Significance
Our work establishes that endogenous regulation of the activity of the tumor-suppressor p53 is a critical component in vertebrate limb regeneration and is required for the process to occur. We show that down-regulation of p53 is required for cell cycle reentry of postmitotic differentiated cells, the critical step in the formation of the progenitors for the process of regeneration. We propose that the absence of tumor-suppressors that stabilize p53 and the presence of dominant-negative isoforms, permit regeneration by negative regulation of p53. This research sheds light onto previously unknown molecular mechanisms critical for limb regeneration and offers a new perspective on the functions of the tumour suppressor p53.
Keywords: myogenesis, chondrogenesis, p73, carcinogenesis
Abstract
Extensive regeneration of the vertebrate body plan is found in salamander and fish species. In these organisms, regeneration takes place through reprogramming of differentiated cells, proliferation, and subsequent redifferentiation of adult tissues. Such plasticity is rarely found in adult mammalian tissues, and this has been proposed as the basis of their inability to regenerate complex structures. Despite their importance, the mechanisms underlying the regulation of the differentiated state during regeneration remain unclear. Here, we analyzed the role of the tumor-suppressor p53 during salamander limb regeneration. The activity of p53 initially decreases and then returns to baseline. Its down-regulation is required for formation of the blastema, and its up-regulation is necessary for the redifferentiation phase. Importantly, we show that a decrease in the level of p53 activity is critical for cell cycle reentry of postmitotic, differentiated cells, whereas an increase is required for muscle differentiation. In addition, we have uncovered a potential mechanism for the regulation of p53 during limb regeneration, based on its competitive inhibition by ΔNp73. Our results suggest that the regulation of p53 activity is a pivotal mechanism that controls the plasticity of the differentiated state during regeneration.
Unlike mammals, which exhibit limited regenerative abilities, the urodele amphibians—or salamanders—are capable of regenerating an extraordinary range of body structures, including ocular tissues, tail, sections of the heart, parts of the nervous system, and entire limbs (1). In salamanders, such as the newt and axolotl, limb regeneration depends on the formation of a blastema, a mound of progenitor cells of restricted potential that arises after amputation (2–4). Following a period of proliferation, blastema cells redifferentiate and restore the structures of the limb.
Extensive evidence indicates that limb regeneration depends on reprogramming of cells in mature limb tissues. Upon amputation, muscle, cartilage, and connective tissue cells underneath the injury site lose their differentiated characteristics and re-enter the cell cycle to give rise to the blastema (5–8). This mechanism has also been observed during zebrafish heart and fin regeneration (9, 10). In contrast, reversals of the differentiated state are rarely observed in mammalian tissues, which led to the suggestion that inability to undergo dedifferentiation could contribute to the failure of regeneration in mammals (11). Despite their significance, the mechanisms underlying regulation of the differentiated state during vertebrate regeneration remain poorly understood.
Recently, the tumor suppressor p53, whose best-characterized functions are in the maintenance of genome stability (12), has been implicated in the suppression of artificial cell reprogramming to pluripotency (13–17) and the promotion of differentiation pathways in mammals (18). In addition, it has been observed that inhibiting p53 disrupts limb regrowth in salamanders (19), although its role in this context has remained unknown. It is possible that p53 could play a role in the regulation of dedifferentiation and redifferentiation events intrinsic to vertebrate regeneration. Our results demonstrate that the regulation of p53 activity is critical for limb regeneration by controlling key cell fate decisions throughout this process.
Results
p53 Activity Is Regulated During Limb Regeneration.
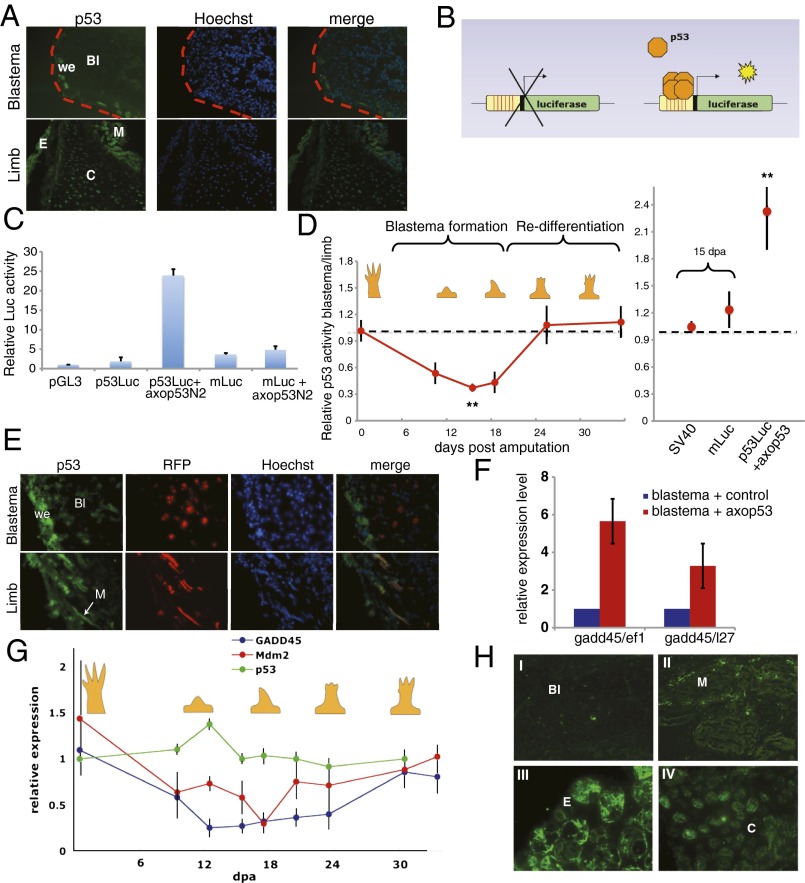
We initially characterized the protein-expression pattern of p53 in both regenerating and intact tissues. The protein levels of p53 were markedly decreased in mid-bud blastemas (Fig. 1A), with the exception of a few glandular Leydig cells underneath the wound epidermis (Fig. S1A). In contrast, the differentiated tissues of the corresponding limb, such as epidermis and muscle, showed strong p53 staining (Fig. 1A), suggesting that the protein levels of p53 in the regenerating blastema are significantly decreased.
