Significance
Here we report the crystal structures of human C-type lectin receptors Mincle (macrophage inducible C-type “calcium-dependent” lectin; CLEC4E) and MCL (macrophage C-type lectin; CLEC4D), both of which are receptors for mycobacterial glycolipid adjuvant cord factor (also called trehalose-6,6′-dimycolate; TDM). Our structural and functional studies clearly reveal the simultaneous recognition of sugar and lipid moieties by Mincle and MCL, distinct from other C-type lectin receptors. Because better adjuvants are desired for enhancing vaccination effects of medical treatments for infectious diseases, cancer, and so forth, these structures provide a framework for the rational design of more effective adjuvants than TDM.
Keywords: X-ray crystallography, innate immunity, mycobacteria, pattern-recognition receptors, myeloid cells
Abstract
Mincle [macrophage inducible Ca2+-dependent (C-type) lectin; CLEC4E] and MCL (macrophage C-type lectin; CLEC4D) are receptors for the cord factor TDM (trehalose-6,6′-dimycolate), a unique glycolipid of mycobacterial cell-surface components, and activate immune cells to confer adjuvant activity. Although it is known that receptor–TDM interactions require both sugar and lipid moieties of TDM, the mechanisms of glycolipid recognition by Mincle and MCL remain unclear. We here report the crystal structures of Mincle, MCL, and the Mincle–citric acid complex. The structures revealed that these receptors are capable of interacting with sugar in a Ca2+-dependent manner, as observed in other C-type lectins. However, Mincle and MCL uniquely possess shallow hydrophobic regions found adjacent to their putative sugar binding sites, which reasonably locate for recognition of fatty acid moieties of glycolipids. Functional studies using mutant receptors as well as glycolipid ligands support this deduced binding mode. These results give insight into the molecular mechanism of glycolipid recognition through C-type lectin receptors, which may provide clues to rational design for effective adjuvants.
Pattern-recognition receptors (PRRs) play important roles in innate immunity. PRRs recognize nucleotides, sugars, lipopolysaccharides, other pathogen components, and self-ligands, and consequently trigger intracellular signaling cascades that initiate innate and adaptive immune responses (1). Among them, Toll-like receptors (TLRs) are well-characterized receptors, in terms of their ligand specificities, ligand-recognition mechanisms, and signaling pathways (2–4). C-type lectin receptors (CLRs) are also a large family of PRRs (5–7). The term “C-type lectin” was introduced to distinguish a group of Ca2+-dependent lectins from other lectins. In the CLRs, two amino acids harboring long carbonyl side chains separated by a proline in a cis conformation coordinate a Ca2+ ion, which forms hydrogen bonds with monosaccharides and determines the binding specificity. CLRs have broad recognition abilities toward not only saccharides but also proteins (5, 7–9). For instance, human NKR-P1 interacts with Lectin-like transcript 1, and some members of the CD94/NKG2 family interact with HLA-E.
Macrophage inducible C-type lectin (Mincle; also called CLEC4E) is a type II transmembrane C-type lectin receptor that is expressed in macrophages, dendritic cells, and monocytes upon stimulation (10). We have reported that Mincle is an FcRγ-coupled activating receptor that recognizes pathogenic fungi and mycobacteria (11–13). Detailed investigations of the ligands of Mincle revealed that Mincle binds glycolipids, such as trehalose-6,6′-dimycolate (TDM) from Mycobacterium tuberculosis, and novel glyceroglycolipids from Malassezia fungus. Malassezia and M. tuberculosis ligands are recognized through the carbohydrate-recognition domain (CRD) in the extracellular region of Mincle (11, 12). The binding of TDM to Mincle leads to the phosphorylation of the immunoreceptor tyrosine-based activation motif (ITAM) in the FcRγ chain, which provides a binding site for the Syk tyrosine kinase. Syk activates the caspase recruitment domain family member 9-mediated NF-κB signaling pathway to promote the expression of TNF and IL-6. A recent report revealed that Mincle plays a nonredundant role in T-cell immune responses to infection by microbes and in the adjuvanticity of mycobacterial cord factor and its synthetic analog, trehalose-dibehenate (14, 15).
Macrophage C-type lectin (MCL; also called CLEC4D) is another C-type lectin receptor expressed in myeloid cells (16, 17). Recently, we found that MCL is also an FcRγ-coupled activating receptor that binds to TDM (15). MCL is distinct from Mincle in the following ways: (i) The expression of Mincle is inducible, whereas MCL is constitutively expressed in myeloid cells; (ii) MCL shows weaker binding affinity to TDM than does Mincle; and (iii) the glutamic acid-proline-asparagine (EPN) motif, a typical glucose/mannose-binding motif, is conserved in Mincle but not in MCL.
We now report the crystal structures of Mincle and MCL, as well as Mincle complexed with citric acid. They have similar overall structures to other typical CLRs, but exhibit characteristic conformations in the vicinity of the Ca2+-binding motif. A patch of hydrophobic amino acids located adjacent to the carbohydrate binding site may likely contribute to the recognition of the fatty acid chain of TDM. The mutational analysis essentially supports this TDM-binding model, and may also explain the different affinities of MCL and Mincle.
Results
Preparation, Crystallization, and Structural Determination of MCL.
The extracellular domain of human MCL (residues 61–215) (Fig. 1) was expressed in Escherichia coli as inclusion bodies, and was refolded in vitro by a dilution method. Ca2+ ions were required in the refolding procedure, and the crude, refolded MCL was purified by sequential gel-filtration chromatography steps (Fig. S1 A and C). The purified MCL was crystallized by the hanging-drop method with 0.1 M bis-Tris propane (pH 6.5), 0.2 M potassium thiocyanate, and 20% (wt/wt) PEG 3350. Crystals of the MCL protein [the space group was I-centered orthorhombic (I222), and the unit-cell parameters were a = 85.19 Å, b = 96.06 Å, c = 104.53 Å] were obtained, and the dataset was collected to the resolution limit of 2.2 Å at the BL32XU beamline at SPring-8 (Harima, Japan) (Table S1). The crystal structure of MCL has 2 α-helices (α1 and α2) and 11 β-strands (β1–β11) (Fig. 2A), which is a typical structural organization of CLRs and partly similar to the solution structure of MCL registered in the Protein Data Bank (PDB) (ID code 2LS8) (Fig. S1E). Two MCL molecules exist in the asymmetric unit. Gel-filtration analysis showed a mixture of the peaks (Fig. S1A), suggesting that MCL may have some conformational variation.
