Significance
The origin of the directional motion in cytoskeleton-based molecular motors has been a major puzzle in spite of the significant advances made by several experimental studies. Especially, an understanding of the unidirectionality of such systems from the structure/energy viewpoint is still lacking. This study attempts to understand the directionality in myosin V, one of the actin-based cytoskeleton motors involved in shuttling cellular cargo, and establishes the previously unknown role of electrostatics in guiding the directional motion.
Keywords: molecular motors, bioenergetics, conformation–chemical coupling, cytoskeletal proteins, intracellular transport
Abstract
Understanding the basis for the action of myosin motors and related molecular machines requires a quantitative energy-based description of the overall functional cycle. Previous theoretical attempts to do so have provided interesting insights on parts of the cycle but could not generate a structure-based free energy landscape for the complete cycle of myosin. In particular, a nonphenomenological structure/energy-based understanding of the unidirectional motion is still missing. Here we use a coarse-grained model of myosin V and generate a structure-based free energy surface of the largest conformational change, namely the transition from the post- to prepowerstroke movement. We also couple the observed energetics of ligand binding/hydrolysis and product release to that of the conformational surface and reproduce the energetics of the complete mechanochemical cycle. It is found that the release in electrostatic free energy upon changing the conformation of the lever arm and the convertor domain from its post- to prepowerstroke state provides the necessary energy to bias the system towards the unidirectional movement of myosin V on the actin filament. The free energy change of 11 kcal is also in the range of ∼2–3 pN, which is consistent with the experimentally observed stalling force required to stop the motor completely on its track. The conformational-chemical coupling generating a successful powerstroke cycle is believed to be conserved among most members of the myosin family, thus highlighting the importance of the previously unknown role of electrostatics free energy in guiding the functional cycle in other actin-based myosin motors.
Myosin constitutes a diverse class of molecular motors that perform mechanical work by producing movement on the actin filaments. These cellular machines tightly couple various steps of the ATP hydrolysis and product release cycle with mechanical force generation on actin and can therefore perform coordinated movements within the cell (1–3). Classical examples of myosin motors constitute the muscle protein that performs a single mechanical force generation cycle and then detaches from the actin filament. These molecules work in large numbers to produce a net directed movement of the actin during muscle contraction (2). Furthermore, living cells have used the basic functional cycle of myosin to generate several other members of the myosin family, some of which produce repetitive mechanochemical movements on the actin filaments. The most studied among these processive motors is myosin V, which is responsible for carrying cargo to different destinations using the intracellular network of actin filaments (4). At the heart of all types of myosin action is its ability to couple large conformational changes with ATP hydrolysis and actin binding/release cycles to eventually perform a unidirectional motion (3). Although some features of the cycle are well understood, one of the most intriguing remaining problems is to unravel the factors that allow myosin V to walk in only one direction toward the “plus end” of actin during its processive motion (this question was nicely articulated in ref. 5). Here, despite having high-resolution structural information and observing the unidirectional myosin movement in several breakthrough experimental studies, the question of why the motor does not show any considerable movement in the opposite direction is still unanswered. This key question about the action of myosin V and related systems is explored in the present work.
Numerous experiments including kinetics and thermodynamic studies (1, 6–8), high-resolution structural studies (9, 10), electron microscopy studies (11), and single-molecule experiments (4, 12, 13) (other relevant studies are discussed in Supporting Information) have advanced our understanding of the action of the system. Apart from ingenious experimental studies that have been able to explore the mechanochemical cycle, there have been theoretical attempts to understand the processes using structure-based or phenomenological modeling approaches in myosin (14–20).
Whereas most of these studies addressed the role of large conformational changes coupled to long-time-scale processes during the cycle, in our view the main open questions are about the structure/energy correlation of the system that is responsible for the unidirectional motion. Progress on this front seems to be more relevant to the central question rather than simulating the dynamical behavior of the different states of myosin, and at present we are not aware of any structure-based model that actually provided an interpretation of the directional motion of myosin [although phenomenological studies and kinetic modeling have been instructive (14, 18, 19)]. Exploring the structure/energy relationship in myosin V presents major challenges, owing in part to the absence of high-quality structural information on all of the intermediate conformations and nucleotide/actin-bound forms. This problem is compounded by the difficulty of capturing the free energy balance between the conformational states of the system using current computer resources, owing to the difficulty of obtaining converging free energies for substantial conformational changes of large and highly charged systems.
To overcome some of the above difficulties, we used our coarse-grained (CG) model (21) that focuses on a consistent description of the electrostatic energy (note that such a physically consistent electrostatic description is not emphasized in most other CG models). Our CG model has been very effective in describing the mechanochemical coupling of F1-ATPase (22) and F0-ATPase (23) as well as the action of other systems (e.g., refs. 24 and 25). Our CG simulations allowed us to reproduce a very reasonable estimate of the change in the conformational free energies of the post- to prepowerstroke states of a single strand of myosin V. After coupling the conformational free energy with the concentration-dependent kinetics of ATP hydrolysis, product release, and acting binding/release cycles, we could produce a semiquantitative description of the processive cycle for the double-headed myosin V that reveals a previously unknown role of electrostatics in guiding the unidirectionality of these molecular motors.
