Significance
CDC5, a DNA binding protein, regulates the growth and immunity of Arabidopsis, however, its functional mechanism remains to be identified. This research reveals that CDC5 acts as a positive transcription factor and a component of the DICER–LIKE1 complex to regulate microRNA (miRNA) accumulation. By identifying a unique regulator and its activities in miRNA biogenesis, this study provides insight on the regulation of miRNA levels. Because CDC5 homologs are present in other organisms, including animals, this study may have a broad effect on the miRNA biogenesis field.
Abstract
CDC5 is a MYB-related protein that exists in plants, animals, and fungi. In Arabidopsis, CDC5 regulates both growth and immunity through unknown mechanisms. Here, we show that CDC5 from Arabidopsis positively regulates the accumulation of microRNAs (miRNAs), which control many biological processes including development and adaptations to environments in plants. CDC5 interacts with both the promoters of genes encoding miRNAs (MIR) and the DNA-dependent RNA polymerase II. As a consequence, lack of CDC5 reduces the occupancy of polymerase II at MIR promoters, as well as MIR promoter activities. In addition, CDC5 is associated with the DICER–LIKE1 complex, which generates miRNAs from their primary transcripts and is required for efficient miRNA production. These results suggest that CDC5 may have dual roles in miRNA biogenesis: functioning as a positive transcription factor of MIR and/or acting as a component of the DICER–LIKE1 complex to enhance primary miRNA processing.
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are ∼22-nucleotide (nt) noncoding RNAs that regulate various biological processes including development, metabolism, and immunity in plants and animals (1–3). miRNAs are generated from primary miRNA transcripts (pri-miRNAs) containing stem–loop structure, whereas siRNAs are derived from long, perfect, double-stranded RNAs (dsRNAs) (1–3). They are associated with members of the Argonaute protein family to repress gene expression at posttranscriptional and/or transcriptional levels (1–3). In addition to miRNAs, plants encode two major classes of siRNAs: siRNAs derived from repeated DNAs (ra-siRNAs) and transacting siRNAs (ta-siRNAs) (4–6).
Studies in Arabidopsis have established the framework of miRNA biogenesis in plants (1–3). In Arabidopsis, pri-miRNAs are primarily transcribed by DNA-dependent RNA polymerase II (Pol II), with assistance from the mediator complex and the transcription factor Negative on TATA less2 (NOT2) (7, 8). After transcription, pri-miRNAs are processed by an RNase III enzyme called DICER–LIKE1 (DCL1) to miRNA precursors and then to mature miRNAs (9, 10). The efficient processing of pri-miRNA requires SERRATE (SE; a zinc finger protein), TOUGH (an RNA-binding protein), and a dephosphorylated HYPONASTIC LEAVES1 (HYL1; a double-stranded RNA binding protein) that form a complex with DCL1 (11–18). SE and HYL1 also promote the processing accuracy of pri-miRNAs (19). Four other proteins, DAWDLE (DDL; an RNA binding protein), Cap-binding protein 20, Cap-binding protein 80, and NOT2, which are associated with the DCL1 complex (8, 20–22), also function in miRNA biogenesis. Recent studies also reveal that the correct localization of DCL1 requires NOT2 and MODIFIER OF SNC1, 2 (an RNA binding protein) (8, 23). In addition, the accumulation of a subset of miRNAs requires a proline-rich protein named SICKLE (24).
The cell division cycle 5 (CDC5) protein is a conserved protein in animals, plants, and fungi (25). It was first isolated from Schizosaccharomyces pombe as a cell cycle regulator. CDC5 is considered a putative transcription factor, as it is a MYB (a transcription factor)-related protein (26–28). In human and yeast, CDC5 has been shown to act as a component of spliceosome to participate in mRNA splicing (29, 30). In Arabidopsis, CDC5 binds DNA and is required for development and immunity to bacteria infection (31, 32). However, how CDC5 functions in Arabidopsis is unclear.
Here, we show that CDC5 plays important roles in the biogenesis of miRNAs and siRNAs in Arabidopsis. CDC5 interacts with both Pol II and the promoters of genes encoding miRNAs (MIR). As a consequence, impairment of CDC5 reduces the MIR promoter activity and the occupancy of Pol II at the MIR promoter. In addition, CDC5 is associated with the DCL1 complex and is required for efficient miRNA production. On the basis of these results, we propose that CDC5 positively regulates the transcription and/or processing of pri-miRNAs.
Results
CDC5 Is Required for the Accumulation of miRNAs and siRNAs.
