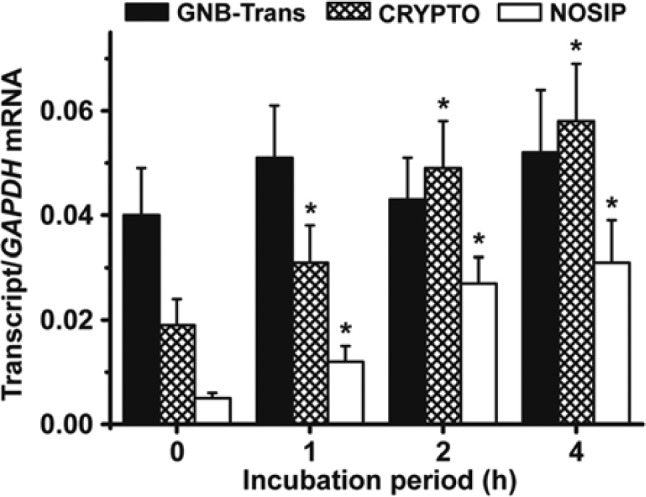
Effect of light on the steady-state expression of the genes encoding for sponge GNB-Trans (SDGNB-Trans), cryptochrome (SDCRYPTO), and nitric oxide synthase–interacting protein (NOSIP) (SDNOSIP-r). Six-day-old primmorphs, kept for the last 3 days in the dark, were exposed to light for 0, 1, 2, or 4 hr. Then, RNA was isolated and quantitative real-time PCR was used to determine the steady-state expression levels of SDGNB-Trans (closed bars), SDCRYPTO (hatched bars), SDNOSIP-r (open bars), and SDGAPDH. The latter ones were used as reference for normalization. Each data point represents the messenger RNA (mRNA) level of the respective expressed gene normalized to the amount of GAPDH transcripts, and results are means ± SD (five experiments per time point); p<0.05.
Co-immunoprecipitation Analysis
Based on the data summarized in the previous paragraph, it is reasonable to assume that the steady-state transcript level of GNB-Trans is not extensively and significantly different in the dark or during light exposure. Therefore, we selected this molecule as a “reference protein” for co-immunoprecipitation.
Extracts were prepared from 6-day-old primmorphs that were kept for the last 3 days in the dark. The particle-free supernatants were obtained and subjected to A/G agarose beads and coated with anti–GNB-Trans antibodies. After incubation, the beads were collected by centrifugation, the proteins were eluted from them, and the polypeptides were identified by using sponge anti–GNB-Trans antibodies (PoAb-r-GNB-Trans_SUBDO), anti-cryptochrome antibodies (PoAb-r-CRY_SUBDO), or anti-NOSIP antibodies (PoAb-r-NOSIP-r_SUBDO). As expected, all extracts from primmorphs, kept for 6 days (last 3 days in the dark), or from aggregates, cultivated in the dark (time 0) or exposed for 1 or 2 hr under light (time 1 or 2 hr), gave strong signals for wild-type GNB-Trans (Fig. 10, upper panel). In contrast, only faint signals were seen with both anti-cryptochrome (Fig. 10, middle panel) and anti-NOSIP sera (Fig. 10, lower panel) at time 0; increasing antigen signals for cryptochrome and NOSIP were seen after exposure of the primmorphs for 1 hr and were even stronger after 2 hr of white light.
Figure 10.
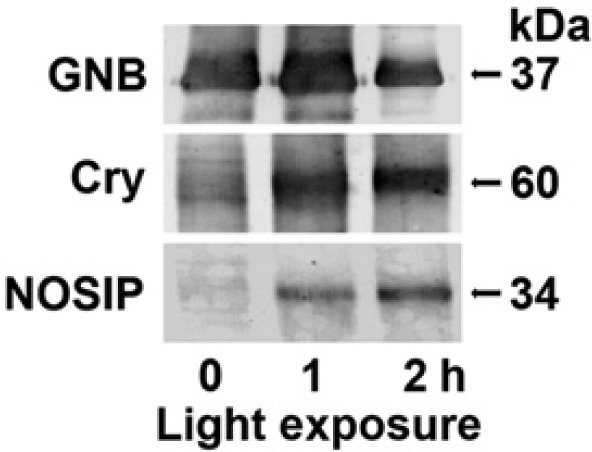
Co-immunoprecipitation of wild-type cryptochrome, nitric oxide synthase–interacting protein (NOSIP), and GNB-Trans by protein A/G agarose beads, coated with anti–GNB-Trans antibodies. Extracts from primmorphs were prepared from either 6-day-old primmorphs, kept for the last 3 days in the dark (time 0), or from those primmorphs that had been exposed for 1 or 2 hr to white light (time 1 and 2 hr). Then the samples were subjected to immunoprecipitation with anti–GNB-Trans beads. After centrifugation, the bound proteins were eluted from the beads and the supernatants were processed by western blotting using the polyclonal antibodies to GNB (upper panel), Cry (middle panel), and NOSIP (lower panel). Further details are given in the Materials and Methods.