II. Background
II.1. The Functional Cycle of a Single Myosin Head.
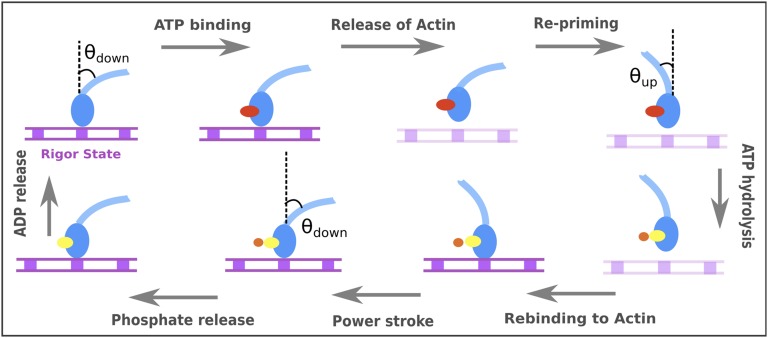
To clarify the nature of the system, we would like to start with a generic description of the functional cycle of a single myosin motor, as understood from scores of experimental and structural findings (1, 3, 9). The cycle is depicted in Fig. 1, which shows that a single myosin molecule can exist predominantly in two distinct conformations, called the “up state” and “down state” (characterized in this work by an angle θup or θdown that the lever axis makes with a tentative axis perpendicular to the actin surface). The cycle in Fig. 1 starts at the “rigor state,” where the lever is in the down state and tightly bound to the actin surface. Binding of ATP leads to release of the myosin head from the actin filament, and after this the lever and the convertor domain changes to the up state from the down state. This conformational reversal, which occurs while myosin is unbound or loosely bound to the actin filament, is often referred as the “repriming” event. The ATP molecule bound to the myosin head undergoes hydrolysis and the products (ADP, Pi) bound head once again rapidly binds actin filaments. The critical “powerstroke” step, or the transition from the up to down state, is thought by some to generate a directional force on the bound actin filament. This step is followed by the release of Pi and then the ADP, after which the system is ready for another cycle. Although current experimental studies cannot tell us directly the molecular origin of the directionality of the single-headed myosin cycle, they provide very useful information when one compiles the relevant energetics estimated from the available kinetic and thermodynamic information (III. Results and Discussion, section III.2).
Fig. 1.
A schematic cycle for a single myosin head (shown in blue). The nucleotide occupancies are denoted in red, yellow, and orange spheres for ATP, ADP, and Pi, respectively. The actin is schematically shown as a track with equally spaced myosin binding sites shown in filled squares. The tight and loose actin-bound states are depicted in dark and light violet, respectively.
II.2. The Unidirectional Processive Runs of the Double-Headed Myosin V.
The double-headed myosin V molecule uses the mechanochemical coupling of the single head to orchestrate repetitive powerstroke cycles that can effectively transport cargo through hundreds of nanometers without detaching completely from the actin filament (4). During the walking process, myosin V takes an average step of about 36 nm on the actin filament and can make several consecutive steps without completely detaching off. The mechanochemical cycle of a single myosin molecule (Fig. 1) holds true for each of the myosin V heads (1), but both cycles occurring in the two heads are coordinated in a fashion so that when one goes through ATP binding, actin detachment, hydrolysis, repriming, and powerstroke cycle in succession the other mostly remains in the ADP bound state tightly bound to the actin filament. The general understanding is that each of the myosin V mechanochemical cycles occurring in one strand is altered differently by the “pulling” or “pushing” force generated on it by the other strand of myosin V, thus leading to an overall coordinated movement of the double stranded molecule over the actin filament (12).
Despite the progress made in understanding the overall myosin V cycle, it is extremely challenging to combine the biochemical and structural findings with a well-defined physical model that reproduces the observed directionality (although it is possible to model the process using phenomenological parameters). In other words, any advance in structure-based understanding of the force generation process must be based on a clear knowledge of the free energy surface. As illustrated in Fig. S1, one must explain why the myosin V steps on the actin filaments predominantly in one direction (to the right, the “plus end”) and not in the opposite direction (to the left). We have only two options for a net directional motion to the right using a qualitatively reasonable free energy surface of the whole process. Either the free energy goes up on the left direction (a thermodynamically controlled mechanism) or the effective barrier to the left direction is higher than that to the right (a kinetically controlled mechanism). In the second case the free energy of the left motion can be as low as that of the right motion. Thus, popular arguments such as “diffusive motion” or “futile lever arm swings” (e.g., ref. 26) are not so effective in understanding the directional motion, which could be studied by constructing a free energy-based model (as in Fig. S1) using structural and kinetics observations. Such an approach should advance our quantitative understanding of the action of myosin.