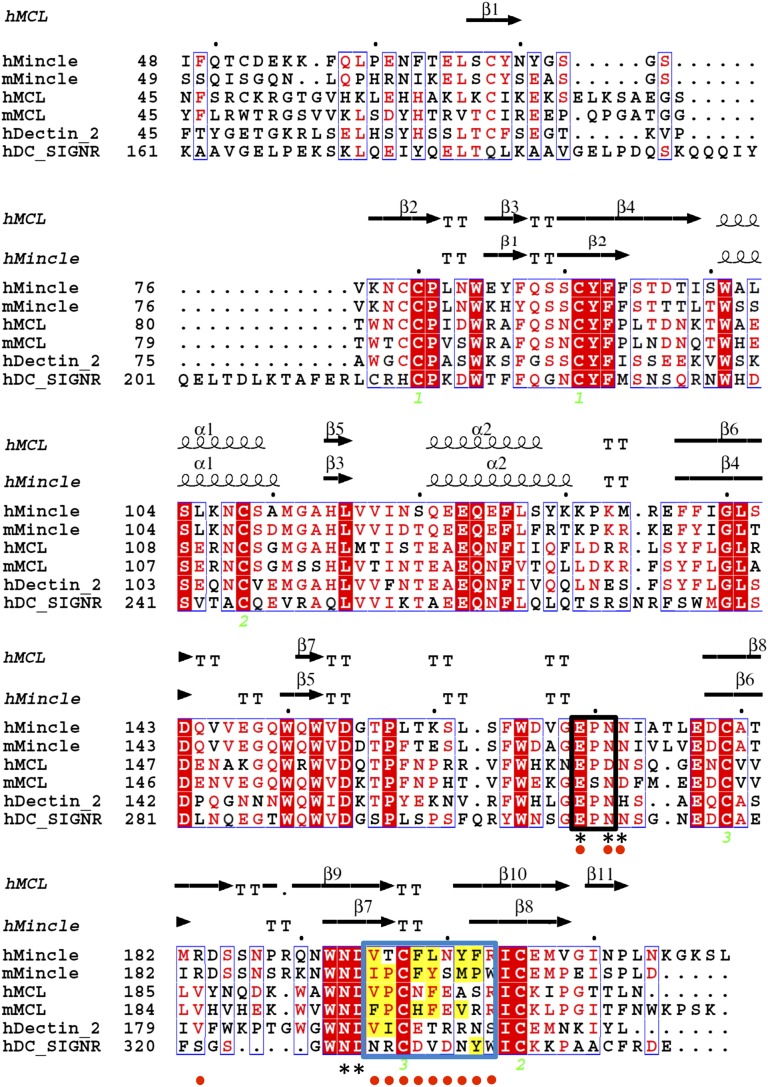
Fig. 1.
Structure-based sequence alignment of Mincle, MCL, DC-SIGNR, and Dectin-2. The sequence alignment of the ectodomains of Mincle, MCL, DC-SIGNR, and Dectin-2 (h, human; m, mouse) is shown, as depicted with ESPript (44). Identical residues are highlighted in red, and similar residues are framed in blue. The secondary structure elements (α, α-helix; β, β-strand; T, turn) of Mincle and MCL are shown above the sequences. The box enclosed by the thick black line indicates EPN motifs, which are usually involved in carbohydrate recognition by C-type lectins. The box enclosed by the thick blue line indicates hydrophobic amino acid loops, and yellow-shaded amino acid residues are hydrophobic residues within Mincle and MCL. The asterisks below the sequences indicate the residues involved in calcium binding in Mincle and MCL. The red-filled circles below the sequences indicate the residues changed to other amino acid residues in the mutational studies. The numbers below the cysteine residues indicate disulfide-bond formation with the cysteine residue with the same number.
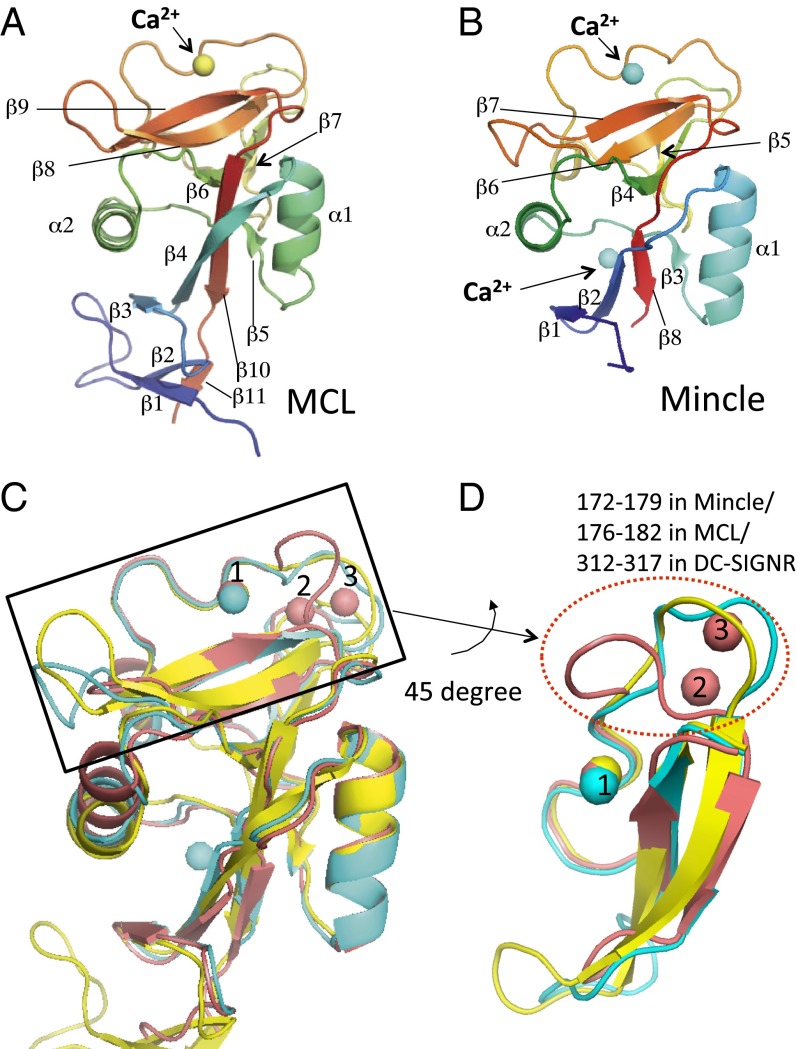
Fig. 2.