From these data, we conclude that GNB-Trans coexists with cryptochrome and NOSIP after the relatively short light exposure of >1 hr.
Association of Cryptochrome, NOSIP, and GNB-Trans to Membranes
To clarify the membrane-binding status of the GNB-Trans, cryptochrome, and NOSIP during different exposure times with white light, the membrane fraction as well as the soluble fraction was prepared from a primmorph extract. The samples were then size-separated by SDS-PAGE and subsequently subjected to western blot analysis. A representative Coomassie brilliant blue–stained SDS-PAGE image from a membrane fraction is shown in Fig. 11A. The membrane and the soluble cytosolic fraction were, after gel size fractionation, blotted onto membranes and reacted with antibodies directed against GNB-Trans, cryptochrome, or NOSIP to detect the respective wild-type proteins GNB-Trans (Fig. 11B), cryptochrome (Fig. 11C) and NOSIP (Fig. 11D).
Figure 11.
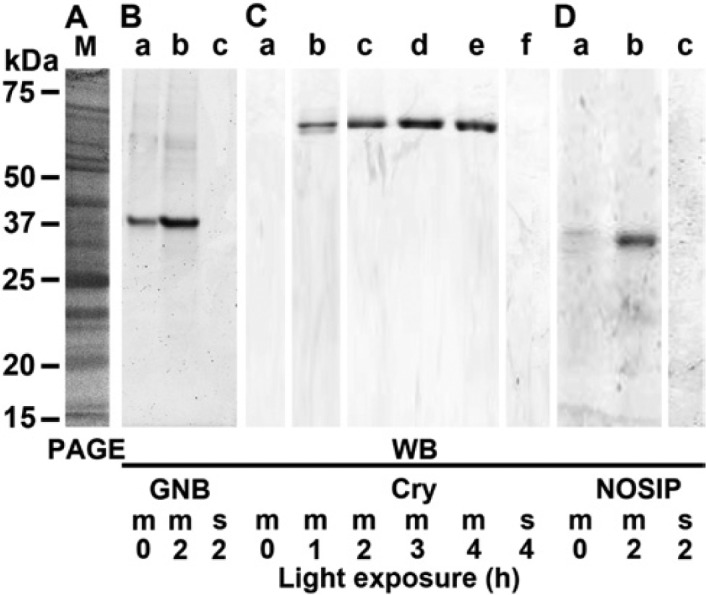
Association of the cell membranes from primmorphs with the wild-type GNB-Trans, cryptochrome, and nitric oxide synthase–interacting protein (NOSIP) protein. The 6-day-old primmorphs, which were kept for the last 3 days in the dark, were then exposed to white light for 0 to 4 hr, as indicated. Then the membrane fraction (m) was separated from the soluble fraction (s) by sucrose step-gradient centrifugation. From both fractions, aliquots of 20 µl were subjected to SDS-PAGE. (A) One sample was stained with Coomassie brilliant blue (M/PAGE). The proteins were then blot-transferred and the membranes were reacted with (B) anti–GNB-Trans, (C) anti-cryptochrome, or (D) anti-NOSIP antibodies to identify the respective proteins, GNB-Trans (GNB), cryptochrome (Cry), and NOSIP, by western blotting.
The data reveals that the membranes from dark-adapted primmorphs showed a distinct signal for the 37.5-kDa GNB-Trans, which increased in intensity in membranes from primmorphs exposed for 2 hr to white light (Fig. 11B, lanes a and b). In contrast, membranes from dark-adapted primmorphs showed no strong signals for cryptochrome (Fig. 11C, lane a) or NOSIP (Fig. 11D, lane a). However, after an exposure time for 2 hr or longer, distinct strong signals were seen for cryptochrome or NOSIP.
Discussion
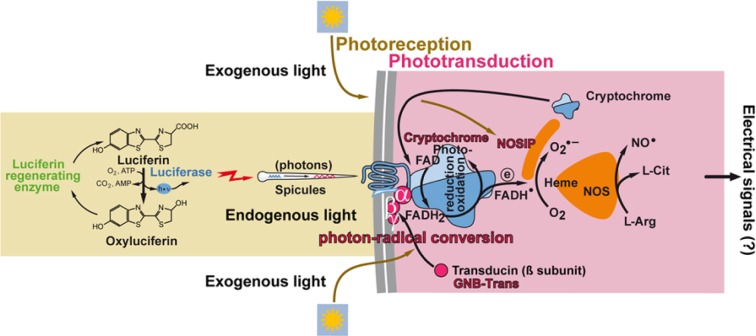
In the present study, we show that, during light exposure of primmorphs from S. domuncula, cryptochrome becomes associated with GNB-Trans and also NOSIP. These three molecules are membrane associated in the light-exposed state. These data support the hypothesis that cryptochrome is causatively involved in both photoreception and phototransduction. A schematic outline of the role of cryptochrome during these processes is sketched in Fig. 12. The property of cryptochrome to act as a molecule involved in photoreception in sponges had been experimentally established in previous studies (Müller et al. 2010; Rivera et al. 2012). The cofactor within the cryptochrome FAD undergoes reduction to FADH2 during light exposure. A fine, more detailed analysis of whether this process is restricted only to exogenous daylight as proposed (Müller et al. 2010; Rivera et al. 2012) or is also proceeding during endogenous luciferase-generated light emission (Müller et al. 2010) remains to be studied.