Fig. 1.
p53 activity is regulated during limb regeneration. (A) Representative longitudinal section of a blastema at 15 dpa or its corresponding limb, stained with antibodies against axolotl p53 and Hoechst 33258. (B) Schematic representation of the p53 luciferase reporter assay. (C) Luciferase activity assay in axolotl AL1 cells 48 h after transfection with the indicated constructs. Luciferase level was normalized to the level of Renilla luciferase, and expressed in relation to the pGL3 control vector. (D) Luciferase activity assay at different stages of limb regeneration. p53Luc was electroporated alongside a Renilla luciferase control vector into both limbs of an axolotl (0 dpa, p53Luc+axop53), or into a blastema and its contralateral limb (10, 15, 18, 25, 35 dpa; SV40, mLuc). Luciferase activity was normalized to the level of Renilla luciferase and expressed relative to the activity of a single intact limb. SV40 is a luciferase expression vector without p53 binding sites (**P < 0.01). (E) Representative longitudinal section of a blastema or limb following electroporation of pRFP-N2 and staining with antibodies against axolotl p53 and Hoechst 33258. (F) qRT-PCR analysis of Gadd45 levels in blastemas, normalized to Ef1-α or L-27, 72 h after electroporation with the indicated vectors. (G) qRT-PCR analysis of Gadd45, Mdm2, and p53 expression levels in blastemas at different stages of regeneration relative to a normal limb, normalized to those of Ef1-α. Similar results were obtained normalizing to L27. (H) Representative images of blastemal (I), muscle (II), epidermal (III), and cartilage (IV) cells stained with antibodies against axolotl Gadd45. Values are given as means ± SEM, n = 6 in C and F; n = 8 in D and G. Bl, blastema mesenchyme; C. cartilage E, epidermis; M, muscle; we, wound epidermis. (Magnification: A, 20×; E, 40×; H, I and II, 40×; H, III and IV, 60×.)
Because p53 is regulated both by protein stabilization and by modulating its activity (20, 21), we assessed the net activity of p53 during regeneration using a p53 reporter (p53Luc), in which firefly luciferase is driven by 13 consensus p53 binding sites (Fig. 1B). The activity of the p53 reporter increased 20-fold upon overexpression of axolotl p53 in an axolotl cell line, AL1 (Fig. 1C). This effect was both dose-dependent (Fig. S1B) and specific, as a reporter in which the p53 binding sites are mutated (mLuc) did not respond to p53 overexpression (Fig. 1C). The reporter, along with a control plasmid, was introduced by electroporation into blastemas at different stages of regeneration and into their contralateral intact limbs, and the relative luciferase activity was quantified. The level of p53 activity declined up to threefold concomitant with the formation of the blastema, and recovered to that of the contralateral limb during the redifferentiation phase (Fig. 1D). As expected, the expression of the p53 mutant reporter or an SV40 promoter-driven luciferase vector did not result in differences in luciferase activity between limb and mid-bud blastemas (Fig. 1D). By delivering red fluorescent protein (RFP) constructs into limbs and blastemas under similar conditions to those used for the p53 reporter, we noted that only mesenchymal cells in the blastema, limb muscle, and limb connective tissue cells can be successfully electroporated, but not epidermis or cartilage (Fig. 1E). This finding suggests that the observed changes in p53 activity reflect differences between the mesenchymal elements of regenerating and intact limbs.
In parallel, we analyzed changes in the expression levels of two p53-regulated genes, growth arrest and DNA damage 45 (gadd45) and mouse double minute 2 (mdm2) (21), which are also p53 targets in the axolotl (Fig. 1F). Quantitative real-time PCR (qRT-PCR) analysis revealed a decrease in the expression levels of Gadd45 and Mdm2 between the early (9 d postamputation, dpa) and late (18 dpa) bud stages, corresponding to the period of blastema formation, followed by a return to the initial levels upon redifferentiation (Fig. 1G). No changes in the levels of p53 mRNA were observed, suggesting that p53 regulation takes place at the posttranscriptional level (Fig. 1G). Additionally, we observed a significant decrease of Gadd45 protein levels in mesenchymal blastemal cells compared with mesenchymal limb tissues, such as muscle and cartilage (Fig. 1H), strongly supporting the observations made using the p53 reporter. These data suggest that p53 activity is tightly regulated during limb regeneration in the mesenchymal compartment, being down-regulated during the stage of blastema formation and up-regulated to its original level during the redifferentiation phase.
Modulation of p53 Activity Is Key to Achieve Proper Regeneration.
