Significance
Molecular mechanisms responsible for differences in longevity between animal species are largely unknown. Here we show that the longest-lived rodent, the naked mole-rat, has more accurate protein translation than the mouse. Furthermore, we show that the naked mole-rat has a unique fragmented ribosomal RNA structure. Such cleaved ribosomal RNA has been reported for only one other species of mammal. This article suggests the importance of protein translation in aging and provides insight into the mechanisms of longevity.
Keywords: aging, NMR
Abstract
The naked mole-rat (Heterocephalus glaber) is a subterranean eusocial rodent with a markedly long lifespan and resistance to tumorigenesis. Multiple data implicate modulation of protein translation in longevity. Here we report that 28S ribosomal RNA (rRNA) of the naked mole-rat is processed into two smaller fragments of unequal size. The two breakpoints are located in the 28S rRNA divergent region 6 and excise a fragment of 263 nt. The excised fragment is unique to the naked mole-rat rRNA and does not show homology to other genomic regions. Because this hidden break site could alter ribosome structure, we investigated whether translation rate and amino acid incorporation fidelity were altered. We report that naked mole-rat fibroblasts have significantly increased translational fidelity despite having comparable translation rates with mouse fibroblasts. Although we cannot directly test whether the unique 28S rRNA structure contributes to the increased fidelity of translation, we speculate that it may change the folding or dynamics of the large ribosomal subunit, altering the rate of GTP hydrolysis and/or interaction of the large subunit with tRNA during accommodation, thus affecting the fidelity of protein synthesis. In summary, our results show that naked mole-rat cells produce fewer aberrant proteins, supporting the hypothesis that the more stable proteome of the naked mole-rat contributes to its longevity.
Naked mole-rats (NMRs; Heterocephalus glaber) are extreme longevity outliers compared with other rodents of their size. Although only slightly larger than a laboratory mouse, the NMR lives an order of magnitude longer, with a maximum recorded lifespan of more than 30 y for both reproductive and nonreproductive castes (1). It was also noted that NMRs are markedly resistant to the increased frailty that accompanies aging in most metazoans. Furthermore, despite observations of many aged individuals, no neoplasia has been reported in the NMR (2, 3).
NMR cells have been experimentally shown to be much more resistant to oncogenic transformation than mouse cells (4, 5). The mechanism of tumor resistance is mediated by hypersensitivity of NMR cells to contact inhibition triggered by hyaluronan of extremely high molecular mass (5–7). Although these studies explain why the animals are tumor-resistant, they do not explain their overall longevity and their ability to maintain somatic integrity until extreme age. Previous studies have indicated that the NMR is able to tolerate high intracellular levels of oxidized proteins and oxidized lipids (8–10). Furthermore, the NMR is notably able to maintain proteomic integrity (as evidenced by lower protein unfolding after treatment with urea) until very late in its lifespan without an age-dependent increase in protein ubiquitination, such as observed in short-lived mice (11).
Multiple signaling pathways implicated in aging and cancer converge on factors that modify the rate of translation initiation and elongation (12–15). One of the main functions of the mTOR pathway is the promotion of protein translation in response to nutrients. Modulating the mTOR pathway genetically or pharmacologically, using the inhibitor rapamycin, has been shown to extend lifespan in yeast, Caenorhabditis elegans, Drosophila, and mice (14). The exact mechanism by which modulating protein translation via inhibition of mTOR extends lifespan is unclear. Two major explanations that were proposed are global reduction in mRNA translation, leading to better maintenance of protein homeostasis, or differential translation of specific mRNAs beneficial for longevity and stress resistance (reviewed in ref. 13).
Proper protein folding and stability are heavily implicated in aging. Overexpression of heat shock transcription factors and chaperones that aid protein folding can extend lifespan in Drosophila melanogaster (16) and C. elegans (17, 18). In bacteria, aging occurs by asymmetrical segregation of protein aggregates (19), and a similar mechanism has been described in yeast (20). The formation of aggregates as a result of mistranslated polypeptides has been implicated in several aging-related diseases such as amyotrophic lateral sclerosis, Huntington’s disease, and Alzheimer’s disease (21). A major factor determining proteome quality is translational fidelity of the ribosome. Severe disruptions of translational fidelity have been shown to cause accumulation of aggregates and pathological neurodegeneration (22).
The connection between translation and aging led us to examine NMR ribosomes for longevity-promoting phenotypes such as altered translation rate and translational fidelity. We describe a unique ribosomal DNA insertion, which is processed after rRNA transcription and breaks the NMR 28S rRNA into two distinct fragments. We assayed ribosomal translation rate (combined initiation and elongation) by measuring bulk incorporation of [35S]-cystein/methionine into nascent polypeptide chains. We found that NMR and mouse skin fibroblasts have roughly equal rates of translation. By using firefly luciferase reporters with a variety of mutations, including amino acid substitutions or stop codons that required misincorporation or frameshift to occur to reactivate the luciferase, we found that primary NMR fibroblasts have a fourfold lower rate of amino acid misincorporation than primary mouse fibroblasts. These results suggest that NMR cells produce fewer aberrant proteins and support the hypothesis that the NMR’s stable proteome is a contributor to its longevity.