Figure 12.
Schematic outline of the light–cryptochrome photoreception/phototransduction system in sponges. It is established that sponges can react to exogenous and, very likely, also endogenous light via the cryptochrome photoreceptor. Spicules might act as waveguides for the transmission of photons. Bound to cell membranes and likely in concert with the G protein β (GNB-Trans), cryptochrome transforms photon signals into spin-correlated FADH-superoxide radical pairs (FADH• and O2•−). This complex is linked with the nitric oxide synthase (NOS) system via the nitric oxide synthase–interacting protein (NOSIP) protein. Finally, it is proposed that a further signal transformation occurs in the NOSes under generation of electrical signals. The verification of the latter reaction, the generation of electrical signals, remains to be experimentally approached.
Alike previously reported for cryptochrome (Muller et al. 2010; Rivera et al. 2012), also the deduced protein GNB-Trans and NOSIP, described in the present study, share the highest similarities to the related mammalian polypeptides. As expected (Seack et al. 1998), the GNB-Trans protein, the newly described β subunit of transducin, likewise shows the characteristic six Trp-Asp (WD) repeat profile segments (Gilman 1987). Also, the sponge NOSIP protein in the phylogenetic tree forms the basis for all metazoan NOSIP proteins with its characteristic U-box family domain (Functional RNA Project Databases 2012). These two sponge proteins, GNB-Trans and NOSIP, are substantially accumulated around the spicules within S. domuncula, suggesting a functional role in this area.
The role of cryptochrome in phototransduction in sponges is proposed in the present study. All metazoan cryptochromes are localized intracellularly in the nucleus, or they shuttle between the cytoplasm and nucleus to exert their circadian transcription loop (reviewed in Sancar 2000) or also possibly in cell membranes (Solov’yov et al. 2010). Cryptochrome is described as a molecule that shuttles in a controlled structure-associated manner between the “soluble” and the “membranous” state, allowing a radical pair-based magnetoreceptor/magnetotransduction reaction (Solov’yov et al. 2010) or signal transduction (Partch et al. 2005). In turn, cryptochrome, as the “second photoactive pigment” in sponges, should preferentially be coupled with cell membrane-associated photoreceptor(s), such as rhodopsin, that might transduce the light signal through intermediate molecules to cryptochrome (Sancar 2000). Systems that couple and convert light-absorbing molecules (photoreception) to signal transduction or with chemical energy reactions (phototransduction) are rare in nature (Presti and Delbrück 1978). Earlier studies have suggested that cryptochrome undergoes a direct conformational rearrangement upon light exposure (Partch et al. 2005). In vertebrates, transducin, as a heterotrimeric G protein, is expressed in retina rods and cones and mediates phototransduction (Lerea et al. 1986). The data given here underscore that cryptochrome is linked to the GNB-Trans molecule, a switching molecule functionally acting during light signal exposure of mammalians (Danciger et al. 1992). In mammals, (retina) transducin is linked to the photoreceptors, the rods and cones, that convert light (electromagnetic radiation), or photons, into signals that cause a change in cell membrane potential, resulting in phototransduction (Hecht et al. 1942). In sponges, no evidence is available that indicates the existence of any opsin membrane-bound G protein–coupled receptors (Shichida and Matsuyama 2009). It is well established that, in mammalian eyes, light is captured by opsin. After absorption of photons, G protein/transducin is activated, a reaction during which the activated Gα is bound to the cyclic nucleotide phosphodiesterase, causing a modulation of the cGMP-gated channels and, in turn, a change in a hyperpolarization of membranes. In view of data that indicate that, at least in yeast, G proteins are not always coupled membrane receptors (Cismowski et al. 1999), we hypothesize that, in sponges, the GNB-Trans molecule is contributing to the conformational change of the cryptochrome (Kondoh et al. 2011).
As outlined earlier, FAD undergoes photoreduction, and during the subsequent reoxidation process of FADH2 to FAD, most likely an intermediate spin-correlated FADH-superoxide radical pair (FADH• and O2•−) (Weber et al. 2010; Müller and Ahmad 2011) is formed. Hence, during this photochemical reaction, a conversion of photons to radical pairs takes place. Data support the view that the NOS system is contributing with the heme domain, existing within the NOS molecule, to the regeneration of FAD. During this process, molecular oxygen is taking up an electron.
In 1990, NO• had been proposed to comprise a novel signal transduction mechanism for transcellular communication (Ignarro 1990) and mediate recruitment of cells to a functional unit. At present, the effect of NO• has been implicated in only a few reaction mechanisms related to those described during neurotransmission or neuromodulation (reviewed in Benarroch 2011).
