Significance
Many secondary-active transporters use a sodium gradient to translocate their substrate along with a sodium ion or ions across the membrane. In contrast to other, closely related transporters, the carnitine transporter CaiT does not depend on an ion gradient. We show here that the positively charged amino acid sidechain arginine 262 (R262) in CaiT replaces the sodium ion required by other transporters. Mutating R262 in CaiT makes substrate binding and transport sodium dependent. Modeling studies reveal that R262 adopts various orientations in different conformational states of the CaiT transport cycle. We propose that this oscillation of R262 mimics sodium binding and dissociation that is crucial for triggering conformational changes resulting in substrate translocation.
Keywords: secondary-active transport, substrate/product antiport, sodium-dependent transport, membrane transport mechanism, membrane protein structure
Abstract
Most secondary-active transporters transport their substrates using an electrochemical ion gradient. In contrast, the carnitine transporter (CaiT) is an ion-independent, l-carnitine/γ-butyrobetaine antiporter belonging to the betaine/carnitine/choline transporter family of secondary transporters. Recently determined crystal structures of CaiT from Escherichia coli and Proteus mirabilis revealed an inverted five-transmembrane-helix repeat similar to that in the amino acid/Na+ symporter LeuT. The ion independence of CaiT makes it unique in this family. Here we show that mutations of arginine 262 (R262) make CaiT Na+-dependent. The transport activity of R262 mutants increased by 30–40% in the presence of a membrane potential, indicating substrate/Na+ cotransport. Structural and biochemical characterization revealed that R262 plays a crucial role in substrate binding by stabilizing the partly unwound TM1′ helix. Modeling CaiT from P. mirabilis in the outward-open and closed states on the corresponding structures of the related symporter BetP reveals alternating orientations of the buried R262 sidechain, which mimic sodium binding and unbinding in the Na+-coupled substrate symporters. We propose that a similar mechanism is operative in other Na+/H+-independent transporters, in which a positively charged amino acid replaces the cotransported cation. The oscillation of the R262 sidechain in CaiT indicates how a positive charge triggers the change between outward-open and inward-open conformations as a unifying critical step in LeuT-type transporters.
The carnitine/γ-butyrobetaine antiporter CaiT belongs to the betaine/carnitine/choline transporter family of secondary transporters that transfer substrates containing a quaternary ammonium group (1, 2) in and out of the cell. In Escherichia coli and other enterobacteria, such as Proteus mirabilis, carnitine is taken up by CaiT and converted to γ-butyrobetaine via the reaction intermediate crotonobetaine (3–5), which serves as an external electron acceptor under anaerobic growth conditions (4). Biochemical studies of E. coli CaiT (EcCaiT) have shown that it is a constitutively active, Na+/H+-independent antiporter (6).
Crystal structures of CaiT from P. mirabilis (PmCaiT) and E. coli (EcCaiT) were recently determined with and without bound substrate (7, 8). These structures revealed a trimeric assembly of CaiT, as previously found (9). The protein was in an inward-facing conformation with two substrate molecules bound per EcCaiT monomer: one in the central transport site and another in an external binding site (7). Fluorescent binding assays with the protein reconstituted into liposomes indicated that substrate binding was cooperative. This suggested a regulatory role for the external binding site, which was proposed to increase substrate affinity and initiate substrate transport (7). Strikingly, the crystal structures revealed that CaiT adopts a fold similar to that of the LeuT-type transporters (7, 10, 11). This places it in the amino acid–polyamine-organocation (APC) superfamily (12), which shares a conserved architecture of two inverted repeats of five transmembrane (TM) helices each, implying common mechanistic principles. Among the APC transporters, the leucine transporter LeuT from the neurotransmitter/sodium symporter family (10), the betaine transporter BetP from the betaine/choline/carnitine transporter family (13), the benzyl-hydantoin transporter Mhp1 of the nucleobase/cation symport 1 family, and the Na+/galactose symporter vSGLT of the solute/sodium symporter family are substrate/sodium symporters (14, 15). Although the substrate to sodium stoichiometry varies for each Na+-dependent LeuT-type transporter, most of them possess a conserved sodium-binding site (Na2 site) at which the binding and dissociation of a sodium ion is proposed to facilitate structural changes that lead to substrate transport (16–18). Although an additional sodium-binding site (Na1 site) exists in transporters such as LeuT and BetP, the position or even the presence of this site is not strictly conserved among the Na+-dependent LeuT-type transporters (14, 15, 19–21).
