Significance
Fungal and oomycete pathogens cause devastating diseases in crop plants and facilitate infection by delivering effector molecules into the plant cell. The secreted effector protein AvrM from flax rust, a fungal pathogen that infects flax plants, internalizes into host cells in the absence of the pathogen, binds to phosphoinositides, and is recognized directly by the resistance protein M in flax to initiate effector-triggered immunity. We describe the crystal structure of AvrM and identify functionally important surface regions in the protein, which advances our understanding of the molecular mechanisms underlying how effectors enter host cells and how they are detected by the plant immune system.
Keywords: innate immunity, plant cell internalization, plant disease resistance, avirulence protein, lipid binding
Abstract
Fungal and oomycete pathogens cause some of the most devastating diseases in crop plants, and facilitate infection by delivering a large number of effector molecules into the plant cell. AvrM is a secreted effector protein from flax rust (Melampsora lini) that can internalize into plant cells in the absence of the pathogen, binds to phosphoinositides (PIPs), and is recognized directly by the resistance protein M in flax (Linum usitatissimum), resulting in effector-triggered immunity. We determined the crystal structures of two naturally occurring variants of AvrM, AvrM-A and avrM, and both reveal an L-shaped fold consisting of a tandem duplicated four-helix motif, which displays similarity to the WY domain core in oomycete effectors. In the crystals, both AvrM variants form a dimer with an unusual nonglobular shape. Our functional analysis of AvrM reveals that a hydrophobic surface patch conserved between both variants is required for internalization into plant cells, whereas the C-terminal coiled-coil domain mediates interaction with M. AvrM binding to PIPs is dependent on positive surface charges, and mutations that abrogate PIP binding have no significant effect on internalization, suggesting that AvrM binding to PIPs is not essential for transport of AvrM across the plant membrane. The structure of AvrM and the identification of functionally important surface regions advance our understanding of the molecular mechanisms underlying how effectors enter plant cells and how they are detected by the plant immune system.
Filamentous eukaryotic microbes such as fungi and oomycetes cause devastating diseases in many economically important crop plants, including rice, corn, wheat, soybean, and potato. During infection, oomycetes and biotrophic fungal pathogens establish a physical interaction with their host through specialized feeding structures, known as haustoria, which facilitate secretion of a vast array of effector proteins to overcome plant defenses and promote host colonization (1–3). The effectors are generally highly divergent even among related species, and generally lack similarity to proteins currently available in the databases, making it difficult to predict biological function from sequence alone. Some of the effectors accumulate in the plant intercellular space (apoplast), whereas others are translocated into the host cell. At present, the host targets and virulence mechanisms of fungal and oomycete effectors are poorly understood. A subset of the plant-translocated effectors called avirulence (Avr) proteins are recognized directly or indirectly and with high specificity by plant disease resistance (R) proteins (4–6). This recognition event leads to activation of effector-triggered immunity (ETI), which usually includes rapid localized host cell death at the site of infection—termed the hypersensitive response (HR)—and results in the plants being immune to pathogen infection.
The question of how fungal and oomycete effector proteins are transported across the host-cell membrane is currently a topic of considerable interest and controversy. Delivery of effectors into host cells by eukaryotic pathogens has been demonstrated in several plant pathogen systems (7–12), and, in some cases, fungal and oomycete host-cell translocation can occur in the absence of the pathogen (7, 12–16), suggesting that a host-encoded transport machinery is responsible for internalization of the effectors. Conserved short N-terminal motifs such as RXLR and LXLFLAK have been identified in oomycete-effector protein families (17), and they have been shown to be both necessary and sufficient for plant-cell entry (9, 16, 18). Although the N-terminal regions of some fungal effectors including AvrL567 and AvrM from flax rust are required for plant-cell entry (7), conserved uptake signals have so far not been identified in fungal pathogens. RXLR motifs in oomycete effectors, and RXLR-like motifs in some fungal effectors, have been reported to be required for host-cell translocation and binding to phosphatidylinositol 3-phosphate (PI3P). Based on these findings, Kale et al. (15) proposed that oomycete and fungal effector proteins are translocated into the host cytoplasm after binding to PI3P associated with the external face of the cell, in a process that involves lipid raft-mediated endocytosis. However, this view has recently been challenged by reports showing that the C-terminal effector domain of AVR3a from Phytophthora infestans, and not the N-terminal region harboring the RXLR motif, is required for binding to PI3P (19–21). There is also an ongoing debate in the literature about the significance of the RXLR motifs in host-cell translocation (22, 23).
The interaction between flax and the fungal pathogen Melampsora lini (flax rust) has been adopted as a model system for studying plant–fungal rust interactions. Several M. lini effector genes have been identified (AvrL567, AvrM, AvrP123, and AvrP4) and they encode small, secreted proteins that are recognized by TIR-NB-LRR (Toll-interleukin receptor, nucleotide binding, leucine-rich repeat) R proteins inside the plant cell (24–27). The AvrM effector is delivered into host cells during infection, and interacts directly with the M resistance protein (7, 28). AvrM has no significant sequence similarity to proteins of known structure and its internalization mechanism, virulence functions, and host targets are unknown. Six naturally occurring variants (AvrM-A to AvrM-E and avrM) have been identified. AvrM-A to AvrM-D are recognized by M and induce HR, whereas AvrM-E and avrM evade recognition (26). The C-terminal region of AvrM-A (residues 106–343) forms a structured and protease-resistant domain that can dimerize in plants and in solution (28). On transient expression in planta, secreted AvrM internalizes into the plant cell cytosol and deletion studies have revealed that residues 123 to 153 are necessary and sufficient for this internalization (7), whereas the C-terminal region (residues 225–343) is required for M-dependent ETI (28). AvrM-A can also bind to negatively charged phospholipids including PI3P, but the role of PIP binding in host-cell translocation is not clear, and the lipid-binding sites do not overlap with the region identified for AvrM uptake (29).