In our attempts to look for a tentative reason for the directionality we must focus on the asymmetric structural elements in the system. There are several ways of introducing structural asymmetry in the myosin V molecule as it walks over the actin filament, but the most obvious among them is the asymmetry in the interactions arising owing to the θdown to θup conformational change in the lever and the connected convertor domain, whereas other options are much less likely to contribute significantly to the asymmetry in the forward and backward motion (discussed in Supporting Information and below). Thus, we will focus here on understanding the energetics of the change in θ and its relationship to the directional motion in myosin V.
III. Results and Discussion
III.1. CG Modeling of the Conformational Landscape of Single-Headed Myosin V.
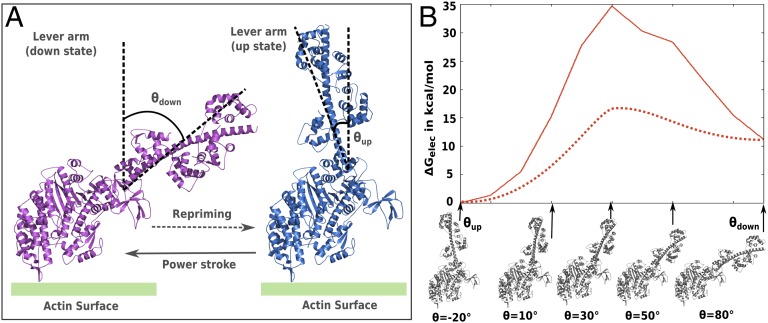
As discussed above, our working hypothesis is that the most important factor contributing to the functional directionality is due to the asymmetry in the energies of the θup and θdown conformations. To explore this hypothesis, we have to understand the conformational free energy landscape of moving from θup to θdown states, and such an attempt is done here using the known pre- and postpowerstroke structures of myosin (9) (Fig. 2A and Supporting Information give a description of myosin V modeling using known structural considerations). The free energy contributions of the protein side chains were evaluated for each of the generated intermediates between the post- and prepowerstroke states using our CG model (21) and the resulting free energy surface is shown in Fig. 2B, whereas the relevant energetic contributions are summarized in Table S1. It should be noted that the effect of the nucleotides and/or actin binding were not incorporated in this calculations and will be included in the analysis below using the experimental information (this is a reasonable strategy because we found that the corresponding energies of the chemical steps do not contribute towards establishing the unidirectionality). As seen from the conformational free energy map of Fig. 2B, the system has an inherent asymmetry, where the energy at θup (prepowerstroke) is almost 11 kcal/mol lower than θdown (postpowerstroke) and the key source for the energy difference between the beginning and end points is of electrostatic origin (Table S1). The residues contributing maximally to the electrostatics free energy change between the pre- and postpowerstroke states are shown in Fig. S2 and discussed in the SI Text. Apparently, Fig. 2B shows that there is a significant rise in free energy as the system tries to shuffle between the two conformations. However, whereas the energy difference between the two conformations is reasonable (because they have been modeled using high-resolution structural data), the actual barrier is probably much lower than that estimated here (shown as a dotted line in Fig. 2B). This is so because we have not attempted to obtain the least energy path along the conformational route, but rather arrived at a quantitative understanding of the energy at the end points, which is the most important part in exploring the reason behind the unidirectionality. A better job of producing an exact path based on converged conformational free energies could be achieved using computationally costly sampling approaches or moving the system in directions implied by the normal modes, as attempted in previous studies (16, 17).
Fig. 2.
(A) Structural model of myosin V lever arm movement used for the current study. The key angles θup and θdown that the lever arm makes with a tentative axis perpendicular to the actin surface characterizes the conformational change during the pre- and postpowerstroke. The tentative actin surface is denoted in green. (B) The calculated CG electrostatic free energy surface (solid red line) and scaled free energy surface (dotted red line) for moving from pre- to postpowerstroke conformation of a single myosin V molecule is shown. The structures on the x axis represent the initial, intermediate, and final states between the θup and θdown conformation. The angular value only provides an estimate of the progression of the conformational reaction coordinate along the path.
III.2. The Energetics of Ligand Binding/Release and Catalysis for Single-Headed Myosin V.