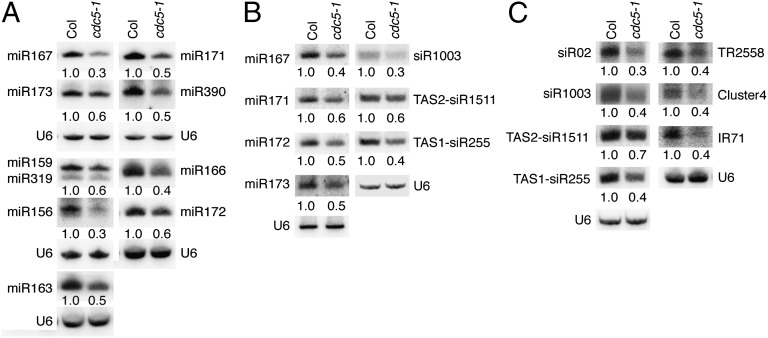
In cdc5-1, a T-DNA insertion disrupts the expression of CDC5, resulting in multiple developmental defects such as smaller plant size, altered leaf shape, late flowering, and sterility (31, 32). We reasoned that cdc5-1 might impair miRNA accumulation, as the alteration in miRNA levels often causes pleiotropic developmental defects (33, 34). We thus performed Northern blot analysis to examine miRNA abundance in inflorescences of cdc5-1 and Columbia-0 (Col; wild-type control). The levels of all examined miRNAs (miR166/165, miR167, miR159/319, miR390, miR171, miR172, miR173, miR156, and miR163) were reduced in cdc5-1 compared with those in Col (Fig. 1A and Fig. S1A). A CDC5-YFP transgene driven by the CDC5 promoter (pCDC5::CDC5-YFP) fully restored miRNA levels (Fig. S2A), demonstrating that cdc5-1 is responsible for the reduction of miRNA abundance. In addition, cdc5-1 exhibited a similar effect on the levels of several examined miRNAs in leaves as those in inflorescences (Fig. 1B and Fig. S1B). We also tested the effect of cdc5-1 on the accumulation of endogenous siRNAs, including two ta-siRNAs, TAS1-siR255 and TAS2-siR1511, and five siRNAs derived from ra-siRNAs, siR02, siR1003, cluster 4, IR71, and TR2588. The levels of these siRNAs were lower in cdc5-1 than those in Col (Fig. 1C and Fig. S1C).
Fig. 1.
cdc5-1 reduces the accumulation of miRNAs and siRNAs. (A) miRNA abundance in inflorescences of cdc5-1 and Col. (B) miRNA abundance in leaves of cdc5-1 and Col. (C) siRNA abundance in inflorescences cdc5-1 and Col. Col: wild-type control of cdc5-1. U6: spliceosomal RNA U6. Small RNAs were detected by Northern blot. After Northern blot, the radioactive signals were detected with phosphor imager and quantified with ImageQuant (V5.2). To determine relative abundance of small RNAs in cdc5-1, the amount of a miRNA or siRNA in cdc5-1 was normalized to U6 RNA and compared with that in Col. Value of miRNAs or siRNAs in Col was set as 1. The number below cdc5-1 indicated the relative abundance of miRNAs or siRNAs, which is the average value of three repeats. P < 0.05; except for siR255 in Fig. 1C (t test). For miR159/319, the upper band was miR159 and the lower band was miR319.
We next examined the effects of cdc5-1 on miRNA and ta-siRNA function by analyzing the expression levels of their targets, using quantitative RT-PCR (qRT-PCR). The transcript levels of several examined targets (ARF8, CUC1, MYB65, PPR, SPL6, SPL10, and ARF3) were moderately increased in cdc5-1 relative to those in Col (Fig. S2B).
CDC5 Regulates the Transcription of MIR.
We next performed qRT-PCR to examine the levels of seven pri-miRNAs (pri-miR158a, pri-miR159a, pri-miR167a, pri-miR171a, pri-miR172a, pri-miR172b, and pri-miR173) in Col and cdc5-1. The levels of these pri-miRNAs decreased in cdc5-1 compared with those in Col (Fig. 2A). The reduced levels of pri-miRNAs and miRNAs in cdc5-1 can result from impaired transcription of pri-miRNAs. Alternatively, it can be a consequence of reduced stability and rapid degradation of pri-miRNAs by nonspecific nucleases, degradation of mature miRNAs, or enhanced posttranscriptional processing of pri-miRNAs in cdc5-1. To test these alternative explanations, we first determined whether CDC5 regulates MIR transcription by examining the effect of cdc5-1 on the expression of a GUS reporter gene driven by the MIR172b promoter (pMIR172b::GUS) (20). We have previously used this system to determine the function of DDL in regulating MIR transcription (20). If CDC5 is indeed a positive transcription regulator of MIR, cdc5-1 will reduce the expression levels of GUS. We crossed cdc5-1 with a Col transgenic line, which contains the pMIR172b::GUS transgene (20). In the second (F2) generation, we obtained CDC5+ (CDC5/CDC5 or CDC5/cdc5) and cdc5-1 genotypes containing pMIR172b::GUS. GUS staining on these plants revealed that the GUS activity was lower in cdc5-1 than that in CDC5+ (Fig. 2B). qRT-PCR analysis confirmed that the GUS mRNA levels in cdc5-1 were reduced relative to those in CDC5+(Fig. 2C).
