Significance
22q11 deletion syndrome (22q11DS) is a chromosome disorder that frequently accompanies psychiatric conditions such as schizophrenia. However, it remains elusive how the chromosomal microdeletion causes the mental manifestation. Here we show that a 22q11DS mouse model has deficits in the development of interneurons and hippocampal dentate gyrus and that DiGeorge syndrome critical region gene 8 (Dgcr8), a microprocessor of microRNA and one of the genes in 22q11, underlies these neurodevelopmental abnormalities. Dgcr8 regulates Chemokine receptor 4/Chemokine ligand 12 (Cxcr4/Cxcl12; Sdf1) signaling, which is indispensable for interneuron and dentate gyrus development. Finally, we observe decreased expression of CXCL12 in olfactory neurons from sporadic schizophrenia. Given the increased risk of 22q11DS in schizophrenia, the overall study suggests that CXCR4/CXCL12 signaling may represent a common downstream mediator in the pathophysiology of schizophrenia.
Keywords: hippocampus, dentate gyrus
Abstract
22q11 deletion syndrome (22q11DS) frequently accompanies psychiatric conditions, some of which are classified as schizophrenia and bipolar disorder in the current diagnostic categorization. However, it remains elusive how the chromosomal microdeletion leads to the mental manifestation at the mechanistic level. Here we show that a 22q11DS mouse model with a deletion of 18 orthologous genes of human 22q11 (Df1/+ mice) has deficits in migration of cortical interneurons and hippocampal dentate precursor cells. Furthermore, Df1/+ mice show functional defects in Chemokine receptor 4/Chemokine ligand 12 (Cxcr4/Cxcl12; Sdf1) signaling, which reportedly underlie interneuron migration. Notably, the defects in interneuron progenitors are rescued by ectopic expression of Dgcr8, one of the genes in 22q11 microdeletion. Furthermore, heterozygous knockout mice for Dgcr8 show similar neurodevelopmental abnormalities as Df1/+ mice. Thus, Dgcr8-mediated regulation of microRNA is likely to underlie Cxcr4/Cxcl12 signaling and associated neurodevelopmental defects. Finally, we observe that expression of CXCL12 is decreased in olfactory neurons from sporadic cases with schizophrenia compared with normal controls. Given the increased risk of 22q11DS in schizophrenia that frequently shows interneuron abnormalities, the overall study suggests that CXCR4/CXCL12 signaling may represent a common downstream mediator in the pathophysiology of schizophrenia and related mental conditions.
The 22q11.2 deletion syndrome (22q11DS) is frequently associated with major mental conditions, such as schizophrenia (SZ) (1). Some reports have indicated that 22q11DS might account for up to 1–2% of subjects diagnosed with SZ (2, 3). All of the genes, except one, in the human 22q11.2 locus exist on mouse chromosome 16, although the organization is different (4). This has facilitated the generation of mouse models of 22q11DS, which carry different-size hemizygous deletions of the 22q11-related region (5–8). These mouse models include Df1/+ and LgDel/+ mice: The former has a deletion from Es2 to Ufd1l, whereas the latter has a deletion from Idd to Hira. A recent study using the LgDel/+ mouse model showed that the hemizygous deletion of the 22q11-related region led to delayed migration of interneurons, altered distribution of parvalbumin (PV)-positive interneurons (9), and reduced Chemokine (C-X-C motif) receptor 4 (Cxcr4) expression known to play a role in interneuron migration (10), although it remains to be determined whether Cxcr4 signaling is impaired or not in this model mouse. Given that changes in PV-positive interneurons occur in the pathology of SZ (11, 12), these reports are intriguing. Nonetheless, the mechanism and clinical evidence that link these phenotypic changes are unclear.
DiGeorge syndrome critical region gene 8 (Dgcr8) is one of the genes in the 22q11-related region, and has been proposed to be responsible, at least in part, for psychiatric manifestations (13). Dgcr8 heterozygous knockout mice show working memory deficits and sensory information-processing deficits (6, 14), which are also seen in SZ patients. However, it remains elusive how the deficit of this specific molecule can underlie these behavior changes.
Here we show that another mouse model of 22q11DS, Df1/+ mice, which have a shorter deletion of the 22q11-related region, also have abnormal interneuron migration. Using Df1/+ and Dgcr8 heterozygous knockout mice, we directly demonstrate that interneuron progenitors show deficits in Cxcr4/Chemokine (C-X-C motif) ligand 12(Cxcl12) signaling, and that Cxcr4-dependent hippocampal dentate gyrus development is also affected. Furthermore, the decreased preference of Df1/+ interneuron progenitors for Cxcl12 could be rescued by overexpression of Dgcr8, suggesting the involvement of Dgcr8-regulated microRNA (miRNA) in this deficit. Finally, we provide evidence that Cxcl12 is down-regulated in the olfactory epithelium from SZ patients.
Results
Df1/+ Mice Show Interneuron Migration Deficits.