A small number of LeuT-type transporters, namely, AdiC (arginine/agmatine antiporter), ApcT (broad-specificity amino acid transporter), and CaiT, are Na+-independent (6, 22–25). The crystal structure of ApcT revealed that a lysine residue (K158) occupies a position equivalent to the Na2 site in LeuT. This lysine is proposed to undergo a protonation/deprotonation event that leads to conformational changes facilitating substrate transport (25). In CaiT, a methionine residue (M331) occupies a position equivalent to Na1 in LeuT, whereas a positively charged arginine residue (R262) occupies the Na2 site (7). Previously, we have shown that mutating M331 reduces transport activity but does not induce Na+ dependence in CaiT (7). Here we report that point mutations of R262 render CaiT inactive. Strikingly, the transport activity was partially restored by the addition of sodium, thus making these mutants Na+-dependent. Unlike wild-type, the transport activity of R262 mutants increased by 30–40% when a membrane potential was applied, suggesting that Na+ and substrate were cotransported. To find out whether and how the mutation affects the Na2 site, we determined the crystal structure of CaiT R262E. Although we did not find any major changes in the mutant protein, comparison with the substrate-bound EcCaiT wild-type protein revealed that the γ-butyrobetaine substrate adopts a different orientation at the central binding site, in which it directly interacts with the unwound part of TM1′. (To make the nomenclature consistent, we adopt the same helix numbering as in LeuT. Because CaiT has two extra helices at the N terminus in comparison with LeuT, TM3 in CaiT corresponds to TM1 in LeuT and is denoted as TM1′, and the remainder of the CaiT helices follow suit.) Because R262 is known to play a role in stabilizing the unwound part of TM1′ (7), we propose that R262 is crucial for substrate binding, similar to Na+ in the Na2 site of LeuT and BetP (17, 19). Indeed, our fluorescent binding assays using R262 mutants showed markedly decreased substrate affinity. Modeling CaiT in various conformations with BetP as a template revealed that R262 undergoes an oscillatory movement, contributing to different hydrogen bond networks in each conformation. We suggest that this movement of the positively charged R262 sidechain in CaiT mimics Na+ binding/unbinding in Na+-dependent LeuT-type transporters and plays a central role in the transport mechanism.
Results
The Conserved Na2 Site in LeuT-Type Transporters Is Replaced by R262 in CaiT.
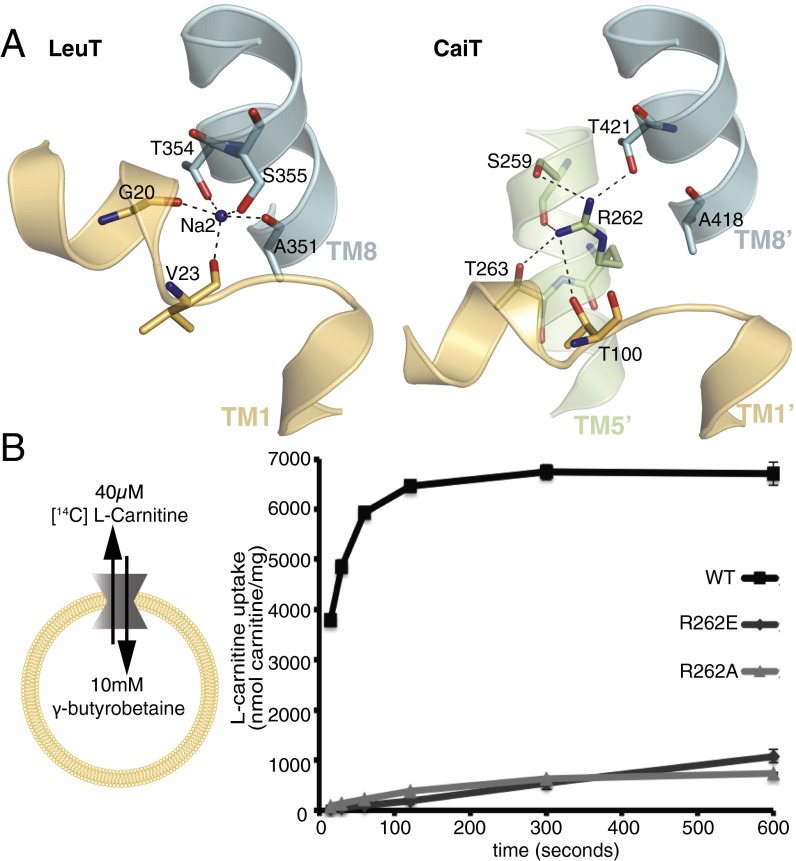
The Na2 site has been identified in several of the LeuT-type transporters including LeuT, vSGLT, MhP1, and BetP (10, 13–15, 18, 20, 21) by structural and biochemical approaches (Fig. S1). Comparing the structures of CaiT and LeuT, we found that a positively charged sidechain of R262 in TM5′ occupies the Na2 position in CaiT (7) (Fig. 1A). Akin to CaiT, the proposed H+-coupled broad-specificity amino acid transporter ApcT also contains a positively charged amino acid residue (K158) in the Na2 site (Fig. S1). This K158 residue was calculated to have a pKa 3–4 U below the normal pKa of a lysine residue (25), and hence was proposed to undergo protonation/deprotonation cycles that facilitate substrate transport. A similar analysis in CaiT revealed that the pKa of the R262 sidechain (model pKa 12.5) in its microenvironment is predicted to be reduced by around two pKa units (26), which is not indicative of protonation/deprotonation under physiological conditions. Previous studies have found that transport in CaiT is independent of an electrochemical proton gradient (6); however, the exact experimental conditions were not described. We therefore measured the carnitine uptake activity of PmCaiT in a wide pH range from 5 to 11, both within and outside the proteoliposomes. Surprisingly, CaiT was maximally active when the pH was adjusted to 7 both inside and outside the proteoliposomes (Fig. S2). This indicates that R262 in CaiT does not undergo a protonation/deprotonation cycle, as K158 in ApcT was proposed to do, which would point to a different mechanism of substrate transport in the latter.
Fig. 1.