To understand the molecular basis of plant-cell translocation, PIP binding and AvrM:M interaction, we solved crystal structures of the C-terminal domains of two different AvrM variants, AvrM-A and avrM. AvrM-A and avrM share 93% amino acid sequence identity in the C-terminal shared region, and also differ by a 62-aa internal deletion in avrM and a 34-aa deletion at the C terminus of AvrM-A. Both structures reveal a unique L-shaped helical fold and form a dimer with an unusual nonglobular shape. Surprisingly, the structures contain two four-helix repeats that share some similarity to the overall architecture of the WY domain in oomycete effectors (30). Analysis of the structure combined with site-directed mutagenesis reveals that a conserved hydrophobic surface patch is required for pathogen-independent internalization, but that PIP binding is surface charge-dependent and not required for this process. Mapping of AvrM-A deletion mutants demonstrates that M recognizes the C-terminal coiled-coil domain of AvrM-A, and mutational analysis of single polymorphic residues in AvrM-A and avrM suggest that multiple contact points are required for the M:AvrM interaction.
Results and Discussion
AvrM Structure.
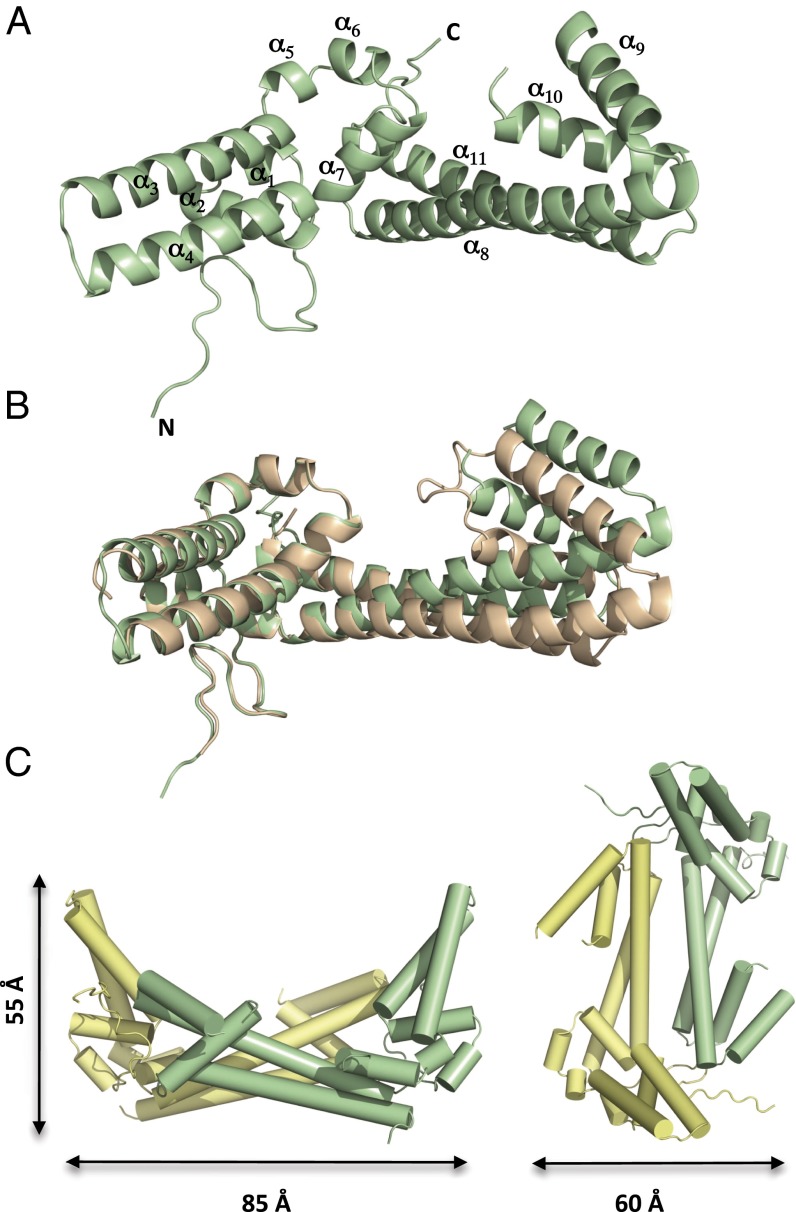
We determined the crystal structures of the C-terminal domain shared between AvrM-A (residues 103–343) and avrM (residues 46–280). The crystals of AvrM-A and avrM contain four and eight molecules in the asymmetric unit, respectively. Both structures have an α-helical fold and adopt an L-shaped structure consisting of a 20-aa residue loop at the N terminus, followed by 11 α-helices (Fig. 1A). Helix pairs α3–α4 and α8–α11 form antiparallel coiled coils that are related to each other by a rotation of ∼90°, resulting in the characteristic elongated L shape. At the tip of the C-terminal coiled-coil domain, two additional helices (α9 and α10) have been inserted and are connected by a flexible loop. Overall, the AvrM-A and avrM structures are similar to each other, with an rms distance (rmsd) of 1.48 Å for 185 structurally equivalent Cα atoms, but there are significant differences in the conformation of the α8 and α11 helices (Fig. 1B). In chains C and G of AvrM-A, the α9 and α10 helices have undergone rotations of ∼35° and 50° as a result of differences in the crystal packing arrangement (SI Appendix, Fig. S1A), suggesting that the tip of the C-terminal coiled-coil domain is flexible and can exist in several conformations.