Fig. 2.
cdc5-1 reduces the promoter activity of MIR. (A) The transcript levels of various pri-miRNAs in inflorescences of cdc5-1 and Col determined by qRT-PCR. The abundance of pri-miRNAs in cdc5-1 was normalized to that of UBQUITIN5 (UBQ5) and compared with that in Col. Value of Col was set to 1. SD of three technical replications was shown as error bars. (B) The levels of GUS in CDC5+ and cdc5-1 harboring pMIR172b::GUS. CDC5+:CDC5/CDC5, or CDC5/cdc5-1. Twenty plants containing GUS were analyzed for each of CDC5+ and cdc5-1 genotypes. An image for each genotype is shown. (C) The transcript levels of GUS driven by MIR172b promoter in CDC5+ and cdc5-1. GUS transcript levels were determined by qRT-PCR. The GUS mRNA levels in cdc5-1 were normalized to UBQ5 and compared with those in CDC5+. *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
CDC5 Is Required for Pol II Occupancy at the Promoter of MIR.
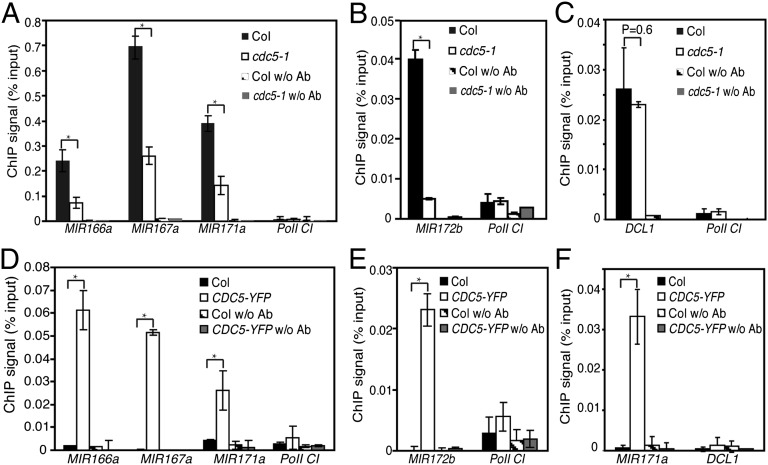
To confirm that CDC5 is a positive transcription factor of MIR, we monitored the occupancy of RNA Pol II at the promoters of MIR166a, MIR167a, MIR171a, and MIR172b in cdc5-1 and Col by chromatin immunoprecipitation (ChIP), using an antibody against the second largest subunit of Pol II (RPB2), as described by Kim et al. (7). We included a no-antibody ChIP as a negative control. After ChIP, the MIR166a, MIR167a, MIR171a, and MIR172b promoter fragments were examined by qPCR. As previously reported (7), the promoter regions of these four MIRs, but not Pol II C1 (a genomic fragment between genes At2g17470 and At2g17460) (7), were enriched in RPB2 immunoprecipitates relative to the no-antibody control in Col. cdc5-1 reduced the occupancy of Pol II at these regions relative to Col (Fig. 3 A and B). Because the transcript levels of DCL1 are not affected in cdc5-1 (described in CDC5 Is Associated with the DCL1 Complex), we also included it as a negative control for the ChIP assay. The association of Pol II with DCL1 promoter did not significantly change in cdc5-1 (Fig. 3C). These data further supported that CDC5 positively regulates MIR transcription in Arabidopsis. However, we cannot rule out the possibility that CDC5 also regulates the transcription of some protein-coding genes.
Fig. 3.
CDC5 is required for the recruitment of Pol II to MIR promoters. (A–C) The occupancy of Pol II at various promoters detected by ChIP using anti-RBP2 antibody in cdc5-1 and Col. (D–F) The association of CDC5 with various promoters detected by ChIP using anti-YFP antibody in plants containing pCDC5::CDC5-YFP. DNA copurified with CDC5 or Pol II was analyzed with qRT-PCR. The intergenic region between At2g17470 and At2g17460 (Pol II C1) that is not occupied by Pol II was used as a negative control. ChIP with no antibodies was performed as another control. Means and standard derivations of three technical repeats are presented, and three biological replicates gave similar results. Please note that the results of Pol II C1 in RBP2 ChIP (A and B) and in CDC5 ChIP (D and E) were showed twice, respectively, for control purposes. *P < 0.05 (t test).
CDC5 Interacts with MIR Promoters.
Because CDC5 is a putative MYB domain-containing transcription factor and has a DNA binding activity (27), we next examined whether CDC5 binds the promoter of MIRs. We performed ChIP, using an antibody against YFP on the cdc5-1 complementation line containing pCDC5::CDC5-YFP (Fig. S1A) and Col. qPCR analysis showed that MIR166a, MIR167a, MIR171a, and MIR172b promoter fragments were enriched in the CDC5–YFP complex, but not in the Col and no-antibody controls (Fig. 3 D and E). In addition, CDC5 did not bind the promoter of DCL1 (Fig. 3F). These results suggested that CDC5 is associated with MIR promoters.