Results
NMR 28S rRNA Has an Atypical Pattern.
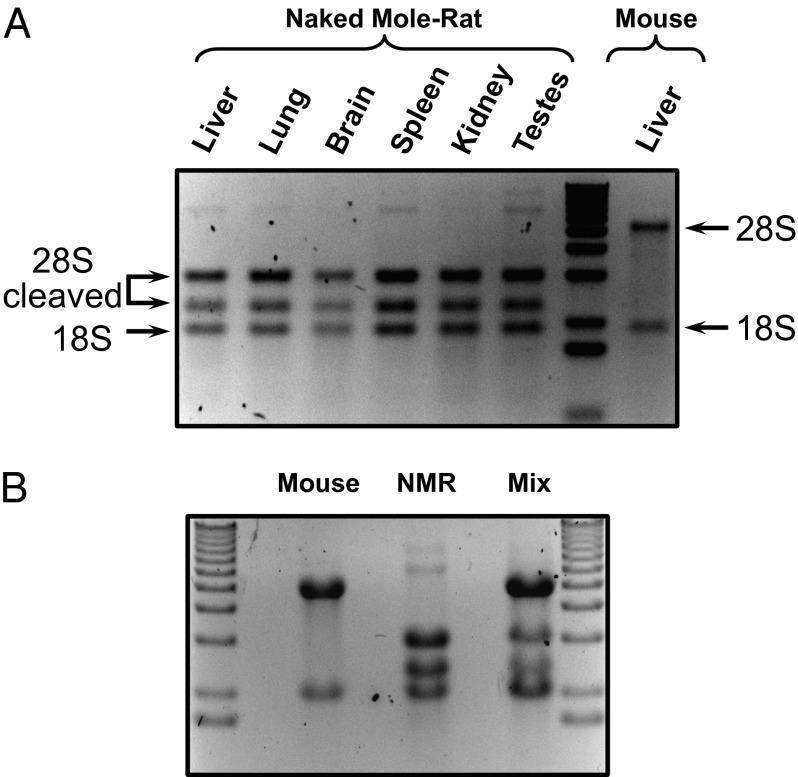
During routine extraction of RNA, we noticed that the 28S ribosomal RNA of the NMR was processed into two fragments of unequal size. NMR RNA extracts from several tissues (liver, lung, brain, spleen, kidney, and testis) exhibited an unusual rRNA band pattern, whereas mouse RNA extracts did not (Fig. 1A). We observed this pattern after using two different methods of RNA extraction in extracts prepared from tissues and cultured cell lines (Fig. S1A). We also extracted RNA from other nonmodel rodents (Fig. S1B) and found the break to be unique for the NMR.
Fig. 1.
NMR rRNA shows an unusual fragmented pattern. (A) 28R rRNA from multiple organs of the NMR appears as two fragments. Arrows indicate 28S fragments and an 18S band. (B) The fragmentation of NMR 28S rRNA is not caused by degradation during RNA extraction. Either NMR and mouse RNA were extracted from fibroblasts independently or the cells from both species were mixed together before extraction. RNA was separated on 1% denaturing agarose gel.
To confirm that the break was not a result of RNase contamination, we combined NMR cells with mouse cells and simultaneously extracted RNA after homogenizing them. When independently extracted, the fibroblast RNA showed three bands and two bands for NMR and mouse, respectively. In the mixed sample, all bands could be seen, indicating that no nuclease contamination was cleaving the mouse rRNA (Fig. 1B). This NMR 28S rDNA was undergoing specific processing in vivo.
Mapping of the 28S rRNA Cleavage Sites.
To determine the site and sequence of the NMR 28S rRNA breakage point, we first designed primers to conserved 28S rDNA sequence from the mouse genome. We estimated the location of the breakpoint region by noting that the sizes of the two fragments (2.5 and 3 kb) were only slightly different, meaning the break would be within ∼1 kb of the center. This allowed us to amplify a central 1-kb fragment of genomic NMR 28S rDNA. This fragment included 28S divergent region 6 (D6) that exhibited a 118-nt unique insertion compared with the mouse sequence flanked by regions of weak homology with the mouse D6 (Fig. 2). Using the 1-kb sequence and a publically available NMR coding DNA sequence, we designed primers to perform 5′ RACE and 3′ RACE. After aligning with the NMR 28S rDNA sequence, we identified the two cleavage sites and found that a 263-nt fragment flanked by 5-nt direct repeats 5′-CGGAC-3′ was cleaved out in the 28S rRNA, leaving behind the fragmented 28S rRNA. The 263-nt fragment included the 118-nt insertion that was not homologous to the mouse sequence (Fig. 2). The ribosomal DNA loci are repeated thousands of times, and from our RNA extractions, most of the products seemed to be cleaved. We used a bioinformatics approach to test whether the identified cut sites were conserved among all of the 28S rDNA loci in the NMR. From whole-genome shotgun reads from two different NMR genome sequences [Broad Institute (http://www.ncbi.nlm.nih.gov/nuccore/AHKG00000000) and Kim et al. (23)], the cut sites were found to be conserved among all of the rDNA loci (Fig. S2 and Table S1), which confirmed that the majority of 28S rRNAs are cleaved in the NMR.