Structures of MCL and Mincle, and structural comparison with DC-SIGNR. (A and B) Cartoon models of the overall structures of MCL (A) and Mincle (B). Secondary structure elements are shown. Gradient rainbow color from blue to red indicates N- to C-terminal. The yellow and cyan spheres represent Ca2+ ions in MCL and Mincle, respectively. (C and D) Overall structures (C) and putative ligand binding sites (D) of MCL (yellow), Mincle (cyan), and DC-SIGNR (pink) are shown. (D) Close-up view of the black box in C. Yellow, blue, and pink spheres represent Ca2+ ions in Mincle, MCL, and DC-SIGNR, respectively. The red dotted oval indicates the large structural differences of loops among these CLRs (Results).
Preparation, Crystallization, and Structural Determination of Mincle.
Using a similar refolding method to that for MCL, we also prepared the extracellular domain of Mincle (residues 74–219) (Fig. 1). The expression, refolding, and purification were successful. However, the crystallization was not successful, because the refolded Mincle was not sufficiently soluble at high concentrations. To improve the protein solubility, we performed site-directed mutagenesis and changed the hydrophobic amino acids presumably located on the surface of the Mincle protein to hydrophilic amino acids, as found in the corresponding residues of MCL. Among them, the mutant with the substitution of isoleucine to lysine at residue 99 (I99K mutant) formed good crystals by the hanging-drop method under two conditions. One is 1 M lithium chloride, 0.1 M citric acid (pH 4), and 20% (wt/vol) PEG 6000, and the other is 0.2 M NH4SO4, 0.1 M bis-Tris (pH 5.5), 25% (wt/vol) PEG 3350. These diffraction data were collected to the resolution limit of 1.3 and 1.35 Å at the BL5A and BL17A beamlines at High Energy Accelerator Research Organization (KEK) (Tsukuba, Japan), respectively. Both crystals have the same space group, primitive trigonal systems (P31), and similar unit-cell parameters (Table S1). Mincle exhibits a typical CLR fold, as shown in Fig. 2B. The asymmetric unit contained one molecule of Mincle, and no physiologically important packing was detected. This is consistent with the gel-filtration analysis showing that Mincle behaves as a monomer, although the eluted time is later than the expected one, likely due to the affinity of Mincle to the glucose-based dextran resin of the Superdex column (Fig. S1 B and D).
Structural Comparison Between Mincle, MCL, and Other C-Type Lectins.
MCL and Mincle superimposed well on each other [root-mean-square deviation (rmsd) 1.5 Å for 124 Cα atoms] (Fig. 2 A and B and Fig. S2B). However, Mincle has two calcium ions, whereas MCL has only one. A DALI analysis (18) indicated that MCL and Mincle share high homology with mouse collectin (PDB ID code 2OX9) [rmsd of 1.3 Å for 120 Cα atoms, 28% identity (MCL); 1.07 Å for 118 Cα atoms, 35% identity (Mincle)] (19) and DC-SIGNR (PDB ID code 1K9J) [rmsd of 1.7 Å for 114 Cα atoms, 37% identity (MCL); 1.3 Å for 121 Cα atoms, 45% identity (Mincle)], which has been extensively studied as an entry receptor of HIV (20, 21). Because collectin recognizes fucose-based oligosaccharides, rather than glucose- or mannose-based ones, we chose to compare the structural features of MCL, Mincle, and DC-SIGNR (Fig. 2 C and D). The entire structures and positions of the amino acid residues in the putative CRD are similar. Specifically, the positions of the Ca2+ ions (site 1) are the same among the three proteins. The EPN motif (residues 169–171 in Mincle) is often observed in C-type lectins, and contributes to carbohydrate recognition via a Ca2+ ion-mediated binding network (Fig. 3 A and B). In contrast, the glutamic acid-proline-aspartic acid (EPD) motif of MCL (residues 173–175) is an unusual sequence among C-type lectins (6) (Fig. 1). However, the Ca2+ ion and other amino acids involved in carbohydrate recognition are located in this region, as in other C-type lectins (Figs. 2 and 3 A–C). These results indicated that Mincle and MCL recognize carbohydrates through these motifs in slightly different but similar ways.