Role of R262 in substrate uptake by CaiT. (A) Comparison of the Na2 site in LeuT and CaiT. In LeuT, Na2 is coordinated by sidechains as well as backbone carbonyl interactions from TM1 and TM8, whereas in CaiT the Na2 position is occupied by R262 with similar hydrogen bond interactions to TM1′ and TM8′. (B) l-carnitine uptake by PmCaiT wild-type and R262 mutants. Proteoliposomes were preloaded with 10 mM cold γ-butyrobetaine, and counterflow uptake activity of 40 μM [14C] l-carnitine was measured. All data points are the averages of three independent experiments, with error bars indicating the SD of the measurements.
To understand the role of R262 in the CaiT transport mechanism, we changed this residue into a neutral alanine (R262A) or a negatively charged glutamate (R262E). These mutations were introduced into the P. mirabilis CaiT (PmCaiT), as the better resolution of the PmCaiT wild-type structure allowed more detailed comparison. The R262A and R262E mutants were successfully expressed, purified, and reconstituted into liposomes. The gel filtration profiles of the mutants show that both mutants are trimers, similar to the wild-type protein, and remain stable throughout the purification procedure (Fig. S3). The proteoliposomes were preloaded with 10 mM γ-butyrobetaine and assayed for uptake of [14C] l-carnitine (Fig. 1B). Although the wild-type protein showed robust uptake, this was drastically reduced in the R262A and R262E mutants, indicating the importance of R262 for CaiT transport activity (Fig. 1B).
Mutating R262 Renders CaiT Na+-Dependent.
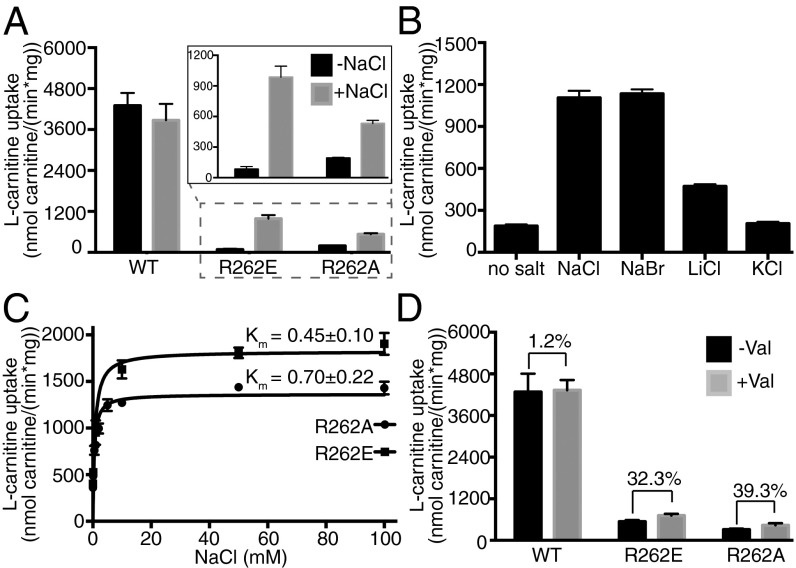
Because the positively charged R262 sidechain was proposed to replace the Na2 ion in CaiT (7), we investigated whether the activity of mutants R262E and R262A could be rescued by adding NaCl to the transport assay. Indeed, addition of 50 mM NaCl to the reaction buffer increased the uptake activity of R262E and R262A mutants by nearly 12-fold and threefold, respectively (Fig. 2A).
Fig. 2.
Substrate uptake by R262 mutants is Na+-dependent. (A) Initial rates of l-carnitine uptake by proteoliposomes with or without 50 mM NaCl. (Inset) Uptake activities of R262E and R262A. (B) Na+-specific uptake by R262E. Initial uptake activity rates were measured in the absence of salt or in the presence of 50 mM NaCl, NaBr, LiCl, or KCl. (C) The Na+ dependence of R262 mutants can be saturated. Initial uptake rates by R262 mutants were measured, whereas the Na+ concentration was increased from 0.1 to 100 mM. (D) Initial [14C] l-carnitine uptake rates of wild-type, R262E, and R262A proteoliposomes in the absence or presence of valinomycin. Proteoliposomes were preloaded with 50 mM KCl, and 50 mM NaCl and 1 µM valinomycin were added to the external buffer to create an inside-negative potential. A 30–40% increase in uptake activity is observed for both R262A and R262E in the presence of a membrane potential. All data points are the averages of three independent experiments, with error bars indicating the SD of the measurements.
To ensure transport is specifically stimulated by sodium ions, uptake assays were performed with 50 mM NaCl, NaBr, LiCl, or KCl in the external medium. Although NaCl and NaBr increased the [14C] l-carnitine uptake activity of the R262E mutant to similar levels, KCl did not stimulate uptake activity significantly (Fig. 2B). The increase in uptake in the presence of LiCl was about 40% compared with NaCl. This indicates that uptake is, in fact, preferentially stimulated by sodium ions and that the protein can accommodate Li+ better than K+. Further uptake assays were carried out under a range of different osmotic conditions to exclude the possibility of an increased uptake resulting from liposome shrinkage caused by NaCl in the external buffer (27) (Fig. S4). We also characterized the transport kinetics of these mutants in the presence of Na+ and found that in comparison with wild-type, they have lower Km and Kcat (Fig. S5).
We asked whether the Na+ dependence of these mutants could be saturated. Uptake kinetics using increasing concentrations of NaCl outside the proteoliposomes revealed an apparent Km (Na+) of 0.45 ± 0.10 mM for R262E and 0.70 ± 0.22 mM for R262A (Fig. 2C). This indicates slightly higher Na+ affinities of the CaiT mutants than of the Na+-dependent symporter BetP (19).