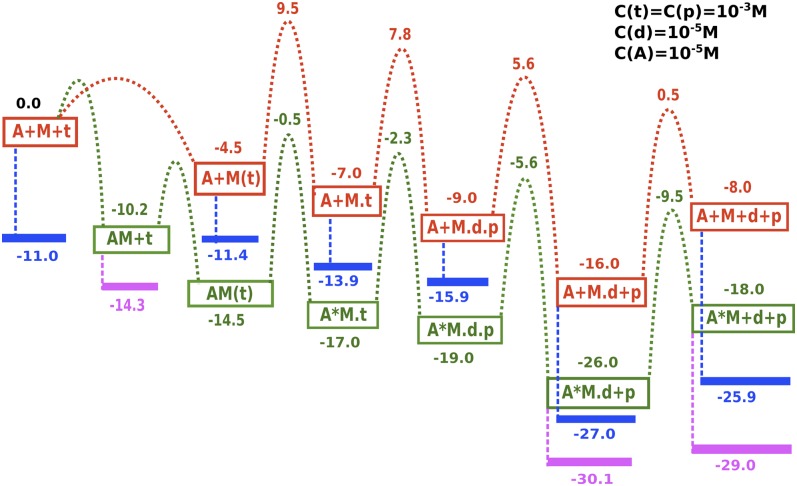
Although the trend displayed in Fig. 2B provides an insight about the asymmetry between the θup and θdown conformations, it is essential to consider the other energy contributions coming from the binding/release and catalytic steps to arrive at a complete picture of the cycle. Here, it is done by collecting the relevant kinetic and thermodynamic experimental results and building a consensus free energy diagram of the single myosin V cycle (Table S2 and Fig. S3). Adding the effect of the physiological concentrations to the standard-state energies obtained in Fig. S3, we arrive at the free energy surface of Fig. 3 for the chemical and actin binding/release steps of myosin V. The concentration effects have been calculated for values near physiological conditions indicated in the figure. The fact that the total free energy change after the release of ADP is around 8 kcal allowed us to provide a rough estimate of the free energy release in some of the steps such as hydrolysis and Pi release, which are not easily apparent from the kinetics experiments alone.
Fig. 3.
The free energy diagram of the chemical steps for the single strand of myosin V. The figure uses experimental kinetic parameters tabulated in Table S2 and Fig. S3 and adds the concentration effect to the standard Gibbs free energies obtained from the rate equations. Myosin, Actin, ATP, ADP, and Pi are denoted by M, A, t, d, and p, respectively. The pathways denoted in red and green are for myosin V in actin-free or actin-bound forms, respectively. The effect of concentrations is highlighted in blue and violet for the myosin V in actin-free or actin-bound forms, respectively. Note that this cycle alone (even with concentration effects) cannot be used to establish the directionality in a walking myosin V molecule because all actin-bound myosin states are much lower in energy than the actin-free states, and the system will not revert to the actin-free state without the correct coupling of the conformational free energy.
The kinetics and thermodynamics information given in Fig. 3 for the single-headed myosin does not provide a complete picture about the myosin V walking cycle. As seen from Fig. 3, the energetics of the free myosin is always higher compared with the actin-bound form [because the binding of actin is always a fast and downhill process (1)]. This implies that for a functional cycle the conformational energy must be coupled to the chemical and binding energies to allow the ATP bound myosin head to remain predominantly in the actin-free form and to revert back to the actin-bound form upon ATP hydrolysis and product release. The reversal of myosin head from actin-free to actin-bound state is the most important part of the cycle because this is necessary for a successful powerstroke generation. Likewise, the reversal of actin-bound myosin head to the actin-free state after ATP binding is crucial for the repriming of the cycle. Multiple studies have attempted to understand this problem and have led to several hypotheses based on structural and kinetic modeling (9, 14, 26), but a clear quantitative basis of the conformation/chemical coupling leading to the myosin V processive cycle has not yet been provided.
III.3. Modeling the Energetics Behind the Unidirectional Myosin V Walking Cycle.