CDC5 Interacts with Pol II.
The association of CDC5 with MIR promoters and the reduced Pol II occupancy at MIR promoters in cdc5-1 suggest that CDC5 may positively regulate MIR transcription by promoting the recruitment of Pol II to their promoters, which predicts a potential CDC5–Pol II interaction. Thus, we tested the association of CDC5 with Pol II through reciprocal coimmunoprecipitation (co-IP). We extracted proteins from the cdc5-1 complementation line expressing pCDC5::CDC5-YFP and a Col control expressing a YFP transgene. IP was performed with either anti-YFP antibody or anti-RPB2 antibody. Western blots detected RPB2 in the CDC5–YFP complex and CDC5–YFP in the RPB2 immunoprecipitates, respectively (Fig. 4 A and B). In contrast, the interaction between YFP and RPB2 were not detected (Fig. 4 A and B). In addition, protein G beads without antibody failed to pull down either CDC5–YFP or RPB2. Both CDC5 and Pol II bind DNAs, suggesting that the CDC5–Pol II interaction may depend on DNA. However, DNase I treatment during IP had no obvious effect on CDC5–Pol II interaction (Fig. 4C). These results suggested that the CDC5–Pol II association might be DNA-independent.
Fig. 4.
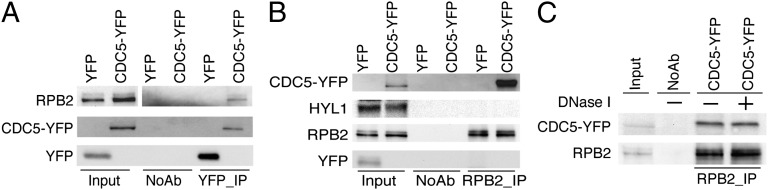
CDC5 interacts with Pol II. (A and B) Co-IP between CDC5–YFP and Pol II. (C) Co-IP between CDC5–YFP and Pol II is DNA-independent. Protein extracts isolated from inflorescences of plants containing CDC5–YFP or YFP were used to perform IP, using either Anti-YFP or Anti-RBP2. YFP, CDC5–YFP, and RBP2 were detected by Western blot, using anti-YFP antibody and anti-RPB2, respectively, and labeled on the left side of the picture. Two percent of input proteins were used for RPB2, whereas 20% input proteins were used for YFP and DCL1–YFP, respectively.
CDC5 Is Required for the Accumulation of miR162 in an in Vitro Assay.
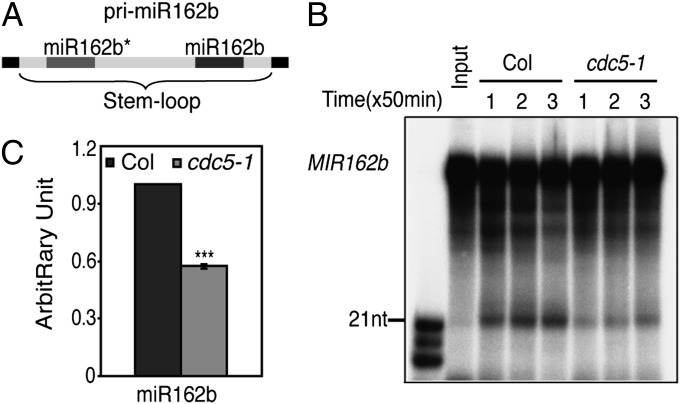
We next asked whether CDC5 could function after MIR transcription by examining the effect of cdc5-1 on the accumulation of miRNAs in an in vitro processing assay (13, 35). In this in vitro processing assay, a portion of pri-miR162b that contains the predicted stem–loop of miR162b with 6-nt arms at each end (MIR162b; Fig. 5A) was used. A radioactive-labeled MIR162b probe was generated by in vitro transcription under the presence of [α-32P] UTP. Radioactive-labeled MIR162b was then incubated with protein extracts from young flower buds of either cdc5-1 or Col, respectively. After reactions were stopped at 50, 100, and 150 min, RNAs were extracted and resolved on a denaturing polyacrylamide gel. The protein extracts of cdc5-1 generated less miR162b than that of Col (Fig. 5B). Quantitative analysis at the 100-min point showed that the abundance of miR162 in cdc5-1 was ∼50% that in Col (Fig. 5C). These results suggested that CDC5 might have roles in miRNA biogenesis other than acting as a MIR transcription factor.