Fig. 1.
Crystal structures of AvrM-A and avrM proteins. (A) Ribbon diagram showing the overall structure of avrM. The elongated L-shaped molecule consists of an N-terminal loop region followed by 11 α-helices. (B) Superposition of the avrM (green) and AvrM-A (wheat) structures. Structural differences are observed in the α8 and α11 helices of the coiled-coil domain. (C) In the crystal structures of AvrM, the C-terminal coiled-coil domains of two molecules (green and yellow) interact to form a four-helix bundle.
Database searches using the AvrM structures revealed only low-significance similarities to other proteins containing antiparallel coiled-coil and hairpin domains, including viral fusion and envelope proteins, seryl tRNA synthethase, and the repressor of primer protein. The α-helical coiled coil is a ubiquitous protein:protein interaction motif found in diverse proteins exhibiting a broad range of different biological functions (31). In AvrM, the C-terminal antiparallel coiled-coil domain has an important role in mediating self-association as an extensive interface between these domains is observed in the crystal structures of both AvrM variants. The dimer has a nonglobular arc-shape with overall dimensions of 85 × 60 × 55 Å, and a total of 1,950 Å2 of the surface area of each monomer is buried at the dimer interface (Fig. 1C), which is consistent with the protein being a stable dimer in solution (28). In the dimer, two C-terminal coiled-coil domains interact to form a four-helix bundle stabilized by hydrophobic and electrostatic interactions (SI Appendix, Fig. S1 B and C). A similar arrangement of two antiparallel coiled-coil domains is observed in the structures of box C/D RNA protein complexes (32). In these complexes, the coiled-coil domains play an important scaffolding role, and are required for correct positioning of other domains and interacting proteins for binding and modification of RNA. Although differences in the remaining part of the structures suggest a role for AvrM in RNA processing is unlikely, it is possible that the C-terminal coiled-coil domain of AvrM exerts a similar scaffolding role. The N-terminal coiled-coil domain does not seem to be involved in self-association based on the crystal structures, and could possibly mediate interactions with host proteins or other host components.
Examination of the electrostatic surface potential of AvrM revealed that the charge is distributed unevenly (SI Appendix, Fig. S2C). The outer surface of the arc shaped dimer has a continuous positive charge encompassing both the N- and C-terminal coiled-coil domains, whereas the inner surface has negative and neutral surface patches.
Structural Conservation in Poplar Rust Homologs.
AvrM shares sequence similarity with eight proteins of unknown function from the fungal pathogen Melampsora larici-populina, responsible for a rust disease in poplar. The sequence identity varies between 26% and 34%, and there are only 16 aa that are strictly conserved among all of them (SI Appendix, Fig. S3). The signal peptide (residues 1–28 in AvrM-A/avrM), and the uptake region (residues 123–153 in AvrM-A and residues 61–91 in avrM) are similar in all of these proteins, and this suggests that they may all be effector proteins transported into the host cell. Several of the residues involved in stabilizing the overall fold of AvrM are conserved in the poplar-rust homologs, which may suggest that maintaining the characteristic shape is important for function (SI Appendix, Figs. S2 A, B, and D and S3). The C-terminal region and the region between the signal sequence and the start of the AvrM-A and avrM structures is highly variable in sequence and length in the poplar rust homologs, and they also have sequence insertions ranging from 18 to 83 residues in the N-terminal coiled-coil domain. The overall low sequence identity between the poplar rust homologs and the presence of highly diverse sequence insertions may suggest they target similar but distinct components inside the host cell and that the structural fold has the ability to rapidly diverge to adapt the existing virulence functions or gain new ones, and avoid recognition by host R proteins.
AvrM Is a Repeat Protein.
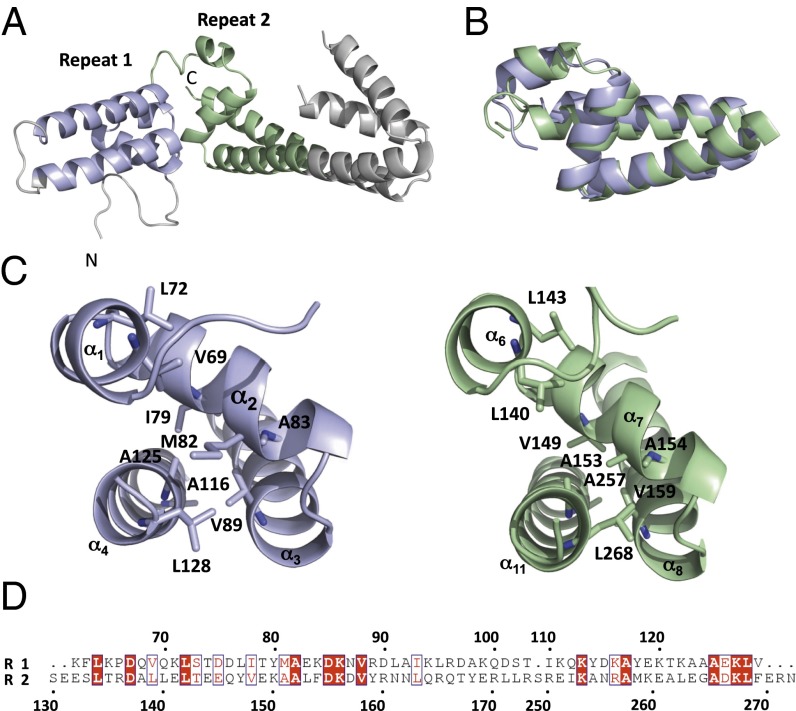
Analysis of the structures revealed that the two antiparallel coiled-coil/hairpin domain regions of AvrM are a part of a structural repeat that is not evident in the sequence. Helices α1 to α4 (residues 124–191 in AvrM-A and residues 62–129 in avrM) resemble helices α6 to α8 and α11 (residues 192–236/313–335 in AvrM-A and residues 130–175/250–272 in avrM), with an rmsd of 2.2 Å for 61 structurally equivalent Cα atoms (Fig. 2 A and B). A structure-based sequence alignment shows 20% identical residues (Fig. 2D) and several of these residues are hydrophobic and involved in stabilizing the overall fold (Fig. 2C). The three last helices of the repeats form a three-helix bundle, in which the first helix is crossing over and stabilizing the conformation of the second and third helices, which resembles the overall architecture of the three-helix WY domain identified in the structures of the oomycete RXLR effectors PexRD2, AVR3a11, and ATR1 (20, 33, 34) (SI Appendix, Fig. S4). The topology of the AvrM repeats is reversed compared with the WY domains, and the structures can therefore not be superimposed directly (rmsd > 7 Å). The WY domain fold can accommodate highly diverse protein sequences, and the presence of similar subdomains in AvrM and related homologs with high sequence diversity may suggest that these three-helix bundle motifs are a common architecture in effector proteins from diverse pathogens, and are well suited to evolutionary changes to avoid host immune recognition and adapt to multiple host targets for enhancement of pathogen virulence.