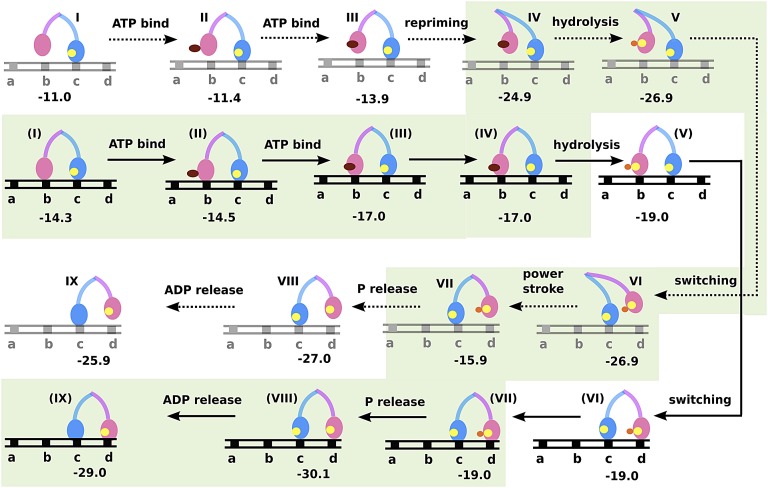
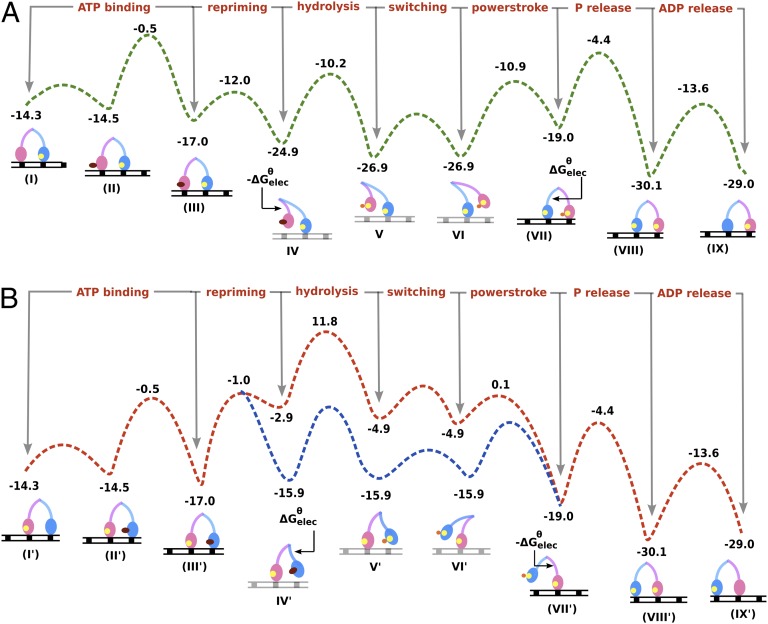
At this stage, we can finally combine the information obtained from the conformational landscape (Fig. 2) with the energetics of the chemical steps of myosin V single head (Fig. 3) to arrive at a feasible functional cycle for the double-headed molecule. Fig. 4 shows such an analysis where the leading leg is ADP-bound and strongly attached to actin as the lagging leg covers its journey starting from the rigor state, to repriming, moving past the leading strand and rebinding actin at a new position. The energetics for each subsequent step is shown for both actin-free (steps I to IX) and actin-bound [steps (I) to (IX)] lagging strand. We have analyzed both actin-bound and -free forms to be able to compare similar states of myosin V and arrive at a least free energy-based description of the actual functional cycle. This approach clearly captured the energetics involved in shifting of the population from actin-bound to actin-unbound states for ATP-bound myosin V [steps (III) to IV] and vice versa after ATP hydrolysis [steps VI to (VII)]. The analysis shown in Fig. 4 describes the functional cycle when we follow the least energy path, and this energetically feasible pathway is highlighted in green. The result elucidates a previously unknown role of the electrostatic conformational free energy in guiding myosin V through its directional cycle. The release of the conformational free energy upon repriming (−11 kcal) makes the actin-free state lower in energy than the actin-bound state [compare steps IV and (IV) and also note that this would not have happened when one only considers the kinetics in Fig. 3]. This leads to the stabilization of the ATP-bound lagging myosin head in the actin-free form. After ATP hydrolysis, commencement of the powerstroke leads to a gain in electrostatic free energy and this makes the actin-free form much less favorable than the actin-bound form [compare steps (VII) and VII], finally leading to product release and the establishment of the rigor state.
Fig. 4.
The free energy landscape for myosin V walking toward the actin “plus end” (obtained by combining information from Fig. 3 and Fig. 2). The states denoted I to IX are when myosin V lagging strand (pink) is in the actin-free state, while the states denoted (I) to (IX) are for both myosin V strands bound to actin. The nucleotide occupancies are denoted in deep red, yellow, and orange spheres for ATP, ADP, and Pi, respectively. The actin is shown as a black or gray track with equally spaced myosin binding sites denoted as a, b, c, and d, where moving in a → d indicates the “plus end” direction. The least free energy path highlighted in green reveals a downhill path as myosin walks from b → d. It also highlights the role of electrostatic energy released during repriming in stabilizing the actin-free ATP bound form [states (IV) to IV] and the energy gained through powerstroke in stabilizing the actin-bound form [states VII to (VII)] of the pink strand.
The free energy surface for the functional cycle along with the tentative barriers calculated from Figs. 2 and 3 is shown in Fig. 5A, showing a downhill path as myosin V moves to the right direction (“plus end” of actin). The large stabilization energy arising from the post- to prepowerstroke change during each 36-nm walking step guides the system consistently toward the correct direction. It is also worth mentioning that the free energy change that guides the directional cycle also provides an idea of the stall force (∼2–3 pN) required to stop the myosin from moving forward. It is also possible that the high electrostatic barrier for the lever arm swing during powerstroke is around 16 kcal (at par with the ADP release step and much lower than that estimated in Fig. 2, solid curve) and actually the uphill conformational barrier might be the reason behind the tightly coupled ADP release step. Also, note that the actual barrier involved in the process of “switching” the lagging strand from its lagging position to the next actin binding site can be low and is not estimated in this study. It is assumed from other studies that this movement can be more akin to a random search of the lagging head under the constraint of the strongly actin-bound leading strand (27).