Fig. 2.
Structure of the NMR 28S rRNA and localization of break sites. (A) Overall structures of the 28S rRNA in mouse, NMR, and Ctenomys. The mouse 28S rRNA has no cleavage sites. The D6 region of the NMR 28S rRNA contains a unique 118-nt insertion (shaded box). The two cleavage sites then excise a 263-nt fragment. Ctenomys is the only other known mammal with fragmented rRNA. The D6 region of the Ctenomys 28S rRNA contains a unique 106-bp insertion (shaded box) with the cleavage site located within this unique sequence. Thick black lines represent the 28S rRNA, the white box represents the D6 region, the arrows denote the cleavage sites, and the size of the D6 region and the distance between the two cleavage sites are indicated. (B) The alignment of the NMR and mouse 28S rDNA sequences. The box highlights the D6 region, the arrows indicate the cleavage sites, and hyphens indicate the gap.
NMR Translation Rate Does Not Differ from the Mouse.
We hypothesized that the fragmented 28S rRNA might cause alternate assembly or structure of the NMR ribosome, with detectable consequences for protein synthesis rate or fidelity. To investigate whether the NMR ribosome had a different translation rate than the mouse ribosome, we implemented an assay based on measuring [35S]-cystein/methionine incorporation into nascent protein. As the physiological temperature of the NMR is 32 °C, rather than 37 °C, and NMR fibroblasts are usually cultured at 32 °C, we tested the translation rate at both temperatures for both mouse and NMR cells. Mouse and NMR fibroblasts were given a pulse of [35S]-cystein/methionine for 30 min, cultured at 32 °C and 37 °C, total cell proteins were extracted, and the amount of radioactive isotope incorporation was measured by a scintillation counter.
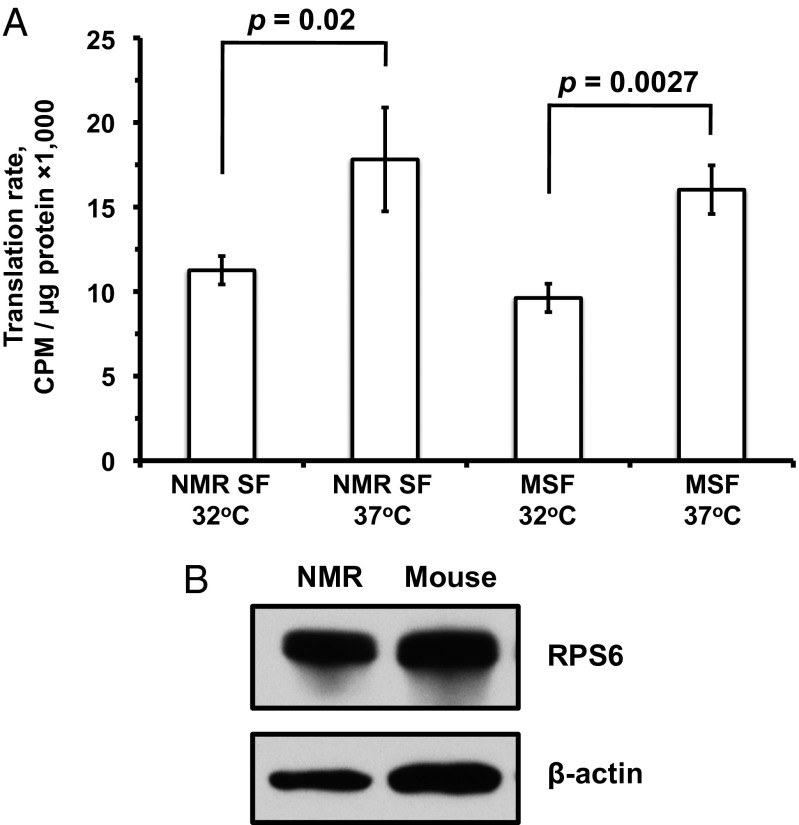
The translation rate was found to be altered twofold by shifting the temperature by 5° in both species (Student t test, NMR, P = 0.02; mouse, P = 0.0027), but no significant differences were seen between the NMR and mouse cells when they were temperature-matched (Fig. 3A). To confirm that the protein quantification was accurate and that mouse and NMR fibroblasts have similar amounts of ribosomes, we performed Western blots on ribosomal protein S6 (RPS6). No large differences were seen in the overall amount of ribosomal protein or loading controls from mouse and NMR fibroblasts (Fig. 3B). Taken together, these results show that the NMR translation rate is equivalent to that of the mouse when they are temperature-matched, and that the cells express similar levels of a ribosomal protein.
Fig. 3.
NMR and mouse have similar translation rates. (A) [35S]-cystein/methionine incorporation assay performed on growing fibroblasts showing NMR translation occurs at roughly equal speed to that of mouse fibroblasts. The assays were performed at 32 °C and 37 °C for both mouse and NMR cells. NMR SF are primary NMR skin fibroblast and MSF are primary mouse skin fibroblast. (B) Western blot against RPS6, a protein component of the mature ribosome (Upper) and β-actin as loading control (Lower).