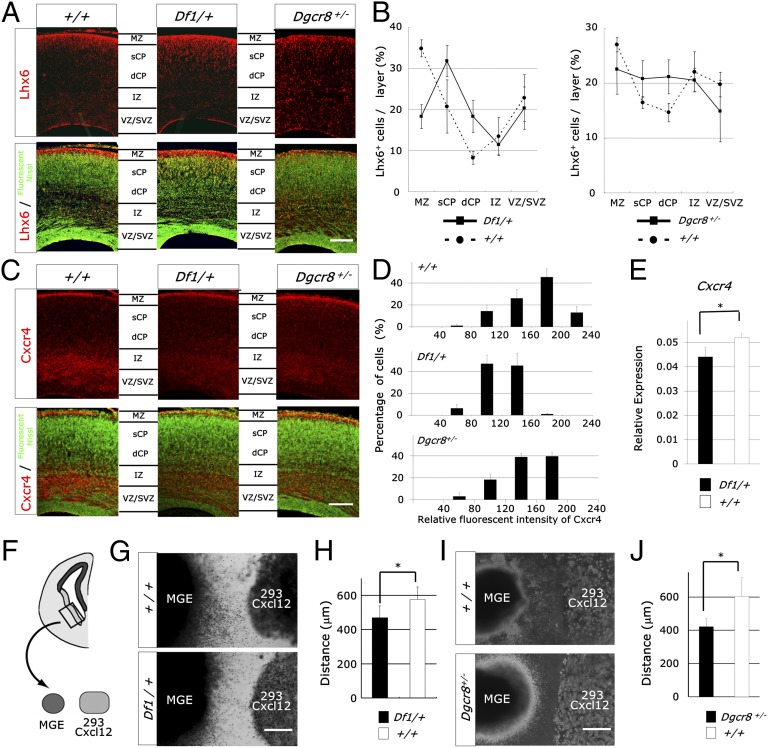
To determine which genes are responsible for interneuron migration deficits, we examined Df1/+ mice, which have a shorter deletion compared with LgDel/+ mice (Fig. S1A). Immunohistochemical studies of Lhx6 and Gad67 showed that the distribution of interneurons was altered at embryonic day E18.5 (E18.5), with a reduced number in the marginal zone and an increased number in the deep cortical plate [Lhx6, genotype × layer interaction, F4,16 = 8.81, P = 0.0006 (n = 3 embryos); Gad67, genotype × layer interaction, F4,16 = 5.50, P = 0.0056 (n = 3 embryos) (ANOVA)] (Fig. 1 A and B and Fig. S1 B and C). Furthermore, the number of PV-positive interneurons was decreased in the medial prefrontal cortex of 1-mo-old Df1/+ mice [control mice, 1.05 ± 0.24 × 104 cells per mm3; Df1/+ mice, 7.74 ± 0.61 × 103 cells per mm3; P = 0.040 (n = 4–6 mice) (Student t test)]. Taken together, these data suggest that at least one of the 18 genes deleted in Df1/+ mice directly underlie interneuron abnormalities.
Fig. 1.
Microdeletion of the 22q11-related region reduced the Cxcl12-induced chemotaxis of MGE-derived cells. (A–D) Immunofluorescence for Lhx6 (red; A), Cxcr4 (red; C), and fluorescent Nissl (green) of coronal sections of E18.5 Df1/+, Dgcr8+/−, and control cerebral cortices. Quantification of the distribution of marker-positive cells per layer (B) and the relative fluorescence intensity of Cxcr4 per cell (D). Values are mean ± SD. dCP, deep cortical plate; IZ, intermediate zone; MZ, marginal zone; sCP, superficial cortical plate; VZ/SVZ, ventricular zone/subventricular zone. (Scale bars, 200 μm.) (E) Quantitative real-time RT-PCR of Cxcr4 in the E15.5 Df1/+ cortex. Values have been normalized to β-actin abundance. Values are mean ± SD. *P = 0.025. (F–J) Schematic of the experimental design (F). MGE explants of E13.5 Df1/+ (G), Dgcr8+/− (I), and control embryos were cultured in Matrigel adjacent to Cxcl12-expressing 293T cell aggregates for 60 h. (Scale bars, 400 μm.) The distance migrated by the farthest 20 MGE-derived cells was measured (H and J). Values are mean ± SD. *P = 0.0079 (H), *P = 0.012 (J).
Medial Ganglionic Eminence-Derived Interneuron Progenitors in Df1/+ Mice Aberrantly Respond to Cxcl12.
Previous studies have demonstrated that Cxcr4/Cxcl12 and Neuregulin/ErbB4 signaling are crucial for cortical interneuron distribution (15–18). Immunohistochemical studies showed that Cxcr4 expression is decreased in the cortex of E18.5 Df1/+ embryos [genotype, F1,4 = 19.50, P = 0.012 (n = 3 embryos) (ANOVA)] (Fig. 1 C and D), which was also reported in LgDel/+ mice (10). Furthermore, quantification of the relative fluorescence intensity of Cxcr4 per cell suggests that each cell expresses less Cxcr4 (Fig. 1E). The reduction of Cxcr4 expression was also confirmed by real-time RT-PCR [P = 0.025 (Student t test) (n = 3 E15.5 embryos)] (Fig. 1F). In contrast, Neuregulin/ErbB4 signaling-related genes were not affected in Df1/+ medial ganglionic eminence (MGE) and cortex (Fig. S2).