Fig. 5.
(A) The free energy surface for the functional path when myosin walks toward the actin “plus end” is shown in green. (B) The free energy surface for the reverse walking scheme is shown in red. The figure is based on Fig S4. An alternative route is represented using blue dashed lines that designate the likely possibility that the reverse motion can also be achieved through a combination of the powerstroke in blue myosin strand and repriming in pink myosin strand to minimize the large ΔGθelec contribution. However, this lower energy surface (blue) is still higher than the functional surface (green). The electrostatic free energy for the conformational contribution (ΔGθelec) affecting the repriming and powerstroke of the specific myosin strand are also indicated in the appropriate steps.
The significance of the above finding in demonstrating the unidirectionality of myosin V is further clarified in Fig. 5B and Fig. S4. Here we show that the motion in the opposite direction is much less feasible owing to the higher energies (at steps IV′, V′ and VI′) leading to higher effective barrier for the whole process. Fig. S4 shows an analysis similar to that in Fig. 4, but instead we try to take the leading strand through the ATP binding, repriming, hydrolysis, powerstroke, and product release steps and thus attempt to move the myosin V in the opposite direction. The path shaded in green in Fig. S4 is further shown in Fig. 5B and highlights the high energy cost associated with such a reverse walking scheme. This high energy originates from the fact that the leading strand is already in the θup state and thus cannot release the electrostatic energy upon repriming that can lead to the ATP bound actin-free state of myosin. Fig. 5 also highlights the important role of the conformational free energy (electrostatic) in dictating the directionality of the myosin V run on actin. Such an analysis identifies the energetics responsible for the unidirectional myosin V motion toward the “plus end” of actin filament for a significant stretch of time (5). It is important to note that the directionality is primarily enforced once we successfully couple the conformational free energy with the chemical steps. The effect of physiological concentrations of the ligands on the standard free energies or the energy released upon ADP and Pi release do not lead to the switching between actin-bound and actin-free forms in the mechanochemical cycle of myosin. Thus, the chemical steps alone probably cannot provide the basis for a successful powerstroke cycle and, more importantly, they cannot lead to the overall directional motion.
Trying to assess the “fidelity” of moving in one direction predicted by the landscapes of Fig. 5 is in principle straightforward. What is needed is a Langevin dynamics treatment with a proper friction coefficient determined by our renormalization treatment. However, a careful treatment should consider the least energy path motions of the lever arm described by the changing θ as well as the binding and release of the ligands (see our work on F1-ATPase in ref. 22). At present, because we are mainly interested in understanding the unidirectionality, we may get a “back of the envelope” estimate by considering the rate of the competing processes shown in Fig. 5 A and B. This can be done by comparing the competing rates of moving from state (I) to (IX) in the correct direction and state (I′) to (IX′) in the backward direction. Because in both cases we do not have any significant return from the states of ADP release [(IX) or (IX′)], which are strongly downhill, we can consider a very simplified model with
 |
where 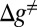 is the difference between the highest point and the initial point in the landscape for each forward- or backward-moving cycle. The proportionality constant
is the difference between the highest point and the initial point in the landscape for each forward- or backward-moving cycle. The proportionality constant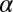 is introduced because there are ways to move without introducing the full contribution from
is introduced because there are ways to move without introducing the full contribution from 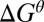 during the highest points in the free energy profile. Obviously, Eq. 1 gives a clear advantage in moving forward, which is what we like to establish in this work. Nevertheless, combining the analysis by a more careful time-dependent renormalization study (with detailed landscape for the flexibility of the arms and the effect of different loads as well as ligand concentrations) should allow one to narrow down the determination of the exact
during the highest points in the free energy profile. Obviously, Eq. 1 gives a clear advantage in moving forward, which is what we like to establish in this work. Nevertheless, combining the analysis by a more careful time-dependent renormalization study (with detailed landscape for the flexibility of the arms and the effect of different loads as well as ligand concentrations) should allow one to narrow down the determination of the exact 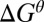 .
.