On the basis of our earlier finding that showed that, in sponges, NO• is synthesized during exposure to UV light (Müller, Ushijima, et al. 2006), a process that is inhibited by the NO-specific scavenger PTIO, we investigated the effect of light on NO• production. The data revealed that the overall NO• production in sponge cells is not dependent on light. However, the level of NO• can be significantly suppressed by the NOS inhibitor L-NMMA. The latter result prompted us to study the level of NOS in the presence and absence of light. In this series of experiments, a strong increase in NOS activity was observed. The NOS system is known to affect the migration properties, especially of endothelial cells (Murohara et al. 1999), and also directs distinct morphogenetic processes (e.g., during angiogenesis) (Kashiwagi et al. 2005). Therefore, we studied the effect of the NOS inhibitor L-NMMA on the formation of primmorphs of S. domuncula cells. During this process, the cells undergo proliferation and differentiation (Müller et al. 1999) to 3D cell aggregates, comprising a channel system (Wiens et al. 2003). The time-lapse analyses show that those processes of cell differentiation and migration in primmorphs are significantly inhibited by this NOS inhibitor.
Quantitative PCR studies revealed that, during the initial phase of light exposure, a strong upregulation of the genes encoding for cryptochrome and NOSIP occurs, while the expression of GNB-Trans remains unaffected. From earlier studies with sponge systems, it is known that cryptochrome is an inducible gene (Müller et al. 2010; Rivera et al. 2012). The same property has been described for the mammalian NOSIP (Yu et al. 2012). On the basis of this result, we performed co-immunoprecipitation experiments by applying sponge-specific antibodies against cryptochrome, NOSIP, and GNB-Trans and could identify that these three proteins become associated in response to light exposure. This association is significant, since (almost) no cryptochrome or NOSIP is seen to be associated with GNB-Trans during the dark phase. The functional interaction between these three molecules during light exposure is also supported by the demonstration that, only in the presence of light, can cryptochrome and NOSIP be detected in the membrane fraction; the cytosolic cryptochrome as well as NOSIP fraction cannot be detected with the methods applied here. This finding corroborates our hypothesis that cryptochrome, as a photoreceptor, also acts as a molecule that comprises a role in phototransduction (Fig. 12).
In conclusion, previous studies have established that sponge tissue reacts both to exogenous daylight and to endogenous light, emitted after chemical reaction and likely also via luciferin/luciferase (Müller et al. 2010; Rivera et al. 2012), with an increased expression of cryptochrome (Fig. 12). According to the available data, it must be proposed that the photons are transmitted in both cases via the spicules to the cryptochrome photoreceptor, a process during which FAD undergoes reduction to FADH2. During the subsequent reoxidation phase, FADH-superoxide radical pairs (FADH• and O2•−) are assumed to be formed. Based on the proposed functional parallelism between cryptochrome expression and NO• production, a close relationship between the reoxidation of FADH2 and an interaction between cryptochrome and NOS is proposed. Since cryptochrome is coexpressed and co-localized with NOSIP, it is plausible to assume that NOSIP acts as a linker between cryptochrome and NOS. This view would imply that cryptochrome acts as a photoreceptor, perhaps in conjunction with GNB-Trans, under conversion of photons to FADH-superoxide radical pairs. The latter process might proceed together with the heme domain of NOS. Interdomain electron transfer processes are key steps in NO• synthesis by coupling reactions between the NOS domains (Feng et al. 2010). This transfer is facilitated by calmodulin, which controls the intersubunit binding of FMN to heme. Heme is a switchpoint during the synthesis of NO• from arginine/N-hydroxy-l-arginine under consumption of oxygen and NADPH and the release of l-citrulline (see Sabat et al. 2013). Since NO, as a diffusible molecule, can act as a retrograde diffusible messenger in the brain and can promote long-term potentiation through glutamatergic transmission, it appears to be promising to study the interrelationship between NO• production and channel gating, resulting in depolarization via Nav channels followed by activation of cell Cav channels as described (Figueroa et al. 2007) (Fig. 12). The next step in our research is the identification of the NOS(es) in the S. domuncula system.
Footnotes
Sequences: The following sequences have been deposited (EMBL/GenBank): the cDNA for the S. domuncula nitric oxide synthase–interacting protein (SDNOSIP-l gene) under the accession number HF678443 and the cDNA from S. domuncula transducin β-like protein (SDGNB-Trans) under the accession number HF678444.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: W.E.G.M. is a holder of an ERC Advanced Investigator Grant (no. 268476 “BIOSILICA”). This work was supported by grants from the German Bundesministerium für Bildung und Forschung (project “Center of Excellence BIOTECmarin”), the Deutsche Forschungsgemeinschaft (SCHR 277/10-2), the European Commission (Seventh Framework Programme, Marie-Curie Initial Training Network “BIOMINTEC,” grant no. 215507; and Industry-Academia Partnerships and Pathways “CoreShell,” grant no. 286059), the International Human Frontier Science Program, and the Public Welfare Project of Ministry of Land and Resources of the People’s Republic of China (grant no. 201011005-06).