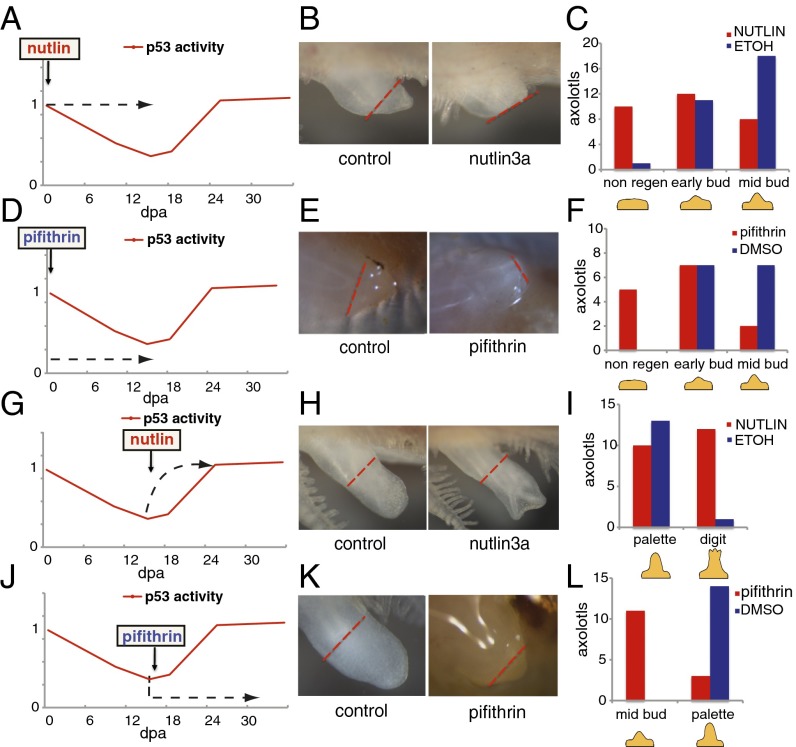
To investigate the biological relevance of modulating p53 activity, we took advantage of two well-characterized compounds: α-pifithrin, a p53 inhibitor (22), and nutlin3a, a p53 stabilizer, which disrupts the p53–Mdm2 interaction (23). Both compounds act as p53 modulators in AL1 cells, as they induce the predicted changes in the expression levels of Gadd45 and Mdm2 (Fig. S2). To evaluate the outcome of disrupting the normal pattern of p53 activity during limb regeneration, we treated axolotls with α-pifithrin and nutlin3a during key stages of this process (Fig. 2).
Fig. 2.
Regulation of p53 activity is necessary for limb regeneration. Schematic representation of axolotl treatment programs with nutlin3a (A and G) or α-pifithrin (D and J) during the blastema formation or redifferentiation phases of limb regeneration. Dotted arrow indicates the duration of each treatment and how they affect p53 activity. (B, E, H, and K) Representative images of blastemas 10 d after the treatment indicated in A, D, G, and J, and the equivalent experiment using vehicle alone (control). (C, F, I, and L) The number of axolotls in each stage of regeneration at the end of the corresponding treatment as indicated in A, D, G, and J.
Stabilization of the p53 level at the time of blastema formation, when it normally decreases, led to an impairment of the regeneration process (Fig. 2 A–C and Fig. S3). The defects induced by nutlin3a treatment were reversible and correlated with the levels of Gadd45 up-regulation in individual animals (Fig. S2). Furthermore, the defects were not caused by an increase in apoptosis, as no change in the number of apoptotic cells was observed between treatments (Fig. S2). This finding suggests that down-regulation of p53 activity is required for blastema formation. Nonetheless, treatment with α-pifithrin during this period also led to an impairment in regeneration (Fig. 2 D–F and Fig. S3) that was reversible and correlated with the inhibition of Gadd45 (Fig. S2), suggesting that a minimum level of p53 activity is required for blastema formation. In addition, neither nutlin nor α-pifithrin treatment lead to changes in expression of the p53 family members p63 and p73 (Fig. S2). Taken together, these observations support the existence of upper and lower thresholds of p53 activity, above or below which blastema formation is impaired.
Next, we analyzed the effects of p53 stabilization on the redifferentiation phase. Interestingly, nutlin3a treatment resulted in a detectable acceleration of regeneration. Blastemas exposed to constant nutlin3a from the mid-bud stage reached the digit stage ∼3 d earlier than their control counterparts (Fig. 2 G–I and Fig. S3), suggesting that p53 could play a role during the redifferentiation phase. To test this theory further, α-pifithrin was administered from the mid-bud stage of regeneration onwards, when p53 activity increases to its normal levels. Whereas the blastemas from control treated axolotls progressed to the palette stage after 10 d of treatment, α-pifithrin–treated blastemas remained at the mid-bud stage (Fig. 2 J–L and Fig. S3), suggesting that p53 activity is required for redifferentiation. In support of this finding, the overexpression of the p53 dominant-negative construct DDp53, from the mid-bud stage onwards, resulted in delayed and defective regeneration (see also Fig. 5 F–H and Fig. S7).
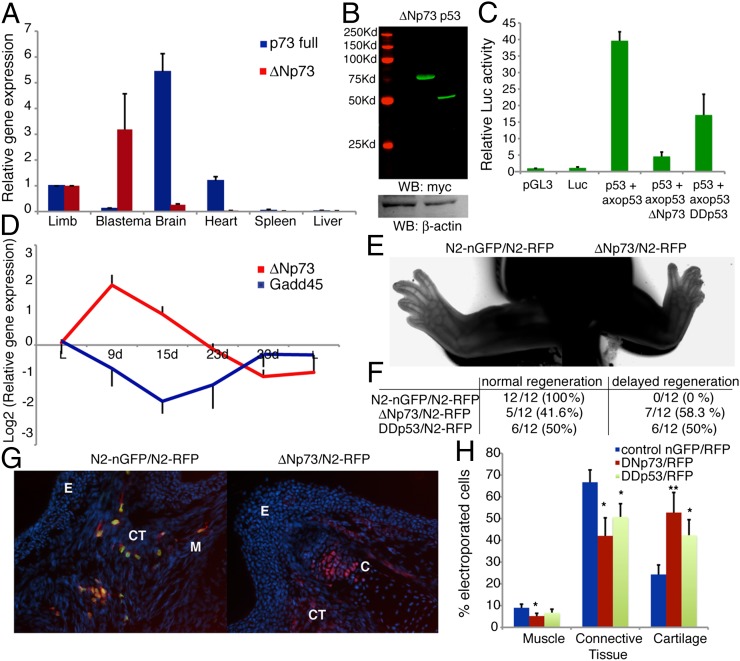
Fig. 5.