NMR Has Higher Translational Fidelity than the Mouse.
We next examined whether altered ribosomal structure might affect the overall translation fidelity of proteins. To assay mouse and NMR cells for translational fidelity, we constructed a set of firefly luciferase mutant reporters. In these reporters, a key amino acid in the luciferase protein (lysine 529) was mutated to completely destroy luciferase activity (24), and only if the wrong amino acid were incorporated into the nascent polypeptide would enzymatic activity be restored. The reporters included all triplet positions, premature stop (third codon mutation at the beginning of the protein), and both positive and negative frameshifts by nucleotide insertion and deletion, respectively.
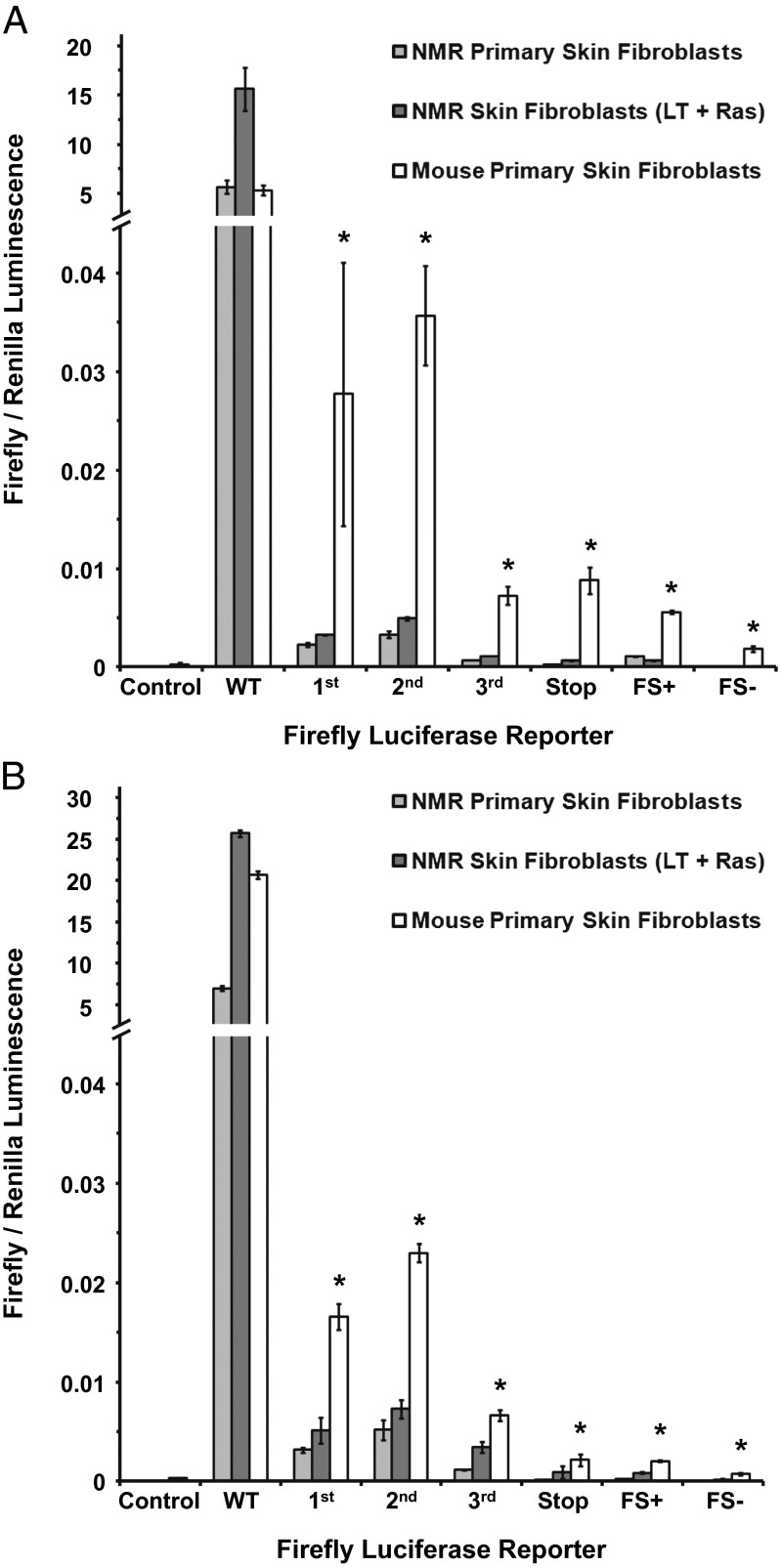
The luciferase assay indicated that NMR cells had higher protein translation fidelity than mouse cells. This was particularly noticeable when mouse and NMR cells were grown at 32 °C, as the NMR cells had ∼10-fold lower misincorporation frequency for the lysine triplet mutants, and these differences were highly statistically significant for all reporters (Student t test, P < 0.005) (Fig. 4A). At 37 °C, NMR cells had approximately a fourfold lower misincorporation; this was also statistically significant for all reporters (Student t test, P < 0.05) (Fig. 4B).