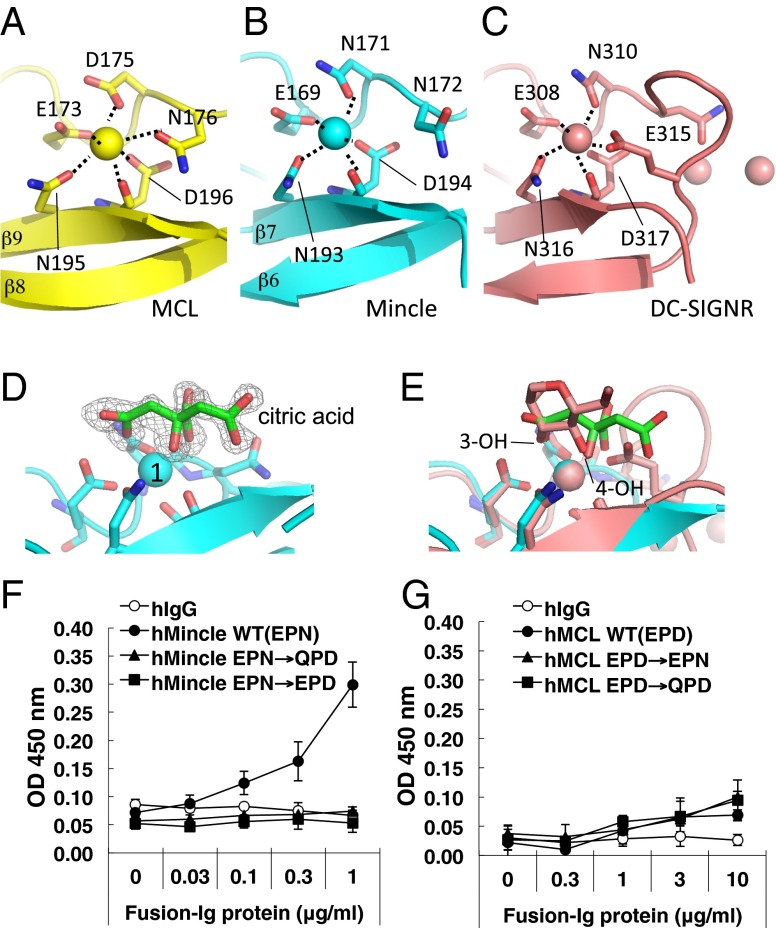
Fig. 3.
Structural comparison of the putative ligand binding sites in MCL, Mincle, and DC-SIGNR, and in vitro binding assays of the Mincle and MCL mutants. (A–C) Close-up views of the putative ligand binding sites of MCL (yellow) (A), Mincle (cyan) (B), and DC-SIGNR (pink) (C) are shown. The amino acid residues involved in and close to Ca2+ ion binding are shown as stick models. Interactions with Ca2+ ions are shown as black dotted lines. (D) Composite omit map (2Fo − Fc) for citric acid in Mincle. The electron-density map is contoured at 1.0σ, and the resolution is 1.3 Å. The citric acid is shown with the O atoms colored red and the C atoms in green. Putative amino acids involved in Ca2+ binding are depicted by sticks. (E) The superimposed structures of Mincle (cyan; same as D) and DC-SIGNR (pink) are shown. The stick model indicates the mannose (the O atoms are colored red and the C atoms are in pink) in the DC–SIGNR complex. (F and G) Mincle–Ig, mutated Mincle–Ig, or hIgG (F) and MCL–Ig, mutated MCL–Igs, or hIgG (G) were incubated with plate-coated TDM. Bound proteins were detected by anti–hIgG-HRP. Error bars indicate ± standard deviation of three independent experiments.
The regions surrounding the Ca2+-bound sites in MCL and Mincle are distinctly different from those in DC-SIGNR. In DC-SIGNR, two additional bound Ca2+ ions are observed close to the site (red, 2 and 3) (Fig. 2C), and they stabilize the typical protein conformation of the C-type lectins (6). The Ca2+ (site 2 and 3) ions push the loop (residues 312–317) close to the Ca2+ (site 1) ion (Fig. 2D, red dotted oval). In contrast, the corresponding loops in MCL and Mincle are located far from the Ca2+ (site 1) ion. The asparagine/aspartate residues just following the EPD/EPN sequences are conserved (Fig. 1). The directions of the asparagines in MCL (residue 176) and Mincle (residue 172) are different from those in other CLRs, such as DC-SIGNR (Fig. 3 A–C). The asparagine in DC-SIGNR is used to bind the Ca2+ (site 2) ion, and therefore the side chain faces the opposite direction of the Ca2+ (site 1) ion. In contrast, neither Mincle nor MCL coordinates Ca2+ (site 2 and 3) ions, and their asparagine side chains extend in different directions compared with other C-type lectins.
Calcium Binding and Ligand Recognition.
In the crystals of Mincle grown in 1 M lithium chloride, 0.1 M citric acid (pH 4), and 20% (wt/vol) PEG 6000, a strong electron density in addition to that of the Mincle protein was observed close to the Ca2+ (site 1) ion and matched a citric acid molecule (Fig. 3D). The superimposition of the amino acids in the Ca2+ ion-binding regions of the ligand complex structures of Mincle and DC-SIGNR revealed the well-conserved locations of the oxygen atoms of the ligands citrate and mannose (equatorial 3- and 4-OH groups), respectively (Fig. 3E). Because the chemical property of the sugar moiety is different from citric acid, we cannot simply compare the recognition modes, but these data may support the idea that Mincle can use this Ca2+ ion to bind nucleophiles for the sugar moieties of TDM and Malassezia ligands, in essentially the same manner as generally observed in CLRs including DC-SIGNR.
The calcium binding site in human and mouse Mincle includes the EPN motif, well-conserved in the mannose-recognizing C-type lectins, as described above. We examined whether the EPN motif in Mincle is involved in direct TDM recognition using soluble Mincle protein (Mincle–Ig). Mincle–Ig (MincleWT), but not control Ig, selectively bound to plate-coated TDM, as previously reported (11, 12). This recognition was shown to require the EPN motif, as the binding was eliminated by introducing a mutation of EPN into glutamine-proline-aspartic acid (QPD), a putative galactose-recognition sequence (MincleQPD) (22). Substitution of the EPN motif into MCL-type EPD (MincleEPD) also impaired the binding capacity, although their reactivities to anti-hIgG were comparable (Fig. 3F and Fig. S3A). These data suggested that EPN in human Mincle is indispensable for TDM recognition, as previously shown in mouse Mincle (11, 12). In contrast, the direct binding of MCL to TDM was much weaker than that of Mincle (Fig. 3G), consistent a the previous report that MCL recognizes TDM with less affinity than Mincle (15). Mutation of the EPD sequence of MCL into QPD (MCLQPD) did not have a large impact on TDM binding in higher concentrations. Unexpectedly, however, the EPD-to-EPN mutation in MCL, which was expected to coordinate the Ca2+ ion location well and facilitate carbohydrate binding, did not improve the affinity for TDM (Fig. 3G). These results suggested that the TDM binding site of MCL might be distinct from that used by Mincle. Furthermore, the side chain of Arg183 in Mincle is in a suitable position to interact with the hydroxyl groups of TDM, based on the crystal structure of Mincle (23) (Fig. 4A). This arginine residue of Mincle is well-conserved from fish to mammals. In contrast, the valine (Val186) at the corresponding position of human MCL is conserved among placentalia; however, its side chain cannot reach the putative carbohydrate recognition site (Fig. 4B). To verify the role of Arg183 in TDM recognition, we introduced the R183V mutation into Mincle and tested its function in an NFAT-GFP reporter assay. This mutation reduced NFAT-GFP activity in the reporter cell assay, suggesting that Arg183 of Mincle is crucially involved in ligand recognition (Fig. 4C).