ΔNp73 acts as a p53 dominant negative and is up-regulated during blastema formation. (A) qRT-PCR analysis of axolotl ΔNp73 or full-length p73 in several tissues relative to their levels in normal limbs, normalized to Ef1-α. (B) Western blot analysis of myc-tagged proteins and β-actin in AL1 cell extracts 48 h posttransfection with axolotl ΔNp73-myc or p53-myc constructs. (C) Luciferase activity assay in AL1 cells 48 h posttransfection with the indicated vectors, normalized to the level of Renilla luciferase and expressed relative to the activity of pGL3 control vector. (D) qRT-PCR analysis of axolotl ΔNp73 and Gadd45 expression levels in regenerating blastemas relative to a normal limb, normalized to those of Ef1-α. (E) Regenerated limbs, 3.5 wk after coelectroporation of pair-matched mid-bud blastemas with either N2-nGFP or ΔNp73 alongside N2-RFP. (F) Quantification of limbs exhibiting normal or delayed regeneration 3.5 wk following electroporation of the indicated constructs. (G) Representative longitudinal sections of regenerated limbs. Note characteristic cell foci in cartilage of ΔNp73-expressing cells. (Magnification: 30×.) (H) Percentage of electroporated cells incorporated into muscle, connective tissue and cartilage following electroporation as indicated in F. Values represent the mean ± SEM, n = 4 in A–C; n = 6 in D and H (*P < 0.05, **P < 0.01). E, epidermis; C cartilage; CT, connective tissue; M, muscle.
Although these observations indicate that the regulation of its activity is essential for limb regeneration, the specific functions of p53 during this process remained obscure. To gain insights into this problem, we investigated the biological functions of salamander p53. In mammals, p53 participates in the maintenance of genome stability by regulating gene expression upon DNA injury, leading to cell cycle arrest, apoptosis, or senescence (12). To determine whether this result is also true for salamander p53, we exposed axolotl AL1 or newt (Notophthalmus viridescens) A1 cells to UV damage. This process resulted in p53 stabilization and up-regulation of Gadd45 (Fig. S4), suggesting that salamander p53 responds to UV-induced DNA damage much like its mammalian counterpart. In addition, an increase in p53 protein levels led to a decrease in the proportion of cells in S-phase, implying that the role of p53 as a negative cell cycle regulator is conserved in salamanders. Thus, the genome-protective functions of p53 are conserved in salamanders and could be important during regeneration.
Regulation of p53 Activity Is Essential for Myogenesis.
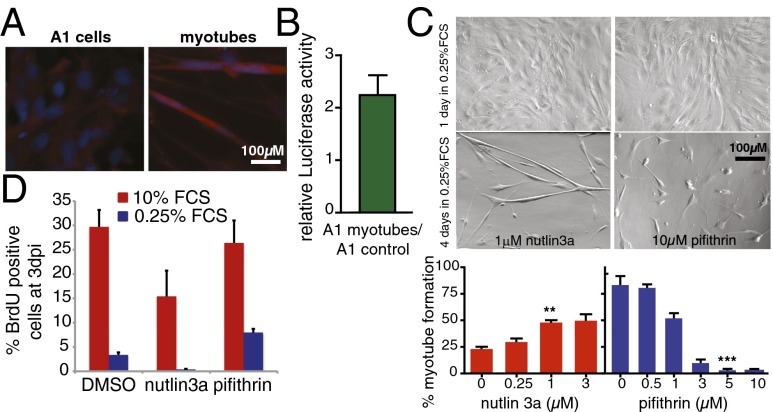
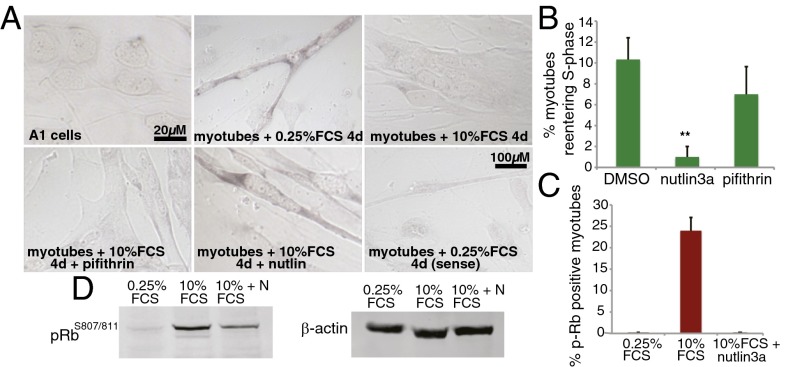
The requirement for p53 at later stages of limb regeneration suggests that p53 could be involved in cell differentiation processes intrinsic to the redifferentiation phase. To test whether this is a function of salamander p53, we used an amphibian model of myogenesis, the A1 cell line, which can be induced to form myotubes following a 4-d period in 0.25% FCS (24). Staining with p53 antibodies showed increased protein levels of p53 in myotubes compared with undifferentiated cells (Fig. 3A). In addition, the relative levels of p53 activity were increased 2.5-fold in newly formed myotubes (Fig. 3B). To determine whether such changes are relevant for myotube formation, we studied the effects of α-pifithrin or nutlin3a treatment during the induction of myogenesis. Treatment with the p53 stabilizer nutlin3a increased the formation of myotubes in 2% FCS, a less-favorable condition for myogenesis (Fig. 3C). In contrast, administration of α-pifithrin abrogated myotube formation in a reversible (Fig. S5) and dose-dependent manner (Fig. 3C). Notably, p53 inhibition did not preclude cell cycle withdrawal before myogenesis (Fig. 3D). Taken together, these results demonstrate that p53 activity is required for myotube differentiation in salamander cells.