References
- Achike FI, Kwan CY. 2003. Nitric oxide, human diseases and the herbal products that affect the nitric oxide signalling pathway. Clin Exp Pharmacol Physiol. 30:605–615 [DOI] [PubMed] [Google Scholar]
- Aizenberg J, Weaver JC, Thanawala MS, Sundar VC, Morse DE, Fratzel P. 2005. Skeleton von Euplectella sp.: structural hierarchy from nanoscale to the macroscale. Science. 309:275–278 [DOI] [PubMed] [Google Scholar]
- Akarid K, Sinet M, Desforges B, Gougerot-Pocidalo MA. 1995. Inhibitory effect of nitric oxide on the replication of a murine retrovirus in vitro and in vivo. J Virol. 69:7001–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano F, Noda T. 1995. Improved detection of nitric oxide radical (NO•) production in an activated macrophage culture with a radical scavenger, carboxy PTIO and Griess reagent. FEBS Lett. 368:425–428 [DOI] [PubMed] [Google Scholar]
- Andrew BL, Part NJ. 1972. Properties of fast and slow motor units in hind limb and tail muscles of the rat. Q J Exp Physiol Cogn Med Sci. 57:213–225 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. 2000. The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol. 10:R132–R134 [DOI] [PubMed] [Google Scholar]
- Benarroch EE. 2011. Nitric oxide: a pleiotropic signal in the nervous system. Neurology. 77:1568–1576 [DOI] [PubMed] [Google Scholar]
- Borden KL, Freemont PS. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 6:395–401 [DOI] [PubMed] [Google Scholar]
- Bordner AJ. 2009. Predicting protein-protein binding sites in membrane proteins. BMC Bioinformatics. 10:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer F, Pfannkuchen M, Baltz A, Hauser T, Thiel V. 2008. Light inside sponges. J Exp Marine Biol Ecol. 367:61–64 [Google Scholar]
- Cattaneo-Vietti R, Bavestrello G, Cerrano C, Sara A, Benatti U, Giovine M, Gaino E. 1996. Optical fibres in an Antarctic sponge. Nature. 383:397–398 [Google Scholar]
- Chiou WF, Sung YJ, Liao JF, Shum AY, Chen CF. 1997. Inhibitory effect of dehydroevodiamine and evodiamine on nitric oxide production in cultured murine macrophages. J Nat Prod. 60:708–711 [DOI] [PubMed] [Google Scholar]
- Cismowski MJ, Takesono A, Ma C, Lizano JS, Xie X, Fuernkranz H, Lanier SM, Duzic E. 1999. Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat Biotechnol. 17:878–883 [DOI] [PubMed] [Google Scholar]
- Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT. 2000. Current protocols in protein science. Chichester, UK: John Wiley; p. 2.0.1–2.8.17 [Google Scholar]
- Compton S, Jones CG. 1985. Mechanism of dye response and interference in the Bradford protein assay. Anal Biochem. 151:369–374 [DOI] [PubMed] [Google Scholar]
- Danciger M, Chakraborti A, Farber DB, Kozak CA. 1992. Localization of the gene for a third G protein beta-subunit to mouse chromosome 6 near Raf-1. Genomics. 12:688–692 [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. 1978. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Washington, DC: Nat Biomed Res Foundation; p. 345–352 [Google Scholar]
- Dedio J, König P, Wohlfart P, Schröder C, Kummer W, Müller-Esterl W. 2001. NOSIP, a novel modulator of endothelial nitric oxide synthase activity. FASEB J. 15:79–89 [DOI] [PubMed] [Google Scholar]
- Demerec M. 1950. Reaction of populations of unicellular organisms to extreme changes in environment. Am Naturalist. 84:5–16 [Google Scholar]
- Diwan AD, Khan SN, Cammisa FP, Jr, Sandhu HS, Lane JM. 2010. Nitric oxide modulates recombinant human bone morphogenetic protein-2–induced corticocancellous autograft incorporation: a study in rat intertransverse fusion. Eur Spine J. 19:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer J, Schleicher M, Tappe A, Schilling K, Kuner T, Kusumawidijaja G, Müller-Esterl W, Oess S, Kuner R. 2004. Nitric oxide synthase (NOS)–interacting protein interacts with neuronal NOS and regulates its distribution and activity. J Neurosci. 24:10454–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1993. PHYLIP, ver 3.5. Seattle: University of Washington [Google Scholar]
- Feng C, Fan W, Dupont A, Guy Guillemette J, Ghosh DK, Tollin G. 2010. Electron transfer in a human inducible nitric oxide synthase oxygenase/FMN construct co-expressed with the N-terminal globular domain of calmodulin. FEBS Lett. 584:4335–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa XF, Chen CC, Campbell KP, Damon DN, Day KH, Ramos S, Duling BR. 2007. Are voltage-dependent ion channels involved in the endothelial cell control of vasomotor tone? Am J Physiol Heart Circ Physiol. 293:H1371–H1383 [DOI] [PubMed] [Google Scholar]
- Förstermann U, Münzel T. 2006. Endothelial nitric oxide synthase in vascular disease from marvel to menace. Circulation. 113:1708–1714 [DOI] [PubMed] [Google Scholar]