Most interneurons are generated from the subpallium including the lateral, medial, and caudal ganglionic eminence (19, 20). To directly examine the responsiveness of Df1/+ MGE-derived cells to Cxcl12, we cocultured E13.5 MGE explants obtained from Df1/+ and control embryos with aggregates of 293T cells expressing Cxcl12. This experiment showed the perturbed chemotactic response of Df1/+ MGE-derived cells to Cxcl12 [genotype, F1,15 = 9.37, P = 0.0079 (n = 8–9 embryos) (ANOVA)] (Fig. 1 F–H).
Hippocampal Dentate Precursor Cells in Df1/+ Mice also Show Migration Deficits.
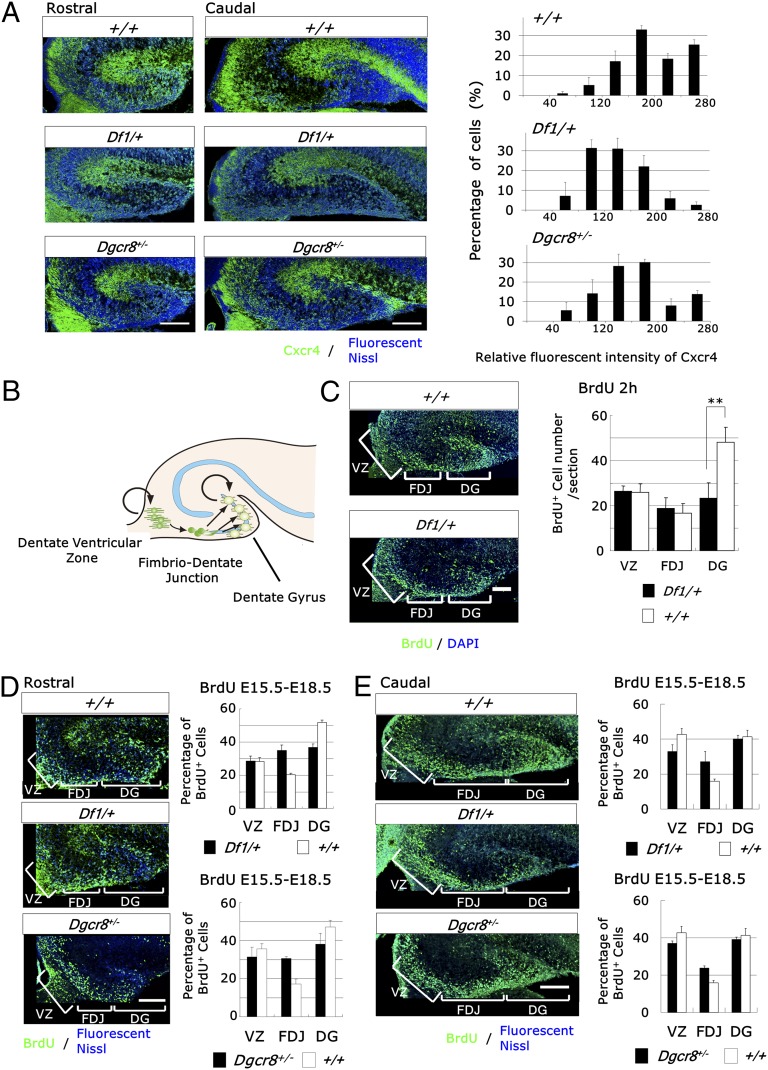
Previous studies indicate that the formation of the hippocampal dentate gyrus (DG) requires Cxcr4/Cxcl12 signaling (21–23). Immunohistochemical and in situ hybridization analysis showed decreased expression of Cxcr4 but not Cxcl12 in the Df1/+ embryonic hippocampus (HP) [genotype, F1,4 = 7.84, P = 0.049 (n = 3 embryos) (ANOVA)] (Fig. 2A and Fig. S3). Furthermore, stereological analysis showed a statistically significant volume reduction in the DG, but not the HP, of perinatal Df1/+ mice [DG, P = 0.0069; HP, P = 0.36 (n = 4 mice) (Student t test)] (Fig. S4A).
Fig. 2.
Df1/+ and Dgcr8+/− hippocampal dentate precursor cells exhibit decreased responses to Cxcl12. (A) Immunofluorescence of the Df1/+, Dgcr8+/−, and control E18.5 hippocampus for Cxcr4 (green) and fluorescent Nissl (blue) (Left). Quantification of the relative fluorescence intensity of Cxcr4 per cell (Right). Values are mean ± SD. (Scale bars, 200 μm.) (B) Schematic representation of the progressive development of dentate neuronal progenitor cells. (C) Representative images of immunofluorescence studies on the Df1/+ and control E18.5 hippocampus for BrdU (green) and DAPI staining of nuclei (blue) (Left). BrdU was administered 2 h before sacrifice. The number of BrdU-positive cells is reduced in the DG but not in the dentate VZ and FDJ of Df1/+ mice (Right). Data are shown as mean ± SD. **P = 0.014. (Scale bar, 100 μm.) (D and E) Representative immunofluorescent images of the Df1/+, Dgcr8+/−, and control E18.5 hippocampus for BrdU (green) and DAPI staining of nuclei (blue) at rostral (D) and caudal levels (E) (Left). BrdU was administered at E15.5 and the mice were killed at E18.5. The distribution of BrdU-positive cells is altered in the Df1/+ and Dgcr8+/− hippocampus (Right). Data are shown as mean ± SD. (Scale bars, 200 μm.)