As stated before in II. Background, section II.2 and in Supporting Information, we can have other potential contributions affecting the overall asymmetry of the system arising from the conformational free energy change. For example, the interaction of myosin head with actin could potentially be different in different parts of actin and can contribute toward establishing the directionality. Although a complete atomistic structure of myosin/F-actin complex is still missing, our current understanding from the experiments suggests that both heads of myosin V bind stereospecifically to F-actin and are separated by ∼36 nm or ∼13 actin monomeric units as it steps on the actin filament during its processive run (28). This reflects the fact that the final and initial states of the acto–myosin complex are similar after each individual cycle, except for the progress in positional coordinates for the overall myosin molecule toward the “plus end.” Although, it is possible that the leading and lagging strands adopt different lever arm geometries (kinked or straight) within the course of the cycle, the focus of our present study is to understand the physical principle behind the directionality of the functional cycle. Armed with a better and more quantitative understanding of the basic functional cycle, one can proceed toward modeling various alterations arising owing to shortened lever arms or effect of pushing and pulling loads on the motor.
Like the acto–myosin interactions, it is also possible to have different contributions arising from myosin interacting with the different forms of the ligand (ATP, ADP and Pi, only ADP, or ligand-free) during the cycle. These ligand-specific interactions definitely contribute toward the overall free energy of the functional cycle. However, comparison of the ATP-bound, ADP-bound, or ligand-free myosin head shows very little deviance in the overall structure. Alterations of side chain arrangements and certain loop regions in and around the nucleotide-binding regions have a potential to affect θup and θdown states in an asymmetric way. However, considering the small number of structural changes compared with the large shifts in the lever arm and the adjacent convertor domain during the powerstroke generation (9), it is most probable that the main deciding factor toward asymmetry arises from their interaction with the rest of the conserved myosin head domains.
IV. Concluding Remarks
Providing a structure/energy basis of the powerstroke generation and the conformational–chemical coupling is crucial to elucidate the nature of the processive directional movement in translational molecular motors. The present work has advanced the understanding of the origin of the unidirectionality of the myosin V motor by providing a structure-based landscape of the factor that is most likely responsible for the directionality. Combining the free energy surface of the most important large-scale conformational change occurring in the myosin motors with that of the ATP hydrolysis and the myosin/actin binding energy provides a semiquantitative understanding of the overall function. Furthermore, it provides a clear free energy-based conceptual basis of the occurrence of different key states during the myosin V processive movement and, most importantly, provides a basis for the unidirectionality shown during its processive movement along the actin filament. Our study highlights that the change in the electrostatic free energy of the lever arm and convertor region during the repriming leads to the stabilization of the lagging myosin V strand in the actin-free state, while the powerstroke leads to an overall increase in the conformational free energy that once again stabilizes the ADP bound myosin V strand in an actin-bound form, thus guiding the overall functional directionality toward the actin “plus end.”
Our result indicates that the free energy change involved in the lever arm swing of myosin V lagging strand biases the landscape so that the molecular motor predominantly moves in the correct direction while taking several steps along the actin filaments. It is estimated that the most significant contributor toward the unidirectional motion is due to the change in electrostatic interaction energy between the drastically different lever arm positions (θup and θdown) and the fairly conserved myosin head domain. Obviously, a more detailed study that will explore the effects due to ligand occupancy should help in fine-tuning the overall result. However, a similar trend in the energetics of the two conformational states will most likely prevail and dominate the unidirectional movement.
Another aspect of our current study is the consideration of the effect of cellular ligand concentrations in altering the relative free energies of the key states during the functional cycle. Although it has been shown in III. Results and Discussion that the concentration effect is not the origin of the directionality in myosin action, our study indicates that the motors can only be active within a certain range of ligand concentrations. Thus, our theoretical approach provides a realistic modeling platform for cellular systems that are actually dependent on environmental factors such as ligand concentrations. Overall, the quantitative knowledge of the free energy surface underlying the myosin V cycle provides further opportunities to explore the motor function under different experimental conditions. For example, the effect of loads on the conformational barrier could be simulated by our renormalization approach and thus increase the connection between the theoretical modeling and single-molecule experiments (12, 29).
The concept of Brownian motors is frequently invoked (30) as the underlying theoretical framework for many biological systems. However, once the free energy surface is determined, the motor functionality and the corresponding directional process become clear from the rate constants of individual steps that follows the Boltzmann-based transition-state theory. Thus, although the observation and identification of “diffusive search” is very insightful and potentially can inform us on the ruggedness of the landscape, it does not help in elucidating the origin of the motor action because all biological systems undergo thermal motion on the available configurational space. Similarly, the description of “futile motions” (26) does not tell us much about the origin of the directionality because futile motions occur both in the correct and incorrect direction and what counts is free energy-based preferential selection.
Finally, because the basic functional cycle of the myosin molecules is thought to be conserved among the different members of the family, this current advance in quantifying the principle behind the myosin V directionality is likely to extend our understanding of the other members of the acto–myosin family. This further opens up an opportunity to explore the role of electrostatics in detail and understand the effects of mutations in altering the motor function.