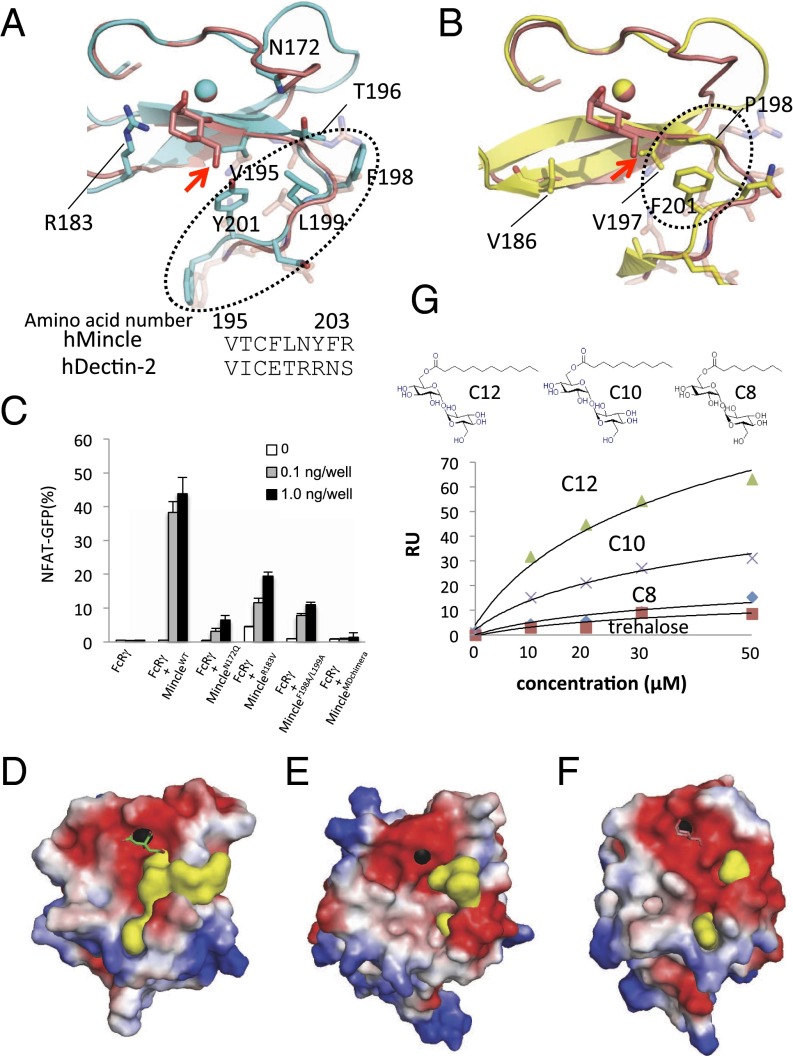
Fig. 4.
Unique amino acid residues in MCL and Mincle, reporter assays of mutant Mincle, and SPR analysis. (A and B) The superimposed structures of Mincle (cyan) and DC-SIGNR (A) and MCL (yellow) and DC-SIGNR (pink) (B) are shown. Arrows indicate the oxygen atom connected with mycolic acid in TDM (mannose binding to DC-SIGNR is shown). Dotted circles indicate the hydrophobic loops found in Mincle and MCL. A sequence comparison between human Mincle and Dectin-2 is shown. (C) Analyses of Mincle and its mutants were performed. Reporter cells expressing human Mincle or its mutants were stimulated with TDM for 18 h. Error bar indicates ± standard deviation of three independent experiments. (D–F) Electrostatic potentials of Mincle (D), MCL (E), and DC-SIGNR (F) are shown. Electrostatic surface potentials were calculated using the program APBS (45) and are represented by PyMOL (www.pymol.org), with the color of the surface potentials in the scale ranging from negatively charged (−4.0 kbT/ec; red) to positively charged amino acids (4.0 kbT/ec; blue). Black spheres are Ca2+ ions. The yellow surface indicates the hydrophobic site. (G) SPR analysis of Mincle and several lengths of acyl chains with trehalose was performed. The C12, C10, and C8 glycolipids used in this experiment are shown. RU, response units.
Taken together, these results strongly suggested that the binding mode of the two OH groups of citric acid to Ca2+ reflects the equatorial 3- and 4-OH groups of mannose and glucose of Mincle/MCL ligands in a similar but slightly different manner from the CLRs (6).
Putative Lipid Recognition Sites.