Fig. 2.
Structural repeat in AvrM. (A) The structures of AvrM contain two four-helical repeats (light blue and green) with similar folds. (B) Superposition of the two AvrM repeats showing the conserved fold [generated by using SSM (37)]. (C and D) Conserved hydrophobic residues stabilize the overall fold of the two repeats. (C) Ribbon diagram of repeat 1 (light blue) and 2 (green) in avrM with the hydrophobic residues highlighted in wireframe. (D) Structure-based sequence alignment of the two repeats. The residues in the ribbon diagrams and in the sequence alignment are numbered according to the avrM sequence.
A Hydrophobic Surface Patch Is Required for Plant-Cell Entry by AvrM.
Previously, we showed that residues 123 to 153 in AvrM-A are necessary and sufficient for mediating internalization of the secreted protein in planta (7). This region has a defined and ordered structure corresponding to part of the N-terminal loop region and α1 and α2 helices (Fig. 3 A–C), and is clearly distinct from the uptake region in RXLR effectors, which is disordered and flexible (20, 33). The internalization region in the flax-rust effector AvrL567 is also completely different in structure to the one in AvrM and corresponds to a region that is partly disordered (residues 27–36) and partly structured (residues 37–50) (24). Kale et al. (15) suggested that the N-terminal region of both AvrM and AvrL567 contained functional RXLR motifs, but our structural analyses show that these motifs are structurally ordered and not disordered as observed for several oomycete effectors. Thus, it is likely that these motifs in AvrM and AvrL567 are not functionally equivalent to the RXLR motif, which is consistent with mutational data on the uptake region in both these proteins (7).
Fig. 3.
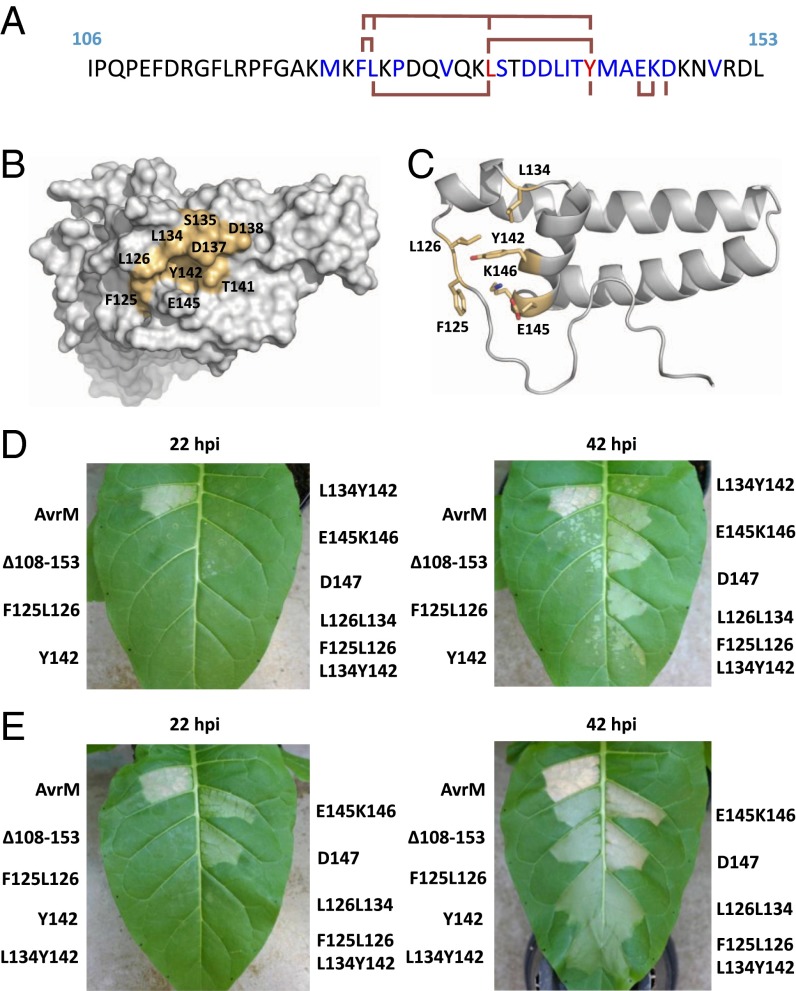
Structural analysis of the AvrM region required for plant-cell entry. (A) Amino acid sequence of AvrM-A (residues 106 to 156). Residues that are conserved or similar in the poplar rust homologs are highlighted in red and blue, respectively. Ala mutants generated in the uptake region are indicated with red lines. (B) Surface representation of the AvrM-A monomer with conserved residues displayed in light orange. (C) Ribbon representation of the molecule in B. The residues that were substituted with Ala and assayed for plant-cell entry are shown in wireframe. (D and E) Agrobacterium-mediated transient expression of AvrM-A, AvrM-A with an internal deletion of aa 108 to 153, and Ala mutants of AvrM-A (F125L126/AA, Y142/A, L134Y142/AA, E145K146/AA, D147/A, L126L134/AA, and F125L126L134Y142/AAAA) in leaves of transgenic tobacco plants (W38) containing the M resistance gene. All the AvrM constructs included the 28-aa signal peptide and a C-terminal cerulean domain. Agroinfiltrations were performed at OD600 of 0.05 (D) and 0.2 (E), and images were prepared 22 and 42 h postinfiltration (hpi).