- Fregly MJ, Blatteis CM. 1996. Environmental physiology. Handbook of Physiology, Sec. 4. Oford, UK: Oxford University Press. p. 157–185 [Google Scholar]
- Functional RNA Project Databases 2012. www.ncRNA.org:database:http://www.ncrna.org/glocal/cgi-bin/hgGene?hgg_gene=uc002pok.1&hgg_prot=Q9Y314&hgg_chrom=chr19&hgg_start=54750779&hgg_end=54775615&hgg_type=knownGene&db=hg18&hgsid=1390157
- Gehring W, Seimiya M. 2010. Eye evolution and the origin of Darwin’s eye prototype. Ital J Zool. 77:124–136 [Google Scholar]
- Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SME, Salerno JC. 2006. Nitric-oxide synthase output state—design and properties of nitric-oxide synthase oxygenase/FMN domain constructs. J Biol Chem. 281:14173–14183 [DOI] [PubMed] [Google Scholar]
- Gilman AG. 1987. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 56:615–649 [DOI] [PubMed] [Google Scholar]
- Gospe SM, Baker SA, Kessler C, Brucato MF, Winter JR, Burns ME, Arshavsky VY. 2011. Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors. J Neurosci. 31:14660–14668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Matsumoto M, Yada M, Nakayama KI. 2004. Interaction of U-box–type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells. 9:533–548 [DOI] [PubMed] [Google Scholar]
- Hecht S, Shlaer S, Pirenne MH. 1942. Energy, quanta, and vision. J Gen Physiol. 25:819–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LH. 1940. Invertebrates: Protozoa through Ctenophora. New York: McGraw-Hill. [Google Scholar]
- Ignarro LJ. 1990. Nitric oxide. A novel signal transduction mechanism for transcellular communication. Hypertension. 16:477–483 [DOI] [PubMed] [Google Scholar]
- Kashiwagi S, Izumi Y, Gohongi T, Demou ZN, Xu L, Huang PL, Buerk DG, Munn LL, Jain RK, Fukumura D. 2005. NO mediates mural cell recruitment and vessel morphogenesis in murine melanomas and tissue-engineered blood vessels. J Clin Invest. 115:1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M. 2011. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol. 413:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasko A, Schröder HC, Perović S, Steffen R, Kruse M, Reichert W, Müller IM, Müller WEG. 1999. Ethylene modulates gene expression in cells of the marine sponge Suberites domuncula and reduces the degree of apoptosis. J Biol Chem. 274:31524–31530 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685 [DOI] [PubMed] [Google Scholar]
- Lakshmi VM, Zenser TV. 2007. 2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potentiates nitrosation of a heterocyclic amine carcinogen by nitric oxide. Life Sci. 80:644–649 [DOI] [PubMed] [Google Scholar]
- LePennec G, Perović S, Ammar MSA, Grebenjuk VA, Steffen R, Müller WEG. 2003. Cultivation of primmorphs from the marine sponge Suberites domuncula: morphogenetic potential of silicon and iron. J Biotechnol. 100:93–108 [DOI] [PubMed] [Google Scholar]
- Lerea CL, Somers DE, Hurley JB, Klock IB, Bunt-Milam AH. 1986. Identification of specific transducin alpha subunits in retinal rod and cone photoreceptors. Science. 234:77–80 [DOI] [PubMed] [Google Scholar]
- Leyhausen G, Schuster DK, Vaith P, Zahn RK, Umezawa H, Falke D, Müller WEG. 1983. Identification and properties of the cell membrane-bound leucine aminopeptidase, interacting with the potential immunostimulant and chemotherapeutic agent bestatin. Biochem Pharmacol. 32:1051–1057 [DOI] [PubMed] [Google Scholar]
- Leys SP, Cronin TW, Degnan BM, Marshall JN. 2002. Spectral sensitivity in a sponge larva. J Comp Physiol A. 188:199–202 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. 2011. Blue-light–dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 D DCT method. Methods. 25:402–408 [DOI] [PubMed] [Google Scholar]
- Miranda KM, Espey MG, Wink DA. 2001. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide. 5:62–71 [DOI] [PubMed] [Google Scholar]
- Müller P, Ahmad M. 2011. Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception. J Biol Chem. 286:21033–21040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WEG, Binder M, von Lintig J, Guo YW, Wang XH, Kaandorp JA, Wiens M, Schröder HC. 2011. Interaction of the retinoic acid signaling pathway with spicule formation in the marine sponge Suberites domuncula through activation of bone morphogenetic protein-1. Biochim Biophys Acta. 1810:1178–1194 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Böhm M, Batel R, de Rosa S, Tommonaro G, Müller IM, Schröder HC. 2000. Application of cell culture for the production of bioactive compounds from sponges: synthesis of avarol by primmorphs from Dysidea avara. J Nat Prod. 63:1077–1081 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Kasueske M, Wang XH, Schröder HC, Wang Y, Pisignano D, Wiens M. 