To determine whether Mincle and MCL use unique amino acids for their interactions with the lipid regions of glycolipids, we verified the characteristics of the surfaces surrounding the putative sites for Ca2+-mediated sugar binding. A series of hydrophobic regions was specifically found in Mincle and MCL but not in other C-type lectins in the vicinity of the putative sugar binding sites (dotted circles in Fig. 4 A and B, yellow surfaces in Fig. 4 D–F, and yellow-shaded amino acid residues in the box enclosed in blue in Fig. 1). The regions are composed of Val195, Thr196, Phe198, Leu199, Tyr201, and Phe202 in Mincle, and Val197, Pro198, and Phe201 in MCL. Mincle has larger hydrophobic areas than MCL, whereas DC-SIGNR has only a much smaller one than both Mincle and MCL (Fig. 4 D–F). If the trehalose part of TDM is placed on the sugar binding site of Mincle, as in the binding mode of mannose to DC-SIGNR, then the mycolic acid attached to the 6-O of the glucose of TDM (Fig. 4 A and B, red arrows) is oriented toward the hydrophobic regions of Mincle and MCL, as described above. To investigate whether the hydrophobic region of Mincle contributes to the recognition of TDM, Ala substitutions of both Phe198 and Leu199 in this region were introduced in reporter cells expressing Mincle (Fig. 4C). The cells expressing the MincleF198A/L199A mutant exhibited reduced NFAT activity in response to TDM. Moreover, we replaced the hydrophobic region of Mincle (residues 195–202) with the corresponding region of another CLR, Dectin-2 (residues 192–199), which lacks the hydrophobic residues (24, 25). The reporter cells expressing this Mincle–Dectin-2 chimeric molecule (MincleMDchimera) (Fig. S4) still retained activity against the anti-Mincle mAb 13D10-H11 (Fig. S3C), which recognizes the conformational epitope on Mincle (Fig. S3D), indicating that the mutation as well as other mutations in this study did not have remarkable effects on overall protein folding and stability on the cell surface. However, the TDM recognition of MincleMDchimera was severely compromised (Fig. 4C). As described above, the Oδ atom of the corresponding residue Asn172 just following the EPN motif in Mincle does not face the Ca2+ ion, which is an unusual type of Ca coordination among C-type lectins (Fig. 3 A and B). Instead, the Nδ atom of Asn172 forms a hydrogen bond with the Oδ atom of Thr196 of the hydrophobic patch (Fig. 4A). The reporter cells expressing the mutant Mincle (MincleN172Q), which has only one additional methylene group, showed reduced NFAT-GFP activity (Fig. 4C). This result may suggest that the N172Q mutation indirectly influences the hydrophobicity of the putative lipid-binding patch via the side chain of Thr196.
To further examine the effect of a set of acyl chains, we performed surface plasmon resonance (SPR) binding assays using a set of trehalose-based glycolipids, which have a single acyl chain with different carbon lengths (C8, C10, C12). These glycolipids have a single and short tail, and thus are expected to be water-soluble while retaining ligand activity. The single acyl chains with trehalose (C10 and C12) bound to Mincle (Fig. 4G and Fig. S5). The affinity of C8 to Mincle is much lower than those of C10 and C12 (Fig. 4G). The crystal structure clearly indicated that the 10-carbon acyl chain with trehalose is reasonably accommodated within the hydrophobic portion in Mincle (Fig. S6).
Discussion
We have determined the crystal structures of the ectodomains of Mincle and MCL, which confirmed that the overall structures of Mincle and MCL are similar to those of other CLRs. Furthermore, we have also solved the crystal structure of Mincle complexed with citric acid, which revealed that the binding mode to citric acid essentially resembles that of glucose/mannose recognition by typical CLRs. We further performed competition binding of glycolipids with citric acid as in Fig. S7, clearly showing that citric acid inhibits glycolipid binding to Mincle, whereas acetic acid does not. Notably, another mannose-binding C-type lectin, codakine, bound similar positions of oxygens of glycerol and glycan in a Ca2+ ion-mediated manner (26). Citric acid is likely accommodated at this position to block the ligands, and hydroxyl groups are likely used following the coordination of Ca2+ ions generally observed in CLRs.
Glycolipids play pivotal roles in innate immunity, as exemplified by the functions of CD1-mediated natural killer T (NKT) cells (27, 28). CD1 family molecules display a variety of glycolipids toward semi-invariant NKT cell receptors to activate NKT cells. Structural analyses of CD1 family proteins have revealed that the lipid parts of glycolipids are deeply accommodated inside the hydrophobic cores of the proteins (29, 30), and thus only the sugar moieties are exposed for recognition by NKT cell receptors (Fig. S6). On the other hand, our present study showed that the putative TDM binding sites of Mincle and MCL include hydrophobic loops uniquely found in Mincle and MCL, which distinguish them from other C-type lectins (Fig. 4 A, B, and D–F). These loops form shallow hydrophobic patches extending from the corresponding position of the 6-OH of glucose in the structure of the mannose complex of DC-SIGNR, which is attached to mycolic acid in the case of TDM (Fig. S6). The mutational study suggested that these CLRs directly recognize the acyl groups of the glycolipid TDM using this shallow hydrophobic region, which is close to the Ca2+ binding site (Fig. 4 D and E). Notably, the SPR binding study using a set of glycolipids clearly showed that the single acyl chain is sufficient for Mincle binding. In addition, importantly, at least a C10 length of the lipid moiety is required (Fig. 4G). These observations might suggest that Mincle recognizes only the sugar-proximal part of the acyl chain of glycolipids. The hydrophobic patch branches out from the potential sugar binding site (downward and to right in Fig. 4D and Fig. S6). These might the sites accommodating the branched acyl chains in mycolic acids, such as TDM and trehalose monomycolate (TMM) (11). The recently discovered ligands of Mincle, which also have branched acyl chains, may interact similarly with TMM and TDM (13). Therefore, the recognition of glycolipids by Mincle and presumably MCL seems to be significantly distinct from those of lipid-recognition proteins, such as CD1 and the Toll-like receptor 4–MD2 complex, which have deep hydrophobic grooves to accommodate the acyl moieties of glycolipids (Fig. S6). Thus, a minimum acyl-chain length is required for glycolipid recognition by CLRs. The unique modes of CLR-glycolipid recognition would be advantageous for host defense responses, because they may allow receptors to recognize these bipolar ligands even within a microbial cell wall or in the micellar form in aqueous solution. Future study of cocrystallization with glycolipids harboring short branched acyl chains, which might have increased binding affinity, would elucidate the lipid-binding modes.