Fig. 3.
p53 activity is essential for myogenesis. (A) Representative image of A1 cells before and 4 d after inducing myotube formation, stained with antibodies against axolotl p53 (red) and Hoechst 33258 (blue). (B) Luciferase activity assay in A1 myotubes (4 d postinduction) relative to that of uninduced A1 cells. A1 cells were transfected with p53Luc and Renilla luciferase. Firefly luciferase level was normalized to the level of Renilla luciferase and expressed as the ratio (A1 myotubes/A1 control), n = 10. (C, Upper) phase contrast micrograph of A1 cells 1 and 4 d after induction of myogenesis. Cells were induced in the presence of 1 µM nutlin3a or 10 µM α-pifithrin. (Lower) Quantification of myotube formation (number of nuclei within myotubes relative to total nuclei expressed as a percentage) in A1 cells were incubated for 4 d in 0.25% FCS in the presence of the indicated doses of nutlin3a or α-pifithrin. (**P < 0.01, ***P < 0.001). (D) Quantification of BrdU+ cells at 3 d after myogenesis induction in the presence of 1 µM nutlin3a, 10 µM α-pifithrin, or DMSO. Values represent the mean ± SEM, n = 6 (C and D).
Postmitotic Cell Cycle Reentry Depends on the Levels of p53 Activity.
Cell cycle reentry of postmitotic differentiated cells is a key event in blastema formation. Given its role in the promotion of differentiation and cell cycle control, the level of p53 activity could be important for this process. Unlike mammalian muscle, salamander myofibers reenter the cell cycle during regeneration. A1 myotubes reenter the cell cycle on serum stimulation (24), providing a valuable model to study factors implicated in this reponse, including p53. In this system, we observed that serum stimulation induced a decrease in the expression of the p53 target gene Gadd45 (Fig. 4A), suggesting that p53 activity is down-regulated under conditions that promote myotube S-phase reentry. To address the importance of such down-regulation, myotubes were treated with 10% FCS along with nutlin3a or α-pifithrin. Stabilization of p53 to equivalent levels present in myotubes before serum stimulation, led to a substantial decrease in the percentage of myotubes in the S-phase (Fig. 4 A and B, and Fig. S6), suggesting that the down-regulation of p53 activity is required for this process. To explore this theory further, we focused on Rb, a critical inducer of irreversible cell cycle withdrawal in mammalian myogenesis (25), the phosphorylation of which is required for cell cycle reentry in salamander myotubes (24). The staining of myotubes with antibodies against phosphorylated RbS807/811 (Fig. 4C and Fig. S6), as well as the analysis of protein extracts (Fig. 4D), revealed a substantial decrease of phosphorylated Rb in myotubes treated with nutlin3a. This finding indicates that p53 stabilization leads to an inhibition of S-phase reentry in differentiated muscle cells by preventing Rb phosphorylation, supporting the view that postmitotic cell cycle reentry depends on the levels of p53.
Fig. 4.
Down-regulation of p53 levels is required for postmitotic cell cycle reentry. (A) In situ hybridization analysis of Gadd45 mRNA expression in A1 cells and myotubes following the indicated treatments. (B) Percentage of myotubes entering S-phase as measured by BrdU incorporation. Myotubes were induced under normal conditions and incubated for 2.5 d in 10% FCS in the presence of DMSO, 1 µM nutlin3a, or 10 µM α-pifithrin (**P < 0.01). (C) Quantification of p-RBS807/811-positive cells. Myotubes were incubated in 0.25% FCS or 10% FCS with 1 µM nutlin3a or vehicle, then fixed and stained as described. (D) Western blot analysis of myotube extracts—including mononucleates—indicating levels of p-RBS807/811 and β-actin. Values from B and C represent the mean ± SEM, n = 6.
Salamander ΔNp73 Acts as a p53 Dominant-Negative and Its Modulation Is Necessary for Limb Regeneration.
To gain insight into the mechanisms behind the regulation of p53 activity, we analyzed the expression patterns of the p53 family members p63 and p73 in regeneration. Whereas p63 levels did not change, we observed a significant up-regulation of p73 in mid-bud blastemas (Fig. S7A). A variety of p73 isoforms have been reported, all of which can compete with active p53 for binding to its DNA targets (26, 27). The ΔNp73 isoform, which lacks the N-terminal transactivation domain (Fig. S7B), has been shown to inhibit p53 functions in mammals by acting as a p53 dominant-negative (28–31). Notably, this form was significantly up-regulated in blastemas compared with differentiated tissues (Fig. 5A) in contrast to full-length p73, which exhibited the inverse expression profile. Absolute mRNA quantification of the p73 isoforms by qRT-PCR indicates that the mRNA levels of ΔNp73 are 10-fold higher than the full-length isoform in mid-bud blastemas, although being barely detectable (<1% of total p73 expression) in limbs. These results suggest a specific role for ΔNp73 in limb regeneration.