Many of the residues in the uptake region of AvrM are conserved in the poplar-rust homologs, including the F125 and L126 residues of the RXLR-like KFLK sequence (but not the K124 and K127 residues), and several residues in α1 and α2 helices. Mapping of these residues onto the structure reveals a conserved surface patch on the exterior side of the helical hairpin (Fig. 3 B and C). The patch has a hydrophobic center consisting of F125, L126, L134, and Y142, and is surrounded by polar and charged amino acids, including S135, D137, D138, T141, and E145. Furthermore, the overall fold of this region of the structure appears to be stabilized by hydrogen bonds between the conserved E145, K146, and D147 residues and the protein backbone. To determine if the conserved surface patch is required for plant-cell entry, we expressed the full-length (secreted) AvrM-A protein with various Ala substitutions in this region in transgenic tobacco (Nicotiana tabacum W38) expressing the M resistance gene (Fig. 3 D and E). The AvrM-A 108–153 deletion mutant (Δ108–153) produced a significantly weaker and delayed HR response compared with wild-type (WT) AvrM-A, which is consistent with previous observations (7). A quadruple Ala mutation of the F125, L126, L134, and Y142 residues (F125L126L134Y142/AAAA) produced an HR response with a comparable phenotype to the Δ108–153 mutant, showing that the cell-entry activity of AvrM depends on these conserved hydrophobic residues. Double Ala mutations of F125 and L126 (F125L126/AA) and L126 and L134 (L126L134/AA) and a single Y142A substitution were also tested; both the F125L126/AA and the Y142A mutants reduced the HR response to similar levels as the Δ108–153 mutant lacking the uptake region, whereas the L126L134/AA mutant showed an intermediate HR phenotype, weaker and delayed compared with WT AvrM-A. Mutations of the conserved E145, K146, and D147 residues resulted in a slightly delayed and weaker HR phenotype compared with WT AvrM-A, but the HR phenotype was significantly stronger than the Δ108–153 deletion and hydrophobic site mutants, suggesting that E145, K146, and D147 are not critical for mediating AvrM internalization. Immunoblot analyses confirmed that all the mutants are expressed (SI Appendix, Fig. S5). Apart from the D147A mutant, all the mutants have a similar, but lower, expression level compared with WT AvrM-A and the Δ108–153 mutant. However, the E145K146/AA mutant induces an HR response that is similar to the D147A mutant, and only slightly weaker than WT AvrM-A, showing that the reduced expression levels are still above the threshold required for producing an HR response in tobacco. We also tested the F125L126L134Y142/AAAA, L126L134/AA, and L134Y142/AA mutations in the context of the nonsecreted AvrM-A protein. Although we observed a slight decrease in the HR induction, this was substantially less than the effect observed for the secreted proteins (SI Appendix, Fig. S6).
Taken together, these results show that the F125 and Y142 residues are most important for efficient AvrM-A internalization, whereas the L126 and L134 residues play a lesser role. The side chains of F125 and Y142 are partially surface-exposed and interact with other residues in the uptake region, which suggests that the structural integrity of this region is important for efficient plant-cell delivery. Interestingly, the K124F125L126K127/AAAA mutant was previously shown to only slightly delay HR (7), whereas the F125L126/AA mutant in this study was equivalent to the 108–153 deletion, indicating that mutation of the positively charged K124 and K127 residues to small hydrophobic Ala residues has a positive effect on AvrM uptake, and suggesting that maintaining a hydrophobic surface potential in this region is important for internalization.
PIP Binding to AvrM Is Charge-Dependent and Is Not Required for Internalization.