Fig. 4.
NMR has higher fidelity of translation than the mouse. Translation fidelity of NMR and mouse skin fibroblasts was quantified as a frequency of amino acid misincorporation into a set of firefly luciferase reporters with a mutated critical lysine 529 (Fig. S3). Misincorporation events can rescue luciferase activity. Luminescence of mutated firefly luciferase was measured and normalized to the luminescence of Renilla luciferase, both under control of the CMV promoter. The positive control (wild-type luciferase) ratio was several orders of magnitude higher and is cropped for visibility. The mutant reporters E - K529E; I - K529I; N - K529N measured misincorporation into the first, second, and third codon positions, respectively; the STOP reporter contained a premature stop codon mutation at amino acid 81; FS+ contained a positive frameshift mutation by nucleotide insertion at position 81; and FS contained a negative frameshift mutation by nucleotide deletion at position 81. Results for the assay are shown for NMR and mouse cells grown at 32 °C (A) and 37 °C (B). Error bars show standard deviation. Asterisks indicate values significantly different between NMR and mouse (P < 0.005) Student t test.
Notably, the NMR cells were able to maintain a high translational fidelity even when forced to grow more quickly. We integrated simian virus 40 (SV40) Large T-antigen (LargeT) and human Ras Glycine 12 to Valine (H-RasV12) mutant protein into wild-type NMR fibroblasts to induce more rapid growth via constant Ras signaling and abrogation of the p53/retinoblastoma protein pRB tumor suppressor pathways to prevent oncogene-induced senescence. The fidelity assays were then repeated on these cells grown at 32 °C and 37 °C. The translation error rate was not significantly different from in the primary NMR fibroblasts at either temperature (Fig. 4 A and B), indicating that even when NMR cells proliferate at a similar rate as mouse cells, they still maintained higher translational fidelity. This result suggests the NMR ribosomes are intrinsically more accurate than the mouse ribosomes.
Discussion
Our study shows an unexpected processing of NMR 28S rRNA into two molecules of 2.5 and 3 kb, corresponding to 3′ and 5′ segments of the rRNA gene. The break occurs within the D6 region and results from two cuts that excise a 263-nt fragment containing a unique 118-nt sequence. This 118-nt sequence bears no significant homology to other known sequences. The cleavage occurs in all NMR organs that we analyzed, as well as in the primary fibroblast cultures.
The excised fragment is flanked by 5-nt direct repeats, which supports that the insert originated from a transposable element. The sequences distal to the cut sites share weak homology with the mouse D6, suggesting that the insert could have also been introduced by homologous recombination. We do not know the mechanism responsible for the cleavage of the NMR 28S rRNA. The cuts could be generated by a ribozyme activity residing within the D6 sequence itself, as was reported for R2 transposons residing within the rDNA locus (25). We tested this possibility by in vitro transcribing 28S rRNA; however, the in vitro transcribed 28S rRNA molecules remained uncleaved. Finally, the cleavage may be mediated by a ribonuclease that recognizes the D6 region.
Fragmented rRNA has been found in Salmonella (26), protozoa (27, 28), a worm (29), and several arthropods (ref. 30 and references therein). However, in vertebrates there is only one other example of a fragmented rRNA, found in rodents of the genus Ctenomys, also known as tuco-tuco (31). In Ctenomys, 28S rRNA is cleaved within the D6 region, but the cleavage occurs with only a single cut site, and sequences surrounding the break share no homology with the NMR sequence (Fig. 2A). Interestingly, Ctenomys is a South American rodent that is phylogenetically distant from the NMR but shares similar ecology. Ctenomys is a subterranean rodent, with some species of the genus leading a social lifestyle. The maximum lifespan in captivity is not determined for Ctenomys, but in the wild, Ctenomys is short-lived, with an average lifespan of 3 y (32).
We found that NMR cells had equivalent translation rates (adjusted for physiological temperature). However, by constructing a set of unique luciferase reporters, we observed that NMR cells have substantially higher translation fidelity than mouse cells, even when transformed with oncogenes to increase their cell cycle speed. NMR cells also displayed higher fidelity of translation for all errors we tested, including misincorporation, stop codon skipping, and frameshifts. A potential caveat of our translation fidelity assay is that active firefly luciferase could be generated not only by incorrect translation but also by incorrect transcription or random mutations of the plasmid DNA during cell growth. This seems unlikely given that if replication or transcription produced active luciferase, we would not see the expected nucleotide position bias that the assay clearly shows (with the third position being the least mistranslated). Furthermore, we would not expect to see such a large bias in favor of insertions over deletions. The misincorporation rate we detect in our assay is also between 1:1,000 and 1:10,000, which is consistent with reports in the literature (24). Thus, we conclude that most of the luminescence from our assay comes from mistranslation, rather than from mistakes at the nucleotide level.