Next, we observed that coexpression of axolotl p53 and ΔNp73, at comparable levels, is able to abrogate p53-mediated transactivation of the p53 reporter in axolotl cells, much like the overexpression of the p53 dominant-negative DDp53 (Fig. 5 B and C), suggesting that axolotl ΔNp73 functions as a p53 dominant-negative. Furthermore, the up-regulation of ΔNp73 precedes the down-regulation of p53 activity during limb regeneration, whereas the decline in ΔNp73 levels coincides with the up-regulation of p53 as the differentiation stage begins (Fig. 5D). Taken together, these results suggest that regulation of p53 activity during limb regeneration may be achieved—at least in part—through modulating ΔNp73 gene expression.
Finally, we probed the biological relevance of the modulation of ΔNp73 by disrupting its expression pattern during axolotl limb regeneration. Electroporation of either DDp53 or ΔNp73 into blastemas led to regeneration delays in more than half of the limbs examined, whereas their contralateral control limbs exhibited normal regeneration (Fig. 5 E and F, and Fig. S7). After electroporation of a ΔNp73GFP fusion protein, ΔNp73 expression was maintained (Fig. S7 C–E). These results suggest that the appropriate level of ΔNp73 expression is an important determinant for normal regeneration. Similarly, these data support our observation that the up-regulation of p53 activity is critical during the redifferentiation phase. Additionally, we observed that the percentage of ΔNp73 electroporated cells incorporated into muscle tissue after regeneration decreased by 40%, whereas the incorporation into cartilage was increased by 60% (Fig. 5 G and H). Similar results were observed for DDp53 electroporated cells (Fig. 5H and Fig. S7). This finding is consistent with the observation that inhibition of p53 interferes with myogenesis. Additionally, the result raises the possibility that p53 acts as a negative regulator of cartilage differentiation. Taken together, these results suggest that the down-regulation of ΔNp73, by relieving its competition with p53, may be controlling cell fate decisions during the redifferentiation phase of limb regeneration.
Discussion
p53 and Blastema Formation.
Our analysis of p53 activity after amputation reveals the existence of a time window when the activity of p53 is down-regulated in the mesenchymal compartment, coinciding with the phase of blastema formation. It is notable that the changes in p53 activity take place within mesenchymal tissues; as in limb regeneration, it is the mesenchymal cells underlying the wound epidermis that re-enter the cell cycle and proliferate to form the blastema (2). Therefore, the spatiotemporal regulation of p53 activity is suggestive of a role in blastema formation.
In axolotls, the stabilization of p53 levels by nutlin3a leads to an impairment or delay in the formation of the limb blastema, suggesting that p53 down-regulation is required for this process. These defects are not caused by induction of apoptosis, indicating that p53 down-regulation is not required for the prevention of programmed cell death but instead plays a different role during blastema formation. Physiological down-regulation of p53 has been previously reported in human embryonic stem cells exposed to hypoxia, where it leads to a state of “enhanced stemness” (32), and during development in Xenopus, where it is required for maintaining ectodermal cells in a pluripotent state (33). Furthermore, inhibition of p53 activity by AurK is crucial for the maintenance of both embryonic and induced pluripotency (34), and during induced pluripotent stem cell induction the reprogramming process itself leads to p53 down-regulation (15). These studies indicate a connection between p53 down-regulation and the reversal of the differentiated state. Significantly, extensive evidence suggests that dedifferentiation is a key mechanism underlying blastema formation in vertebrate regeneration (2, 35), raising the possibility that the essential function of p53 down-regulation is to promote the dedifferentiation of postmitotic, differentiated tissues to give rise to the blastema. The evidence presented in this article supports this hypothesis. Our studies on S-phase reentry in differentiated cells, a key index for the reversal of differentiation (2), demonstrate that this process depends on the levels of p53 activity. Moreover, these results suggest a molecular basis for the impairment of regeneration observed following p53 stabilization during the blastema formation phase.
The dependence of myotube cell cycle reentry on the levels of p53 suggests a mechanism by which the down-regulation of p53 levels contributes to the generation of blastemal cells by facilitating dedifferentiation and cell cycle reentry of differentiated limb cells. This theory is strongly supported by the finding that the joint inactivation of Rb, the inactivation of which by hyperphosphorylation we have shown depends on p53 in salamanders, and alternate reading frame (ARF), the primary function of which is to stabilize p53 (36, 37), induces dedifferentiation followed by cell cycle reentry in differentiated mammalian cells, yielding progenitors with regenerative potential (11). Pajcini et al. have proposed that the absence of ARF in lower vertebrates, including urodeles, could underlie the fundamental differences in regenerative ability between mammals and salamanders. In view of our findings, we propose that the absence of ARF in salamanders could facilitate the negative control of p53 levels, which in turn may increase the plasticity of the differentiated state and generate a regeneration-permissive environment in this species. In this context, it is worth noting that lack of expression of a key p53 target gene, p21, was recently shown to improve healing and ear closure in mice (38), which suggests that the functions of the p53 axis in regeneration could extend to mammalian systems.
Our data also suggest the existence of a threshold of p53 activity below which regeneration cannot take place. An explanation for such a requirement may be found in the role of p53 in the maintenance of genome integrity, which we have shown are conserved in salamanders, as has previously been shown in other amphibians (39). Actively dividing cells within the blastema are likely to encounter replicative challenges that would require the genome-protective activities of p53. We suggest that the requirement for some protective functions during blastema formation could account for our finding that a minimal level of p53 is needed even at this stage.