Fig. 5.
cdc5-1 reduces the accumulation of miR162 in an in vitro processing assay. (A) Schematic diagram of the pri-miR162b fragment (MIR162b) used in the in vitro processing assay. (B) MIR162b processing by protein extracts from cdc5-1 and Col. After reaction, RNAs were extracted, resolved on PAGE gel, and detected with a phosphor imager. (C) Quantification of miR162 production in cdc5-1 relative to Col. The quantitative analysis was performed for the reaction stopped at 100 min, as shown in B. The radioactive signal of miR162 was quantified with ImageQuant (V5.2) and then normalized to input to determine the amount of miR162 produced by cdc5-1 or Col protein extracts (miR162cdc5-1 or miR162Col). The relative level of miR162 produced by cdc5-1 was calculated as miR162cdc5-1 divided by miR162Col. The value of miR162Col was set as 1. The value represents mean of three repeats. ***P < 0.001 (t test).
CDC5 Is Associated with the DCL1 Complex.
To determine how CDC5 acts after MIR transcription, we first examined whether cdc5-1 affected the transcript levels of several known genes involved in miRNA biogenesis, including CBP80, CBP20, DDL, HYL1, DCL1, HEN1, and SE, by qRT-PCR. The expression levels of these genes were slightly increased in cdc5-1 relative to Col (Fig. S3A). Western blot analysis showed that the protein levels of DCL1 and HYL1 were comparable in cdc5-1 to those seen in Col (Fig. S3 B and C).
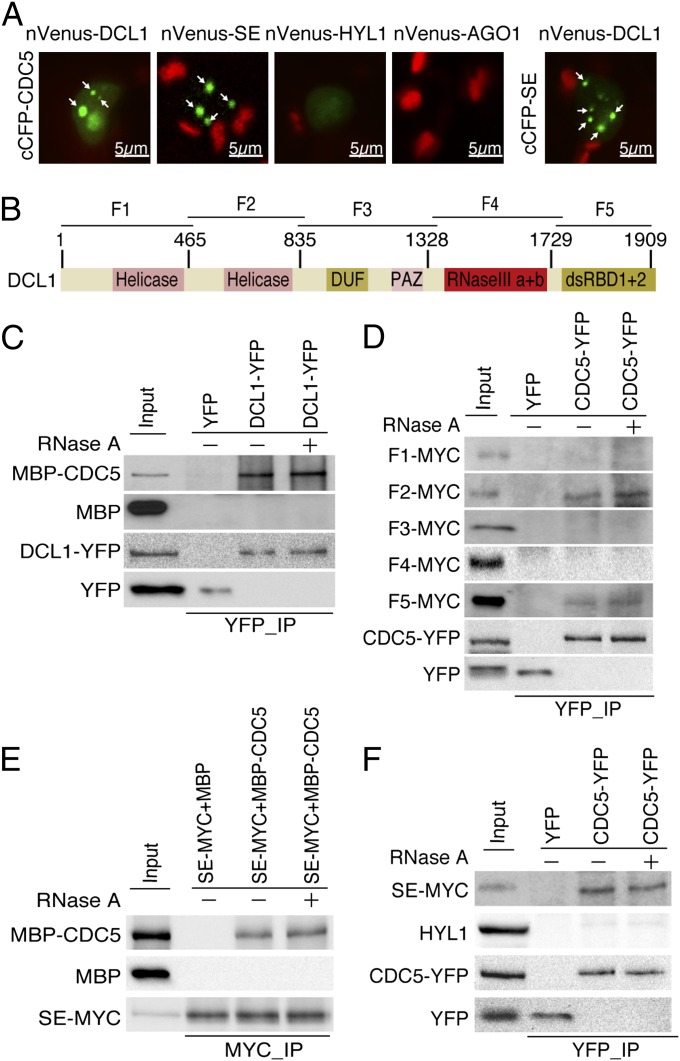
Next we tested the interaction of CDC5 with the DCL1 complex through a bimolecular fluorescence complementation (BiFC) assay. We have previously used this assay to determine the association of TOUGH with the DCL1 complex (13). The protein partners were fused to the N-terminal fragment of Venus (nVenus) or C-terminal fragment of cyan fluorescent protein (cCFP) under the control of a Cauliflower mosaic virus 35S promoter and cointroduced into Nicotiana benthamiana. In this assay, generation of a functional yellow fluorescent protein (YFP) indicates the potential interaction between proteins (36). The CDC5–DCL1, CDC5–SE, and SE–DCL1 (positive control), but not AGO1–CDC5 (negative control), interactions were observed (Fig. 6A). In addition, weak YFP signals were produced from the CDC5–HYL1 pair, indicating either a weak or no interaction between CDC5 and HYL1 (Fig. 6A).
Fig. 6.