Transport by the sodium-coupled symporters LeuT and BetP is known to be electrogenic. Previous work has shown that CaiT is electroneutral, as transport activity is ion-independent (6) and the substrates l-carnitine and γ-butyrobetaine are zwitterionic but uncharged at physiological pH. Because our R262 mutations have made CaiT Na+-dependent, it was interesting to ask whether these mutants are also electrogenic. We addressed this question by monitoring the transport rates of the mutants in the presence of a membrane potential. Proteoliposomes were preloaded with 10 mM γ-butyrobetaine and 50 mM KCl (inside negative), followed by the addition of 1 μM valinomycin and 50 mM NaCl to the external medium. For both mutants, R262A and R262E, addition of valinomycin resulted in a 30–40% increase in uptake activity, whereas there was no effect on the activity of the wild-type (Fig. 2D). Although these data suggest strongly that the R262 mutants are electrogenic, further experiments are required to prove Na+-substrate symport conclusively.
R262 Is Involved in Substrate Binding.
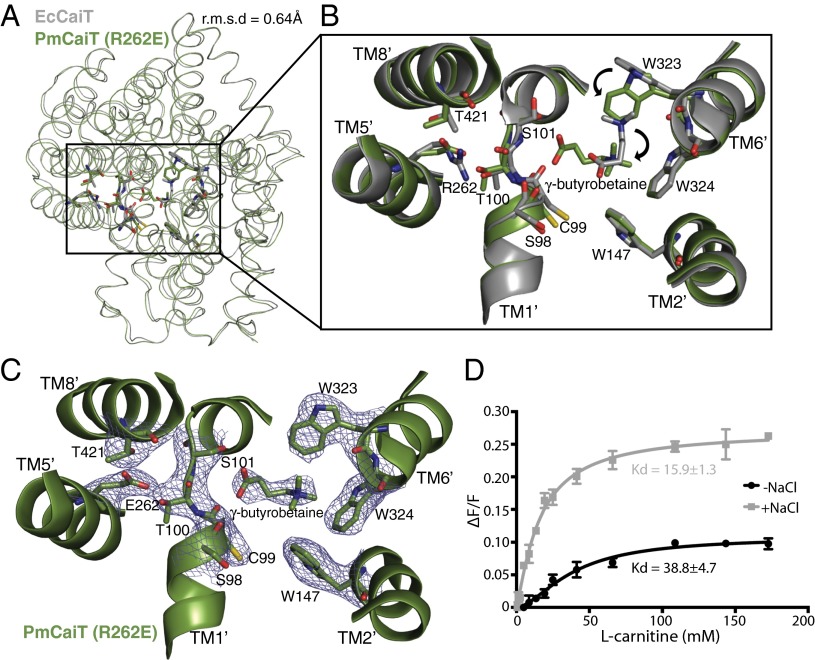
To further our understanding of the Na+-dependence of R262 mutants and the effect of this mutation on the protein conformation, we determined the crystal structure of the PmCaiT R262E mutant. Crystals were grown in the presence of NaCl with the intent of identifying the Na+ binding site in these mutants. The structure was determined at 3.29 Å resolution by molecular replacement, using wild-type PmCaiT as the search model (Table S1). The refined structure is in a substrate-bound inward-open conformation and does not show major structural changes in comparison with the available wild-type structures (rmsd, 0.37 Å for apo PmCaiT and 0.64 Å for substrate-bound EcCaiT) (Fig. 3A), except for the reorientation of the substrate bound at the central binding site (see next paragraph and Fig. 3 B and C). The mutant structure did not reveal any extra density for an ion near the Na2 site, nor did crystallization in the presence of anomalous scatterers such as Tl+ or Rb+ yield an ion-bound structure. We reasoned that the inward-open conformation, which is a sodium-releasing state (17, 18, 20, 28), might preclude ion binding at the Na2 site. This is also evident from the inward-open crystal structures of vSGLT, MhP1, LeuT, and BetP, where the Na2 site is empty (16–18, 20).
Fig. 3.
Substrate binding in CaiT R262 mutants. (A) Superimposition of EcCaiT (chain A) and PmCaiT R262E (PDB codes 2WSW and 4M8J, respectively) reveals an rmsd of 0.64 Å. (B) Central substrate binding site in the PmCaiT R262E structure with a bound γ-butyrobetaine superimposed on the substrate-bound EcCaiT type structure. In the R262E mutant, the substrate has rotated by about 90° compared with the wild-type structure. In addition, W323 has reoriented into a position suitable to accommodate the substrate in this new orientation. (C) 2Fo-Fc map of the central binding site at 1.0 σ. The trimethyl ammonium head group of the substrate is coordinated by the tryptophan box from TM6′ and TM2′, whereas the carboxyl group of the substrate is interacting with the backbone carbonyl of S98 in the unwound TM1′ helix. The glutamate at position 262 establishes a hydrogen bond to the unwound TM1′ helix, which, together with T421 from TM8′, stabilizes the unwound stretch, similar to Arg262 in the wild-type structure. (D) Tryptophan fluorescence-based substrate-binding assay with R262E reconstituted into liposomes. The increase in tryptophan fluorescence with rising l-carnitine concentration was monitored in the presence and absence of 50 mM NaCl. The relative increase in fluorescence was plotted using a one-site binding model to obtain the apparent Kd values for substrate binding. All data points are the averages of three independent experiments; the error bars indicate the SD of the measurements.