2009. Luciferase a light source for the silica-based optical waveguides (spicules) in the demosponge Suberites domuncula. Cell Mol Life Sci. 66:537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WEG, Rothenberger M, Boreiko A, Tremel W, Reiber A, Schröder HC. 2005. Formation of siliceous spicules in the marine demosponge Suberites domuncula. Cell Tissue Res. 321:285–297 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Ushijima H, Batel R, Krasko A, Boreiko A, Müller IM, Schröder HC. 2006. Novel mechanism for the radiation-induced bystander effect: nitric oxide and ethylene determine the response in sponge cells. Mutation Res. 597:62–72 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wang XH, Grebenjuk VA, Korzhev M, Wiens M, Schloßmacher U, Schröder HC. 2012. Nocturnin in the demosponge Suberites domuncula: a potential circadian clock protein controlling glycogenin synthesis in sponges. Biochem J. 448:233–242 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wang XH, Schröder HC, Korzhev M, Grebenjuk VA, Markl JS, Jochum KP, Pisignano D, Wiens M. 2010. A cryptochrome-based photosensory system in the siliceous sponge Suberites domuncula (Demospongiae). FEBS J. 277:1182–1201 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wang XH, Wiens M, Schloßmacher U, Jochum KP, Schröder HC. 2011. Hardening of bio-silica in sponge spicules involves an aging process after its enzymatic polycondensation: evidence for an aquaporin-mediated water absorption. Biochim Biophys Acta. 1810:713–726 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wendt K, Geppert C, Wiens M, Reiber A, Schröder HC. 2006. Novel photoreception system in sponges? Unique transmission properties of the stalk spicules from the hexactinellid Hyalonema sieboldi. Biosensors Bioelectronics. 21:1149–1155 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wiens M, Adell T, Gamulin V, Schröder HC, Müller IM. 2004. Bauplan of urmetazoa: basis for genetic complexity of Metazoa. Intern Rev Cytol. 235:53–92 [DOI] [PubMed] [Google Scholar]
- Müller WEG, Wiens M, Batel R, Steffen R, Schröder HC, Borojevic R, Custodio MR. 1999. Establishment of a primary cell culture from a sponge: primmorphs from Suberites domuncula. Marine Ecol Progr Ser. 178:205–219 [Google Scholar]
- Murohara T, Witzenbichler B, Spyridopoulos I, Asahara T, Ding B, Sullivan A, Losordo DW, Isner JM. 1999. Role of endothelial nitric oxide synthase in endothelial cell migration. Arterioscler Thromb Vasc Biol. 19:1156–1161 [DOI] [PubMed] [Google Scholar]
- Natalio F, Corrales TP, Panthöfer M, Schollmeyer D, Müller WEG, Kappl M, Butt HJ, Tremel W. 2013. Flexible minerals: self-assembled calcite spicules with extreme bending strength. Science. 339:1298–1302 [DOI] [PubMed] [Google Scholar]
- Natalio F, Link T, Müller WEG, Schröder HC, Cui FZ, Wang XH, Wiens M. 2010. Bioengineering of the silica-polymerizing enzyme silicatein-α for a targeted application to hydroxyapatite. Acta Biomater. 6:3720–3728 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB., Jr. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments. Version 1.1.004. cris.com/~ketchup/genedoc.shtml
- Nickel M. 2004. Kinetics and rhythm of body contractions in the sponge Tethya wilhelma (Porifera: Demospongiae). J Exp Biol. 207:4515–4524 [DOI] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgür S, Lee AL, Sancar A. 2005. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 44:3795–3805 [DOI] [PubMed] [Google Scholar]
- Perović S, Krasko A, Prokic I, Müller IM, Müller WEG. 1999. Origin of neuronal-like receptors in Metazoa: cloning of a metabotropic glutamate/GABA-like receptor from the marine sponge Geodia cydonium. Cell Tissue Res. 296:395–404 [DOI] [PubMed] [Google Scholar]
- Presti D, Delbrück M. 1978. Photoreceptors for biosynthesis, energy storage and vision. Plant Cell Environm. 1:81–100 [Google Scholar]
- Pribyl M, Muratov CB, Shvartsman SY. 2003. Long-range signal transmission in autocrine relays. Biophys J. 84:883–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. 2009. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A. 106:1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, Oakley TH. 2012. Blue-light–receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J Exp Biol. 215:1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondard P, Iiri T, Srinivasan S, Meng E, Fujita T, Bourne HR. 2001. Mutant G protein α subunit activated by Gβγ: a model for receptor activation? Proc Natl Acad Sci U S A. 98:6150–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat J, Egawa T, Lu C, Stuehr DJ, Gerfen GJ, Rousseau DL, Yeh SR. 2013. Catalytic intermediates of inducible nitric oxide synthase stabilized by the W188H mutation. J Biol Chem. 288:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs L. 1984. Angewandte Statistik. Berlin, Germany: Springer [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- Sancar A. 2000. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 69:31–67 [DOI] [PubMed] [Google Scholar]