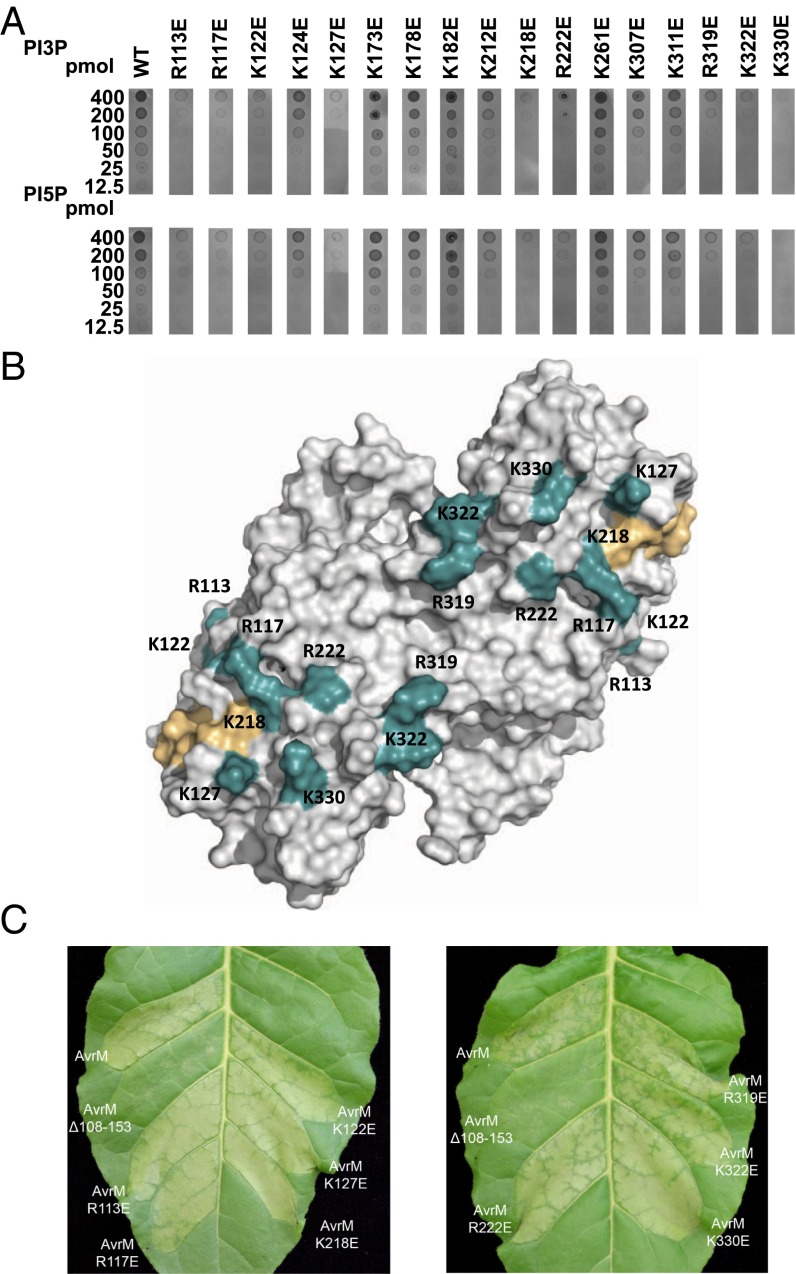
We have previously shown, by using a dot-blot assay, that the C-terminal region of AvrM-A (residues 106–343) can bind to phosphatidyl inositol, phosphatidyl inositol monophosphates, and phosphatidyl serine (PS) (29). The structures of AvrM-A and avrM do not display similarity to any characterized phospholipid-binding domains, and do not contain surface pockets that could provide stereoselective recognition of phospholipid molecules, which is consistent with their ability to bind different types of PIPs and PS. However, AvrM proteins contain several positively charged patches on the surface, with basic side-chain configurations that could mediate binding to head-groups of negatively charged phospholipids. To test this directly, we analyzed the effects of reverse-charge amino acid substitutions in these surface patches on binding to PI3P and phosphatidylinositol 5-phosphate (PI5P). A total of 17 mutants were assayed for PIP binding, and nine of these mutants (R113E, R117E, K122E, K127E, K218E, R222E, R319E, K322E, and K330E) significantly reduced AvrM-A’s ability to bind PI3P and PI5P (Fig. 4A). The largest effect was observed for the K330E mutant, which almost completely abrogated PI3P and PI5P binding to AvrM-A. Seven of these mutations (R117E, K127E, K218E, R222E, R319E, K322E, and K330E) are localized on the base of the arc-shaped dimer, whereas the remaining two mutations (R113E and K122E) map to the N-terminal region of the structure. The R117E, K122E, K127E, and K218E mutants are also close to the conserved hydrophobic surface patch important for AvrM-A internalization (Fig. 4B). Reverse-charge mutations in the N-terminal coiled-coil region (K173E, K178E, and K182E), or the tip of the C-terminal coiled-coil domain (K261E, K307E, and K311E) do not have an effect on AvrM-A’s ability to bind PI3P or PI5P. Overall, PIP recognition by AvrM-A seems best characterized as charge-dependent, and the overall positive surface charge on the outer surface of the arc-shaped dimer needs to be maintained for high-affinity PIP binding.
Fig. 4.
PIP binding to AvrM is charge-dependent and is not required for uptake. (A) Protein-lipid overlay assay of Escherichia coli-expressed GST-AvrM-A and single Glu substitutions of surface exposed Arg and Lys residues. Serial dilutions (400, 200, 100, 50, 25, and 12.5 pmol) of PI3P and PI5P were spotted onto nitrocellulose membranes and incubated with WT or mutant AvrM-A. After rigorous washing, bound proteins were detected by using anti-GST/alkaline phosphatase antibodies. (B) Positions of the reverse-charge mutants (teal) that reduced or abrogated PIP binding (R113E, R117E, K122E, K127E, K218E, R222E, R319E, K322E, and K330E) on the structure of the AvrM-A dimer. The conserved surface patch important for AvrM internalization is shown in light orange. (C) Agrobacterium-mediated transient expression of AvrM-A, AvrM-A with an internal deletion of amino acids 108 to 153 and AvrM-A mutants (shown in B) in leaves of transgenic tobacco plants (W38) containing the M resistance gene. All the AvrM constructs included the 28-aa signal peptide and a C-terminal citrine domain. Agroinfiltrations were performed at an OD600 of 0.4, and images were prepared 29 hpi.