Surprisingly, however, the substrate-bound crystal structure of R262E provided us with unique insights into the role of R262 and the unwound part of helix TM1′ in substrate binding. The glutamate introduced at position 262 forms strong polar interactions with residues in the unwound helix of TM1′, similar to R262 in the wild-type protein, whereas the conformation of TM1′ remains essentially unchanged (Fig. 3B). The structure reveals a different orientation of the bound γ-butyrobetaine, which is rotated by about 90° compared with the γ-butyrobetaine bound in the previously reported substrate-bound E. coli structure (Fig. 3B) (7). In this orientation, the carboxyl group of γ-butyrobetaine forms polar contacts with the unwound part of TM1′ that were not observed in any of the previous CaiT structures. Interactions include the backbone carbonyl of S98, as well as a weak interaction with the sidechain hydroxyl of S101. The trimethyl ammonium head group of γ-butyrobetaine fits perfectly into the tryptophan box constituted by W323, W324, and W147 (Fig. 3C). Notably, the W323 sidechain is rotated by around 90° in contrast to the substrate-bound E. coli structure to accommodate the substrate in the present orientation (Fig. 3B). The interaction between the unwound part of TM1′ and the substrate, together with the known role of R262 in the stabilization of the unwound stretch of helix TM1′ (7), suggests an indirect role for R262 in substrate binding.
To test this, we performed tryptophan fluorescence binding assays with the R262A and R262E mutant proteins incorporated into liposomes (Fig. 3D and Fig. S6). Our binding assays revealed that the apparent substrate binding affinities of the R262 mutants are almost 10-fold lower compared with wild-type (wild-type apparent Kd = 3.63 ± 0.4 mM; R262A apparent Kd = 39.6 ± 4.2 mM; R262E apparent Kd = 38.75 ± 4.7 mM for l-carnitine; Table 1). Remarkably, addition of NaCl increased substrate binding of the R262 mutants (Fig. 3D and Fig. S6), whereas the wild-type protein showed no significant change (Table 1), highlighting the influence of Na+ on substrate binding in R262 mutants. The low affinity of the mutants clearly suggests a role for R262 in substrate binding and explains the effect of the R262 mutations on the transport activity of CaiT.
Table 1.
Dissociation constants (apparent Kd) obtained from tryptophan fluorescence substrate-binding experiments with l-carnitine in the absence and presence of 50 mM NaCl
| Protein | Apparent Kd, mM |
|
| −Na+ | +Na+ | |
| PmCaiT wild-type | 3.63 ± 0.4 | 3.79 ± 0.35 |
| PmCaiT R262E | 38.75 ± 4.7 | 15.9 ± 1.3 |
| PmCaiT R262A | 39.6 ± 4.2 | 16.8 ± 1.2 |
R262 Oscillation Mimics Sodium Binding and Unbinding.
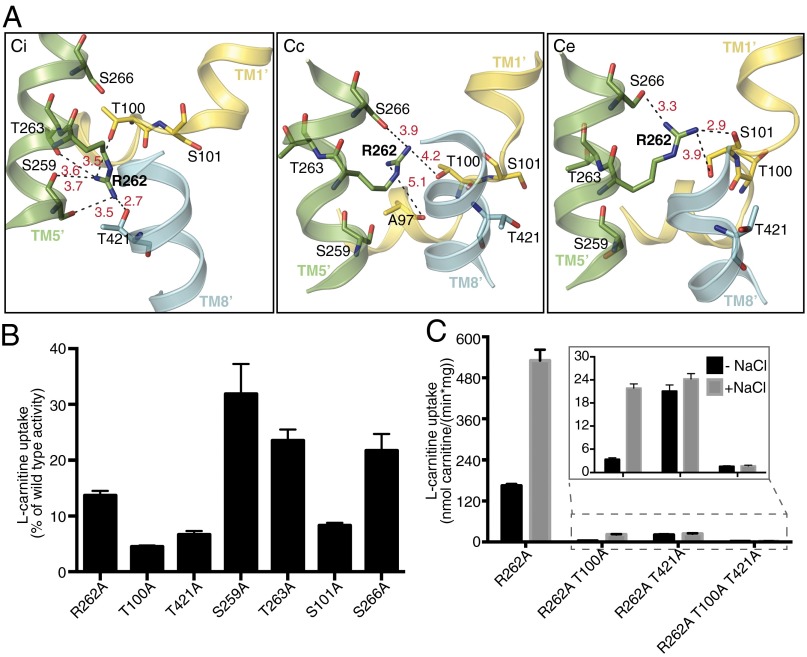
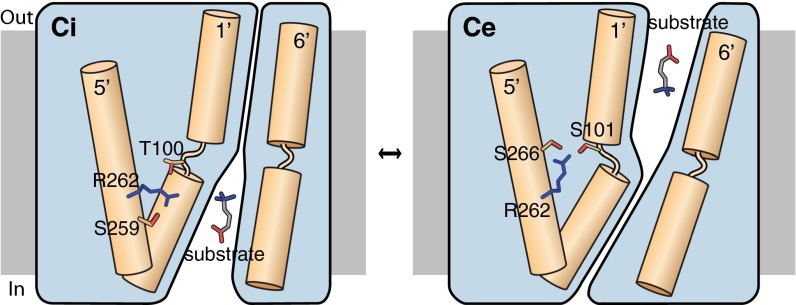
In Na+-dependent transporters, Na+ binding and dissociation facilitates conformational changes, mostly mediated by the unwound, flexible part of TM1′ (16–18). Because the crystal structures of CaiT are all in inward-open conformations with or without bound substrate (Ci and CiS), it is not clear whether a similar mechanism mediated by R262 occurs in CaiT. For a better insight into the dynamics of the Na2 site, we modeled the closed and outward-open conformations of CaiT, using the corresponding BetP structures as templates (18). From these models, it was clear that the core helices forming the substrate-binding site (TM1′, TM2′, TM6′, TM7′) undergo conformational changes that result in the opening or closing of the periplasmic or cytoplasmic pathway. Remarkably, R262 occupies different positions in these modeled conformations, each being stabilized by a separate set of hydrogen bonds (Fig. 4A). In the inward-open conformation, R262 interacts with the sidechains of T100 from TM1′, S259 from TM5′, and T421 from TM8′, as well as the backbone carbonyls of S259 and T263 from TM5′. In contrast, in the modeled closed state, R262 has long-distance interactions with the sidechains of S266 from TM5′ and T100 in the unwound part of TM1′. An additional weak interaction is formed to the backbone carbonyl of A97 in this stretch. In the outward-open conformation, R262 interacts with sidechains of S101 from TM1′ and S266 from TM5′, as well as with the backbone carbonyl of S101. On the basis of these observations, we propose a model, which we refer to as the “arginine oscillation model,” for the CaiT transport mechanism, in which R262 plays a critical role in regulating the flexibility of TM1′ through alternating interactions, thereby inducing the conformational changes in the protein that ultimately result in substrate translocation (Fig. 5).