In addition, it has been shown that a p53/p63-related protein is essential for the proliferation and self-renewal of the neoblast population during homeostasis and regeneration in flatworms (40). Even though it is well established that the mechanisms of regeneration used by invertebrates, like planarians, are quite different from those found in vertebrates (41, 42), it is notable that the complete abrogation of p53-like functions in both settings disrupts regeneration.
p53 and the Redifferentiation Phase.
Our results using p53 modulating compounds, as well as electroporation of DDp53 and ΔNp73, suggest that p53 has an instructive role during the redifferentiation phase. This result is in contrast to mammals, where disruption of p53 has no significant impact on differentiation (43, 44), a result often attributed to the existence of functional redundancy between p53 family members (18). In Xenopus, in which p63 and p73 are not expressed during development (45), inhibition of p53 blocks terminal differentiation (46). Furthermore, regulated expression of a p53 interactor, XFDL156, prevents mesoderm specification in the presumptive ectoderm by inhibiting p53 (33). These reports indicate a role for p53 regulation in differentiation processes during amphibian development and are supportive of our findings.
Our studies on myogenesis provide direct evidence of the role of p53 in cell differentiation in urodeles. We found that both protein and activity levels of p53 increase during myogenesis, consistent with previous reports on the differentiation of mammalian muscle in culture (47, 48). Although stabilization of p53 activity during the induction of myogenesis increases the overall efficiency of the process, inhibition of p53 prevents myotube formation, demonstrating that p53 is essential during muscle differentiation in salamander cells. These findings help explain the effects, identified in our study, of both inhibition and stabilization of p53 during late stages of regeneration. Our observations are supported by previous reports that demonstrate that inhibition or depletion of p53 abrogates myogenesis (49–51). It is notable that during myotube formation the inhibition of p53 does not prevent cell cycle withdrawal but does preclude subsequent stages of differentiation. This finding indicates that p53 acts on the differentiation program independent of its functions in cell cycle regulation.
ΔNp73: A Putative Regulator of p53 Activity.
Although our results have established that the tight regulation of p53 activity is important for limb regeneration, an equally important question is how is such regulation achieved. Even though the protein levels of p53 are decreased in blastema cells, this cannot fully account for the observed changes in p53 activity. Our data strongly suggest that a truncated form of p73, ΔNp73, may play an important role. We show that this form acts as a p53 dominant-negative in the salamander system, as it does in humans (31), that it exhibits an inverse pattern of expression to that of p53 activity throughout regeneration, and that it is up-regulated only in mid-bud blastemas. This finding suggests that ΔNp73 could have a specific role in regeneration, through the targeted competitive inhibition of p53. Critically, we show that overexpression of ΔNp73 during the redifferentiation phase leads to defects in regeneration that are similar to those induced by overexpression of DDp53 and, although milder (likely because of less than 100% transfection efficiency), are reminiscent of the regeneration defects caused by pifithrin-mediated inhibition of p53. Furthermore, ΔNp73 electroporation causes a decrease in the proportion of cells differentiating into muscle, consistent with both our observations using the p53 inhibitor during myogenesis, as well as a previous study describing the inhibition of myogenesis by ΔNp73 (28).
Interestingly, it has been shown that overexpression of ΔNp73 increases human induced pluripotent stem cell reprogramming efficiency (52), mirroring the effect of down-regulating p53 by siRNA (17). Thus, it is notable that ΔNp73 has been shown to abrogate functions of p53 that are relevant to regeneration. Interestingly, in contrast to their effect on muscle cells, ΔNp73 and DDp53 have a positive effect on cartilage formation, suggesting that p53 might be a negative regulator of this process. Supporting this hypothesis, a recent study has demonstrated that down-regulation of Mdm2, leading to p53 stabilization, inhibits chondrogenesis (53).
A detailed understanding of the molecular mechanisms underlying vertebrate regeneration is required if we are ever to stimulate such a process in humans. Herein we have identified the regulation of p53 activity as a central mechanism behind salamander limb regeneration. Moreover, we have uncovered functions of salamander p53 in the maintenance of the differentiated state, which provide insights into the role played by this protein in the regeneration of complex structures.
Methods
Procedures for care and manipulation of newts and axolotls used in this study were carried out in compliance with the Animals (Scientific Procedures) Act 1986, approved by the United Kingdom Home Office. Axolotls were allowed to regenerate at 20 °C, and limbs or blastemas collected and processed for immunohistochemistry, qRT-PCR, or luciferase assays. Firefly/Renilla luciferase activity was measured using the Dual Luciferase Kit (Promega). Electroporation was carried out by administration of 10 electric pulses (duration: 100 ms, voltage: 80 V/cm) using a SD9 electroporation device (ETL). Full experimental procedures used in this study are available in SI Methods. See Tables S1–S3 for lists of primers used in cloning and RT-PCR, and for antibodies and reagents used.
Supplementary Material
Acknowledgments
We thank N. Shaikh and A. Kumar for technical advice; A. Joerger for help with bioinformatics analysis of p53; and S. Roy and T. Hupp for providing reagents. This work was supported by a Medical Research Council Programme Grant and a Research Professorship (to J.P.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310519110/-/DCSupplemental.