During development, neuronal precursors of dentate granule cells migrate to the subpial zone of the DG from the dentate ventricular zone (dVZ) through the fimbrio-dentate junction (FDJ) (Fig. 2B). The precursors in the subpial zone then proliferate and generate a large number of DG granule cells during the first month of postnatal life (21, 22). We first analyzed cell proliferation following a single injection of BrdU at E18.5 2 h before sacrifice. The number of BrdU-positive cells was 50% lower in Df1/+ mice compared with control mice specifically in the DG but not in the dVZ or the FDJ [genotype, F1,8 = 9.82, P = 0.014 (n = 4–6 embryos) (ANOVA)] (Fig. 2C). The decreased proliferation in the E18.5 DG could be caused by the decreased generation of dentate precursors in the dVZ or by a migration defect from the dVZ to the DG. To distinguish between these two possibilities, we examined the generation of dentate precursors in earlier developmental stages using BrdU. No difference was observed in the number of BrdU-positive cells in the dVZ at E15.5 and E16.5 (Fig. S5 A and B). Additionally, neither Wnt3a nor Lef1 was affected in Df1/+ mice, although Wnt signaling is known to be essential for the proliferation of dentate precursors (Fig. S5 C and D) (24). These results support the hypothesis that the generation of dentate precursors is intact in Df1/+ mice.
We next examined the effects of 22q11-related region deficiency on the migration of dentate precursors. To trace the migrating dentate precursors, we performed a BrdU pulse experiment. We injected BrdU at E15.5 and performed analysis at E18.5. A drastic decrease in the number of BrdU-positive cells was observed in the DG of Df1/+ mice compared with control mice, but not in the dVZ and the FDJ [genotype × region interaction, F2,8 = 40.26, P = 0.0001 (n = 3 embryos) (ANOVA)] (Fig. 2D). The altered distribution of BrdU-positive cells was also observed at a more caudal level [genotype × region interaction, F2,8 = 8.21, P = 0.012 (n = 3 embryos) (ANOVA)] (Fig. 2E). These results suggest that the migration of dentate precursors to the DG may be delayed. Furthermore, immunohistochemical analysis of Nestin and Prox1 in the DG of Df1/+ mice showed widely dispersed Nestin-positive precursors in the DG (Fig. S6). In contrast, Nestin-positive dentate precursor cells and Prox1-positive granule cells formed different layered organizations (Fig. S6). Taken together, these data show that haplodeficiency of the 22q11-related region caused the migration deficits of dentate precursors to the DG subpial zone.
Hippocampal Dentate Precursor Cells in Df1/+ Mice also Aberrantly Respond to Cxcl12.
To directly examine whether Cxcr4/Cxcl12 signaling deficits cause DG developmental abnormalities, we performed a transwell migration assay using postnatal day 0 (P0) Df1/+ DG-derived cells and Cxcl12. Dose–response studies showed a typical biphasic response in control DG-derived cells, with a peak at 0.1 μg/mL of Cxcl12. In contrast, Df1/+ DG-derived cells showed a lower chemotactic response to Cxcl12, with a peak at 1 μg/mL [dose × genotype interaction, F3,39 = 4.03, P = 0.014 (n = 7–8) (ANOVA)] (Fig. S7A). Finally, to test whether the migration of Df1/+ dentate precursors is reduced in a more physiological condition, we cocultured E17.5 dVZ explants with aggregates of Cxcl12-expressing 293T cells in Matrigel matrices. Df1/+ dVZ-derived cells exhibited a decreased response to Cxcl12 compared with control dVZ-derived cells [genotype, F1,10 = 5.51, P = 0.041 (n = 5–7 embryos) (ANOVA)] (Fig. S7B). The migration of control dVZ-derived cells is Cxcr4-dependent, because a Cxcr4 inhibitor, AMD3100, inhibited migration [treatment, F2,6 = 20.18, P = 0.0022 (n = 3 embryos) (ANOVA)] (Fig. S8 A and B). Thus, microdeletion of the 22q11-related region caused Cxcr4/Cxcl12 signaling deficits in dentate precursor cells.
Dgcr8 Rescued the Reduced Responsiveness of Df1/+ MGE-Derived Cells to Cxcl12.