Fig. 4.
Arginine oscillation model. (A) Orientation of R262 in three different conformations of PmCaiT. The inward-open conformation (CiS) is the published CaiT structure (7), whereas the closed (Cc) and outward open (Ce) conformations are modeled on the crystal structures of BetP in the corresponding conformations. R262 sidechain oscillates, making and breaking a number of hydrogen bonds, while the transporter moves from the inward-open via the closed state to the outward-open state and then back to the inward-open state. Black dashed lines indicate hydrogen bonds, with bond distances in Å shown in red. (B) Residues forming hydrogen bonds to R262 in the wild-type structure or the two models were mutated to alanine, and transport activity was measured in the presence of 50 mM NaCl. The initial rates of uptake were plotted as a percentage of wild-type activity. (C) Uptake activity of TM1′ unwound helix mutants in combination with the R262A mutation in the presence or absence of 50 mM NaCl. Uptake activities of R262A/T100A, R262A/T421A, and R262A/T100A/T421A are shown in the inset. All data points in B and C are the averages of three independent experiments, with error bars indicating the SD of the measurements.
Fig. 5.
Schematic representation of the arginine oscillation model as a mechanism of substrate transport in CaiT. In the inward-open conformation (Ci), R262 interacts with T100 from the unwound part of TM1′, as well as with S259 from TM5′. Binding of substrate near the partly unwound TM1′ of CaiT triggers reorientation of R262, which now coordinates with S101 from TM1′ and S266 from TM5′. This is accompanied by the flexing of TM1′ and coordinated movements of nearby helices, ultimately resulting in the closing of cytoplasmic pathway and substrate release into the periplasm (Ce).
On the basis of the previous CaiT crystal structures, the residues coordinating R262 during its proposed oscillation were not expected to have a functional role in substrate transport. Therefore, we reasoned that measuring the transport activity of mutants in which these residues had been changed could lend support to the arginine oscillation model. Toward this end, the residues interacting with R262 through their sidechains in the various CaiT conformations (Fig. 4A) were mutated to alanines. These single mutants were reconstituted into liposomes, and their carnitine uptake activity was measured. In agreement with our model, all mutants showed a significantly reduced transport activity compared with wild-type (Fig. 4B). It is plausible that in the R262 mutants, some of these residues coordinate the Na+, which would contribute to their Na+ dependence. To test this, we combined these mutants with R262A. Although R262A/T100A was still Na+-dependent, the overall transport activity was much lower than that of R262A alone. This implies that the sidechain of T100 is not crucial for coordinating the Na+ but still plays a role in substrate translocation. However, R262A/T421A completely lost its sodium dependence, suggesting a critical role for T421 in the coordination of Na+ in the R262 mutants (Fig. 4C).
Interestingly, T421 from TM8′ is highly conserved in other LeuT-type transporters (Fig. S7), where it is directly involved in Na2 coordination. Mutation of the corresponding residue T467 in BetP affected betaine uptake activity and resulted in significantly increased Km and Kd in comparison with wild-type (19). This unveils yet another common feature between the Na+-independent CaiT and the Na+-dependent transporters. Although these experiments suggest that T100, S101, S259, T263, S266, and T421 are involved in coordinating R262 by stabilizing it in various positions in the course of the transport cycle, it still remains to be seen how exactly this changes the protein conformation. Crystal structures of CaiT in the remaining conformations are needed to provide structural evidence for the arginine oscillation model.
Discussion
Altering the Electrostatic Component in CaiT.