CDC5 interacts with the DCL1 complex. (A) BiFC analysis of CDC5 with DCL1, SE, HYL1, and AGO1. Respective pairs of cCFP (cCFP–CDC5, cCFP–SE) and nVenus (nVenus–DCL1, nVenus–HYL1, nVenus–SE, and nVenus–AGO1) fused proteins were coinfiltrated into N. benthamiana leaves. Yellow fluorescence (green in image) signals were examined at 48 h after infiltration by confocal microscopy. Arrow indicates the BiFC signal. The red spot was inflorescence from chlorophyll. Thirty nuclei were examined for each pair, and an image is shown. (B) Schematic diagram of DCL1 domains and truncated DCL1 fragments used for protein interaction assay. (C) Coimmunoprecipitation between CDC5 and DCL1. The protein pairs in the protein extracts were indicated by the labels on the left side of and on top of the picture. DCL1–YFP/YFP and MBP–CDC5/MBP were detected by Western blot, using anti-YFP and anti-MBP, respectively, and labeled on the left side of the picture. One percent input protein was used for MBP–CDC5 and MBP. Twenty percent input proteins were used for DCL1–YFP and YFP. (D) Coimmunoprecipitation between CDC5 with the helicase and dsRNA binding domains of DCL1. Truncated DCL1 proteins fused with a MYC tag at their N terminus were expressed in N. benthamiana leaves. The protein pairs in the protein extracts were indicated by the labels on the left side of and on top of the picture. Anti-MYC antibody was used to detect MYC fusion proteins in Western blots. Labels on the left side of picture indicate proteins detected by Western blot. Five percent input proteins were used for MYC tagged proteins, whereas 20% inputs were used for DCL1–YFP and YFP. Please note only one IP picture was shown for CDC5–YFP and YFP, respectively. (E and F) Coimmunoprecipitation between CDC5 and SERRATE (SE). The protein pairs in the protein extracts were indicated by the labels on the left side of and on top of the picture. Proteins detected by Western blot were indicated on the left side of the picture. Two percent of input proteins were used for SE–MYC. Twenty percent inputs proteins were used for MBP and YFP tagged proteins.
We performed a co-IP assay to confirm the BiFC results. The DCL1–YFP fusion protein and YFP were expressed in N. benthamiana, respectively, whereas recombinant CDC5 fused with a maltose-binding protein epitope tag at its N terminus (MBP–CDC5) and MBP were expressed in Escherichia coli BL21 (13). Next, anti-YFP antibody conjugated with protein G agarose beads was incubated with the protein mixture containing MBP–CDC5/DCL–YFP, MBP–CDC5/YFP, or MBP/DCL1–YFP to capture the DCL1–YFP or YFP complex. We were able to detect MBP–CDC5, but not MBP, in the DCL1–YFP complex (Fig. 6C). In contrast, YFP did not pull down either MBP or MBP–CDC5 (Fig. 6C). In addition, RNase A treatment did not impair the CDC5–DCL1 interaction, although it abolished an RNA-mediated AGO4–FDM1 interaction (Fig. 6C and Fig. S3D). These results indicated that the CDC5–DCL1 interaction may not be RNA-mediated.
We further determined the protein domains of DCL1 that mediate the DCL1–CDC5 interaction. Five different DCL1 fragments named F1 (aa 1–468, covering amino terminus to helicase domain 1), F2 (aa 465–840; helicase domain 2), F3 (aa 835–1330; domain of unknown function and Piwi/Argonaute/Zwille domain), F4 (aa 1328–1700; RNase IIIa+IIIb domains), and F5 (aa 1729–1909; dsRNA binding domains I+II) were expressed in N. benthamiana, respectively, as described (37; Fig. 6B). CDC5–YFP was able to pull down F2 (Helicase domain 2) and F5 (dsRNA binding domains I+II), but not other fragments (Fig. 6D).
We next examined the interactions of CDC5 with SE and HYL1. CDC5 and SE, but not CDC5 and HYL1, were able to pull down each other, which was not affected by RNase A treatment (Fig. 6 E and F). In addition, the interactions among controls were not detected (Fig. 6 E and F). The interaction of CDC5 with SE and DCL1 suggested that CDC5 is a component of the DCL1 complex. However, we did not detect the HYL1–CDC5 interaction (Fig. 6F). This was not unexpected, as CDC5 may be weakly associated with the DCL1 complex, or its association with HYL1 may need bridge proteins. In fact, NOT2 has been shown to interact with DCL1 and SE, but not HYL1 (8).
Discussion
In conclusion, we show that CDC5, a MYB-related and evolutionarily conserved protein, is an important player in miRNA biogenesis. This is evidenced by reduced levels of pri-miRNAs and less accumulation of miRNAs in cdc5-1. Impairment of CDC5 function causes both immunity and pleiotropic development defects, which agrees with the crucial roles of miRNAs in regulating multiple biological processes (31, 32). However, it is possible that the regulation of genes other than small RNAs by CDC5 also contributes to the observed phenotypes in cdc5-1.