The production of NO and IL-6 by bone marrow-derived macrophages (BMMφ), which express Mincle and MCL, was reportedly changed by stimulation with several lengths of acyl chains, revealing the importance of acyl-chain length (31). Fungal glycolipids, recently identified as Mincle ligands, have more complicated and branched lipid moieties. The structural and functional data presented here showed that Mincle and MCL probably require an acyl chain longer than 10 carbons for glycolipid recognition, thus clearly providing important clues for the design of better adjuvants than TDM.
The present study indicated that Mincle has a higher affinity for TDM than MCL, which is consistent with our in vitro binding study (Fig. 3 F and G). The crystal structures of MCL and Mincle clearly revealed that MCL has a smaller hydrophobic patch next to the putative Ca2+-mediated sugar binding site compared with that in Mincle. The different sizes of these hydrophobic sites might explain the affinity differences of the two CLRs observed in the binding data.
Typical CLRs that simply recognize sugars, such as DC-SIGNR and CEL-IV, exhibit remarkably low affinities (Kd ∼mM) (32, 33). They require multiple valencies of sugar ligands to mediate signaling. However, SPR analysis revealed that Mincle seemed to show higher affinity, suggesting that it can detect small numbers of glycolipids on fungal surfaces. On the other hand, MCL showed much lower affinity than Mincle but essentially the same ligand specificity. It is plausible that MCL-mediated signaling requires multiple valencies of glycolipid ligands. Therefore, Mincle and MCL may play distinct roles in physiological events.
CLRs often form homodimers or heterodimers on the cell surface. As described above, the multivalent ligands on the bacterial surface likely induce the multimerization of CLRs (either monomeric or dimeric structures), which may mediate efficient signaling. The recent report by Lobato-Pascual et al. demonstrated that Mincle and MCL form a disulfide-linked heterodimer associated with the FcRγ chain (34). The heterodimeric complex formation between Mincle and MCL through the N-terminal β-strand and/or stalk regions, as previously reported for maltose-binding protein (35), for efficient recognition/signaling would be an intriguing issue to be addressed.
Materials and Methods
Plasmid Construction.
E. coli expression plasmids encoding the partial extracellular domain of human Mincle (residues 74–219), pET22-Mincle, and the extracellular domain (residues 61–215) of human MCL, pET22-MCL, were constructed (details are available in SI Materials and Methods) (36).
To improve the solubility and crystallization of Mincle, we synthesized, purified, and crystallized several mutated Mincle proteins. Among them, the I99K mutant was produced with a high yield and generated good crystals.
Preparation of Recombinant Proteins.
pET22-Mincle, pET22-Mincle I99K, and pET22-MCL plasmids were transformed into E. coli strain BL21(DE3) pLysS, and the protein was obtained as inclusion bodies. The protein was solubilized in a buffer containing 6 M guanidine⋅HCl, 50 mM MES (pH 6.5), 100 mM NaCl, and 10 mM EDTA for 12 h at 4 °C. One hour after the addition of DTT (10 mM), the solubilized proteins were slowly diluted into 1 L of buffer containing 0.1 M Tris⋅HCl (pH 8.5), 1 M l-arginine, 2 mM EDTA, 6.3 mM cystamine, 3.7 mM cysteamine, and 0.1 mM phenylmethylsulfonyl fluoride. The refolding mixture was purified by gel-filtration chromatography. The buffer was finally exchanged to 20 mM Tris⋅HCl (pH 8.0), with 5 mM CaCl2 for crystallization.
Crystallization and Structure Determination.
Crystals of purified Mincle I99K and MCL were grown at 20 °C [reservoir solutions: 1 M lithium chloride, 0.1 M citric acid, pH 4, 20% (wt/vol) PEG 6000, and 0.1 M bis-Tris propane, pH 6.5, 0.2 M potassium thiocyanate, 20% (wt/wt) PEG 3350, respectively] by the hanging-drop vapor-diffusion method. Crystals were equilibrated in a cryoprotectant consisting of reservoir solution supplemented with 16% (vol/vol) glycerol. X-ray data were collected on beamlines BL32XU in SPring-8 and BL5A in KEK. The data were processed with HKL2000 (37) or XDS (38). The structure was solved by molecular replacement with Phaser (39), using CD69 as the search model (PDB ID code 1FM5). Several rounds of model building in Coot (40) and refinement in PHENIX (41) were performed. The final refinement statistics are provided in Table S1. The coordinates for the refined Mincle, Mincle–citrate complex, and MCL structures have been deposited in the Protein Data Bank (ID codes 3WH3, 3WH2, and 3WHD, respectively).
Binding Assay Using Ig-Fusion Proteins.
The MCL–Ig and Mincle–Ig fusion proteins were prepared as described previously. Briefly, the C terminus of the extracellular domain of human MCL (residues 42–215), human Mincle (residues 46–219), or their mutants was fused to the N terminus of the hIgG1 Fc region. The Ig-fusion proteins were incubated with 0.2 μg per well of plate-coated TDM or plate-coated anti-human IgG, and the bound proteins were detected by using HRP-labeled anti-human IgG.
Reporter Assay.
Reporter cells were prepared as described previously (11, 36). Briefly, 2B4-NFAT-GFP reporter cells were transfected with FcRγ, together with Mincle and the mutants. The reporter cells were stimulated with various concentrations of plate-coated TDM or anti-human Mincle antibody (13D10-H11). The activation of NFAT-GFP was monitored by flow cytometry.
SPR Analysis.
SPR analysis was performed similarly as described previously for other cell-surface receptors (42). Briefly, Mincle and MCL were each dissolved in 10 mM sodium acetate (pH 4) containing 5 mM CaCl2. SPR experiments were performed with a BIAcore T3000 (GE Healthcare). All of the proteins were covalently immobilized on a CM5 sensor chip by amine coupling (GE Healthcare). β2-Microglobuline was used as a negative control protein. All glycolipids (C12, C10, and C8), and trehalose as a negative control, were injected in 10 mM Hepes (pH 7.4) containing 150 mM NaCl and 5 mM CaCl2 with or without 5% (vol/vol) dimethyl sulfoxide. The data were analyzed using BIAevaluation software, version 4.1 (GE Healthcare).