In this study we have shown that, and how, the positively charged residue R262 in CaiT functionally substitutes for a Na+ in the Na2 binding site. The Na2 site is known to be structurally and functionally important in the Na+-dependent transporters LeuT and BetP (10, 18). The Na+-dependent nature of the CaiT mutants R262A and R262E suggests the necessity of a positive charge at the Na2 site for substrate binding and transport. Interestingly, a direct correlation between the electrostatic interactions of an amino acid sidechain and ion-dependence or independence of transport has been observed in the GABA (γ-aminobutyric acid) transporter GAT-1 as well as in LeuT and Tyt1 (29). Although the eukaryotic neurotransmitter:sodium symporters including GAT-1 are chloride-dependent, their bacterial counterparts LeuT, Tyt1, and TnaT are chloride-independent. Introduction of a glutamate near the putative Na1 binding site in GAT-1 renders this transporter chloride-independent, whereas the reciprocal mutations make LeuT and Tyt1 chloride-dependent (29–31). Similarly, replacement of an aspartate by threonine transformed the light-driven proton pump bacteriorhodopsin into a chloride pump (32). We now show that the electrostatic charge in the Na2 site of a LeuT-type transporter can be changed from that of a positively charged amino acid sidechain to a sodium ion. In contrast, mutating M331 that substitutes for the sodium ion in the Na1 site does not make CaiT Na+-dependent (7).
Our data indicate that CaiT may have evolved into a Na+-independent transporter from an ancestral Na+-dependent transporter. A positively charged sidechain in the Na2 site would have been an advantage in making CaiT independent of a second substrate such as Na+ or H+. Sodium independence makes sense for the biological role of CaiT, which is to carry out substrate/product antiport of l-carnitine and γ-butyrobetaine down their concentration gradients (6). Therefore, an additional dependence on a Na+ gradient would be unnecessarily restrictive without offering a selective advantage. In contrast, this dependence would be a selective disadvantage, as a Na+ gradient sufficient to drive transport would not normally be available in the intestinal tract, which is the natural habitat of both E. coli and P. mirabilis.
Implications for the Transport Mechanism.
The predicted high pKa of the R262 sidechain suggested that its amino group is not deprotonated during the outward-open to inward-open transition of the transporter. Moreover, our transport assays confirm a previous report that CaiT is not H+-dependent (Fig. S2). It was therefore interesting to see whether there is a mechanism in CaiT that mimics the process of Na+-binding and unbinding that would induce conformational changes through the unwound part of TM1′. The changing hydrogen bond network centered on R262 in the inward-open crystal structure and in our models of the closed and outward-open states suggest an oscillatory movement of the arginine sidechain. We propose that this oscillating mechanism simulates Na+ binding and unbinding during the transport cycle, thus triggering substrate release, as in LeuT (17, 33), vSGLT (16), and BetP (18) (Fig. 5). Functionally, CaiT does not depend on an ion gradient, as it is involved in l-carnitine/γ-butyrobetaine transport and caters to the cells’ need for l-carnitine during anaerobic growth or nutrient starvation. Moreover, as pointed out earlier, a Na+ gradient is not normally available in the native environment of enterobacteria such as E. coli or P. mirabilis. There is a further advantage in a tethered positive charge replacing the functionally equivalent, but more mobile, Na2: Unlike BetP or LeuT, which are symporters, CaiT is an antiporter, and as such can switch between different conformational states only in the substrate-bound form, whereas in the symporters, the inside-open to outside-open transition has to take place in the absence of either substrate or Na+. The energy barrier for these transitions in CaiT is likely to be similar in both directions, as they are both substrate-mediated. This would call for a symmetrical event such as R262 oscillation to control the flexibility of the unwound part of TM1′ in both transport directions, in contrast to the Na+-dependent symporters, where Na+-mediated conformational changes occur only during the outward-open to inward-open transition.
Substrate-induced alternating access for antiporters such as GlpT (34) or AdiC (35) has been proposed to be mediated either by a single substrate binding site or two different sites, respectively. In CaiT, substrate binding is likely to trigger the conformational change by interacting with the unwound region of TM1′. This would affect the hydrogen bond donors available to R262, which in turn would respond by reorienting itself accordingly. This oscillation of R262 is accompanied by the flexing of the unwound part of TM1′, as well as the movement of the surrounding TM helices, thus opening and closing the cytoplasmic pathway (Fig. 5).
In our R262 mutants, the arginine-mediated, substrate-induced conformational changes fail to take place, even though the substrate can bind, albeit with lower affinity (Fig. 3D, Fig. S6, and Table 1). This explains the stabilization of a differently oriented substrate in our crystal structure of PmCaiT R262E, a likely intermediate step in the transport cycle, where the carboxyl group of the substrates interacts with the partly unwound TM1′ helix (Fig. 3 B and C).
In summary, our biochemical and structural characterization of CaiT and the role of R262 in its transport mechanism illustrates how CaiT evolved to be ion-independent, whereas the mode of conformational changes remains largely conserved across the large APC transporter family, with a few exceptions such as AdiC. Our results demonstrate that a positively charged amino acid can assume the functional role of the sodium ion in the alternating access transport mechanism. Similar alternating conformations of a charged amino acid sidechain simulating ion binding or dissociation could be operative in other ion-independent transporters.