References
- 1.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 2.Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat Rev Mol Cell Biol. 2002;3(8):566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- 3.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460(7251):60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 4.Stewart R, et al. Comparative RNA-seq analysis in the unsequenced axolotl: The oncogene burst highlights early gene expression in the blastema. PLOS Comput Biol. 2013;9(3):e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay ED, Fischman DA. Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev Biol. 1961;3:26–59. doi: 10.1016/0012-1606(61)90009-4. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Velloso CP, Imokawa Y, Brockes JP. Plasticity of retrovirus-labelled myotubes in the newt limb regeneration blastema. Dev Biol. 2000;218(2):125–136. doi: 10.1006/dbio.1999.9569. [DOI] [PubMed] [Google Scholar]
- 7.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci USA. 1993;90(15):7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen TP. Stability of chondrocyte differentiation and contribution of muscle to cartilage during limb regeneration in the axolotl (Siredon mexicanum) J Exp Zool. 1968;167(1):49–78. doi: 10.1002/jez.1401670105. [DOI] [PubMed] [Google Scholar]
- 9.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knopf F, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20(5):713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7(2):198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vousden KH. p53: Death star. Cell. 2000;103(5):691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 13.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3(5):475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31(9):1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 19.Villiard E, et al. Urodele p53 tolerates amino acid changes found in p53 variants linked to human cancer. BMC Evol Biol. 2007;7:180. doi: 10.1186/1471-2148-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274(51):36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 21.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 22.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285(5434):1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 23.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka EM, Gann AA, Gates PB, Brockes JP. Newt myotubes reenter the cell cycle by phosphorylation of the retinoblastoma protein. J Cell Biol. 1997;136(1):155–165. doi: 10.1083/jcb.136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7(5):597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 26.Brandt T, Petrovich M, Joerger AC, Veprintsev DB. Conservation of DNA-binding specificity and oligomerisation properties within the p53 family. BMC Genomics. 2009;10:628. doi: 10.1186/1471-2164-10-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X. Tied up in loops: Positive and negative autoregulation of p53. Cold Spring Harb Perspect Biol. 2010;2(5):a000984. doi: 10.1101/cshperspect.a000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cam H, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10(4):281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Pozniak CD, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289(5477):304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 30.Stiewe T, Theseling CC, Pützer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: Implications for tumorigenesis. J Biol Chem. 2002;277(16):14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 31.Stiewe T, Zimmermann S, Frilling A, Esche H, Pützer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002;62(13):3598–3602. [PubMed] [Google Scholar]
- 32.Das B, et al. HIF-2α suppresses p53 to enhance the stemness and regenerative potential of human embryonic stem cells. Stem Cells. 2012;30(8):1685–1695. doi: 10.1002/stem.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasai N, Yakura R, Kamiya D, Nakazawa Y, Sasai Y. Ectodermal factor restricts mesoderm differentiation by inhibiting p53. Cell. 2008;133(5):878–890. doi: 10.1016/j.cell.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Lee DF, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11(2):179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odelberg SJ. Unraveling the molecular basis for regenerative cellular plasticity. PLoS Biol. 2004;2(8):E232. doi: 10.1371/journal.pbio.0020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96(12):6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1(1):20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 38.Bedelbaeva K, et al. Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci USA. 2010;107(13):5845–5850. doi: 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bensaad K, Rouillard D, Soussi T. Regulation of the cell cycle by p53 after DNA damage in an amphibian cell line. Oncogene. 2001;20(29):3766–3775. doi: 10.1038/sj.onc.1204492. [DOI] [PubMed] [Google Scholar]
- 40.Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137(2):213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockes JP, Kumar A, Velloso CP. Regeneration as an evolutionary variable. J Anat. 2001;199(Pt 1-2):3–11. doi: 10.1046/j.1469-7580.2001.19910003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21(1):172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 44.White JD, Rachel C, Vermeulen R, Davies M, Grounds MD. The role of p53 in vivo during skeletal muscle post-natal development and regeneration: Studies in p53 knockout mice. Int J Dev Biol. 2002;46(4):577–582. [PubMed] [Google Scholar]
- 45.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7(3):165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 46.Wallingford JB, Seufert DW, Virta VC, Vize PD. p53 activity is essential for normal development in Xenopus. Curr Biol. 1997;7(10):747–757. doi: 10.1016/s0960-9822(06)00333-2. [DOI] [PubMed] [Google Scholar]
- 47.Cerone MA, et al. p53 is involved in the differentiation but not in the differentiation-associated apoptosis of myoblasts. Cell Death Differ. 2000;7(5):506–508. doi: 10.1038/sj.cdd.4400676. [DOI] [PubMed] [Google Scholar]
- 48.Soddu S, et al. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol. 1996;134(1):193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzaro G, Bossi G, Coen S, Sacchi A, Soddu S. The role of wild-type p53 in the differentiation of primary hemopoietic and muscle cells. Oncogene. 1999;18(42):5831–5835. doi: 10.1038/sj.onc.1202962. [DOI] [PubMed] [Google Scholar]
- 50.Porrello A, et al. p53 regulates myogenesis by triggering the differentiation activity of pRb. J Cell Biol. 2000;151(6):1295–1304. doi: 10.1083/jcb.151.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirai H, et al. Post-mitotic role of nucleostemin as a promoter of skeletal muscle cell differentiation. Biochem Biophys Res Commun. 2010;391(1):299–304. doi: 10.1016/j.bbrc.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Y, Cheng Z, Yang Z, Zheng J, Lin T. DNp73 improves generation efficiency of human induced pluripotent stem cells. BMC Cell Biol. 2012;13:9. doi: 10.1186/1471-2121-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Song J, Jin EJ. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J Biol Chem. 2010;285(35):26900–26907. doi: 10.1074/jbc.M110.115105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.