We also tested the ability of the nine mutants that significantly reduced PIP binding to enter plant cells after secretion. Transient expression of these mutants fused to YFP-citrine in W38::M tobacco leaves produced a similar HR response compared with WT AvrM-A (Fig. 4C). Fluorescence microscopy confirmed the cytosolic localization of these proteins (SI Appendix, Fig. S7), suggesting that PIP binding is not required for translocation of AvrM-A into plant cells. This is consistent with our previous data, which showed that a subregion of AvrM sufficient to direct uptake of a fusion protein into plant cells did not bind to PI3P and PI5P (29).
The C-Terminal Coiled-Coil Domain Interacts with M.
Previous truncation studies identified amino acids 206 to 335 as the minimal region that could induce a necrotic response equal to the WT protein and interact directly with M in yeast (28). This region corresponds to the C-terminal coiled-coil domain region of AvrM, and contains 11 of the 13 polymorphic residue positions observed in the AvrM alleles (SI Appendix, Fig. S8). The side chains of seven of these polymorphic residues in AvrM-A/avrM are highly solvent-exposed, and are accessible for mediating direct interactions with the M resistance protein (SI Appendix, Fig. S9), and they localize to the inner (K232/R170, T259/N197, P280/L217) and outer surfaces of the arc-shaped dimer (K226/Q164, E316/K253, K326/E263, and K333/E270). Previous studies have shown that single, double, and triple mutations of E316K, K326E, and K333E in AvrM-A, and K253E, E263K, and E270 in avrM do not lead to loss or gain of recognition of AvrM-A and avrM, respectively, suggesting that these positions in AvrM-A are not involved in direct interaction with M, or that additional positions also are required for the interaction (28). To test if the remaining polymorphic residues positions are critical for recognition by M, we mutated them individually in AvrM-A and in an avrM variant lacking the 34-aa C-terminal extension (avrM-CT-Δ34), and tested their ability to produce an HR in W38::M tobacco leaves. All the mutants produced a similar HR to WT AvrM-A or avrM-CT-Δ34 (SI Appendix, Fig. S10), showing that mutation of individual polymorphic residue positions is not sufficient to produce a gain or loss of recognition phenotype. Overall, our results suggest that the M:AvrM interaction has multiple contact points, which is consistent with the L6:AvrL567 interaction in flax/flax rust and the RPP1:ATR1 interaction in Arabidopsis/Hyaloperonospora arabidopsidis (24, 34).
Conclusions
We report the crystal structures of two naturally occurring variants of AvrM, revealing a dimeric protein with an unusual nonglobular shape, each polypeptide consisting of a tandem duplicated four-helix motif related to the WY domain core in oomycete effectors. Our functional analysis reveals that a hydrophobic surface patch conserved between both variants is required for internalization into plant cells, whereas the C-terminal coiled-coil domain mediates interaction with M through multiple contact points.
Overall, our structural and functional data on AvrM localization and PIP binding do not support the proposed hypothesis for the general role of extracellular PI3P in plant-cell entry by oomycete and fungal effector proteins (15). Furthermore, the structural differences between the uptake region in RXLR effectors, AvrL567 and AvrM suggest that fungal effector proteins may use different transport mechanisms for plant-cell translocation. The ability of fungal and oomycete effectors to enter the host cell independently of the pathogen suggests that they may hijack a plant-derived membrane-transport system or have autonomous membrane-crossing properties. Interestingly, the Yersinia pestis effector protein YopM, which is normally injected into mammalian host cells by the type-III secretion system, was recently shown to also have cell-penetrating properties, with either of the two N-terminal α-helices of YopM capable of autonomous translocation across the cell membrane (35). The structural properties of AvrM, including positive and hydrophobic surface patches required for negatively charged phospholipid binding and uptake, indicate that AvrM has intrinsic membrane-binding properties (7). Interestingly, the region shown to be involved in plant-cell entry (residues 122–153 in AvrM-A) coincides well with the first of the two four-helix repeat motifs observed in the AvrM structures. The similarity in structure and sequence between the two repeats (Fig. 2) may suggest that AvrM, similar to YopM, has two redundant regions capable of mediating host-cell translocation, which may explain why the AvrM-A Δ108–153 mutant only delays HR in transient assays in tobacco. The ability of AvrM and other fungal and oomycete effectors to bind negatively charged phospholipids such as PIPs may be a consequence of selection for a particular surface charge distribution required for membrane recruitment rather than a requirement for a specific PIP-related uptake mechanism. Importantly, the transient expression assay does not distinguish between uptake of the AvrM protein after secretion into the apoplast, or entry into the cytosol from an internal vesicular structure before secretion. We have not observed M gene-dependent HR after infiltration of the purified AvrM protein into tobacco or flax leaves. Secretion of the protein into the sealed compartment of the extrahaustorial matrix during infection may be important for host delivery, possibly for protection from apoplastic proteases or to allow accumulation of high local protein concentrations to drive endocytosis. It will be important to test mutant derivatives of AvrM in transgenic rust lines to determine whether the results of the in planta assays truly reflect the mechanism of effector delivery during infection. A further understanding of the molecular mechanisms involved in effector translocation across the host membrane cells may provide us with the rationale to block effector uptake and function, which has the potential to provide new therapeutic agents for disease control.
Materials and Methods
Cloning, Protein Production, and Purification.
Standard methods were used as described previously (36) and in SI Appendix, SI Materials and Methods.
Crystallization and Structure Determination.