Note Added in Proof.
While this paper was under revision, Feinberg et al. (43) reported the crystal structure of bovine Mincle complexed with trehalose, whose binding mode is similar to that of human Mincle for glycolipids we proposed here.
Supplementary Material
Acknowledgments
We thank Dr. S. Kita, M. Nagata, and M. Shiokawa for their kind help with data collection. We also thank the beamline staff at the Photon Factory (Tsukuba, Japan) and SPring-8 for technical help during the X-ray data collection. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, and the Ministry of Health, Labor and Welfare of Japan (to K.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3WH3, 3WH2, and 3WHD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312649110/-/DCSupplemental.
References
- 1.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: Shaping immune responses. Nat Rev Immunol. 2009;9(7):465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: Regulation through compartmentalization. Nat Rev Immunol. 2009;9(8):535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choe J, Kelker MS, Wilson IA. Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309(5734):581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 5.Kuroki K, Furukawa A, Maenaka K. Molecular recognition of paired receptors in the immune system. Front Microbiol. 2012;3:429. doi: 10.3389/fmicb.2012.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272(24):6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 7.Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancho D, Reis e Sousa C. Sensing of cell death by myeloid C-type lectin receptors. Curr Opin Immunol. 2013;25(1):46–52. doi: 10.1016/j.coi.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyake Y, Ishikawa E, Ishikawa T, Yamasaki S. Self and nonself recognition through C-type lectin receptor, Mincle. Self Nonself. 2010;1(4):310–313. doi: 10.4161/self.1.4.13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto M, et al. A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J Immunol. 1999;163(9):5039–5048. [PubMed] [Google Scholar]
- 11.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206(13):2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamasaki S, et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc Natl Acad Sci USA. 2009;106(6):1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa T, et al. Identification of distinct ligands for the C-type lectin receptors Mincle and Dectin-2 in the pathogenic fungus Malassezia. Cell Host Microbe. 2013;13(4):477–488. doi: 10.1016/j.chom.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Schoenen H, et al. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J Immunol. 2010;184(6):2756–2760. doi: 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyake Y, et al. C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38(5):1050–1062. doi: 10.1016/j.immuni.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Arce I, Martínez-Muñoz L, Roda-Navarro P, Fernández-Ruiz E. The human C-type lectin CLECSF8 is a novel monocyte/macrophage endocytic receptor. Eur J Immunol. 2004;34(1):210–220. doi: 10.1002/eji.200324230. [DOI] [PubMed] [Google Scholar]
- 17.Graham LM, et al. The C-type lectin receptor CLECSF8 (CLEC4D) is expressed by myeloid cells and triggers cellular activation through Syk kinase. J Biol Chem. 2012;287(31):25964–25974. doi: 10.1074/jbc.M112.384164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg H, Taylor ME, Weis WI. Scavenger receptor C-type lectin binds to the leukocyte cell surface glycan Lewis(x) by a novel mechanism. J Biol Chem. 2007;282(23):17250–17258. doi: 10.1074/jbc.M701624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg H, Mitchell DA, Drickamer K, Weis WI. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294(5549):2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 21.Geijtenbeek TB, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 22.Drickamer K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature. 1992;360(6400):183–186. doi: 10.1038/360183a0. [DOI] [PubMed] [Google Scholar]
- 23.Fujihashi M, Peapus DH, Kamiya N, Nagata Y, Miki K. Crystal structure of fucose-specific lectin from Aleuria aurantia binding ligands at three of its five sugar recognition sites. Biochemistry. 2003;42(38):11093–11099. doi: 10.1021/bi034983z. [DOI] [PubMed] [Google Scholar]
- 24.Robinson MJ, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J Exp Med. 2009;206(9):2037–2051. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Gourdine JP, et al. High affinity interaction between a bivalve C-type lectin and a biantennary complex-type N-glycan revealed by crystallography and microcalorimetry. J Biol Chem. 2008;283(44):30112–30120. doi: 10.1074/jbc.M804353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 28.Moody DB, Besra GS. Glycolipid targets of CD1-mediated T-cell responses. Immunology. 2001;104(3):243–251. doi: 10.1046/j.1365-2567.2001.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat Immunol. 2002;3(8):721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 30.Koch M, et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6(8):819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 31.Khan AA, et al. Long-chain lipids are required for the innate immune recognition of trehalose diesters by macrophages. ChemBioChem. 2011;12(17):2572–2576. doi: 10.1002/cbic.201100451. [DOI] [PubMed] [Google Scholar]
- 32.Probert F, Whittaker SB, Crispin M, Mitchell DA, Dixon AM. Solution NMR analyses of the C-type carbohydrate recognition domain of DC-SIGNR protein reveal different binding modes for HIV-derived oligosaccharides and smaller glycan fragments. J Biol Chem. 2013;288(31):22745–22757. doi: 10.1074/jbc.M113.458299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatakeyama T, et al. Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif. J Biol Chem. 2011;286(12):10305–10315. doi: 10.1074/jbc.M110.200576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobato-Pascual A, Saether PC, Fossum S, Dissen E, Daws MR. Mincle, the receptor for mycobacterial cord factor, forms a functional receptor complex with MCL and FcεRIγ. Eur J Immunol. August 26, 2013 doi: 10.1002/eji.201343752. [DOI] [PubMed] [Google Scholar]
- 35.Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2(12):1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 36.Yamasaki S, et al. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol. 2008;9(10):1179–1188. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132. [DOI] [PubMed]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabata S, et al. Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J Biol Chem. 2008;283(14):8893–8901. doi: 10.1074/jbc.M709793200. [DOI] [PubMed] [Google Scholar]
- 43.Feinberg H, et al. Mechanism for recognition of an unusual mycobacterial glycolipid by the macrophage receptor Mincle. J Biol Chem. August 19, 2013 doi: 10.1074/jbc.M113.497149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.