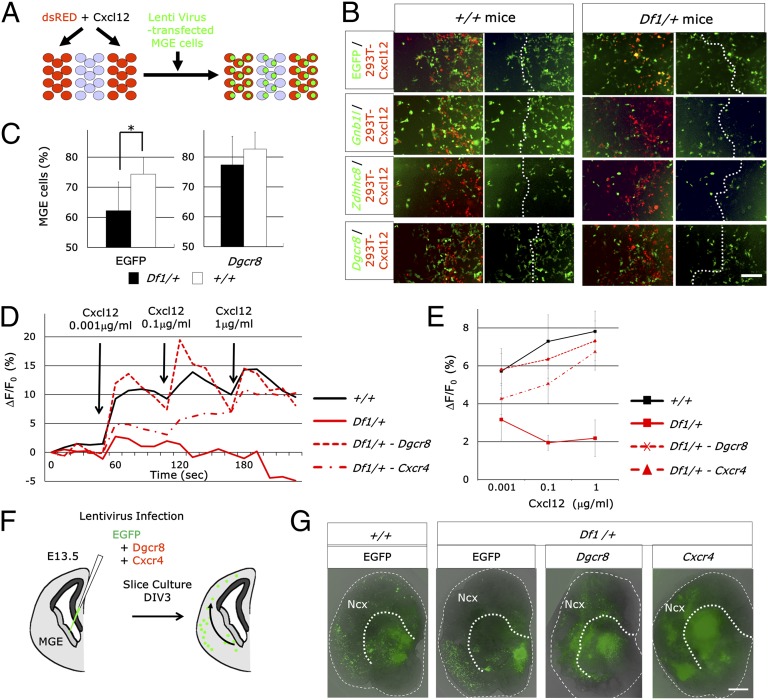
To identify the gene, or genes, in the 22q11-related region that is responsible for the migration deficits observed in Df1/+ mice, we performed a stripe choice assay. Dissociated E13.5 MGE cells obtained from Df1/+ and control mice were infected by EGFP-expressing lentivirus and then given a chance to migrate on top of alternating stripes of 293T cells nontransfected or transfected with DsRed and Cxcl12 (Fig. 3A). Their final position was identified by fluorescence of EGFP 48 h later. Df1/+ MGE-derived cells showed a decreased preference for Cxcl12-expressing cells compared with nontransfected cell stripes [genotype, F1,18 = 5.55, P = 0.03 (n = 4–7 embryos) (ANOVA)] (Fig. 3 B and C). For rescue experiments, we used lentiviral vectors to express a 22q11-related gene—Gnb1l, Zdhhc8, or Dgcr8—with EGFP. Only Dgcr8 significantly restored the decreased preference of Df1/+ MGE-derived cells for Cxcl12 [treatment, F1,18 = 10.158, P = 0.0051 (n = 4–7 embryos) (ANOVA)] (Fig. 3B). To test whether Cxcr4-mediated signaling is normalized by Dgcr8 or Cxcr4 overexpression, we measured the Cxcl12-induced increase in intracellular calcium ([Ca2+]i) by Rhod3 Ca2+ imaging experiments (Fig. 3 D and E). We applied various concentrations of Cxcl12 to E13.5 Df1/+ MGE-derived neural progenitor cells. The [Ca2+]i response to Cxcl12 was significantly decreased in Df1/+ neural progenitors compared with control neural progenitors [genotype, F1,8 = 12.82, P = 0.0072 (n = 5 embryos) (ANOVA)]. Both Dgcr8 and Cxcr4 rescued this deficit in [Ca2+]i response to Cxcl12 of Df1/+ MGE-derived neural progenitor cells [genotype, F2,15 = 6.27, P = 0.011 (n = 5–7 embryos) (ANOVA); Dgcr8, P = 0.0032; Cxcr4, P = 0.0283 (Fisher's Least Significant Difference test)].
Fig. 3.
Chemotactic deficits of Df1/+ MGE-derived cells are restored by lentivirus-mediated reintroduction of Dgcr8. (A) Schematic of the experimental design. MGE-derived cells from Df1/+ and control embryos were infected by GFP-expressing lentivirus and plated on top of alternating stripes of 293T cells nontransfected and transfected with DsRed and Cxcl12. The distribution of MGE-derived cells was assessed 48 h later. (B) Distribution of EGFP-positive MGE-derived cells from E13.5 Df1/+ and control embryos. For rescue experiments, Gnb1l, Zdhhc8, or Dgcr8 with EGFP was introduced by lentivirus. The dotted lines indicate the boundary between nontransfected and transfected 293T cells. (Scale bar, 200 μm.) (C) Quantification of the percentage of MGE-derived cells on Cxcl12-expressing 293T cells. Values are mean ± SD. *P = 0.03. (D and E) Representative example of [Ca2+]i response after Cxcl12 addition in Df1/+ and control MGE-derived neuronal progenitors. For rescue experiments, Dgcr8 or Cxcr4 with EGFP was introduced by lentivirus. Averaged data for the concentration-dependent effect of Cxcl12 on [Ca2+]i responses (E). Error bars represent the SEM. (F and G) Schematic of the experimental paradigm used to analyze the effects of Dgcr8 or Cxcr4 overexpression on the migration deficits of MGE-derived interneurons from Df1/+ mice (F). Brain slice cultures were prepared from Df1/+ and control E13.5 embryos. Dgcr8 or Cxcr4 with EGFP was introduced by lentivirus (G). Dotted lines represent the pallial/subpallial boundary. Note the decreased migration from the MGE to the neocortex (Ncx) in Df1/+ mice and the partial rescue of this migration deficit by Dgcr8 and Cxcr4 72 h after infection [3 days in vitro (DIV3)]. (Scale bar, 300 μm.)
Finally, to examine whether Dgcr8 or Cxcr4 overexpression is sufficient to rescue the migration deficits of Df1/+ MGE-derived interneurons, we used a slice tissue culture assay. Brain slices were prepared from E13.5 embryos of Df1/+ and wild-type mice. Lentiviruses, which express GFP with or without Dgcr8 or Cxcr4, were injected into the MGE (Fig. 3F). Three days after control GFP-expressing lentiviral infection, a decreased number of GFP+ cells were observed in the cortex of Df1/+ brain slices. In contrast, either Dgcr8 or Cxcr4 overexpression at least partially rescued the number of GFP-positive cells in the cortex of Df1/+ brain slices (Fig. 3 F and G). Taken together, our data suggest that interneuron migration deficits in Df1/+ mice are caused by Cxcr4/Cxcl12 signaling deficits, and we show that their decreased responsiveness to Cxcl12 can be rescued by the overexpression of Dgcr8.