On the basis of studies of CDC5 homologs in other organisms, the roles of plant CDC5 in transcription have been speculated (31, 32). This study provides direct evidence to support the theory that CDC5 is a positive transcription factor. Tthat CDC5 does not bind the DCL1 promoter and that cdc5-1 does not significantly affect the occupancy of Pol II at the DCL1 promoter suggest that CDC5 may not be a general transcription factor. Rather, it may affect the expression of MIRs. It is possible that CDC5 can also act as a transcription factor for some protein-coding genes. Given that CDC5 interacts with Pol II and Pol II binds DNA promoter sequences, it is possible that the DNA amplified by qPCR in the CDC5 ChIP was bound to Pol II, rather than to CDC5. However, this seems not to be the case, as MIR promoters, but not the Pol II-dependent DCL1 promoter, were predominately enriched in the CDC5 ChIP. Lack of CDC5 in cdc5-1 reduces MIR promoter activity and the occupancy of Pol II at MIR promoters, suggesting that CDC5 may have a direct role in promoting the transcription of MIR by recruiting Pol II to their promoters. It is possible that CDC5 also contributes to Pol II activity through its interaction with Pol II. However, cdc5-1 does not significantly affect DCL1 transcript levels, as well as the occupancy of Pol II at its promoter, suggesting that the CDC5–Pol II interaction by itself maybe not sufficient to regulate the Pol II activity. Whether the CDC5–Pol II interaction is required for the regulation of MIR transcription needs to be further investigated.
CDC5 may also have a role in promoting miRNA maturation. This is unlikely to be caused by the reduced transcription of key genes involved in miRNA biogenesis, as their transcript levels are slightly increased in cdc5-1. Rather, CDC5 may act as a component of the DCL1 complex to enhance pri-miRNA processing efficiency on the basis of the association of CDC5 with the DCL1 complex and the fact that cdc5-1 reduces the production of miR162 in vitro. CDC5 interacts with the helicase and dsRNA binding domains of DCL1, which regulate DCL1 activity (10, 38). Structure studies have revealed that the interaction of human dicer with other proteins can cause dicer conformational change, and therefore improve its activity (39). Thus, it is possible that CDC5 may regulate pri-miRNA processing through its interaction with DCL1.
In summary, our study reveals that CDC5 can positively regulate processing and/or transcription of pri-miRNAs. It is unlikely that CDC5 regulates the transcription of all MIRs, as it is not a general transcription factor. Thus, CDC5 may only regulate some pri-miRNAs at both transcriptional and posttranscriptional levels. However, CDC5 may have a general role in regulating pri-miRNA processing, as it acts as cofactor of DCL1. In addition, CDC5 is predominantly expressed in the proliferating cells (32), suggesting that CDC5 may have cell-specific activities on miRNA accumulation. CDC5 is required for the accumulation of ra-siRNAs and ta-siRNAs. It is unclear whether CDC5 has a direct role in ta-siRNA biogenesis, as the generation of ta-siRNAs requires miRNAs. On the basis of the function of CDC5 in the miRNA pathway, CDC5 may have two contributions, which are not mutually exclusive, to the production of ra-siRNAs. First, it may affect Pol IV activity, which is thought to produce the precursor RNAs of ra-siRNAs. Second, it may regulate the DCL3 activity that generates 24-nt ra-siRNAs from long dsRNAs. Clearly, these two possibilities need to be examined in the near future.
Materials and Methods
Plant Materials.
The cdc5-1 (SAIL_207_F03) mutant that is in the Columbia genetic background was obtained from the Arabidopsis Biological Resources Center (31, 32). Transgenic line harboring pMIR172b::GUS (20) was crossed to cdc5-1. In F2 generation, CDC5+ (CDC5/CDC5 and CDC5/cdc5-1) and cdc5-1 containing pMIR172b::GUS were identified by genotyping of cdc5-1 and GUS.
RNA Analysis.
Northern blot analysis of small RNAs and qRT- PCR analysis of pri-miRNA and miRNA target transcription levels were performed as described (13).
Plasmid Construction.
A ∼5.2-Kb genomic DNA covering the CDC5 coding region and promoter was PCR amplified from Col genomic DNA and cloned to pMDC204 to generate the pCDC5::CDC5-YFP construct. A full-length CDC5 cDNA was amplified by RT-PCR and ligated to pMAL-c5x (New England Biolabs) to produce the MBP-CDC5 plasmid.CDC5 cDNA was cloned into pSAT4-C-CFP. The CDC5-C-CFP fragment was then released by I-SceI restriction enzyme digestion and subsequently cloned into the pPZP-ocs-bar-RCS2-2 vector. SE cDNA was amplified by RT-PCR and cloned into pEarleyGate203 vector to generate the SE-MYC construct. The truncated DCL1 (F1 to F5)-MYC plasmids were obtained from the laboratory of Y. Adam Yuan (National University of Singapore, Singapore) (12). The primers used for plasmid construction are listed in Table S1.
Plant Complementation.
The pCDC5::CDC5-YFP plasmid was transformed into CDC5/cdc5-1. The transgenic plants were selected using hygromycin resistance. In T2 generation, cdc5-1 harboring pCDC5::CDC5-YFP was identified by genotyping of YFP and cdc5-1.