Pérez et al. reported that the NMR proteome has higher levels of cysteine than the mouse proteome and that the proteins are resistant to unfolding stressors such as 1 M urea. These results are consistent with high levels of translation fidelity (11). According to our results, the mouse proteome would have two- to 10-fold higher levels of proteins with misincorporated amino acids relative to the NMR. These misincorporated amino acids could increase the number of proteins that are unstable under challenging conditions, such as urea treatment. Pérez et al. also report that the NMR does not show an age-dependent increase in protein ubiquitination. This is consistent with the high fidelity of protein translation in the NMR. More recently, it has been reported that NMR proteins maintain a higher level of activity after carbonylation and oxidation than the equivalent mouse proteins (33). Again, it is possible that fewer amino acid misincorporations allow proteins to tolerate more stochastic improper modifications and oxidative damage, and thus retain tertiary structure and activity.
Given the remarkable longevity of the NMR, it is possible that increased translational fidelity plays a role in its longevity. The “error catastrophe theory of aging” first proposed by Orgel in 1963 (34) and explored in Escherichia coli by Edelmann and Gallant in 1977 posits that early defects in translational fidelity lead to a vicious cycle of progressively more severe defects in protein homeostasis (35). Differences in translational fidelity may play an important role in determining lifespan. Many age-related disorders are caused by protein aggregation (21), and problems with translational fidelity can lead to the accumulation of misfolded protein and the formation of inclusion bodies (22). The proteome is maintained by various mechanisms, all of which show an age-dependent decline (36). During heat stress, chaperones are up-regulated, but this mechanism fails with age (37, 38). Proteasome activity also declines with age (39). Aging also leads to a decline in macroautophagy, which eliminates misfolded or incorrectly glycosylated proteins from the cell (40). It is possible that when proteome maintenance mechanisms fail, it allows the aberrant mistranslated proteins to accumulate. Species, such as the NMR, with a lower overall level of mistranslated proteins would thus be buffered against some of the decline of old age, given that they have fewer misfolded proteins to begin with.
The main source of mistranslated proteins in E. coli is discrimination of noncognate or near-cognate tRNAs during translation (24). The ribosome structure itself confers at least some of the fidelity of the process, as shown by hyperaccurate ribosomes (41), and its sequence and structure (42) may be altered by natural selection in species that evolve long lifespans. Indeed, in E. coli, there is an observed evolutionary tradeoff between ribosomal speed and accuracy (41). Because the NMR in vitro translation system currently cannot be reconstituted from purified components, we are unable to directly test whether 28S cleavage is responsible for improved translation fidelity in the NMR. The cleavage leads to deletion of the major part of the D6 region corresponding to a helix 45 of 23S rRNA in E. coli and leaves two fragments of 28S rRNA disconnected. This may change the folding or dynamics of the large ribosomal subunit, altering the rate of GTP hydrolysis by elongation factor 1A and/or interaction of the large subunit with tRNA during accommodation, thus affecting the fidelity of protein synthesis. In particular, in bacterial ribosome, helix 45 of 23S rRNA is connected to the L11-stalk (L12 stalk in mammals), which is formed by helices 42, 43, and 44 through coaxial stacking with helices 40 and 41. Thus, the deletion of helix 45 may affect dynamics of the L11 stalk, which directly interacts with the elbow of the aminoacyl-tRNA in the intermediate (A/T) state of tRNA accommodation (43).
In summary, we have shown that NMR has a unique form of 28S rRNA that is processed in two fragments. The processing occurs by two cuts within the D6 region. Furthermore, we demonstrate that NMR has higher fidelity of translation than the mouse. We hypothesize that the unique structure of the NMR 28S rRNA may contribute to the higher fidelity of translation, which in turn may contribute to exceptional longevity of the NMR.
Materials and Methods
RNA Extraction and Denaturing Gel Electrophoresis.
RNA from rodent tissues and cells was extracted either by RNEasy Kit (Qiagen), according to the manufacturer’s instructions, or by TriReagent extraction. TriReagent extraction was performed by mixing cells or tissues with TriReagent, rupturing in a glass homogenizer, and centrifuging at high speed (>10,000 × g) at 4 °C on a tabletop centrifuge. The aqueous phase was then washed with isopropanol to precipitate RNA and then washed with ethanol, dried, and resuspended in RNase-free water.
Denaturing agarose gels were prepared in double-distilled H2O and 20× Mops buffer diluted to 1× concentration. Formaldehyde was added to a final concentration of 2% (vol/vol). RNA samples were denatured in formaldehyde/Mops buffer at 80 °C for 5 min and then immediately placed on ice for at least 2 min. Samples were loaded and run at ∼100 V for 2–3 h.
rRNA Break Mapping.
The unbroken 28S rRNA was estimated to be ∼5 kbp, and because the two fragments were close in size, we estimated that the break probably happened within 1,000 bp of the middle. 5′ RACE primers specific to highly conserved regions downstream of the putative 28S midpoint were designed. A 5′/3′ second-generation RACE kit (Roche) was used to obtain a fragment that mapped from the conserved region to the 5′ break point. Primers were designed that mapped upstream of the putative breakpoint, and 3′ RACE was performed by ligating an adenylated linker to the 3′ end of the ribosomal RNA. PCR was then used to extend from the upstream conserved site to the start point of the break. Primer sequences are shown in Table S2.