Changes in Downstream Targets of Dgcr8 in the Df1/+ MGE.
miR-200a and miR-224 have been reported to increase Cxcr4 expression (25, 26). Thus, we examined the expression of these miRNAs in the MGE of E13.5 Df1/+ mice by quantitative real-time RT-PCR. miR-200a was decreased in the Df1/+ MGE, whereas miR-224 was not affected [miR-200a, P = 0.026; miR-224, P = 0.93 (n = 3 embryos) (Student t test)] (Fig. S9A). To determine whether loss of miR-200a causes Cxcr4 down-regulation in MGE-derived neural progenitors, we used LNA miRNA inhibitors. MGE-derived neural progenitors were transfected with anti–miR-200a or anti–miR-224. Anti–miR-200a but not anti–miR-224 down-regulated Cxcr4 expression 3 d after transfection compared with a negative control inhibitor (Fig. S9B). The decreased level of miR-200a may be partially responsible for the Cxcr4/Cxcl12 signaling deficits in Df1/+ mice.
Defects in Migration of MGE-Derived Cells and Dentate Precursor Cells in Mice of Dgcr8 Haploinsufficiency.
To examine whether Dgcr8 haploinsufficiency affects the development of cortical interneurons and the DG, we analyzed the neurodevelopment of Dgcr8 heterozygous mice. Immunohistochemical analysis of Lhx6 and Gad67 revealed that the distribution of interneurons was also altered in the E18.5 Dgcr8+/− cortex [Lhx6, layer × genotype interaction, F4,24 = 4.21, P = 0.01 (n = 4 embryos); Gad67, layer × genotype interaction, F4,16 = 5.74, P = 0.0046 (n = 3 embryos) (ANOVA)] (Fig. 1 A and B and Fig. S1 B and C). Cxcr4 expression is also decreased in the E18.5 Dgcr8+/− cortex [genotype, F1,4 = 19.50, P = 0.012 (n = 3) (ANOVA)] (Fig. 1 C and D). E13.5 Dgcr8+/− MGE-derived cells also showed decreased responsiveness to Cxcl12 when cocultured with Cxcl12-expressing 293T cells [genotype, F1.7 = 11.34, P = 0.011 (n = 3–6 embryos) (ANOVA)] (Fig. 1 I and J).
Like the Df1/+ mice, the volume of the DG of P0 Dgcr8+/− mice is decreased [P = 0.036 (n = 6 mice) (Student t test)] (Fig. S4B). A BrdU pulse experiment showed migration deficits of dentate precursor cells in the Dgcr8+/− HP [genotype × region interaction; rostral, F2,8 = 9.77, P = 0.0071 (n = 3 embryos); caudal, F2,8 = 9.42, P = 0.0079 (n = 3 embryos) (ANOVA)] (Fig. 2 D and E), which was also confirmed by ectopic Nestin-positive precursors (Fig. S6). E17.5 Dgcr8+/− dVZ-derived cells exhibited a decreased response to Cxcl12 [genotype, F1,4 = 9.19, P = 0.039 (n = 3 embryos) (ANOVA)] (Fig. S7C), although the reduced Cxcr4 expression in the E18.0 Dgcr8+/− HP was not statistically significant [genotype, F1,4 = 3.57, P = 0.13 (n = 3 mice) (ANOVA)] (Fig. 2A). Taken together, these data suggest that Dgcr8 is essential for the normal migration of cortical MGE-derived cells and hippocampal dentate precursor cells.
CXCL12 Is Reduced in the Neuronal Layers of the Olfactory Epithelium in Patients with SZ.