V. Methods
The present work uses a CG model (21) that describes the main chains by an explicit model and represents the side chains by a simplified united atom model. The model has a unique treatment of the electrostatic energy including the self-energy that gives its reliable features. The details of our model are given elsewhere (24) and it is also discussed in Supporting Information. The details of the kinetics and thermodynamics data used to calculate the standard free energies are provided in Supporting Information.
Supplementary Material
Acknowledgments
We thank the University of Southern California High Performance Computing and Communication Center for computational resources. This work was supported by National Science Foundation Grant MCB-0342276 and National Institutes of Health Grant R01-AI055926.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317641110/-/DCSupplemental.
References
- 1.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16(1):61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 4.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400(6744):590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 5.Kinosita K, Jr, Ali MY, Adachi K, Shiroguchi K, Itoh H. How two-foot molecular motors may walk. Adv Exp Med Biol. 2005;565:205–218, discussion 218–219, 379–395. doi: 10.1007/0-387-24990-7_16. [DOI] [PubMed] [Google Scholar]
- 6.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96(24):13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41(26):8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 8.Trybus KM, Krementsova E, Freyzon Y. Kinetic characterization of a monomeric unconventional myosin V construct. J Biol Chem. 1999;274(39):27448–27456. doi: 10.1074/jbc.274.39.27448. [DOI] [PubMed] [Google Scholar]
- 9.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23(23):4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442(7099):208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 12.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci USA. 2005;102(39):13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300(5628):2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 14.Lan G, Sun SX. Dynamics of myosin-V processivity. Biophys J. 2005;88(2):999–1008. doi: 10.1529/biophysj.104.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Ma L, Yang Y, Cui Q. Mechanochemical coupling in the myosin motor domain. II. Analysis of critical residues. PLOS Comput Biol. 2007;3(2):e23. doi: 10.1371/journal.pcbi.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tehver R, Thirumalai D. Rigor to post-rigor transition in myosin V: Link between the dynamics and the supporting architecture. Structure. 2010;18(4):471–481. doi: 10.1016/j.str.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Navizet I, Lavery R, Jernigan RL. Myosin flexibility: Structural domains and collective vibrations. Proteins. 2004;54(3):384–393. doi: 10.1002/prot.10476. [DOI] [PubMed] [Google Scholar]
- 18.Craig EM, Linke H. Mechanochemical model for myosin V. Proc Natl Acad Sci USA. 2009;106(43):18261–18266. doi: 10.1073/pnas.0908192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinczewski M, Tehver R, Thirumalai D. Design principles governing the motility of myosin V. Proc Natl Acad Sci USA. 2013;110:E4059–E4068. doi: 10.1073/pnas.1312393110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng W. Coarse-grained modeling of conformational transitions underlying the processive stepping of myosin V dimer along filamentous actin. Proteins. 2011;79(7):2291–2305. doi: 10.1002/prot.23055. [DOI] [PubMed] [Google Scholar]
- 21.Messer BM, et al. Multiscale simulations of protein landscapes: Using coarse-grained models as reference potentials to full explicit models. Proteins. 2010;78(5):1212–1227. doi: 10.1002/prot.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Warshel A. Electrostatic origin of the mechanochemical rotary mechanism and the catalytic dwell of F1-ATPase. Proc Natl Acad Sci USA. 2011;108(51):20550–20555. doi: 10.1073/pnas.1117024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee S, Warshel A. Realistic simulations of the coupling between the protomotive force and the mechanical rotation of the F0-ATPase. Proc Natl Acad Sci USA. 2012;109(37):14876–14881. doi: 10.1073/pnas.1212841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dryga A, Chakrabarty S, Vicatos S, Warshel A. Realistic simulation of the activation of voltage-gated ion channels. Proc Natl Acad Sci USA. 2012;109(9):3335–3340. doi: 10.1073/pnas.1121094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rychkova A, Mukherjee S, Bora RP, Warshel A. Simulating the pulling of stalled elongated peptide from the ribosome by the translocon. Proc Natl Acad Sci USA. 2013;110(25):10195–10200. doi: 10.1073/pnas.1307869110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Málnási-Csizmadia A, Kovács M. Emerging complex pathways of the actomyosin powerstroke. Trends Biochem Sci. 2010;35(12):684–690. doi: 10.1016/j.tibs.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beausang JF, Shroder DY, Nelson PC, Goldman YE. Tilting and wobble of myosin V by high-speed single-molecule polarized fluorescence microscopy. Biophys J. 2013;104(6):1263–1273. doi: 10.1016/j.bpj.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker ML, et al. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405(6788):804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 29.Dryga A, Warshel A. Renormalizing SMD: The renormalization approach and its use in long time simulations and accelerated PMF calculations of macromolecules. J Phys Chem B. 2010;114(39):12720–12728. doi: 10.1021/jp1056122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Astumian RD. Making molecules into motors. Sci Am. 2001;285(1):56–64. doi: 10.1038/scientificamerican0701-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.