Tissue Culture.
NMR fibroblasts were grown at 32 °C (in vivo body temperature, unless otherwise indicated) 5% CO2, 3% O2 on treated polystyrene culture dishes (Corning) in eagle's minimum essential medium (EMEM) media (ATCC) supplemented with 15% FBS (Gibco), nonessential amino acids, sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco). Mouse cells were grown in identical conditions except at 37 °C, unless otherwise indicated.
Translation Rate Assay.
Cells were grown to ∼75% confluence, and media was removed and incubated for 15 min with cysteine/methionine-free media to deplete intracellular pools. The media was then removed, and cells were incubated for 30 min in media containing [35S]-methionine/cystein. Radioactive media was then removed, and cells were incubated with 1.5 mL ice-cold lysis buffer and scraped from the plate, using a disposable plastic scraper. The cells were then stored at −80 °C overnight. Trichloroacetic acid (TCA) precipitation was used the following day to pellet proteins, and the pellet was washed with acetone. Lowry assay was performed to determine protein concentration. Radioactivity was evaluated by scintillation counter (Beckman).
Western Blot.
Proteins samples were run through 15% SDS/PAGE and transferred to nitrocellulose via the TransBlot Turbo system (BioRad). The membrane was cut and blotted separately for ribosomal protein RPS6 and beta-actin (loading control). The membrane was incubated with primary antibody overnight at 4 °C and with secondary antibody for 1 h at room temperature. Antibodies used were Santa Cruz biotechnology SC-47778 (beta-actin) and Cell Signaling 2317 (RPS6).
Plasmid Construction and Transfection.
pGL3 wild-type luciferase plasmid (Promega) was mutated as indicated by site-directed mutagenesis, using a QuikChange kit (Agilent). Several clones were picked, miniprepped, and sent for sequencing to confirm proper mutagenesis. Correct plasmids were prepped using EndoFree Maxi prep kit (Qiagen). Cells were thawed and grown for two passages before being harvested. Cells were counted, and 1 × 106 cells were transfected using Nuclefector transfection (Lonza) with program U-020 in human dermal fibroblast (NHDF) solution with 5 µg reporter plasmid [mutant, wild-type-positive control, or hypoxanthine-guanine phosphoribosyltransferase (HPRT) negative control] and 0.1 μg Renilla luciferase plasmid (transfection/promoter control). Cells were then pipetted onto 10 cm Corning treated plates and allowed to grow for 24 h at the indicated temperature. Mutation sites within Luciferase gene are shown in Fig. S3.
Luciferase Assay.
A Promega Dual-Luciferase assay kit was used to perform analysis on transfected cells. Cells were harvested, counted, and lysed, using the provided buffer. Extracts were thoroughly mixed with LAR-II reagent, and luminescence was measured. The sample was then mixed with STOP&GLO solution (halting firefly luciferase and starting Renilla luciferase), and luminescence was measured. Ratio of firefly to Renilla was used as an indicator of translational fidelity.
Assembly of 28S rRNA Sequence from Illumina Raw Reads.
First, we mapped the paired-end reads to the mouse 28S DNA sequence, using Bowtie2 (44), which allows mismatches and small indels/gaps. Then Velvet (45) was used to assemble the reads that are mapped to the mouse 28S DNA sequence. The set of contigs with the largest N50 were selected. However, it is hard to obtain the reads from divergent regions by this method. To find scaffolds that the cover NMR 28S gene, we mapped our contigs to all of the scaffolds obtained by genome-wide assembly. Finally, we find one scaffold that matched our contigs the most.
Divergence Analysis of the Cleavage Sites in the D6 Region.
To assess the divergence/conservation at the two cutting sites in D6 among all of the copies of 28S in NMR, we used the following strategy: Given a single sequenced copy of NMR D6 and its flanking regions, the conserved 50-bp sequences immediately upstream to the start of D6 and downstream to the end of D6 were used as the target for the 5′ and 3′ end cutting sites, separately. We mapped the target to the reads with the maximum 3 mismatch (i.e., we take the sequencing short reads as the references and do alignment for the target). After we get the matched reads, if the read covers the cutting site, we save the subsequence around the cutting site. We kept ±15 bp from the 5′ end cutting site and ±18 bp from the 3′ end cutting site, by which we centered the cutting site in the subsequences around the cutting sites. Finally, the multiple sequence alignment program Clustal Omega (46) was used to align the subsequences and the nucleotide frequency was counted based on the multiple sequence alignment result. Fig. S2 shows the divergence analysis strategy. Table S1 shows that nucleotide frequency at the two cutting sites. The cutting sites were highly conserved among the 28S rRNA copies.
Supplementary Material
Acknowledgments
We thank members of the T. Eickbush and G. Culver laboratories at the University of Rochester for useful discussion and suggestions. This work was supported by the National Institutes of Health and Ellison Medical Foundation grants (to V.G.) and by Life Extension Foundation grants (to V.G. and A.S).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313473110/-/DCSupplemental.