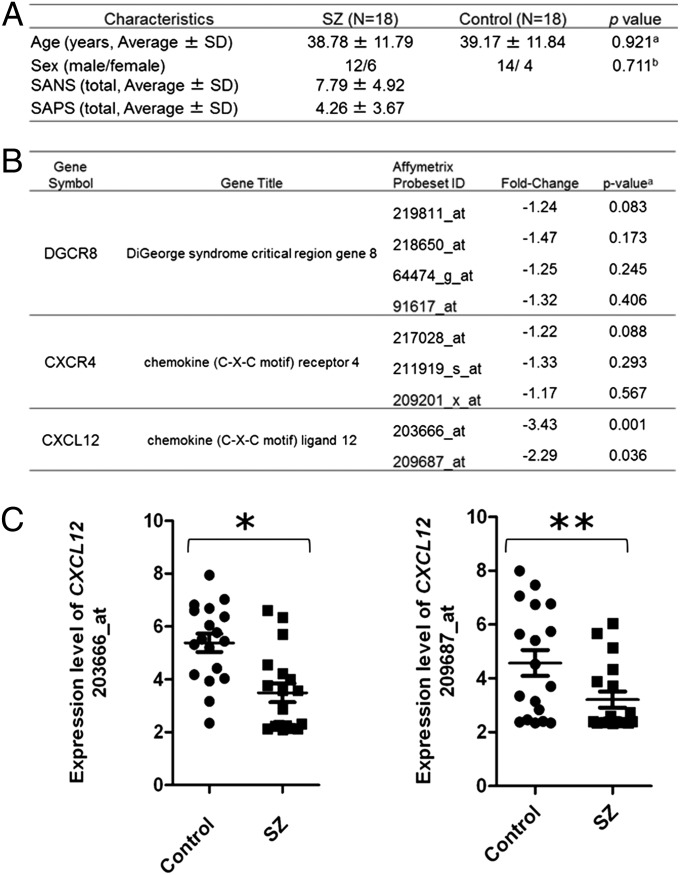
The above data suggest the significance of Dgcr8 and its downstream Cxcr4/Cxcl12 signaling in the developmental abnormalities in a 22q11DS mouse model. Next, we examined whether this molecular pathway might be altered in neuronal cells in patients with SZ. It is very difficult to obtain neurons or neuronal cells from living subjects. However, via nasal biopsy combined with laser-capture microdissection, we can obtain neuronal layers of the olfactory epithelium (27). By using this methodology, we compared the expression of CXCR4, CXCL12, and DGCR8 in the neuronal layers of the olfactory epithelium between normal controls and sporadic cases of SZ. The demographic summary is presented in Fig. 4A. We observed a significant reduction in the expression of CXCL12 in patients with SZ compared with normal controls, whereas no difference was observed in the expression of CXCR4 or DGCR8 between these two groups (Fig. 4 B and C). A significant difference in the expression of CXCL12 in sporadic cases of SZ compared with matched normal controls suggests that the CXCR4/CXCL12 pathway may contribute to the pathophysiology of SZ and be disturbed in a substantial subgroup diagnosed with SZ.
Fig. 4.
CXCL12 is decreased in the olfactory neuronal layer in patients with SZ. (A) Demographics of human subjects in this study. a, ANOVA; b, Fisher’s exact test; SANS and SAPS, Scales for the Assessment of Positive and Negative Symptoms, respectively. (B) Microarray analysis of DGCR8, CXCR4, and CXCL12 expression in the laser-capture microdissected neuronal layer of the olfactory epithelium from control and SZ subjects. (C) The expression level of CXCL12 for each control (circles) and SZ (squares) subject. The results of each probe are shown separately and correspond to the Affymetrix probeset ID shown in B. *P = 0.001, **P = 0.036.
Discussion
The main findings of the present study are as follows: We demonstrated that the haplodeletion of 18 orthologs of the human genes in the 22q11.2 region (Df1/+ mice) causes defects in cortical interneuron migration and hippocampal dentate precursor migration and shows functional abnormalities in Cxcr4/Cxcl12 signaling. This study shows a direct link between the 22q11 microdeletion and these developmental abnormalities via a Cxcr4/Cxcl12 signaling deficiency. With multiple lines of evidence, we proved that Dgcr8 in the 22q11 region might play, at least in part, a role as an upstream master regulator. In parallel, in microdissection of human neuronal tissues via nasal biopsy, we showed that CXCL12 expression was significantly decreased in sporadic SZ patients compared with normal controls. Although it is unlikely, we acknowledge that the CXCL12 signal may also come from nonneuronal cells, such as macrophages and sustentacular cells.
Our findings are consistent with a recent study by Meechan et al. (10), in which the disturbances in the placement of PV interneurons and the reduced expression of Cxcr4 were reported in another mouse model of 22q11DS (LgDel/+ mice). In the present study, by using rescue experiments of Df1/+ interneuron migration, we could pin down the pivotal role of Dgcr8-mediated miRNA regulation in the downstream phenotypes of 22q11 deletion, including Cxcr4/Cxcl12 functional deficits. miRNA-mediated regulation can buffer increases or reductions in gene dosage (28, 29). Haploinsufficiency of Dgcr8 might disrupt miRNA-mediated buffering effects, and uncovers the effects of 22q11 microdeletion as well as biological or environmental perturbations. This study will open a window to study Cxcr4/Cxcl12 functional deficits more mechanistically in the context of 22q11DS, in particular in the link with Dgcr8.
Although 22q11DS accounts for only a very small subset of SZ, we found significant reduction of CXCL12 levels, but failed to observe changes in DGCR8 levels, in sporadic SZ patients. Of note, in 22q11DS mouse models, Cxcr4 is affected. We believe that these two positive (changes in either Cxcr4 or CXCR12) and negative (no change in DGCR8) results are equally important to explore a possible link between 22q11DS and SZ. The positive results of CXCL12 suggest that Cxcr4/Cxcl12 signaling deficits observed in 22q11DS mouse models may have some relevance in SZ pathophysiology. DGCR8 may underlie an upstream etiology for aberrant CXCR4/CXCL12 signaling deficits in 22q11DS-associated mental manifestation (including SZ-like disturbances). However, this specific etiology may not account for most cases of sporadic SZ. This working hypothesis fits with a well-appreciated notion that SZ is caused by multiple etiologies but has some levels of commonality in the pathophysiology. The proposal from the present study that CXCR4/CXCL12 may underlie a common pathophysiology of SZ may have translational potential—for example, aiding biomarker cultivation in SZ and related conditions.