Methods
PmCaiT wild-type and mutant proteins were purified and reconstituted into liposomes, as described previously (6, 7). Uptake of [14C] l-carnitine was measured using proteoliposomes preloaded with 10 mM cold γ-butyrobetaine (6, 7). Tryptophan fluorescent assays were performed with protein reconstituted into liposomes (6, 7). R262E diffraction data were collected at the Swiss Light Source. The structure was solved by molecular replacement using PHASER (36), with the wild-type protein as the search model [Protein Data Bank (PDB) code 2WSW], and the refinement was performed using Python-based Hierarchical Environment for Integrated Xtallogrpahy (PHENIX) (37). Homology modeling of CaiT was performed using Modeler 9.11 (38), with BetP Structures (PDB codes 4DOJ and 4AIN) as templates. Detailed materials and methods are available in SI Methods.
Supplementary Material
Acknowledgments
We thank the Swiss Light Source Staff for help with the X-ray data collection, Özkan Yildiz for help with structure determination and discussions, Christine Ziegler for comments on the manuscript, and Paolo Lastrico for help with the model drawing. S.K. acknowledges a PhD fellowship from the International Max Planck Research School on Biological Membranes. This work was funded by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4M8J).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309071110/-/DCSupplemental.
References
- 1.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64(2):354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17(4):163–176. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seim H, Löster H, Claus R, Kleber H-P, Strack E. Formation of γ-butyrobetaine and trimethylamine from quaternary ammonium compounds structure-related to l-carnitine and choline by Proteus vulgaris. FEMS Microbiol Lett. 1982;13(2):201–205. [Google Scholar]
- 4.Seim H, Löster H, Claus R, Kleber H-P, Strack E. Stimulation of the anaerobic growth of Salmonella typhimurium by reduction of L-carnitine, carnitine derivatives and structure-related trimethylammonium compounds. Arch Microbiol. 1982;132(1):91–95. doi: 10.1007/BF00690825. [DOI] [PubMed] [Google Scholar]
- 5.Kleber H-P. Bacterial carnitine metabolism. FEMS Microbiol Lett. 1997;147(1):1–9. doi: 10.1111/j.1574-6968.1997.tb10212.x. [DOI] [PubMed] [Google Scholar]
- 6.Jung H, et al. CaiT of Escherichia coli, a new transporter catalyzing L-carnitine/gamma -butyrobetaine exchange. J Biol Chem. 2002;277(42):39251–39258. doi: 10.1074/jbc.M206319200. [DOI] [PubMed] [Google Scholar]
- 7.Schulze S, Köster S, Geldmacher U, Terwisscha van Scheltinga AC, Kühlbrandt W. Structural basis of Na(+)-independent and cooperative substrate/product antiport in CaiT. Nature. 2010;467(7312):233–236. doi: 10.1038/nature09310. [DOI] [PubMed] [Google Scholar]
- 8.Tang L, Bai L, Wang W-H, Jiang T. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol. 2010;17(4):492–496. doi: 10.1038/nsmb.1788. [DOI] [PubMed] [Google Scholar]
- 9.Vinothkumar KR, Raunser S, Jung H, Kühlbrandt W. Oligomeric structure of the carnitine transporter CaiT from Escherichia coli. J Biol Chem. 2006;281(8):4795–4801. doi: 10.1074/jbc.M508993200. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl—dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 11.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807(2):167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Wong FH, et al. The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol. 2012;22(2):105–113. doi: 10.1159/000338542. [DOI] [PubMed] [Google Scholar]
- 13.Farwick M, Siewe RM, Krämer R. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol. 1995;177(16):4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322(5902):709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turk E, et al. Molecular characterization of Vibrio parahaemolyticus vSGLT: a model for sodium-coupled sugar cotransporters. J Biol Chem. 2000;275(33):25711–25716. doi: 10.1074/jbc.M003127200. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468(7326):988–991. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez C, Koshy C, Yildiz Ö, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012;490(7418):126–130. doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 19.Khafizov K, et al. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. Proc Natl Acad Sci USA. 2012;109(44):E3035–E3044. doi: 10.1073/pnas.1209039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimamura T, et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328(5977):470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321(5890):810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419(6908):715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 23.Gong S, Richard H, Foster JW. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol. 2003;185(15):4402–4409. doi: 10.1128/JB.185.15.4402-4409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reig N, et al. Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: a model for APC transporters. J Biol Chem. 2007;282(18):13270–13281. doi: 10.1074/jbc.M610695200. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325(5943):1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61(4):704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 27.Rübenhagen R, Morbach S, Krämer R. The osmoreactive betaine carrier BetP from Corynebacterium glutamicum is a sensor for cytoplasmic K+ EMBO J. 2001;20(19):5412–5420. doi: 10.1093/emboj/20.19.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Tajkhorshid E. Ion-releasing state of a secondary membrane transporter. Biophys J. 2009;97(11):L29–L31. doi: 10.1016/j.bpj.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449(7163):726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Substrate-dependent proton antiport in neurotransmitter:sodium symporters. Nat Chem Biol. 2010;6(2):109–116. doi: 10.1038/nchembio.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kantcheva AK, et al. Chloride binding site of neurotransmitter sodium symporters. Proc Natl Acad Sci USA. 2013;110(21):8489–8494. doi: 10.1073/pnas.1221279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki J, et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269(5220):73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 33.Caplan DA, Subbotina JO, Noskov SY. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophys J. 2008;95(10):4613–4621. doi: 10.1529/biophysj.108.139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301(5633):616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 35.Kowalczyk L, et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc Natl Acad Sci USA. 2011;108(10):3935–3940. doi: 10.1073/pnas.1018081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Eswar N, Eramian D, Webb B, Shen M-Y, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.