References
- 1.Buffenstein R. The naked mole-rat: A new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60(11):1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 2.Lewis KN, Andziak B, Yang T, Buffenstein R. The Naked Mole-Rat Response to Oxidative Stress: Just Deal with It. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): A retrospective survey of lesions in a zoo population. Vet Pathol. 2013;50(4):607–621. doi: 10.1177/0300985812471543. [DOI] [PubMed] [Google Scholar]
- 4.Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9(4):626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seluanov A, et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci USA. 2009;106(46):19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian X, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499(7458):346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azpurua J, Seluanov A. Long-lived cancer-resistant rodents as new model species for cancer research. Frontiers Genet. 2013;3:319. doi: 10.3389/fgene.2012.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulbert AJ, Faulks SC, Buffenstein R. Oxidation-resistant membrane phospholipids can explain longevity differences among the longest-living rodents and similarly-sized mice. J Gerontol A Biol Sci Med Sci. 2006;61(10):1009–1018. doi: 10.1093/gerona/61.10.1009. [DOI] [PubMed] [Google Scholar]
- 9.Andziak B, et al. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell. 2006;5(6):463–471. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 10.Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5(6):525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 11.Pérez VI, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106(9):3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25(48):6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 13.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaeberlein M, Kennedy BK. Hot topics in aging research: Protein translation and TOR signaling, 2010. Aging Cell. 2011;10(2):185–190. doi: 10.1111/j.1474-9726.2010.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrow G, Samson M, Michaud S, Tanguay RM (2004) Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J 18(3):598–599. [DOI] [PubMed]
- 17.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15(2):657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 19.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105(8):3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budovsky A, Fraifeld VE, Aronov S. Linking cell polarity, aging and rejuvenation. Biogerontology. 2011;12(2):167–175. doi: 10.1007/s10522-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 21.Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36(12):2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 23.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479(7372):223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13(1):87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30(13):3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgin AB, Parodos K, Lane DJ, Pace NR. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60(3):405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 27.Schnare MN, Gray MW. Sixteen discrete RNA components in the cytoplasmic ribosome of Euglena gracilis. J Mol Biol. 1990;215(1):73–83. doi: 10.1016/S0022-2836(05)80096-8. [DOI] [PubMed] [Google Scholar]
- 28.Boer PH, Gray MW. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988;55(3):399–411. doi: 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 29.van Keulen H, Mertz PM, LoVerde PT, Shi H, Rekosh DM. Characterization of a 54-nucleotide gap region in the 28S rRNA gene of Schistosoma mansoni. Mol Biochem Parasitol. 1991;45(2):205–214. doi: 10.1016/0166-6851(91)90087-m. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara H, Ishikawa H. Molecular mechanism of introduction of the hidden break into the 28S rRNA of insects: Implication based on structural studies. Nucleic Acids Res. 1986;14(16):6393–6401. doi: 10.1093/nar/14.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melen GJ, Pesce CG, Rossi MS, Kornblihtt AR. Novel processing in a mammalian nuclear 28S pre-rRNA: Tissue-specific elimination of an ‘intron’ bearing a hidden break site. EMBO J. 1999;18(11):3107–3118. doi: 10.1093/emboj/18.11.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowak R. Walker's Mammals of the World. Baltimore: John Hopkins Univ Press; 1999. [Google Scholar]
- 33.De Waal EM, et al. Elevated protein carbonylation and oxidative stress do not affect protein structure and function in the long-living naked-mole rat: A proteomic approach. Biochem Biophys Res Commun. 2013;434(4):815–819. doi: 10.1016/j.bbrc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci USA. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edelmann P, Gallant J (1977) On the translational error theory of aging. Proc Natl Acad Sci USA 74(8):3396–3398. [DOI] [PMC free article] [PubMed]
- 36.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arslan MA, Csermely P, Soti C. Protein homeostasis and molecular chaperones in aging. Biogerontology. 2006;7(5-6):383–389. doi: 10.1007/s10522-006-9053-7. [DOI] [PubMed] [Google Scholar]
- 38.Shpund S, Gershon D. Alterations in the chaperone activity of HSP70 in aging organisms. Arch Gerontol Geriatr. 1997;24(2):125–131. doi: 10.1016/s0167-4943(96)00745-5. [DOI] [PubMed] [Google Scholar]
- 39.Baraibar MA, Friguet B. Changes of the proteasomal system during the aging process. Prog Mol Biol Transl Sci. 2012;109:249–275. doi: 10.1016/B978-0-12-397863-9.00007-9. [DOI] [PubMed] [Google Scholar]
- 40.Cuervo AM, et al. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy. 2005;1(3):131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 41.von Ahsen U. Translational fidelity: Error-prone versus hyper-accurate ribosomes. Chem Biol. 1998;5(1):R3–R6. doi: 10.1016/s1074-5521(98)90080-1. [DOI] [PubMed] [Google Scholar]
- 42.Vila-Sanjurjo A, et al. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc Natl Acad Sci USA. 2003;100(15):8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2012;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.