- Schröder HC, Boreiko A, Korzhev M, Tahir MN, Tremel W, Eckert C, Ushijima H, Müller IM, Müller WEG. 2006. Co-expression and functional interaction of silicatein with galectin: matrix-guided formation of siliceous spicules in the marine demosponge Suberites domuncula. J Biol Chem. 281:12001–12009 [DOI] [PubMed] [Google Scholar]
- Schröder HC, Krasko A, Gundacker D, Leys SP, Müller IM, Müller WEG. 2003. Molecular and functional analysis of the (6-4) photolyase from the hexactinellid Aphrocallistes vastus. Biochim Biophys Acta. 1651:41–49 [DOI] [PubMed] [Google Scholar]
- Seack J, Kruse M, Müller WEG. 1998. Evolutionary analysis of G-proteins in early metazoans: cloning of α- and β-subunits from the sponge Geodia cydonium. Biochim Biophys Acta. 1401:93–103 [DOI] [PubMed] [Google Scholar]
- Shichida Y, Matsuyama T. 2009. Evolution of opsins and phototransduction. Phil Trans R Soc B. 364:2881–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Underwood BA, Auldridge M, Loucas HM, Shibuya K, Schmelz E, Clark DG, Klee HJ. 2004. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of Petunia flowers. Plant Physiol. 136:3504–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solov’yov IA, Mouritsen H, Schulten K. 2010. Acuity of a cryptochrome and vision-based magnetoreception system in birds. Biophys J. 99:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SpongeBase 2010. [Accessed August 15, 2010]. https://octavia.vk.medizin.uni-mainz.de/login.cgi
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 466:720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi H, Han YH, Ichikawa K. 2003. A kinetic study of 3-chlorophenol enhanced hydroxyl radical generation during ozonation. Water Res. 37:4924–4928 [DOI] [PubMed] [Google Scholar]
- Wang XH, Schloßmacher U, Schröder HC, Müller WEG. 2013. Biologically-induced transition of bio-silica sol to mesoscopic gelatinous flocs: a biomimetic approach to a controlled fabrication of bio-silica structures. Soft Matter. 9:654–664 [Google Scholar]
- Wang XH, Schröder HC, Wang K, Kaandorp JA, Müller WEG. 2012. Genetic, biological and structural hierarchies during sponge spicule formation: from soft sol-gels to solid 3D silica composite structures. Soft Matter. 8:9501–9518 [Google Scholar]
- Wang XH, Wiens M, Schröder HC, Hu S, Mugnaioli E, Kolb U, Tremel W, Pisignano D, Müller WEG. 2010. Morphology of sponge spicules: silicatein a structural protein for bio-silica formation. Adv Biomater Adv Eng Mater. 12:B422–B437 [Google Scholar]
- Wang Y, Elion EA. 2003. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol Biol Cell. 14:2543–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Glick MC, Nass MK. 1966. Membranes of animal cells, I: methods of isolation of the surface membrane. J Cell Physiol. 68:269–287 [Google Scholar]
- Weber S, Biskup T, Okafuji A, Marino AR, Berthold T, Link G, Hitomi K, Getzoff ED, Schleicher E, Norris JR. 2010. Origin of light-induced spin-correlated radical pairs in cryptochrome. J Phys Chem B. 114:14745–14754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyrer S, Rützler K, Rieger R. 1999. Serotonin in Porifera? Evidence from developing Tedania ignis, the Caribbean fire sponge (Demospongiae). Memoirs Queensland Museum. 44:659–665 [Google Scholar]
- Wiens M, Batel R, Korzhev M, Müller WEG. 2003. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol. 206:3261–3271 [DOI] [PubMed] [Google Scholar]
- Wiens M, Korzhev M, Perovic-Ottstadt S, Luthringer B, Brandt D, Klein S, Müller WEG. 2007. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol Biol Evol. 24:792–804 [DOI] [PubMed] [Google Scholar]
- Wiens M, Koziol C, Hassanein HMA, Batel R, Müller WEG. 1998. Induction of gene expression of the chaperones 14-3-3 and HSP70 induced by PCB 118 (2,3′,4,4′,5-pentachlorobiphenyl) in the marine sponge Geodia cydonium: novelbiomarkers for polychlorinated biphenyls. Mar Ecol Prog Ser. 165:247–257 [Google Scholar]
- Wiens M, Wang XH, Unger A, Schröder HC, Grebenjuk VA, Pisignano D, Jochum KP, Müller WEG. 2010. Flashing light signaling circuit in sponges: endogenous light generation after tissue ablation in Suberites domuncula. J Cell Biochem. 111:1377–1389 [DOI] [PubMed] [Google Scholar]
- Yu X, Zhong Y, Zhu Z, Wu T, Shen A, Huang Y. 2012. Increased expression of nitric oxide synthase interacting protein (NOSIP) following traumatic spinal cord injury in rats. J Mol Histol. 43:661–668 [DOI] [PubMed] [Google Scholar]