ChIP Assay.
ChIP was performed as described by Kim et al. (7). Three biological replicates were performed. Anti-RPB2 and anti-GFP and GFP variants antibodies (Clontech) were used for immunoprecipitation. qPCR was performed on DNAs copurified with Pol II or CDC5, using primers listed in Table S1.
Co-IP Assay.
For the Pol II–CDC5 co-IP, protein extracts from plants expressing pCDC5::CDC5-YFP or YFP were incubated with anti-GFP (and GFP variants; Clontech) antibodies or anti-RBP2 coupled to protein G-agarose beads for 4 h at 4 °C. After five-time washing, the proteins in the immunoprecipitates were subjected to Western blot analysis, using anti-GFP antibody and anti-RBP2 antibody, respectively. For the interactions of CDC5 with components of the DCL1 complex, MBP-CDC5 and MBP were expressed in BL21 and extracted, following the manufacturer’s protocol (New England Biolabs), whereas DCL1-YFP, truncated DCL1-MYC (F1–F5), SE-MYC, and YFP were expressed in N. benthamiana (20). HYL1 and CDC5–YFP were obtained from inflorescences of Col and plants expressing pCDC5::CDC5-YFP, respectively. Anti-GFP (and GFP variants) and anti-MYC antibodies were used to capture and detect corresponsive YFP- and MYC-tagged proteins, respectively. Anti-HYL1 and anti-MBP antibodies (New England Biolabs) were used to detect HYL1 and MBP-tagged proteins, respectively, in Western blot.
Dicer Activity Assay.
MIR162b was prepared by in vitro transcription under the presence of [α-32P] UTP. In vitro dicer activity assay was performed according to Qi et al. (35) and Ren et al. (13). Radioactive signals were quantified with ImageQuant (5.2).
BiFC Assay.
Paired cCFP and nVenus constructs were coinfiltrated into N. benthamiana leaves. After 48 h, yellow fluorescence signals and chlorophyll auto fluorescence signals were exited at 488 nm and detected by confocal microscopy (Fluoview 500 workstation; Olympus) with a narrow barrier filter (BA505–525 nm).
Supplementary Material
Acknowledgments
We thank Dr. Y. Adam Yuan (National University of Singapore) for providing the DCL1 plasmids (F1–F5). Support for this work was received by National Science Foundation Grant MCB-1121193 (to B.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310644110/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579(26):5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez F. Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 2006;11(9):460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22(5):268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Herr AJ, Baulcombe DC. RNA silencing pathways in plants. Cold Spring Harb Symp Quant Biol. 2004;69:363–370. doi: 10.1101/sqb.2004.69.363. [DOI] [PubMed] [Google Scholar]
- 7.Kim YJ, et al. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011;30(5):814–822. doi: 10.1038/emboj.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, et al. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12(17):1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104(13):5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manavella PA, et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151(4):859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Ren G, et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(31):12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48(9):1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 15.Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14(4):346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 16.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101(4):1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47(6):841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 18.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7(10):1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105(29):9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(25):8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, et al. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013;23(5):645–657. doi: 10.1038/cr.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan X, et al. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci USA. 2012;109(44):18198–18203. doi: 10.1073/pnas.1216199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohi R, et al. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol Cell Biol. 1998;18(7):4097–4108. doi: 10.1128/mcb.18.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohi R, et al. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 1994;13(2):471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirayama T, Shinozaki K. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93(23):13371–13376. doi: 10.1073/pnas.93.23.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein HS, Coughlin SR. A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. J Biol Chem. 1998;273(8):4666–4671. doi: 10.1074/jbc.273.8.4666. [DOI] [PubMed] [Google Scholar]
- 29.Burns CG, Ohi R, Krainer AR, Gould KL. Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc Natl Acad Sci USA. 1999;96(24):13789–13794. doi: 10.1073/pnas.96.24.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald WH, Ohi R, Smelkova N, Frendewey D, Gould KL. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol Cell Biol. 1999;19(8):5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palma K, et al. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21(12):1484–1493. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Z, et al. AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 2007;17(9):815–828. doi: 10.1038/cr.2007.71. [DOI] [PubMed] [Google Scholar]
- 33.Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126(23):5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 34.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12(12):2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19(3):421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J Am Chem Soc. 2000;122(23):5658–5659. [Google Scholar]
- 37.Machida S, Yuan YA. Crystal structure of Arabidopsis thaliana Dawdle forkhead-associated domain reveals a conserved phospho-threonine recognition cleft for dicer-like 1 binding. Mol Plant. 2013;6(4):1290–1300. doi: 10.1093/mp/sst007. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Axtell MJ, Fedoroff NV. The helicase and RNaseIIIa domains of Arabidopsis DCL1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 2012;159(2):748–758. doi: 10.1104/pp.112.193508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau PW, et al. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19(4):436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.