Materials and Methods
The generation of Df1/+ 22q11DS model mice was previously described (30). XG058 ES cells (BayGenomics; http://baygenomics.ucsf.edu) were used to generate Dgcr8+/− mice. Mice were maintained on a C57Bl6 genetic background for at least 11 generations. Mouse colonies were maintained in accordance with the protocols approved by the Committee on Animal Research at the Research Institute, Shiga Medical Center.
Olfactory epithelium tissues were obtained by nasal biopsy as previously described (27). Full details of subjects and clinical assessment and the analysis of the microarray are presented in SI Materials and Methods.
Full descriptions of the volumetric measurement of the hippocampus, as well as descriptions of BrdU labeling analysis, immunohistochemical analysis, in situ hybridization, chemotaxis assay, explant coculture, stripe choice assay, slice culture, neural progenitor cell culture, Ca imaging, miRNA real-time RT-PCR, miRNA knockdown studies, and real-time RT-PCR are detailed in SI Materials and Methods.
Statistical significance was assessed by unpaired Student t test. All data, unless stated otherwise, are expressed as mean ± SD. For comparisons of more than two groups, one- or two-way repeated-measures analysis of variance (ANOVA) followed by Fisher’s LSD test was used. A probability of less than 5% (P < 0.05) was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 25670154 (to K. Tanigaki), the NOVARTIS Foundation for the Promotion of Science (K. Tanigaki), Uehara Memorial Foundation (K. Tanigaki), US Public Health Service Grants MH-084018, MH-094268 Silvio O. Conte Center (The Johns Hopkins School of Medicine), MH-069853, MH-085226, MH-088753, and MH-092443, the Stanley, RUSK, and S-R Foundations, the National Alliance for Research on Schizophrenia and Depression, and the Maryland Stem Cell Research Fund (all to A.S.). This project is partly funded by Astellas Pharma (A.S.).
Footnotes
Conflict of interest statement: H.H., K. Tajinda, and N.K. are employees of Astellas Pharma, Inc.
This article is a PNAS Direct Submission. R.J.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312661110/-/DCSupplemental.
References
- 1.Pulver AE, et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis. 1994;182(8):476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Karayiorgou M, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92(17):7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puech A, et al. Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc Natl Acad Sci USA. 1997;94(26):14608–14613. doi: 10.1073/pnas.94.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay EA, et al. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401(6751):379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 6.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40(6):751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 7.Paylor R, Lindsay E. Mouse models of 22q11 deletion syndrome. Biol Psychiatry. 2006;59(12):1172–1179. doi: 10.1016/j.biopsych.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104(4):619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 9.Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci USA. 2009;106(38):16434–16445. doi: 10.1073/pnas.0905696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Cxcr4 regulation of interneuron migration is disrupted in 22q11.2 deletion syndrome. Proc Natl Acad Sci USA. 2012;109(45):18601–18606. doi: 10.1073/pnas.1211507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo TU, Miller JL, Lewis DA. Schizophrenia and the parvalbumin-containing class of cortical local circuit neurons. Am J Psychiatry. 1997;154(7):1013–1015. doi: 10.1176/ajp.154.7.1013. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto T, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimoto S, et al. Selective overexpression of Comt in prefrontal cortex rescues schizophrenia-like phenotypes in a mouse model of 22q11 deletion syndrome. Transcult Psychiatry. 2012;2:e146. doi: 10.1038/tp.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fénelon K, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci USA. 2011;108(11):4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stumm RK, et al. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23(12):5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiveron MC, et al. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci. 2006;26(51):13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Bendito G, et al. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci. 2008;28(7):1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flames N, et al. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44(2):251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 20.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Kataoka H, Coughlin SR, Pleasure SJ. Identification of a transient subpial neurogenic zone in the developing dentate gyrus and its regulation by Cxcl12 and reelin signaling. Development. 2009;136(2):327–335. doi: 10.1242/dev.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagri A, et al. The chemokine SDF1 regulates migration of dentate granule cells. Development. 2002;129(18):4249–4260. doi: 10.1242/dev.129.18.4249. [DOI] [PubMed] [Google Scholar]
- 23.Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci USA. 2002;99(10):7090–7095. doi: 10.1073/pnas.092013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127(3):457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 25.Rosati J, et al. Smad-interacting protein-1 and microRNA 200 family define a nitric oxide-dependent molecular circuitry involved in embryonic stem cell mesendoderm differentiation. Arterioscler Thromb Vasc Biol. 2011;31(4):898–907. doi: 10.1161/ATVBAHA.110.214478. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, et al. MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochem Biophys Res Commun. 2012;425(2):127–133. doi: 10.1016/j.bbrc.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 27.Tajinda K, et al. Neuronal biomarkers from patients with mental illnesses: A novel method through nasal biopsy combined with laser-captured microdissection. Mol Psychiatry. 2010;15(3):231–232. doi: 10.1038/mp.2009.73. [DOI] [PubMed] [Google Scholar]
- 28.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43(3):204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 2010;24(13):1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paylor R, et al. Mice deleted for the DiGeorge/velocardiofacial syndrome region show abnormal sensorimotor gating and learning and memory impairments. Hum Mol Genet. 2001;10(23):2645–2650. doi: 10.1093/hmg/10.23.2645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.