Diffraction-quality crystals of AvrM-A and avrM were obtained by hanging-drop vapor diffusion (36). The structures were solved by SeMet-based single-wavelength anomalous diffraction phasing and refined and validated by using standard macromolecular crystallography methods and software (SI Appendix, SI Materials and Methods and Table S1). The final models of AvrM-A and avrM contain residues 110 to 342 and 46 to 280, respectively. Several of the chains in AvrM-A and avrM have no electron density in the loops connecting helices α3 and α4, or α9 and α10, suggesting that these regions have a disordered or flexible conformation in the crystals. Coordinates and structure factors of AvrM-A and avrM have been deposited in the Protein Data Bank with ID codes 4BJN and 4BJM, respectively.
Lipid-Binding Assay.
Lipids were spotted onto nitrocellulose membranes and incubated with protein solutions, and the lipid–protein interaction was detected as described in SI Appendix, SI Materials and Methods.
Transient in Planta Expression Assays.
Binary expression constructs of AvrM-A and avrM were prepared as described in SI Appendix, SI Materials and Methods. Agrobacterium cultures of strain GV3101-pMP90 containing the binary expression constructs were prepared to an OD600 of 1.0, 0.4, 0.2, or 0.05 in 10 mM Mes (pH 5.6) buffer with 10 mM MgCl2 and 200 μM acetosyringone for infiltration into W38 and W38::M tobacco leaves.
Supplementary Material
Acknowledgments
We thank Brett Collins, Marc Kvansakul, Kim Newell, and the B.K. laboratory members for helpful discussions. This work was supported by Australian Research Council Grants DP120100685 (to B.K., P.N.D., and J.G.E.), and DP1093850 (to A.R.H., D.A.J., P.N.D., and J.G.E.). B.K. is a National Health and Medical Research Council Research Fellow. We acknowledge the use of University of Queensland Remote Operation Crystallisation and X-ray Diffraction Facility and the Australian Synchrotron.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4BJN and 4BJM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307614110/-/DCSupplemental.
References
- 1.Rafiqi M, Ellis JG, Ludowici VA, Hardham AR, Dodds PN. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr Opin Plant Biol. 2012;15(4):477–482. doi: 10.1016/j.pbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012;15(4):483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 3.de Jonge R, Bolton MD, Thomma BP. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr Opin Plant Biol. 2011;14(4):400–406. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 6.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 7.Rafiqi M, et al. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell. 2010;22(6):2017–2032. doi: 10.1105/tpc.109.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khang CH, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22(4):1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450(7166):115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 10.Kemen E, et al. Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol Plant Microbe Interact. 2005;18(11):1130–1139. doi: 10.1094/MPMI-18-1130. [DOI] [PubMed] [Google Scholar]
- 11.Djamei A, et al. Metabolic priming by a secreted fungal effector. Nature. 2011;478(7369):395–398. doi: 10.1038/nature10454. [DOI] [PubMed] [Google Scholar]
- 12.Plett JM, et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21(14):1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Ribot C, et al. The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 2013;74(1):1–12. doi: 10.1111/tpj.12099. [DOI] [PubMed] [Google Scholar]
- 14.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Kale SD, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142(2):284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Dou D, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20(7):1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 18.Schornack S, et al. Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci USA. 2010;107(40):17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaeno T, Shirasu K. The RXLR motif of oomycete effectors is not a sufficient element for binding to phosphatidylinositol monophosphates. Plant Signal Behav. 2013;8(4) doi: 10.4161/psb.23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaeno T, et al. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci USA. 2011;108(35):14682–14687. doi: 10.1073/pnas.1106002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wawra S, et al. Avirulence protein 3a (AVR3a) from the potato pathogen Phytophthora infestans forms homodimers through its predicted translocation region and does not specifically bind phospholipids. J Biol Chem. 2012;287(45):38101–38109. doi: 10.1074/jbc.M112.395129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wawra S, et al. In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate specific autonomous uptake in plant and animal cells. Mol Plant Microbe Interact. 2013;26(5):528–536. doi: 10.1094/MPMI-08-12-0200-R. [DOI] [PubMed] [Google Scholar]
- 23.Tyler BM, et al. Microbe-independent entry of oomycete RxLR effectors and fungal RxLR-like effectors into plant and animal cells is specific and reproducible. Mol Plant Microbe Interact. 2013;26(6):611–616. doi: 10.1094/MPMI-02-13-0051-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CI, et al. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell. 2007;19(9):2898–2912. doi: 10.1105/tpc.107.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodds PN, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103(23):8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18(1):243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16(3):755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catanzariti AM, et al. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact. 2010;23(1):49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan PH, et al. Lipid binding activities of flax rust AvrM and AvrL567 effectors. Plant Signal Behav. 2010;5(10):1272–1275. doi: 10.4161/psb.5.10.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Win J, et al. Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 2012;8(1):e1002400. doi: 10.1371/journal.ppat.1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001;11(2):82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 32.Lin J, et al. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011;469(7331):559–563. doi: 10.1038/nature09688. [DOI] [PubMed] [Google Scholar]
- 33.Boutemy LS, et al. Structures of Phytophthora RXLR effector proteins: A conserved but adaptable fold underpins functional diversity. J Biol Chem. 2011;286(41):35834–35842. doi: 10.1074/jbc.M111.262303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou S, et al. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci USA. 2011;108(32):13323–13328. doi: 10.1073/pnas.1109791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rüter C, Buss C, Scharnert J, Heusipp G, Schmidt MA. A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J Cell Sci. 2010;123(pt 13):2190–2198. doi: 10.1242/jcs.063016. [DOI] [PubMed] [Google Scholar]
- 36.Ve T, et al. Crystallization and X-ray diffraction analysis of the C-terminal domain of the flax rust effector protein AvrM. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(pt 12):1603–1607. doi: 10.1107/S1744309111037675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.